Abstract
Deregulation of the hedgehog (HH) pathway results in overexpression of the GLI target BCL2 and is an initiating event in specific tumor types including basal cell carcinoma of the skin. Regulation of the HH pathway during keratinocyte differentiation is not well understood. We measured HH pathway activity in response to differentiation stimuli in keratinocytes. An upregulation of suppressor of fused (SUFU), a negative regulator of the HH pathway, lowered HH pathway activity and was accompanied by loss of BCL2 expression associated with keratinocyte differentiation. We used in vitro and in vivo models to demonstrate that ΔNp63α, a crucial regulator of epidermal development, activates SUFU transcription in keratinocytes. Increasing SUFU protein levels inhibited GLI-mediated gene activation in suprabasal keratinocytes and promoted differentiation. Loss of SUFU expression caused deregulation of keratinocyte differentiation and BCL2 overexpression. Using in vivo murine models, we also provide evidence of GLI-mediated regulation of the TP63 pathway. p63 expression appears essential to establish an optimally functioning HH pathway. These observations present a regulatory mechanism by which SUFU acts as an interacting node between the HH and TP63 pathways to mediate differentiation and maintain epidermal homeostasis. Disruption of this regulatory node can be an important contributor to multistep carcinogenesis.
Keywords: SUFU, TP63, BCL2, keratinocyte, differentiation, non-melanoma skin cancer
Epidermal homeostasis necessitates a balance between proliferative cells and keratinocyte differentiation.1 This balance is regulated by the action of characteristic developmental pathways acting sequentially through specific transcription factor modules.2 These pathways preserve epidermal homeostasis by their ability to regulate the spatiotemporal expression of downstream target proteins. Deregulation of these pathways has been shown to directly contribute to the formation of tumors, including basal cell carcinomas (BCCs).2, 3
A homolog of TP53, TP63, has an essential role in development. Murine models lacking TP63 show extensive defects in epidermal development and die at birth.4, 5 Alternate promoter usage gives rise to two different classes of p63 protein characterized by the presence or absence of an N-terminal transactivation domain (TA) and ΔN, respectively. C-terminal splicing results in isoforms with three distinct C-termini (α, β, and γ). The six distinct p63 isoforms have diverse transcriptional and functional activities. TAp63 is expressed early during embryogenesis and is critical for the maintenance of stem cells in the epidermis.6, 7 ΔNp63α, the predominant p63 isoform expressed in epidermal keratinocytes, has a crucial role in epidermal morphogenesis and is the focus of this study.8, 9, 10 ΔNp63, identified as a regulator of cell proliferation, also has a role in initiation of epidermal differentiation and stratification.9, 11, 12
Binding of the sonic hedgehog (SHH) ligand to the membrane-bound patched (PTCH) receptor complex results in derepression and activation of the hedgehog (HH) pathway, which enables the effector GLI transcription factor to modulate target gene expression.13 In the absence of receptor-ligand interaction, PTCH inhibits phosphorylated activation of smoothened (SMO). In addition, the HH pathway can be modulated by cytoplasmic suppressor of fused (SUFU), which inhibits nuclear translocation of effector GLI proteins, thereby facilitating GLI processing into their repressor forms.13, 14, 15 An active HH pathway in keratinocytes is associated with cellular proliferation and is necessary for normal epithelial development.16 Deregulation of the pathway due to mutations in PTCH, SMO, and SUFU can be initiating events in specific tumor types.3, 17, 18 In BCCs, constitutive activation of the HH pathway is a frequent event3, 17 that results in disordered proliferation and differentiation, as well as apoptosis resistance through upregulation of the GLI target gene BCL2.19, 20 BCL2, an antiapoptotic member of a signature family of proteins, is often deregulated in cancer.21, 22 In the epidermis, BCL2 protein is confined to basal keratinocytes, the outer root sheath cells of the hair follicle, and is associated with undifferentiated cells having proliferative potential.23 Suprabasal keratinocytes lose expression of BCL2. Very little is known about the mechanisms regulating the HH pathway in differentiating keratinocytes. The goal of the study was to examine potential mechanisms that regulate the HH pathway and its target BCL2 during keratinocyte differentiation.
In the present study, a reduction in HH pathway activity and BCL2 transcript levels along with increased SUFU expression was observed after induction of differentiation in primary epidermal keratinocytes. Targeted overexpression of ΔNp63α resulted in an increase in SUFU protein with consequent downregulation of HH pathway activity in the epidermis of genetically engineered mice. Furthermore, we showed that activation of the HH pathway by loss of SUFU or overexpression of GLI in murine models results in an increase in p63 protein. On the basis of these in vitro and in vivo observations, we postulated a potential mechanism by which differentiation, proliferation, and survival of epidermal keratinocytes could be coordinately regulated by reciprocal TP63/HH pathway interactions.
Results
The HH pathway is downregulated during keratinocyte differentiation
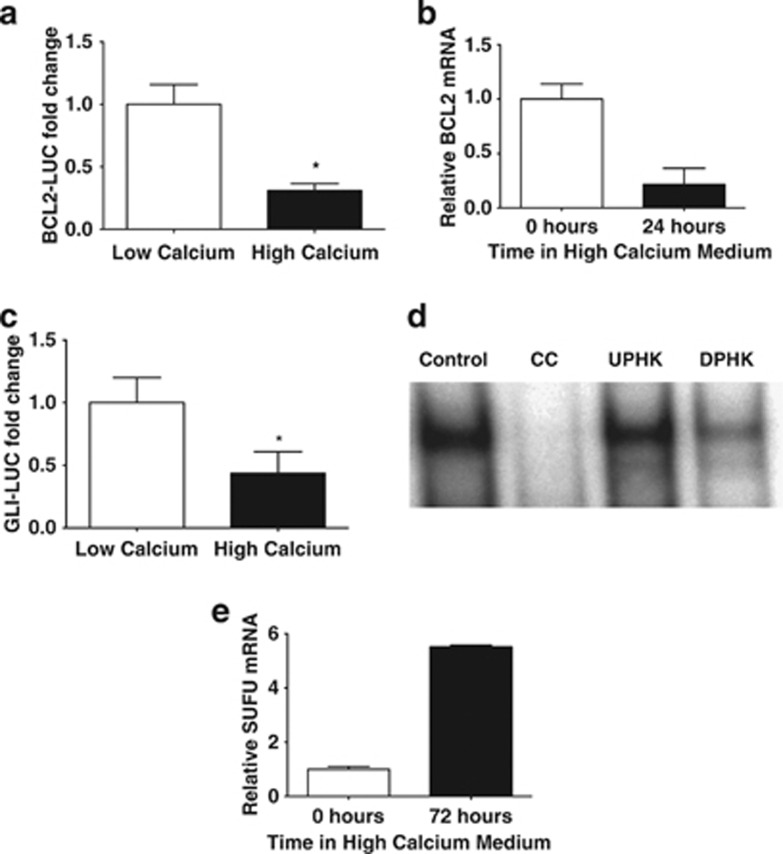
It has been shown that calcium-induced differentiation of primary human keratinocytes (PHKs) results in a selective decrease in BCL2 protein levels24, 25 (Supplementary Figure S1). In order to determine if this decrease in BCL2 protein expression was due to transcriptional changes, luciferase assays using a 2.8-kb BCL2 promoter luciferase (BCL2-LUC) construct were performed in PHKs, which were induced to differentiate by increasing the extracellular calcium concentration in the culture media. Extracellular calcium-mediated differentiation resulted in significant downregulation of BCL2 promoter activity (Figure 1a). The decrease in BCL2 promoter activity was accompanied by a reduction in BCL2 transcript levels as measured by quantitative PCR (qPCR; Figure 1b). In order to determine if the changes in BCL2 transcription corresponded to changes in GLI pathway activity, PHKs were transfected with a GLI luciferase reporter (GLI-LUC). PHKs were grown in regular low calcium medium (0.05 mM) to allow proliferation or high calcium medium (1.5 mM) to induce differentiation. Compared with PHKs grown in low calcium medium, PHKs induced to differentiate in high calcium medium showed a significant repression of the GLI-LUC reporter (Figure 1c) consistent with HH pathway downregulation during differentiation.
