Abstract
Transfer RNA (tRNA) structure, modifications and functions are evolutionary and established in bacteria, archaea and eukaryotes. Typically the tRNA modifications are indispensable for its stability and are required for decoding the mRNA into amino acids for protein synthesis. A conserved methylation has been located on the anticodon loop specifically at the 37th position and it is next to the anticodon bases. This modification is called as m1G37 and it is catalyzed by tRNA (m1G37) methyltransferase (TrmD). It is deciphered that G37 positions occur on few additional amino acids specific tRNA subsets in bacteria. Furthermore, Archaea and Eukaryotes have more number of tRNA subsets which contains G37 position next to the anticodon and the G residue are located at different positions such as G36, G37, G38, 39, and G40. In eight bacterial species, G (guanosine) residues are presents at the 37th and 38th position except three tRNA subsets having G residues at 36th and 39th positions. Therefore we propose that m1G37 modification may be feasible at 36th, 37th, 38th, 39th and 40th positions next to the anticodon of tRNAs. Collectively, methylation at G residues close to the anticodon may be possible at different positions and without restriction of anticodon 3rd base A, C, U or G.
Keywords: tRNAs, Codon, Anticodon, m1G modification
Background
tRNA is an intermediate molecule and it is a class of small RNA being present in Bacteria, Archaea and Eukaryota [1, 2]. Generally it consists of 73 to 93 RNA nucleotides which are involved in the translation process along with the complex of ribosome, mRNA, tRNA with amino acids and co-factors involved in the synthesis of proteins [3]. It has special segments such as amino acid attachment site, TCC loop; variable loop, anticodon loop and D loop thereby forming a tertiary structure designated in the form of a cloverleaf model [4, 5, 6]. In these segments, several modifications occur resulting in posttranscriptional modification of tRNA which influences the tRNA folding, stability and accuracy of translation process [7, 8]. In 1960s, an RNA modification enzyme was first identified by Borek and coworkers (1960) as the tRNA m5U54 methyltransferase and now it is being called as TrmA in Bacteria and Trm2 in Eukarya [9, 10]. In tRNAs, more than 100 modifications have been identified particularly in the highly conserved anticodon stem and loop (ASL) domain base at position 37 which is next to the anticodon site and it is highly prone to modification by tRNA m1G37 methyltransferase (TrmD) which results an addition of methyl group (CH3) to guanosine (G) base next to the anticodon [7, 11, 12]. A modified 1-methylguanosine (m1G37) at position 37 plays a vital role in tRNA function and it acts as a classical frameshift suppressor that leads to the accuracy of translation [13]. The modified nucleosides m1G37 position of tRNA are present in all organisms including the smallest organisms like the Mycoplasma genitalium and this modification occurs in chloroplast and mitochondrial tRNAs as well [7]. The wobble position next to the anticodon of 37th position is frequently modified with a plethora of diverse nucleotides being found at those positions.
A modified m1G37 enhances proofreading in the ribosome and aminoacylated tRNA selection in a tRNA dependent manner [7, 14]. All these suggest that m1G37 had been present in the tRNA of the primitive organisms and it is a part of the minimal set of genes required for life that existed before emergence of these three domains namely the Bacteria, Archaea and eukaryota [15, 16].
tRNA binding protein - TrmD:
TrmD is one of the RNA modification enzymes which catalyze the transfer of a methyl group from S-adenosyl methionine (AdoMet) to the N1 atom of guanosine base at position 37 next to the anticodon of tRNA subsets. TrmD is a member of the TrmH methyltransferase family [6]. Most of this modification occurs in selected amino acid tRNAs for example, tRNALeu; anticodon CAG, GAG, UAG, tRNAPro; anticodon CCG, GGG, UGG and tRNAArg; anticodon CCG [17, 18]. An in vivo method has proven that the three modifications such as pseudouridine (C), 1-methylguanosine (m1G) and 2-methylthio-N6-(4- hydroxyisopentenyl) adenosine (ms2io6A) promote the selection of aa-tRNAs for translation process. Hence, it is believed that one of the modification m1G37 strongly influences selection of tRNAPro and tRNAArg but not much on the selection of tRNALeu such as specific tRNAs [14]. Deficiency of m1G37 in tRNAPro and tRNAArg has reduced the selection of A-site on ribosome-mRNA complex on the translation process [13]. The first crystal structure of Haemophilus influenza TrmD was introduced by Ann et al., (2003) which modifies the m1G37 position in tRNAs. The tRNA anticodon stem loop is possible to make a tight fit on the active site of TrmD on both positions at G36 and G37 [10].
