Abstract
Among New Mexican Hispanic women, breast cancer is detected at a more advanced stage than compared to Non-Hispanic White women. One central factor that has been little studied is the role of critical cytokines. We genotyped incident breast cancer cases and their age-, gender- and smoking-matched controls (N = 40 matched pairs) for 25 single nucleotide polymorphisms (SNPs) in cytokine genes. We measured corresponding serum cytokine levels as well. Five cytokines (IL-1β, IL-5, TNF-α, IL-6 and IL-2) were significantly associated with disease and based on their serum levels, concentrations were higher in the cases than in the controls. Disease odds ratios corresponding to one standard deviation change in log-transformed concentrations of these cytokines were 18.87, 4.10, 3.61, 3.27 and 2.52. Three most statistically significant SNPs were rs2069705, located in the promoter region of the interferon gamma gene (INF-γ); rs2243248, in the promoter of IL-4 (rs2243248); and rs1800925, in the promoter of the IL-13 gene. Increased serum cytokine levels at diagnosis are indicative for immunological alterations and possibly related to genetic susceptibility markers as well. These findings might guide us to understand the presence of SNPs in cytokine genes and serum concentrations among breast cancer patients and potentially in other cancers.
Keywords: Breast cancer, Cytokine, Th1/Th2 balance, Serum cytokine level
1. Introduction
Breast cancer is an important public health problem in New Mexico (NM). It is the most common cancer diagnosed among New Mexican women, and among NM Hispanic women, breast cancer is diagnosed at a more advanced stage and mortality rate is higher compared to Non-Hispanic White women [1]. As New Mexico has the highest proportion of Hispanics of any state in the US (42%), breast cancer survival creates a real challenge for our health care and public health system.
While there are potentially problems with access to care and reduced breast cancer screening among Hispanic women, there may also be biological factors that are important contributors to their poorer prognosis. One biological factor that has been little studied is the role of the immune system, specifically pro- and anti-inflammatory cytokines.
Cytokines are small molecular-weight, regulatory proteins of the immune system secreted by active immune cells (mostly T cells) and other cell types (epithelial cells, endothelial cells, fibroblasts, keratinocytes, etc.) which are part of the immune surveillance system [2–6].
In this study, we attempted to find useful single nucleotide polymorphisms (SNPs) from several cytokine genes that are associated with an increase or decrease in cytokine serum levels among breast cancer cases when compared to healthy controls. Our goal was to determine whether these relationships would be valuable predictors for diagnosis, therapy and treatment responses among breast cancer patients and potentially in other cancers as well [7–9].
Previous studies have explored those common single nucleotide polymorphisms (SNPs) in Th1 and Th2 cytokine genes that can alter gene expression, modulate the balance between Th1/Th2 responsiveness, and influence the susceptibility for cancer [10,11]. Moreover, much is yet to be discovered about the relationship between cytokines and breast cancer. Some data indicates that the presence of variants in IL-6 – 174 promoter gene has been associated with improved disease-free survival of breast cancer patients [12]. One study found that a variant allele in TGF-β1 gene polymorphisms (codon 25 CGG > CCG, rs1800471) in sporadic breast cancer patients was higher among cases than controls [13]; however this was not found in another population [14]. A small study (N = 11 patients) reported that three cytokine levels were associated with Th1/Th2 imbalances among breast cancer patients [15]. In our work we examined the role and interactions of 25 single nucleotide polymorphisms of cytokine genes and their corresponding serum cytokine levels in their association with breast cancer among New Mexican women newly diagnosed with breast cancer.
2. Materials and methods
This project utilized resources of the University of New Mexico (UNM) Expanded Breast Cancer Registry and Tissue Repository (EBCR), which has been collecting specimens and data from breast cancer patients seen at the UNM Cancer Center clinics since February 2006. Participants consented to the donation of serum, plasma and DNA from whole blood, along with DNA extracted from buccal cells and left-over tissue from surgery; a short questionnaire that evaluates the risk factors commonly associated with breast cancer, detailed clinical information and medical record follow-up. Sixty percent of our patients (24 out of 40 women) donated their biological samples at recruitment to our institutional biobanking project. Forty percent of the patients had undergone surgery prior to their blood draw. The average time period to blood donation was 2.3 weeks after surgery, but these women did not start any therapy before their samples have been collected.