Figure 1.
Downregulation of the HH pathway during PHK differentiation. (a) A 2.8-kb BCL2-LUC was transfected into PHKs and cells were grown in low calcium (0.05 mM) or differentiation-inducing media high calcium (1.5 mM) media. A renilla luciferase construct was used to normalize for transfection efficiency. Data are the mean±S.E.M. of three independent experiments in triplicate (*P<0.05). (b) RNA was extracted from PHKs grown in differentiation-inducing media for the indicated time period. qPCR analysis for BCL2 was carried out in triplicate and normalized to GAPD. Data are the mean±S.D. (c) A GLI-LUC was transfected into keratinocytes grown in low calcium (0.05 mM) medium or high calcium (1.5 mM), differentiation-inducing medium. Results are normalized to renilla luciferase readings. Data are the mean±S.E.M. of three experiments (*P<0.05). (d) EMSA assays were carried out using nuclear proteins from undifferentiated PHKs (UPHKS) grown in low calcium media or PHKs induced to differentiate (DPHK) PHK in methylcellulose suspension (MC) for 24 h and were incubated with 32P-labeled GLI consensus-binding site oligonucleotides. The control utilized nuclear extracts from PHK and competed with excess unlabeled cold competitor (CC) GLI consensus oligonucleotide. (e) HaCaT keratinocytes were grown in differentiation-inducing media for the indicated time period. qPCR analysis for SUFU was carried out in triplicate and normalized to GAPD. Data are the mean±S.D.
In order to evaluate changes in GLI-DNA interactions in PHKs during differentiation, electrophoretic mobility shift assays (EMSAs) were performed using protein extracts from PHKs induced to differentiate using methylcellulose suspension.26 A significant reduction was seen in GLI protein binding to a consensus GLI-binding site oligonucleotide in methylcellulose-differentiated PHKs compared with proliferating undifferentiated PHKs (Figure 1d and Supplementary Figure S2). The nuclear translocation of GLI proteins is controlled by SUFU, the negative regulator of the HH pathway.14, 15 Using qPCR, we examined activation of SUFU transcription in differentiated HaCaT keratinocytes. We observed a significant increase in SUFU mRNA levels in keratinocytes induced to differentiate27, 28 (Figure 1e). HaCaT keratinocytes also exhibited a corresponding reduction in BCL2 transcript levels (data not shown). We also observed an increase in SUFU protein expression in PHKs induced to differentiate (Supplementary Figure S3). Together, these observations imply that the GLI pathway is actively downregulated in differentiating keratinocytes secondary to an increase in SUFU transcriptional activity and subsequent reduction in nuclear GLI.
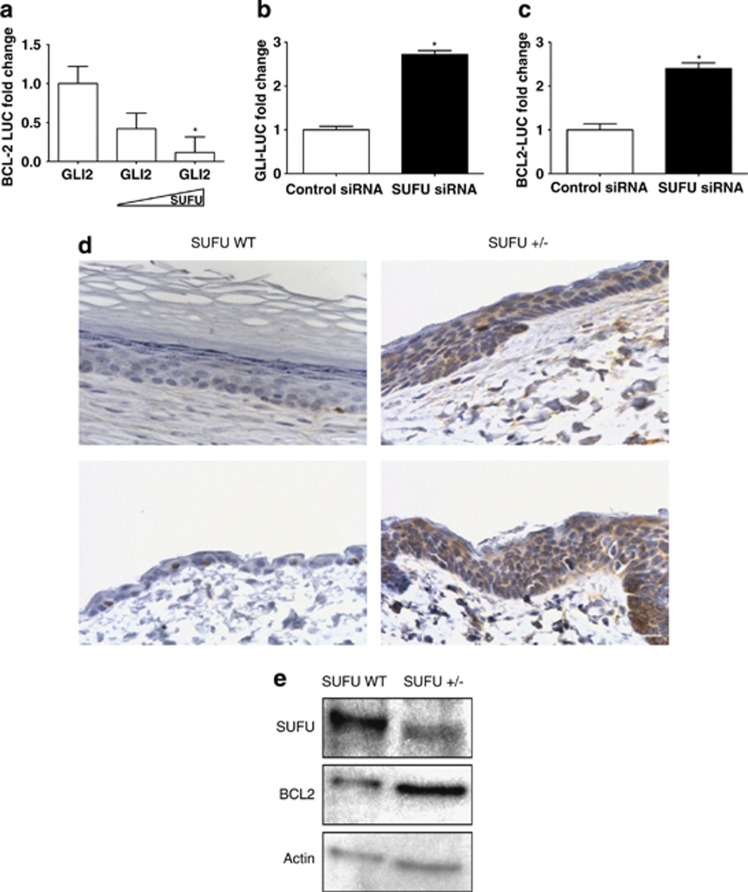
In addition to GLI119 (Supplementary Figure S4), GLI2 is a potent activator of BCL2.20 We confirmed that increasing amounts of SUFU can negate GLI2-mediated activation of the GLI-LUC reporter in PHKs, which has been shown in other systems14 in a dose-responsive manner (Supplementary Figure S5). We sought to determine whether increasing SUFU protein levels could counteract GLI2-dependent activation of BCL2 in PHKs. A GLI2 expression construct was transfected along with a BCL2-LUC construct into PHKs followed by increasing amounts of SUFU expression vector. We observed that cotransfection of the SUFU expression vector into PHKs with GLI2-impeded GLI-mediated activation of the BCL2-LUC in a dose-dependent manner (Figure 2a). This suggests that increasing SUFU levels inhibits the binding of GLIs to the promoters of target genes, thereby preventing their activation during differentiation.
Figure 2.
Increased SUFU levels negate GLI2-mediated transcription. (a) A GLI2 expression construct and increasing amounts of a SUFU expression vector was transfected along with a BCL2-LUC construct into PHKs. Resultant luciferase values were normalized to a renilla luciferase control. Data are the mean±S.E.M. of three experiments (*P<0.05). PHK cells were transfected with control siRNA and SUFU siRNA along with (b) GLI-LUC reporter or (c) BCL2-LUC constructs. Results were normalized to renilla luciferase. Data are the mean±S.E.M. of three experiments (*P<0.05). (d) IHC analysis of BCL2 in multiple skin sections from SUFU WT (left panels) and SUFU+/− (right panels) mice show a higher and more diffuse BCL2 protein expression pattern in the SUFU+/− mice. Scale bar is 20 μm. (e) Western blot analysis of protein lysate from epidermal sections of SUFU WT and SUFU+/− mice demonstrated reduced SUFU expression and increased BCL2 protein expression. β-Actin was used as a loading control
Loss of SUFU disrupts differentiation of keratinocytes
SUFU has been shown to act as a tumor suppressor because of its ability to suppress the HH pathway.18 In order to examine whether loss of SUFU would enhance activity of the HH pathway and its target BCL2 in keratinocytes, BCL2 and GLI reporter activity were assessed using a SUFU knockdown strategy. siRNA-mediated depletion of SUFU in PHKs caused a significant increase in both GLI-LUC (Figure 2b) and BCL2-LUC (Figure 2c) activity compared with controls.