Bacterial TrmD enzyme recognizes the tRNA specifically G36pG37 base nucleotides because at G36th position it might be involved in stabilizing the enzyme-tRNA complex [19]. Aquifex aeolicus TrmD enzyme recognizes and catalyzes the tRNA at anticodon site in the presence of both G36pG37 and A36pG37 nucleotides (20). Mostly purine bases (G or A) at position 37 in tRNA stabilize the codon-anticodon interactions in ribosome attachment. Sequence analysis of more than 3000 tRNA genes from various organisms confirmed that purine base A is found at position 37 in ~80% and purine base G is found in ~20% [20, 21]. TrmD enzyme binds to tRNA subsets even in the absence of GpG residues at positions 36 and 37. This enzyme-tRNA binding complex might be stabilizing the tRNAs. Recently, bacterial m1G37 has been found in tRNAs with G or A base at position 36 [22]. In Archaea and eukaryotes, tRNA (m1G37) methyltransferase called as aTrm5 and eTrm5 respectively, are responsible for the m1G37 modification in all tRNAs [23, 24]. TrmD methylates G residues are nearer to the anticodon in Bacterial, Archaea and Eukaryotes but TrmD, aTrm5 and eTrm5 structures, active sites and types of methylation are quite different among them [25]. In bacteria, m1G37 has been found in the amino acid specific tRNAs which are having anticodons, on the base site, either G36 or A36. For instance, in tRNALeu NAG, tRNAPro NGG, tRNAHis GUG, tRNAArg NCGand tRNAPhe GAA anticodons, the third bases are G36 and A36. In Eukaryotes and Archaea, m1G37 occurs in broad tRNAs subsets with corresponding anticodon base of G36 and A36, tRNALeuNAG, tRNAProNGG, tRNAHisGUG, tRNAGlnNUG, tRNAArgNCG and tRNAPheGAA, tRNASer GGA/UGA/CGA, tRNATyrGUA, tRNACysGCA, tRNATrpCCA, and tRNALeuNAA. Hence, these findings suggest that tRNA anticodon positions at A36 or G36 promotes the m1G37 in all organisms. When there occurs a defect in m1G37 modification, a frameshift mutation may occur which leads to retarded growth in the organisms [23].
Methodology
For the three domains, randomly 11 organisms were selected and their tRNA gene sequences were retrieved from Genomic tRNA database in the FASTA format [26]. BioEdit software has been used for multiple sequence alignments and for identification of the Guanosine base, next to the anticodon in tRNA susbets at different positions such as at 36th, 37th, 38th, 39th and 40th.