2.1. Study population
Cases were selected from the EBCR who were over the age of 18, and willing to provide informed consent, blood samples and a short risk factor questionnaire.
Controls were recruited from advertisements on the UNM campus and included age- and ethnicity- and smoking history-matched women without breast cancer or other malignancies, willing to provide informed consent, blood samples and a short risk factor questionnaire. This project was approved by the UNM Health Science Center Human Research Review Committee (HRRC#:06-362). We summarized population and tumor characteristics in Table 1.
Table 1.
Characteristics of cases (n = 40) and controls (n = 40) in the UNM-EBCR cytokine project.
| Characteristics | Cases Mean (SD) | Controls Mean (SD) |
|---|---|---|
| Age at blood draw | 49.2 (13.6) | 50.2 (10.8) |
| Ethnicity | N (%) | N (%) |
| White | 31 (77.5) | 29 (72.5) |
| Hispanic | 7 (17.5) | 8 (20.0) |
| African-American | 1 (2.5) | 1 (2.5) |
| Native American | 1 (2.5) | 1 (2.5) |
| Refused | 0 (0) | 1 (2.5) |
| Stage at diagnosis | ||
| Stage 0 (DCIS) | 2 (5) | – |
| Stage I | 16 (40) | – |
| Stage II-A | 11 (27.5) | – |
| Stage II-B | 4 (10) | – |
| Stage III-A | 6 (15) | – |
| Stage IV | 1 (2.5) | – |
| Lymph node involvement | ||
| Yes | 10 (25) | – |
| No | 30 (75) | – |
| Cigarette smoking status | ||
| Non-smoker | 38 (95.0) | 36 (90.0) |
2.2. Biological sample collection
2.2.1. Blood collection procedure
During the interview, participants were asked to donate two 9 ml tubes of blood samples. Blood samples were centrifuged for 15 min at 1300g immediately after clotting (at least 30 min but no longer than 60 min). Serum samples were flash-frozen in liquid nitrogen and stored at –80 °C until analysis where they were used to measure cytokine concentrations.
2.2.2. DNA extraction
One Vacutainer tube containing EDTA was used for DNA extraction using Qiagen QIAamp DNA Blood Maxi Kit (Qiagen Inc., Valencia, CA) for up to 350 μg of genomic DNA. Nanodrop was used to measure optical density at 260 and 280 nm and assess DNA yield and quality. The samples were divided into several aliquots, labeled and stored at –80 °C. Average yields using this technique were 125 μg of DNA.
2.3. Laboratory assays
2.3.1. Serum based assays
Serum based assays were carried out using Human Cytokine Antibody Arrays (FAST®Quant Human Th1/Th2 kit, Whatman Inc. Florham Park, NJ) to measure Th1 and Th2 cytokine levels. This assay can determine concentrations of nine cytokines (IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-13, TNF-α and IFN-γ) from as little as 70 μl of sample per patient. This new diagnostic tool conserves valuable sera of patients, limiting required serum use to the minimum. In addition, we measured additional cytokine using monoclonal sandwich ELISA methods to analyze IL-8, IL-12, IL-17, TGF-β1 concentrations (R&D System Inc., Minneapolis, MN). One hundred microliters of serum per participant was used.
2.3.2. DNA genotyping assays
DNA genotyping assays were chosen based on extensive literature review and a candidate gene approach. Because of the absence of a Hispanic/Latino genotype database, the HapMap Central European population database (CEU) was chosen to provide genotype frequencies. The CEU frequencies were used to not only select particular SNPs for the study, but also to serve as a reference to measure the differences between the European and “New Mexican” genotype frequencies. Most of the SNPs selected for this study are promoter SNPs, or single base differences that could change the transcription level of the cytokines by a number of molecular mechanisms. SNPs have been shown to affect the binding affinities for transcription factors that specifically recognize DNA sequences in the promoter [16,17]. This in turn can affect the rates of transcription and ultimately the levels of the cytokine proteins. A primary reason the promoter SNPs were selected as the main class of SNPs is that the serum cytokine levels measured in this study might be a direct consequence of the RNA transcription levels. If these RNA levels are associated with specific SNPs in the promoters of the cytokines, then the genotype of the SNP could predict the serum cytokine levels. After selecting the SNPs, used HapMap (http://hapmap.org/cgi-perl/gbrowse/hapmap), to confirm population frequencies. Genotyping was performed using Applied Biosystem Inc. Taqman assays. Table 2 summarizes information of the SNPs for this project, their identification numbers in NCBI system (rs numbers), their location on chromosomes, their published variant frequencies for Caucasians and the immunological pathways involved.