An immunohistochemical (IHC) assessment of BCL2 protein expression was performed on epidermal sections obtained from SUFU+/− and SUFU wild-type (WT) control littermate mice to evaluate the impact of SUFU deficiency on BCL2 expression in vivo (Figure 2d). BCL2 expression was relatively weak and confined to basal keratinocytes in control mice (Figure 2d). In contrast, BCL2 was present at increased levels in the epidermis of SUFU+/− mice and was no longer confined to basal keratinocytes, but also detectable in the suprabasal layers (Figure 2d). Immunoblots of skin from SUFU WT and SUFU+/− mice showed a decrease in SUFU protein associated with increased BCL2 protein expression (Figure 2e).
In aggregate, the in vivo and in vitro data suggest that induction of SUFU is necessary for the appropriate restriction of BCL2 expression to basal keratinocytes and that loss, or reduction, in SUFU expression in the epidermis is associated with altered differentiation.
ΔNp63α regulates SUFU expression in keratinocytes
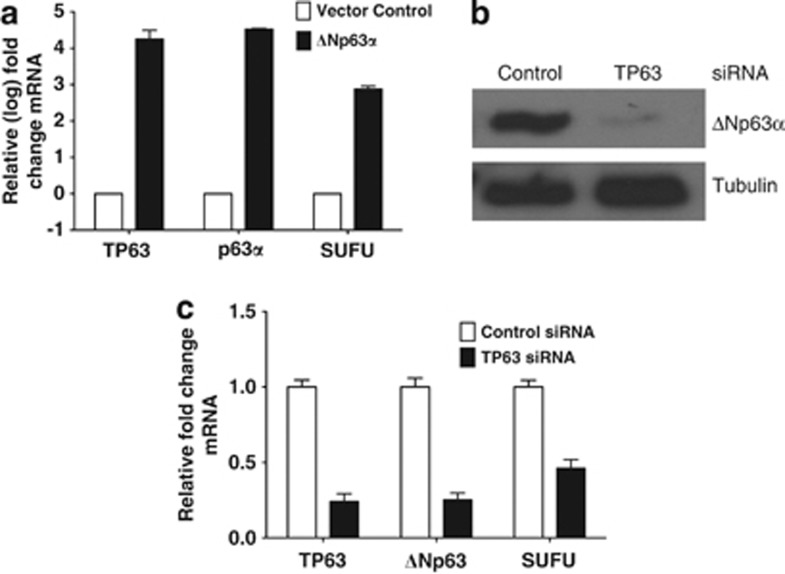
Regulators of SUFU expression during keratinocyte differentiation have not been fully characterized. It is noteworthy that recent studies employing genome-wide ChIP analysis identified SUFU as a target of ΔNp63α.29, 30 The ectopic expression of human ΔNp63α in H1299 cells resulted in an increase in SUFU mRNA as assessed by qPCR (Figure 3a). In order to examine whether loss of ΔNp63α affects SUFU transcriptional activity, a siRNA strategy to knockdown p63 in ME-180 cells was performed. ME-180 cells express only the ΔNp63 isoform30 and p63 knockdown resulted in a significant reduction in the level of ΔNp63α protein (Figure 3b). A significant decline in SUFU mRNA was observed in ΔNp63-depleted ME-180 cells (Figure 3c).
Figure 3.
ΔNp63α regulates SUFU expression. (a) H1299 cells were transfected with vector control or ΔNp63α expression constructs and the level of expression of SUFU, TP63, and p63α transcript was assessed by qPCR. (b) Immunoblot for p63 protein levels in ME-180 cells transfected with control siRNA or TP63 siRNA validated knockdown of p63 protein. α-tubulin was used as a loading control. (c) ME-180 cells were transfected with control siRNA or TP63 siRNA for 48 h. The extracted RNA was examined for SUFU, p63α, and ΔNp63 mRNA levels. qPCR data represent mean±S.D. of three independent experiments carried out in triplicate
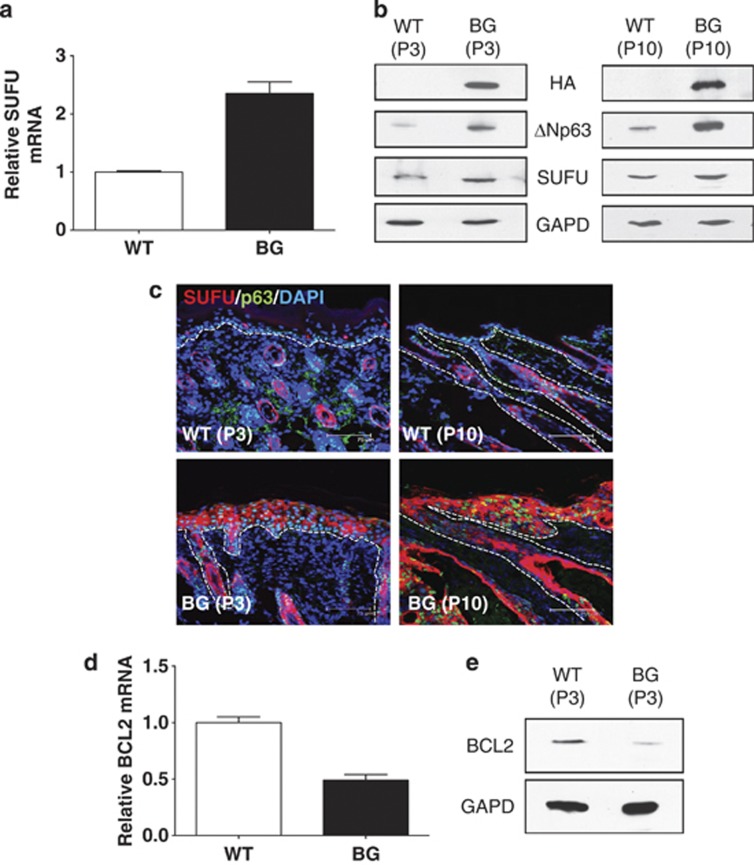
In order to assess the effect of modulating ΔNp63α on SUFU expression in vivo, a transgenic (TG) mouse model in which ΔNp63α was specifically targeted to the basal layer keratinocytes and the outer root sheath of the hair follicles in mouse skin was used (ΔNp63α bigenic mice, ΔNp63α BG).31 qPCR analysis of RNA extracted from the epidermis of ΔNp63α BG mice showed a significant increase in SUFU transcript expression compared with WT littermate controls (Figure 4a). Immunoblot analysis of protein extracts from epidermal tissue demonstrated a corresponding increase in SUFU protein in the skin of ΔNp63α BG mice (Figure 4b). Immunofluorescence microscopy showed that SUFU protein expression was higher and presented a more diffuse distribution in the epidermis of the ΔNp63α BG mice than in WT control mice (Figure 4c). In order to evaluate the effect of ΔNp63α- mediated SUFU activation on BCL2, we assayed both BCL2 transcript and protein levels in the epidermis of ΔNp63α BG and WT mice. We found a significant reduction in BCL2 mRNA (Figure 4d) and protein (Figure 4e and Supplementary Figure S6) levels in keratinocytes obtained from the ΔNp63α BG mice compared with matched control mice.