Results
Guanosine base next to the anticodon:
Previous studies reported that guanosine residue is being present at 37th position next to the anticodon at three different tRNA susbets such as tRNAArg, tRNAPro and tRNALeu [18, 27]. Contradictory to this fact, organisms belonging to Archaea and eukayotes have been shown to contain G residue at different positions (36, 37, 38, 39 and 40) next to the anticodon on various tRNA subsets (tRNAArg, tRNALeu, tRNAPro, tRNAHis, tRNAGln, tRNAPhe, tRNACys, tRNASer, tRNATrp, tRNATry and tRNAGlu) as shown in Table 1 (see supplementary material) and thus the tRNAs codons do not show consistent positions in all tRNA subsets. Even at different positions of codons in tRNAs, the G residues are present next to the anticodon Table 1. E. coli K12 and H. influenzae have only 3 types of tRNA subsets (tRNAArg, tRNAPro and tRNALeu) and both organisms have G residue at 38 positions Table 1. Besides, M. tuberculosis H37Rv, S. aureus and S. pneumoniae D39 revealed 6 types of tRNAs, tRNAArg, tRNAPro and tRNALeu, tRNAHis, tRNAGln and tRNAPhe are having G residue at 37th and 38th positions. A. phagocytophilum has 5 types of tRNA subsets (tRNAArg, tRNAPro, tRNALeu, tRNAHis, and tRNAVal) and its tRNAs have G residue at 37, 38 and 39 positions. Similarly A. aeolicus has 4 types of tRNAs (tRNAArg, tRNAPro, tRNALeu and tRNAHis) and it is holding G residue at the same positions. On the contrary B. Houston-1 is having tRNAs (tRNAGln, tRNAHis, tRNAPro, tRNALeu) with G residues at 36 and 38 positions. Analysis of eight bacterial species, revealed the conserved three tRNA subsets (tRNAArg, tRNAPro and tRNALeu) and to some extent other tRNA sequences also contain G residue close to the anticodon and may be susceptible for m1G modification Table 1. The M. jannaschii and H. sapiens hg19 tRNAs are having numerous tRNA subsets (tRNAArg, tRNALeu, tRNAPro, tRNAHis, tRNAVal, tRNAGln, tRNAPhe, tRNACys, tRNASer, tRNATrp, tRNATry and tRNAGlu) with their G residues at 36, 37, 38, 39 and 40th positions. However, M. jannaschii tRNACys is the only tRNA having G36 position present next to the anticodon. The hg19 of H. sapiens does not have tRNA with G residue at 40th position Table 1. Furthermore, a schematic structure (Figure 1) has been depicted for m1G37 modification at various positions close to the anticodon of tRNAs. The proposed m1G37 modification in tRNAs at G36pG37 positions is shown in Figure 1A. Without m1G37, modification in tRNAs occurs by +1 frameshift mutation in mRNA codons (Figure 1B). Thus the, m1G37 modification is possibly found in tRNAs having A36pG37 segments (Figure 1C), Our analysis of tRNA sequence of these structures confirm that m1G38, m1G39 and m1G40 modifications are possibly present in tRNAs in their respective sites for their functional significances (Figure 1D, E & F).
Figure 1.
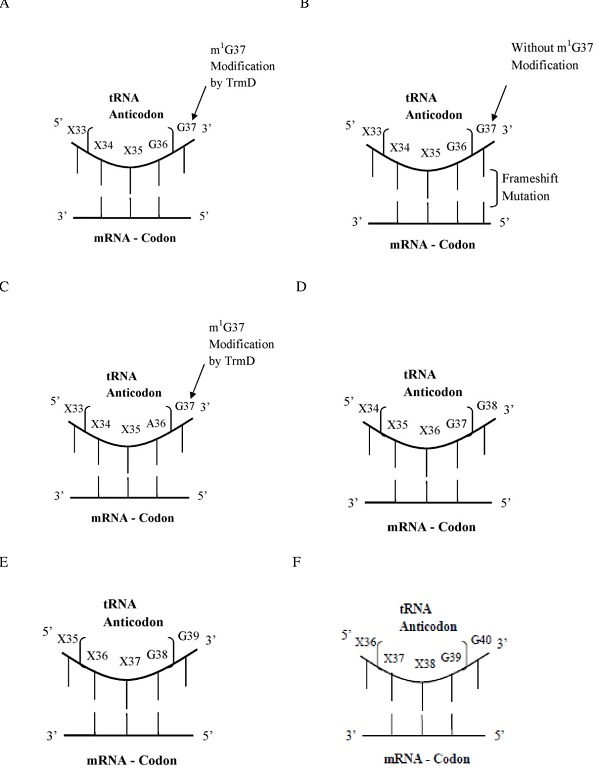
Schematics of m1G modifications by TrmD at diverse positions of guanosine next to the anticodon in tRNAs.