Table 2.
Summary information on single nucleotide polymorphisms used in the UNM-EBCR cytokine project.
| SNP | dbSNP ID | Chromosome location | HapMap frequency data |
Pathway | |||||
|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Frequencies | ||||||||
| IFN-γ –1615 C/T | rs2069705 | 12 | CC | CT | TT | 0.2 | 0.433 | 0.367 | Th1 |
| IL-12 –980 A/G | rs2243113 | 3 | AA | AG | GG | 0.983 | 0.017 | 0 | Th1 |
| IL-1α –889 C/T | rs1800587 | 2 | CC | CT | TT | 0.417 | 0.5 | 0.083 | Th1 |
| IL-1β +3954 C/T | rs1143634 | 2 | CC | CT | TT | 0.589 | 0.375 | 0.036 | Th1 |
| IL-1β –31 C/T | rs1143627 | 2 | CC | CT | TT | 0.133 | 0.467 | 0.4 | Th1 |
| IL-1β –511 A/G | rs16944 | 2 | AA | AG | GG | 0.145 | 0.4 | 0.455 | Th1 |
| IL-2 –384 G/T | rs2069762 | 4 | GG | GT | TT | 0.042 | 0.5 | 0.458 | Th1 |
| IL-6 –597 A/G | rs1800797 | 7 | AA | AG | GG | 0.2 | 0.55 | 0.25 | Th2 |
| IL-8 –204 C/T | rs2227306 | 4 | CC | CT | TT | 0.476 | 0.429 | 0.095 | Th1 |
| IL-8 –251 A/T | rs4073 | 4 | AA | AT | TT | 0.136 | 0.409 | 0.455 | Th1 |
| TNF-α –238 A/G | rs361525 | 6 | AA | AG | GG | 0 | 0.067 | 0.933 | Th1 |
| TNF-α –308 A/G | rs1800629 | 6 | AA | AG | GG | 0 | 0.433 | 0.567 | Th1 |
| TNF-β +252 C/T | rs909253 | 6 | CC | CT | TT | 0.083 | 0.55 | 0.367 | Th1 |
| IL-10 –1082 A/G | rs1800896 | 1 | AA | AG | GG | 0.217 | 0.5 | 0.283 | Th2 |
| IL-10 –3575 A/T | rs1800890 | 1 | AA | AT | TT | 0.137 | 0.49 | 0.373 | Th2 |
| IL-10 –592 A/C | rs1800872 | 1 | AA | AC | CC | 0.087 | 0.304 | 0.609 | Th2 |
| IL-10 –819 C/T | rs1800871 | 1 | CC | CT | TT | 0.636 | 0.273 | 0.091 | Th2 |
| IL-13 +98 C/T | rs20541 | 5 | CC | CT | TT | 0.583 | 0.367 | 0.05 | Th2 |
| IL-13 –1055 C/T | rs1800925 | 5 | CC | CT | TT | 0.653 | 0.304 | 0.043 | Th2 |
| IL-4 –1098 G/T | rs2243248 | 5 | GG | GT | TT | 0 | 0.183 | 0.817 | Th2 |
| IL-4 –524 C/T | rs2243250 | 5 | CC | CT | TT | 0.707 | 0.259 | 0.034 | Th2 |
| IL-5 –745 C/T | rs2069812 | 5 | CC | CT | TT | 0.458 | 0.423 | 0.119 | Th2 |
| IL-17 +18 A/G | rs3819025 | 6 | AA | AG | GG | 0.017 | 0.083 | 0.9 | Th17 |
| TGF-β1 +29 C/T | rs1982073 | 19 | CC | CT | TT | 0.258 | 0.581 | 0.161 | Immunosuppressive |
| TGF-β1 –509 C/T | rs1800469 | 19 | CC | CT | TT | 0.517 | 0.345 | 0.138 | Immunosuppressive |
2.4. Statistical analysis
The comparison of allele frequencies between our population and HapMap Non-Hispanic White population was performed using a Chi-square test. The difference in each of the cytokine levels between matched cases and controls (equivalently, the effect of disease status on each of the cytokine levels) was examined using the paired t-test and the associations of the cytokine levels with the disease status were examined using the conditional logistic regression. The effects of disease status and the genotypes of each of the SNPs on cytokine levels were further analyzed using general linear models in which each pair of matched case and control representing one of the randomized blocks. This technique greatly enhances not only the power of the study but also improved our ability to understand our findings.