Figure 4.
ΔNp63α drives SUFU expression in vivo. (a) qPCR analysis of SUFU mRNA from the epidermis of matched WT and ΔNp63α BG (BG) mice pair. Analysis was performed in triplicate and normalized to B2M. (b) Immunoblot analysis of SUFU protein expression in keratinocytes from two separate matched WT and BG mice pairs (P3, left panel and P10, right panel). GAPD was used as a loading control. (c) Immunflouresence analysis of skin sections from two separate pairs (P3 and P10) of matched WT and BG mice for SUFU protein was carried out. Sections were also costained for DAPI and p63. Analysis of BCL2 mRNA (d) and protein (e) expression in keratinocytes from matched WT and BG mice pair (P3)
HH pathway activation results in increased ΔNp63α expression
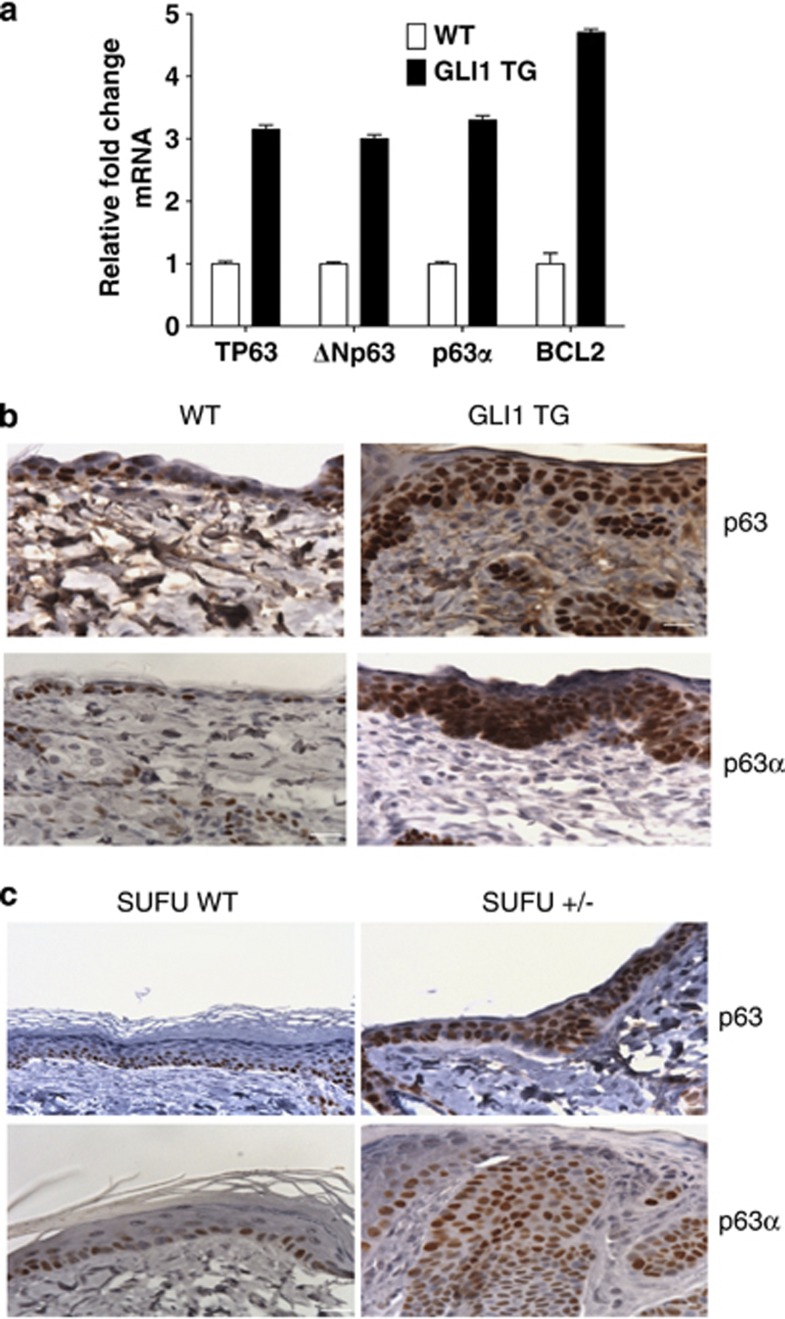
Deficiency of SHH or TP63 results in defects in prostate and epidermal development as observed in genetically engineered mouse models.32, 33, 34, 35 We used a GLI1 TG mouse model (GLI1 TG)36 to assess in vivo activation of ΔNp63α in the epidermis by the HH pathway. qPCR analysis was carried out with primers that measured amounts of the ΔNp63 isoforms or the p63α (ΔN and TA) variants. Analysis of transcripts from the skin of GLI1 TG mice demonstrated an increase in TP63 (pan-p63), ΔNp63, and p63α mRNA relative to control littermate mice as determined by qPCR (Figure 5a). Increases were also observed in transcript levels of the GLI1 target BCL2 (Figure 5a). This suggests an upregulation of ΔNp63α transcription upon GLI1 activation. IHC analysis using an antibody (pan-p63 4A4) to detect all p63 isoforms or only the p63α isoforms demonstrated an increase in p63α protein and the presence of the p63α isoform in the suprabasalar layers of the epidermis in GLI1 TG mice in contrast to WT control littermates (Figure 5b). A similar increase in expression pattern and change in staining pattern for the p63α isoform protein was observed in the SUFU+/− genetically engineered mice when compared with their WT control littermates (Figure 5c). As ΔNp63α is the only α-isoform expressed in mature epidermis, an increase in the expression of p63α isoforms implies an increase in ΔNp63α protein. Together, these observations suggest that deregulation of the HH pathway by either activation of GLI or loss of SUFU in murine models results in an increased expression of ΔNp63α, the predominant p63 isoform, in vivo.
Figure 5.