Distinct tRNAs codons and wobble position:
The various types of amino acid specific tRNAs and its GpG sequences have been listed in Table 1. Eventually the GpG sequence position is defined as the anticodon third residue (wobble position) with NNG carrying the guanosine (N with any one of four bases A, C, G, U) or G. This structural position (GpG) is the hallmark of methylation site being present at G37th position next to the anticodon [10, 28]. Escherichia coli K12 and Haemophilus influenzae have GpG sequences in the anticodon position in the selected tRNAs subsets as in tRNAArg, tRNAPro and tRNALeu. Furthermore Anaplasma phagocytophilum HZ, Aquifex aeolicus, Bartonella henselae Houston-1, Mycobacterium tuberculosis H37Rv, Staphylococcus aureus MRSA252 and Streptococcus pneumoniae have GpG sequence on the additional tRNAs such as tRNAArg, tRNAPro and tRNALeu, tRNAHis, tRNAVal, tRNAGln, tRNAPhe Table 1. Methanococcus jannaschii and Homo sapiens hg19 has GpG sequences on various types of tRNAs has been listed in the Table 1. Consequently it is proposed that guanosine (G) residue can possibly be present next to the anticodon, at the wobble position having G, C, A or U. This structural sequence might be present on different nucleotide numbers such as G35pG36, G36pG37, G37pG38, G38pG39, G39pG40 and G40pG41. Staphylococcus aureus MRSA252 and Streptococcus pneumoniae D39 tRNAPhe have GpG and ApG sequences next to the anticodon. Similarly Archaea Methanococcus jannaschii is having GpG, ApG, CpG structure sequences in the tRNAs. Eukaryote, Homo sapiens hg19 tRNA holds GpG, ApG, UpG residues having positioned next to the anticodon in tRNA subsets. However, in bacterial population (8 species) there is a lack of A, C, U residues at the third position of anticodon in all tRNA susbets. Conversely in Archaea like Methanococcus jannaschii the m1G may possibly be in tRNAs (tRNACysGCA, tRNATryGTA, tRNASerGGA, tRNASerTGA, tRNATrpCCA, tRNALeu TAG/TAA) at A35/37/38/39th positions and tRNAGluTTC at C39 (third base of anticodon) Table 1. Similarly in eukaryotes the m1G is found in tRNAs (tRNACysGCA, tRNATrpCCA, tRNATyrGTA, tRNASer AGA, tRNACys GCA,tRNALeu TAA, tRNAPheGAA, tRNATyrATA, tRNASerAGA, tRNATrpCCA) at the positions of A35/36/37/38 and tRNAArgCCT at C35.
Discussion
Computational analysis of tRNAs in three domains disclosed that m1G37 modification might occur at different positions (36th, 37th, 38th, 39th, or 40th) and the anticodon position is not constant in all the tRNAs. The above modification restricts its location next to the anticodon. Furthermore the m1G37 modification can occur in any of the four possible nuceltodies (G, A, C, U) at the position 36 (anticodon). In the case of E. coli and H. influenza, it has been reported that tRNAs m1G37 modification of G residues occurs at 37th position [22]. The study also revealed that the Escherichia coli K12 and Haemophilus influenzae tRNAs (tRNAArg, tRNAPro and tRNALeu) are having the G residues at 38th position, thus confirming that the modification would be possible at G38 also. Initially it was also believed that all three domains the m1G37 are present in the three tRNA subsets like tRNAArg, tRNAPro and tRNALeu [15, 27]. Analysis of tRNA squences in bacteria, archaea and eukaryota reveals that m1G modification may occur in other amino acid specific tRNA subsets also. Sprinzl et al., (2005) and Goto Ito et al., (2008) reported that A36 base at anticodon is possible in all three domains [21, 23]. For instance, in bacteria, tRNAPheGAA with A36 has been identified and in the same way in archaea, m1G is found in tRNACysGCA with A36. Similarly in eukaryotes the m1G may exist in tRNAPheGAA, tRNASerGGA/UGA/CGA, tRNATyrGUA, tRNACysGCA, tRNATrpGCA, tRNALeuNAA. Besides, the eukayotic tRNAAspGUC contains m1G37 with C base at third position of anticodon. As per the present study, it is evident that in bacterial populations the m1G37 may occur in tRNAs with G residue at 35, 36, 37 or 38 positions. Generally the m1G preferably occurs on the GpG sequence and this implies that the G residue usually occurs at third base of anticodon. In bacterial populations it has been found that the tRNAs is holding GpG sequence in diverse positions Table 1. In Archaea like Methanococcus jannaschii the m1G might as well be in tRNAs having GpG, ApG and CpG sequences. In eukayotes, the m1G might exist in tRNAs containing GpG, ApG or UpG. The sequence UpG is not mentioned in previous report and it is unique as its (m1G37) mechanism can possibly be in tRNAArgCCT (Homo sapiens hg19) at U35 position. Therefore it may be concluded that in Archaea and eukaryotes, the m1G modification might not be restricted to nucleotides G, A, C and U which may be present on the third base of anticodon. Furthermore, the position of m1G can occur at different positions of tRNAs. Nevertheless in Archaea and eukayotes, the m1G might possibly be without restriction for the three tRNA subtypes (tRNAPro, tRNALeu, tRNAArg).