This analysis also provides the evidence for the association between each of the SNP and corresponding cytokine. In analyzing the association of SNP genotype with disease status, binary groups for each of the SNP genotypes were created by combining the heterozygous genotype (e.g. AB) with one of the two possible homozygous genotypes (e.g. AA or BB). As it was not clear whether each SNP had a dominant, recessive, or no effect, for each SNP, heterozygotes were pooled with the one homozygous genotypes that minimize p-value of the test for the association between the binary genotype groups and disease status. Conditional logistic regression was used to examine whether the genotype of each SNP is predictive of disease status. We matched the cases and controls by variables (i.e., smoking status, ethnicity and age) that potentially influence serum cytokine levels, decreasing the likelihood of finding false positive associations by chance. The cytokine levels were log transformed before the statistical tests were performed. Because of the exploratory nature of our study, an alpha of 0.05 was used to decide on statistical significance despite performing multiple comparisons. All statistical analyses utilized R version 2.8.0 statistical software [18,19].
3. Results
3.1. Serum cytokine levels and disease
Of the 13 cytokines observed, five were significantly associated with disease status (paired t-test, p < 0.05). They were IL-1β, IL-5, TNF-α, IL-6 and IL-2, and their concentrations were higher in the cases than in the controls (Table 3). The disease odds ratios corresponding to one standard deviation change in log-transformed concentrations of these five cytokines are 18.87, 4.10, 3.61, 3.27 and 2.52.
Table 3.
Association of cytokine level with disease status.
| Cytokine | Number of matched pairs | Differencea |
Conditional logistic regressionb |
|||
|---|---|---|---|---|---|---|
| Mean | SD | p-Value (paired t-test) | Odds ratio | 95% confidence interval | ||
| IL-17 | 39 | 0.0237 | 0.1533 | 0.3403 | 1.5563 | 0.6145–3.941 |
| IL-12 | 39 | 0.1021 | 0.5453 | 0.2496 | 1.5615 | 0.7212–3.381 |
| IL-8 | 29 | –0.3365 | 1.0115 | 0.0840 | 0.6422 | 0.3819–1.080 |
| TGF-β1 | 36 | –0.0291 | 0.6106 | 0.7766 | 0.9320 | 0.5786–1.501 |
| IL-6 | 20 | 0.9348 | 1.5819 | 0.0160 | 3.2708 | 0.9294–11.51 |
| IL-1β | 20 | 1.3889 | 1.6731 | 0.0015 | 18.8670 | 0.2434–1462 |
| IL-10 | 20 | 0.3766 | 1.7655 | 0.3521 | 1.4208 | 0.6822–2.959 |
| IL-2 | 20 | 0.9947 | 1.9228 | 0.0320 | 2.5238 | 0.9558–6.664 |
| IL-13 | 20 | 0.4624 | 1.7244 | 0.2452 | 1.5417 | 0.7289–3.261 |
| IL-4 | 20 | 0.6766 | 1.8023 | 0.1095 | 1.7454 | 0.8508–3.581 |
| IFN | 20 | 0.5165 | 1.6832 | 0.1860 | 1.9100 | 0.7012–5.203 |
| IL-5 | 20 | 1.0568 | 1.3983 | 0.0031 | 4.1032 | 1.0216–16.48 |
| TNF | 20 | 1.0088 | 1.5774 | 0.0100 | 3.6083 | 0.8504–15.31 |
Difference = the difference of cytokine level (in log scale) between the cases and controls.
Odds ratio = the odds ratio of cases vs. controls for one standard deviation change of cytokine level in log scale.