HH pathway activation results in increased p63 expression. (a) RNA extracted from the epidermis of four matched WT or induced GLI1 TG mice were subjected to qPCR analysis for BCL2, TP63, ΔNp63, and p63α transcript levels. Data is mean±S.D. from a representative matched pair carried out in triplicate normalized to GAPD. (b) Epidermal sections from WT and induced GLI1 TG mice were immunostained using antibodies directed against p63 (upper) and p63α (lower). GLI1 TG mice (right column) show increased expression of p63 compared with their WT (left column) controls. (c) IHC analysis of skin from SUFU WT (left column) and SUFU+/− (right column) carried out using antibodies targeted against p63 (upper) and total p63α (lower). Scale bars are 20 μm
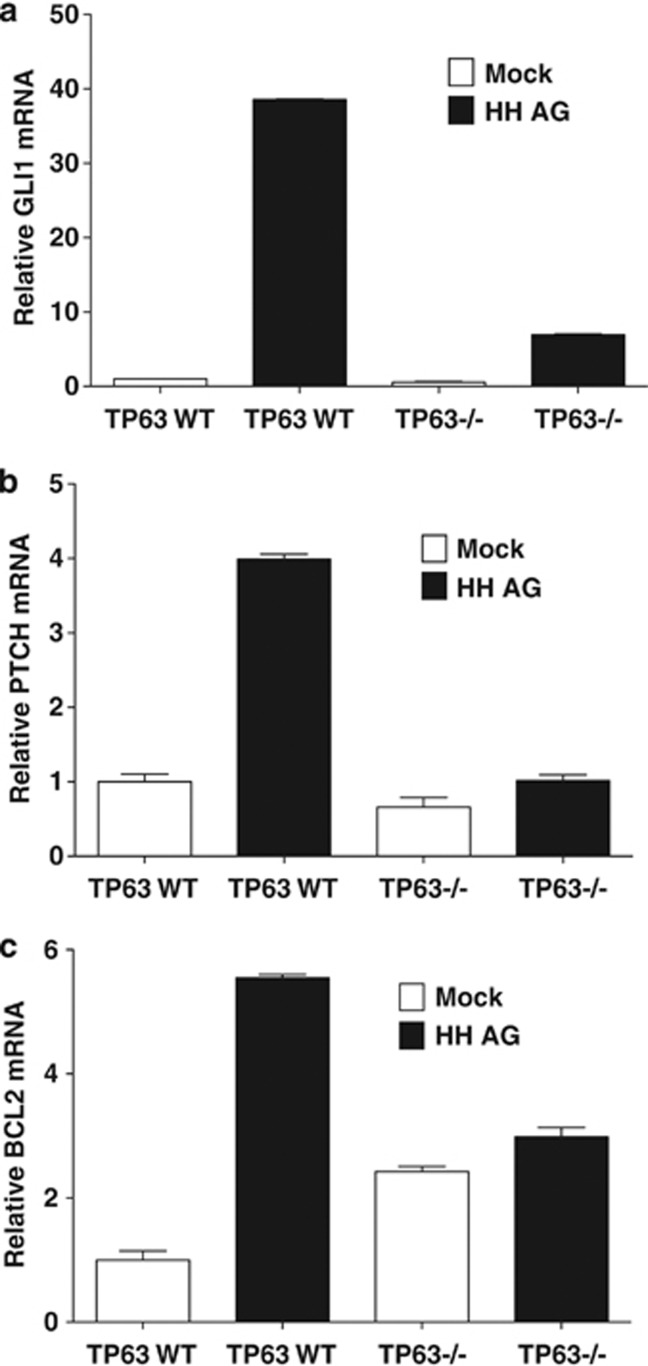
Keratinocytes defective for p63 expression show a hypoproliferative phenotype.12 p63 transcriptionally regulates the expression of SHH, which is a known regulator of cellular proliferative potential.37 Lower levels of SHH protein were observed in TP63−/− mouse embryonic fibroblasts (MEFs).37 In order to examine whether substitution of HH into TP63−/− MEFs would restore the HH pathway, an exogenous HH agonist 1.5 (HH AG)38 was added to TP63 WT and TP63−/− MEFs cultured in vitro. Addition of the HH AG to TP63 WT MEFs resulted in a significant upregulation of the GLI1 and PTCH transcript as expected. In contrast, the induction of these genes was significantly reduced in TP63−/− MEFS (Figures 6a and b). In addition, we found that the HH AG induced a fivefold increase in BCL2 transcript levels in the TP63 WT, but not TP63−/− MEFs (Figure 6c). These findings suggest that lack of p63 severely impairs signal transduction in the HH pathway even after compensation for lowered SHH morphogen in TP63-deficient cells.
Figure 6.
p63 is essential for optimal HH pathway activity. TP63 WT and TP63−/− MEFs were treated with mock or 100 nm HH AG for 20 h and qPCR analysis for (a) GLI1, (b) PTCH, and (c) BCL2 was carried out in triplicate on the purified RNA. Data represent mean±S.D.
Discusssion
The HH and TP63 pathways are essential to epidermal development. Mouse models demonstrate that disruptions in either the HH or TP63 networks have severe consequences in the development of epithelial organs.17, 39, 40, 41 In humans, mutations and alterations in both TP63 and components of the HH pathway are etiological factors for numerous ectodermal dysplasias and epithelial cancers.3, 39, 40, 41, 42
The HH pathway is associated with stem cells and the proliferative compartments of tissues.17 Activating mutations in the HH pathway drive BCL2 expression and proliferation and are a frequent feature of neoplasms including BCC.3, 19, 20, 41 In addition, GLIs and BCL2 are coexpressed in proliferative compartments of the epidermis.20 Although, there are numerous studies demonstrating the mechanisms and mutations, that cause an activation of the pathway and lead to carcinogenesis,3 information is scarce regarding mechanisms by which the HH pathway is suppressed during keratinocyte differentiation.
We found that inducing keratinocytes to differentiate with increased extracellular calcium caused a drop in HH pathway activity and a lowered expression of the target gene BCL2, but not other BCL2 family members. Murine knockout models have shown that only BCL2 among all the BCL2 family members is necessary for normal epidermal development.43 In addition, overexpression of BCL2 by itself in the suprabasalar layers does not affect epidermal development but alters sensitivity to UV-induced apoptosis.44 This suggests that merely altering the ratio of apoptotic/antiapoptotic members by enforced BCL2 expression does not affect epidermal development or spontaneous tumor formation. In BCCs, a mutated HH pathway changes expression patterns of a menu of transcriptional targets involved in cell-cycle regulation, differentiation, and apoptosis, including BCL2. Induction of SUFU expression was responsible for a reduction in GLI binding to target gene promoters. This increase in SUFU expression is necessary to lower HH pathway activity and promote differentiation because partial loss of SUFU results in increased GLI activity, disordered differentiation, and tumor formation in mice.45
Genome-wide ChIP analysis suggests that SUFU is a target of ΔN (amino-terminal truncated protein) p63α, and our preliminary analysis implies that ΔNp63α binds the SUFU promoter at an NF-Y-binding site (data not shown).29, 30 Loss of ΔNp63α resulted in lowered levels of SUFU mRNA. ΔNp63α-mediated HH pathway regulation can also be visualized in in vivo mouse models with targeted overexpression of ΔNp63α to the outer root sheath of the hair follicle. In these mice, a significant change in the cell lineage to a more differentiated state was found with the keratinocytes of the hair follicle expressing differentiation markers K1/K0 and fillagrin not being normally associated with hair follicle keratinocytes.31 Confirmation of the fact that ΔNp63α overexpression leads to significant induction of SUFU that shuts down the HH pathway is obtained from analysis of the epidermis from the ΔNp63α BG mice. We, also observed lowered BCL2 expression levels in the epidermis of ΔNp63α BG mice. In addition, SUFU is a negative regulator of β-catenin,46 and analysis of the ΔNp63α BG mice indicated a suppressed β-catenin/WNT axis.31 This SUFU-mediated downturn in HH and WNT activity results in depletion of stem cells and also hair loss. Interestingly, BCL2 knockout mice also show significant defects in hair follicle cycling.47 Together, these observations suggest that ΔNp63α-mediated overexpression of SUFU restricts HH pathway activation of target genes essential for epidermal cell proliferation and survival.