Supplementary material
Footnotes
Citation:Srinivasan et al, Bioinformation 9(9): 466-470 (2013)
References
- 1.Targanski I, Cherkasova RNA. 2008;14:1095. doi: 10.1261/rna.896108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marck C, Grosjean H. RNA. 2002;8:1189. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crick FH, et al. J Mol Biol. 1968;38:367. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 4.Djelloul M, Denise A. RNA. 2008;14:2489. doi: 10.1261/rna.1061108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazantsev AV, Pace NR. Nat Rev Microbiol. 2006;4:729. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi K, Nureki O. Mol Cells. 2005;30:157. [PubMed] [Google Scholar]
- 7.Gustilo EM, et al. Curr Opin Microbiol. 2008;11:134. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipowsky G, et al. RNA. 1999;5:539. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson JD, Crick FH. Nature. 1953;25:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 10.Watson JD, Crick FH. Nature. 1953;171:964. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 11.Bouadloun F, et al. J Bacteriol. 1986;166:1022. doi: 10.1128/jb.166.3.1022-1027.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JN, Bjork GR. J Bacteriol. 1995;177:6593. doi: 10.1128/jb.177.22.6593-6600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjork GR, et al. FEBS Lett. 1999;452:47. doi: 10.1016/s0014-5793(99)00528-1. [DOI] [PubMed] [Google Scholar]
- 14.Li J, et al. J Mol Biol. 1997;271:209. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- 15.Bjork GR, et al. EMBO J. 2001;20:231. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam JD, et al. Biochemistry. 2011;50:9886. doi: 10.1021/bi2012004. [DOI] [PubMed] [Google Scholar]
- 17.Qian Q, Bjork GR. J Mol Biol. 1997;266:283. doi: 10.1006/jmbi.1996.0789. [DOI] [PubMed] [Google Scholar]
- 18.Li JN, Bjork RNA. 1999;5:395. doi: 10.1017/s1355838299980834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brule H, et al. Biochemistry. 2004;43:9243. doi: 10.1021/bi049671q. [DOI] [PubMed] [Google Scholar]
- 20.Konevega AL, et al. RNA. 2004;10:90. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhling F, et al. Nucleic Acids Res. 2009;37:D159. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn HJ, et al. EMBO J. 2003;22:2593. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto-Ito S, et al. Proteins. 2008;72:1274. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 24.Goto-Ito S, et al. Nat Struct Mol Biol. 2009;16:1109. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan T, Sudarsanam D. Journal of Bioscience & Technology. 2011;6:416. [Google Scholar]
- 26.Lowe TM, Eddy SR. Nucleic Acids Res. 1997;25:955. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjork GR, et al. RNA. 2007;13:1245. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda H, et al. Genes Cells. 2006;11:1353. doi: 10.1111/j.1365-2443.2006.01022.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