The results of our studies utilizing multiple mouse models of GLI activation established HH pathway-mediated activation of ΔNp63α. This is an interesting concept, as it proposes that HH activates ΔNp63α expression, which, in turn, upregulates the GLI repressor SUFU, which then functions as a node in a feed forward inhibitory loop. This mechanism could explain the reduction in ΔNp63α in suprabasal keratinocytes, where GLI is undetectable.48
It has been shown that the β and γ, but not the α-isoforms of TP63, are potent activators of SHH transcription, the loss of which could account for the severe proliferative phenotype of the TP63−/− keratinocytes.12, 37 Our results, however, show that the addition of exogenous HH AG to the TP63−/− MEFs resulted in significantly lowered GLI transcriptional activity compared with control WT MEFs. This suggests that TP63 has other functions besides transcriptional regulation of the SHH ligand in actuating the HH network. In this regard, GLI3 and SMO have been identified as potential targets of p63, and lowered expression of SMO could result in lowered HH pathway activity.30 In addition, TP53 is a negative regulator of target genes of the HH pathway.49, 50 p63 might potentiate HH pathway activity by preventing TP53-mediated inhibition of the HH pathway, as loss of TP53 in keratinocytes results in enhanced expression of SMO.51 This is in agreement with previous studies in which knockdown of TP53 in TP63-ablated keratinocytes can rescue the proliferative deficiency but not the lack of differentiation induction.12 On the basis of our findings, induction of differentiation and inhibition of an active HH pathway by SUFU would require the transcriptional activity of ΔNp63α.
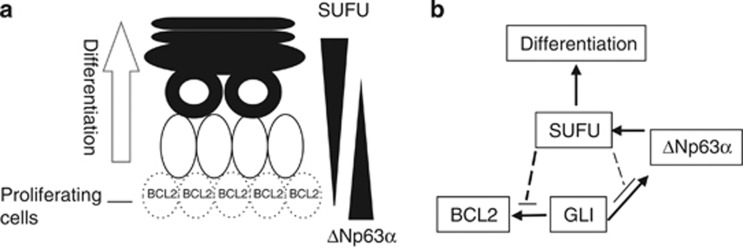
The formation of the adult epidermis requires the multiplication of the stem cell pool and the initiation of differentiation and stratification of keratinocytes. It has been hypothesized that TP63, especially ΔNp63α, is required for stem cell proliferation, and that TP63 deficiency results in defects in the cycling of stem cells.52 Alternatively, it has been suggested that loss of TP63 prevents keratinocytes from entering terminal differentiation and that TP63 is necessary for initiation of differentiation.52 Our study for the first time presents a model demonstrating a potential cross regulation of the HH and TP63 pathways in keratinocytes that maintains epidermal homeostasis by controlling both proliferation and differentiation. On the basis of our findings, we propose a model (Figures 7a and b) in which HH pathway activation in basal keratinocytes results in increased expression of ΔNp63α and BCL2. This increased expression of ΔNp63α is important for cell regeneration and replication because it probably serves to inhibit TP53 activity and promote the HH pathway. Differentiation stimuli cause the cell to detach from the basement membrane and initiate the differentiation program. At, or prior to this point, increased levels of ΔNp63α activate SUFU expression. Accumulation of SUFU protein shuts off the HH pathway by restricting effector GLIs to the cytoplasm. This loss of GLI activity prevents activation of survival/proliferation pathways in differentiated cells including loss of BCL2 expression. This also indirectly affects ΔNp63α expression, a GLI target, whose lowered expression is observed in differentiated keratinocytes.53 Though absolute p63 levels are lower in differentiated cells compared with the keratinocyte stem cells, the levels of phosphorylation of the p63 protein is greatly increased. Suzuki et al.54 suggest that phosphorylation of p63 in keratinocytes in response to calcium occurs before differentiation induced degradation of p63. Our model also suggests a potential explanation for the apparently opposing functions proposed for TP63 by suggesting that TP63 is necessary for cell proliferation by activation of SHH expression and the potentiation of the pathway in the presence of TP53. In addition, ΔNp63α can inhibit GLI by transcriptionally activating SUFU, thereby initiating differentiation.
Figure 7.
Model integrating the TP63 and HH pathways during keratinocyte differentiation. (a) Basal proliferative cells of the epidermis express BCL2. Differentiating suprabasal cells do not express BCL2 and have lowered ΔNp63α, and increased SUFU protein levels. (b) In proliferative cells, an active HH pathway drives ΔNp63α expression, which activates SUFU transcription. The increase in SUFU expression inhibits GLI-mediated activation of target genes, including ΔNp63α, BCL2, and GLI1. Loss of GLI activity restrains proliferation and promotes differentiation of keratinocytes. Further information is available in text
Although our model of TP63/HH crosstalk rather simplistically attempts to explain the dual roles of TP63 in epidermal development, in reality, regulation of epidermal homeostasis is more complicated and involves the interaction of other developmental networks, including the WNT and NOTCH pathways.40 Our study is also limited in that it focuses only on ΔNp63α. This isoform accounts for a majority of TP63 expressed in the epidermis but we cannot exclude the role that the TA, β-, and γ-isoforms might have in tissue development. Studies attempting to investigate the individual roles of the TA and ΔNp63α isoforms of TP63 found that though ΔNp63α can greater compensate for TP63 loss, both isoforms are required for complete epidermal function.6, 55 Loss of TP53 enhances HH driven BCCs by increased SMO expression, a potential p63 target.51 The mechanism by which TP53 affects p63-mediated potentiation of the HH pathway has not been elucidated. TP53 is frequently mutated in BCCs and mutated TP53 can affect TP63 function.56 How mutated TP53 might affect the HH/ΔNp63α crosstalk merits further exploration.
In conclusion, we propose a model for crosstalk between the HH and p63 signaling pathways in keratinocytes. We characterized ΔNp63α as both a transcriptional controller of SUFU and also a target of HH pathway overexpression. Thus, our study might help partially elucidate the complicated dual role that TP63 has in keratinocyte proliferation and differentiation.
Materials and Methods
Cell culture
The H1299 cell line was grown in RPMI medium, and ME-180 cells were grown in McCoys 5a media. HaCaT keratinocytes and the WT and TP63−/− MEFs (ER Flores) were grown in DMEM with glutamine. All the aforementioned media were supplemented with 10% fetal calf serum. PHKs were isolated and propagated as previously described.19 For experiments in which calcium concentrations were changed, KBM-2 media (Cambrex, East Rutherford, NJ, USA) was used with the addition of CaCl2 (Cambrex) to modulate calcium levels in the media. PHKs were placed in KBM-2 medium supplemented with the KGM-2 (Cambrex) growth bullets (comprises BPE, hEGF, insulin, hydrocortisone, epinephrine, transferring, and GA-1000) with 0.05 mM calcium at least 24 h before experiments after washing off the 3T3-J2 feeder cells. 3T3-J2 cells were cultivated in DMEM supplemented with 10% calf serum. Neonatal PHKs (HEKn) obtained from Invitrogen (Carlsbad, CA, USA) and used for protein expression studies were grown as previously described.25 Induction of differentiation of HaCaTs was carried out as previously described.29 Treatment with HH agonist 1.5 (Cellagen Tech, San Diego, CA, USA) was carried out as previously described.57
Transfections with plasmid DNA were carried out using Lipofectamine 2000 (Invitrogen) for H1299/ME-180 cells and Lipofectamine (Invitrogen) or Polyjet (Signagen, Rockville, MD, USA) for PHKs as per the manufacturer's protocols. siRNA transfections irrespective of cell line were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. Lentivirus production and subsequent transduction of PHKs with GLI1 (pLL3.7 GLI1) or control (pLL3.7 EGFP) were carried out as described in Kasper et al.58
Plasmid constructs
The human ΔNp63α expression vector (J Pietenpol, Vanderbilt, TN, USA), GLI reporter,59 GLI2 expression vector (F Aberger), and SUFU expression vector (R Toftgard) were gifts from the respective investigators. The 2.8-kb BCL2-LUC construct was generated as previously described19 and was cloned into the PGL3E luciferase vector (Promega, Madison, WI, USA).
Luciferase assays
Transfections with the promoter luciferase construct and a control renilla luciferase were carried out in triplicate for each experiment. Protein lysates were taken at indicated time points after transfection according to the prescribed protocol. Luciferase assays were carried out as per the manufacturer's instructions using the Dual Luciferase Assay kit (Promega), and luciferase readings were taken on a TD 20/20 luminometer (Turner Design, Sunnyvale, CA, USA). Luciferase readings were normalized to renilla luciferase as indicated. Data is presented as the mean of three independent experiments with error bars representing ±S.E.M.
EMSA and methylcellulose assays
The methylcellulose suspension assay was carried out as previously described.26 EMSAs with the GLI consensus-binding site oligonucleotides were carried out as previously described.19
Quantitative PCR
RNA extracted using the RNeasy Mini kit (Qiagen, Valencia, CA, USA) was used in synthesis of cDNA with reverse transcriptase (Invitrogen). qPCR reactions were set up in triplicate for each probe and condition using Taqman probes (Applied Biosystems, Carlsbad, CA, USA) or primers with SYBR green Mastermix (Applied Biosystems). Results were analyzed using the ΔΔCt method with target gene expression normalized to GAPD, HPRT, or β2 macroglobulin. Data is presented as the mean of the experiments with error bars representing±S.D. Details regarding individual probes and sequences are included in the Supplementary Materials.
Immunofluorescence and IHC
Dorsal skin from WT and ΔNp63 BG animals was dissected and fixed in 10% neutral-buffered formalin, dehydrated, paraffin embedded, and sectioned to 5 μm thickness. Slides were deparaffinized and rehydrated through a graded alcohol series. Antigen retrieval was performed by boiling the slides in a microwave for 20 min in antigen retrieval solution (10 mM sodium citrate, 0.05% Tween-20, pH 6.0). Next, slides were rinsed briefly in phosphate-buffered saline, circled with a PAP pen, and blocked in 5% bovine serum albumin, 0.1% Triton X-100 in phosphate-buffered saline for 1 h. Primary antibodies used at the indicated dilutions were SUFU (1 : 50) (Epitomics, Burlingame, CA, USA) and anti-p63 (clone 4A4; 1 : 50), (Santa Cruz Biotechnology, Dallas, TX, USA). Secondary antibodies used were anti-rabbit IgG Alexa 568 and anti-mouse IgG 488 (1 : 250) (Molecular Probes, Eugene, OR, USA). DAPI was used to stain nuclei. Reagents and protocols from the MOM Basic Kit (Vector Labs, Burlingame, CA, USA) were used. Epidermal tissue obtained from the GLI1 and SUFU TG mice used for IHC were formalin fixed and paraffin embedded. After deparaffinization, the slides were treated in Target Retrieval solution citrate buffer (pH 6.0) (Dako, Carpinteria, CA, USA) and heated in a pressure cooker for 30 min. Slides were processed and probed as previously described.60 Antibodies are listed in the Supplementary Methods.
Genetically engineered mice
All mice were generated in accordance with the guidelines prescribed by the respective institutions. The ΔNp63α BG mice were generated as described.31 Briefly, mouse oocytes from C3Hf/HeRos × C57BL/10 Rospd mice were microinjected with an HA-ΔNp63α DNA construct. PCR genotyping was used to identify the founder lines, which were then crossed with K5tTA mice. The SUFU WT and SUFU+/− mice have been previously described.45 Briefly, a pSufuΔexon1 neo-targeting vector was generated to replace exon1 of SUFU with the neocassette upon homologous recombination in embryonic stem cells. Clones transfected with stable integration of the targeting vector were then isolated. The selected embryonic stem cells were injected into blastoscysts from C57BL/6 mice, which were then implanted into B6CBAF1 recipient female mice. Backcrossing of the male chimeras was carried out with C57BL/6 mice. The GLI1 TG mice used in this study were generated by crossing TREGLI1 mice with K5tTA61 mice as described.36 Briefly, the full-length human GLI1 cDNA was cloned into the tetracycline responsive TRE/pBSA vector. After sequence verification this vector was then microinjeceted into the nuclei of fertilized (C57BL/6J × CBA) F2 oocytes. Transgene positivity was confirmed by PCR genotyping, and the selected mice were crossed with K5tTA mice.61
Microscopy
IHC images were acquired using a Nikon Eclipse 80i (10 × 40 magnification) fitted with a Nikon DS Fi1 color camera all controlled through Nikon Elements software (Nikon Instruments, Melville, NY, USA). Immunoflouresence images were acquired at × 40 magnification and captured using a Nikon digital camera attached to a Nikon FXA fluorescence microscope. Image analysis was performed with Adobe Illustrator software (Adobe Systems, San Jose, CA, USA).
Statistical analysis
Student's t-test was used to calculate significance between treatment groups, and a P-value <0.05 was considered significant.
Acknowledgments
This work was partially supported by NCI grant U01 CA105345. We acknowledge grant support from the Swedish Research Council (to ST) and Swedish Cancer Society (to RT), Estonia Research Council (grant no. 8932), and EMBO Installation grant (to VJ, Austrian Science Fund (project no. P20652) and Austrian Genome Project-Gen AU (to FA)). Also we acknowledge Ahn Hoang and J Havilland for help with IHC, and Henry Adams for advice and guidance in microscopy image acquisition.
Glossary
- HH
hedgehog
- BCC
basal cell carcinoma
- SUFU
suppressor of fused
- SHH
sonic hedgehog
- SMO
smoothened
- PTCH
patched
- TA
transactivation domain
- ΔN
amino-terminal truncated protein
- p63α
α-isoforms of ΔNp63 and TAp63
- PHK
primary human keratinocyte
- IHC
immunohistochemistry
- WT
wild type
- MEF
mouse embryonic fibroblast
- ΔNp63α BG
ΔNp63α bigenic mice
- TG
transgenic
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Fuchs E, Byrne C. The epidermis: rising to the surface. Curr Opin Genet Dev. 1994;4:725–736. doi: 10.1016/0959-437x(94)90140-x. [DOI] [PubMed] [Google Scholar]
- Nagarajan P, Romano RA, Sinha S. Transcriptional control of the differentiation program of interfollicular epidermal keratinocytes. Crit Rev Eukaryot Gene Expr. 2008;18:57–79. doi: 10.1615/critreveukargeneexpr.v18.i1.50. [DOI] [PubMed] [Google Scholar]
- Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8:743–754. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, et al. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- Ghali L, Wong ST, Green J, Tidman N, Quinn AG. Gli1 protein is expressed in basal cell carcinomas, outer root sheath keratinocytes and a subpopulation of mesenchymal cells in normal human skin. J Invest Dermatol. 1999;113:595–599. doi: 10.1046/j.1523-1747.1999.00729.x. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR, et al. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem. 2004;279:1197–1205. doi: 10.1074/jbc.M310589200. [DOI] [PubMed] [Google Scholar]
- Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP, et al. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- Spurgers KB, Chari NS, Bohnenstiehl NL, McDonnell TJ. Molecular mediators of cell death in multistep carcinogenesis: a path to targeted therapy. Cell Death Differ. 2006;13:1360–1370. doi: 10.1038/sj.cdd.4401986. [DOI] [PubMed] [Google Scholar]
- Polakowska RR, Piacentini M, Bartlett R, Goldsmith LA, Haake AR. Apoptosis in human skin development: morphogenesis, periderm, and stem cells. Dev Dyn. 1994;199:176–188. doi: 10.1002/aja.1001990303. [DOI] [PubMed] [Google Scholar]
- Alfandari J, Shnitman Magal S, Jackman A, Schlegel R, Gonen P, Sherman L. HPV16 E6 oncoprotein inhibits apoptosis induced during serum-calcium differentiation of foreskin human keratinocytes. Virology. 1999;257:383–396. doi: 10.1006/viro.1999.9675. [DOI] [PubMed] [Google Scholar]
- Carlsson H, Yhr M, Petersson S, Collins N, Polyak K, Enerback C. Psoriasin (S100A7) and calgranulin-B (S100A9) induction is dependent on reactive oxygen species and is downregulated by Bcl-2 and antioxidants. Cancer Biol Ther. 2005;4:998–1005. doi: 10.4161/cbt.4.9.1969. [DOI] [PubMed] [Google Scholar]
- Flores ER, Lambert PF. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagata Y, Aoki T, Kondo S. Detecting expression of keratins 8/18 in human HaCaT keratinocytes. J Dermatol Sci. 1999;19:139–143. doi: 10.1016/s0923-1811(98)00054-1. [DOI] [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, et al. New p63 targets in keratinocytes identified by a genome-wide approach. Embo J. 2006;25:5105–5116. doi: 10.1038/sj.emboj.7601375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Liu S, Sinha S. Abnormal hair follicle development and altered cell fate of follicular keratinocytes in transgenic mice expressing DeltaNp63alpha. Development. 2010;137:1431–1439. doi: 10.1242/dev.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Desai N, Wang X, Karhadkar SS, Reynon M, Abate-Shen C, et al. Roles for Hedgehog signaling in androgen production and prostate ductal morphogenesis. Dev Biol. 2004;267:387–398. doi: 10.1016/j.ydbio.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Chiang C, Swan RZ, Grachtchouk M, Bolinger M, Litingtung Y, Robertson EK, et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev Biol. 1999;205:1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Dassule HR, Karavanova I, Botchkarev VA, Li J, Danielian PS, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, et al. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M, Jaks V, Are A, Bergstrom A, Schwager A, Svard J, et al. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci USA. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta TM, Kommagani R, Yuan Z, Robbins DJ, Mercer CA, Kadakia MP. p63 overexpression induces the expression of Sonic Hedgehog. Mol Cancer Res. 2006;4:759–768. doi: 10.1158/1541-7786.MCR-05-0149. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky M, Zhang XM, Bottega S, Guicherit O, Wichterle H, Dudek H, et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne T, Brunner HG, van Bokhoven H. p63-associated disorders. Cell Cycle. 2007;6:262–268. doi: 10.4161/cc.6.3.3796. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. p63 in skin appendage development. Cell Cycle. 2007;6:285–290. doi: 10.4161/cc.6.3.3798. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Visinoni AF, Lisboa-Costa T, Pagnan NA, Chautard-Freire-Maia EA. Ectodermal dysplasias: clinical and molecular review. Am J Med Genet A. 2009;149A:1980–2002. doi: 10.1002/ajmg.a.32864. [DOI] [PubMed] [Google Scholar]
- Lippens S, Denecker G, Ovaere P, Vandenabeele P, Declercq W. Death penalty for keratinocytes: apoptosis versus cornification. Cell Death Differ. 2005;12 (Suppl 2:1497–1508. doi: 10.1038/sj.cdd.4401722. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villanueva J, Greenhalgh D, Wang XJ, Bundman D, Cho S, Delehedde M, et al. Human keratin-1.bcl-2 transgenic mice aberrantly express keratin 6, exhibit reduced sensitivity to keratinocyte cell death induction, and are susceptible to skin tumor formation. Oncogene. 1998;16:853–863. doi: 10.1038/sj.onc.1201610. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Zhang X, Liu L, Hui CC, Mainprize TG, Scherer SW, et al. Failure of a medulloblastoma-derived mutant of SUFU to suppress WNT signaling. Oncogene. 2004;23:4577–4583. doi: 10.1038/sj.onc.1207605. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Negishi I, Kuida K, Sawa H, Loh DY. Targeted disruption of Bcl-2 alpha beta in mice: occurrence of gray hair, polycystic kidney disease, and lymphocytopenia. Proc Natl Acad Sci USA. 1994;91:3700–3704. doi: 10.1073/pnas.91.9.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Weinberg WC. p63: defining roles in morphogenesis, homeostasis, and neoplasia of the epidermis. Mol Carcinog. 2007;46:716–724. doi: 10.1002/mc.20337. [DOI] [PubMed] [Google Scholar]
- Brandner S. Nanog, Gli, and p53: a new network of stemness in development and cancer. Embo J. 2010;29:2475–2476. doi: 10.1038/emboj.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Alman B. Protecting the hedgerow: p53 and hedgehog pathway interactions. Cell Cycle. 2010;9:506–511. doi: 10.4161/cc.9.3.10552. [DOI] [PubMed] [Google Scholar]
- Wang GY, Wang J, Mancianti ML, Epstein EH., Jr Basal cell carcinomas arise from hair follicle stem cells in Ptch1(+/−) mice. Cancer Cell. 2011;19:114–124. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon F. p63 and the epithelial stem cell: more than status quo. Genes Dev. 2004;18:465–469. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- King KE, Ponnamperuma RM, Gerdes MJ, Tokino T, Yamashita T, Baker CC, et al. Unique domain functions of p63 isotypes that differentially regulate distinct aspects of epidermal homeostasis. Carcinogenesis. 2006;27:53–63. doi: 10.1093/carcin/bgi200. [DOI] [PubMed] [Google Scholar]
- Suzuki D, Senoo M. Increased p63 phosphorylation marks early transition of epidermal stem cells to progenitors. J Invest Dermatol. 2012;132:2461–2464. doi: 10.1038/jid.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M, Regl G, Eichberger T, Frischauf AM, Aberger F. Efficient manipulation of Hedgehog/GLI signaling using retroviral expression systems. Methods Mol Biol. 2007;397:67–78. doi: 10.1007/978-1-59745-516-9_6. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Assikis VJ, Do KA, Wen S, Wang X, Cho-Vega JH, Brisbay S, et al. Clinical and biomarker correlates of androgen-independent, locally aggressive prostate cancer with limited metastatic potential. Clin Cancer Res. 2004;10:6770–6778. doi: 10.1158/1078-0432.CCR-04-0275. [DOI] [PubMed] [Google Scholar]
- Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.