3.2. Genotype associations
Table 4 shows the association between the binary genotypes of each SNP and disease status determined by conditional logistic regression. The rs2069705 SNP, located in the promoter region of the interferon gamma gene (INF-γ) was significantly associated with the disease status.
Table 4.
Association of genotype with disease status in the UNM-EBCR cytokine project.
| SNP | Description | # pairs | Genotypes | # in 1st group | Odds ratio | 95% CI for odds ratio | p-Value |
|---|---|---|---|---|---|---|---|
| rs2069705 | IFN-γ –1615 C/T | 39 | CC vs. CT and TT | 16 | 5.00 | 1.10–22.82 | 0.038 |
| rs1800587 | IL-1α –889 C/T | 35 | TT vs. TC and CC | 9 | 2.00 | 0.50–8.00 | 0.327 |
| rs1143634 | IL-1β +3954 C/T | 25 | CC vs. CT and TT | 34 | 2.33 | 0.60–9.02 | 0.220 |
| rs1143627 | IL-1β –31 C/T | 36 | CC vs. CT and TT | 25 | 0.67 | 0.24–1.87 | 0.442 |
| rs16944 | IL-1β –511 | 35 | GG vs. AG and AA | 26 | 0.56 | 0.19–1.66 | 0.292 |
| rs2069762 | IL-2 –384 G/T | 35 | GG vs. GT and TT | 9 | 0.25 | 0.03–2.24 | 0.215 |
| rs2243248 | IL-4 –1098 G/T | 34 | TT vs. TG and GG | 58 | 4.00 | 0.85–18.84 | 0.080 |
| rs2243250 | IL-4 –524 C/T | 35 | CC vs. CT and TT | 40 | 1.00 | 0.40–2.52 | 1.000 |
| rs2069812 | IL-5 –745 C/T | 36 | CC vs. CT and TT | 28 | 1.50 | 0.61–3.67 | 0.374 |
| rs1800797 | IL-6 –597 A/G | 38 | GG vs. GA and AA | 30 | 0.75 | 0.26–2.16 | 0.594 |
| rs2227306 | IL-8 –204 C/T | 39 | TT vs. TC and CC | 17 | 0.38 | 0.10–1.41 | 0.147 |
| rs4073 | IL-8 –251 A/T | 39 | AA vs. AT and TT | 22 | 0.71 | 0.23–2.25 | 0.566 |
| rs1800896 | IL-10 –1082 A/G | 38 | AA vs. AG and GG | 20 | 1.80 | 0.60–5.37 | 0.292 |
| rs1800890 | IL-10 –3575 A/T | 39 | TT vs. TA and AA | 10 | 1.67 | 0.40–6.97 | 0.484 |
| rs1800872 | IL-10 –592 A/C | 39 | CC vs. CA and AA | 36 | 1.44 | 0.62–3.38 | 0.396 |
| rs1800871 | IL-10 –819 C/T | 36 | CC vs. CT and TT | 35 | 1.37 | 0.55–3.42 | 0.493 |
| rs2243113 | IL-12 –980 A/G | 37 | AA vs. AG and GG | 68 | 1.00 | 0.14–7.10 | 1.000 |
| rs20541 | IL-13 +98 C/T | 39 | CC vs. CT and TT | 43 | 1.83 | 0.68–4.96 | 0.232 |
| rs1800925 | IL-13 –1055 C/T | 28 | CC vs. CT and TT | 32 | 3.00 | 0.81–11.08 | 0.099 |
| rs3819025 | IL-17 +18 A/G | 38 | GG vs. GA and AA | 59 | 1.33 | 0.30–5.96 | 0.706 |
| rs1982073 | TGF-β1 +29 C/T | 38 | CC vs. CT and TT | 12 | 0.71 | 0.23–2.25 | 0.566 |
| rs1800469 | TGF-β1 –509 C/T | 38 | TT vs. TC and CC | 4 | 0.33 | 0.03–3.20 | 0.341 |
| rs361525 | TNF-α –238 A/G | 38 | GG vs. GA and AA | 69 | 0.67 | 0.11–3.99 | 0.657 |
| rs1800629 | TNF-α –308 A/G | 39 | GG vs. GA and AA | 50 | 0.80 | 0.32–2.03 | 0.638 |
| rs909253 | TNF-β +252 C/T | 38 | CC vs. CT and TT | 23 | 1.71 | 0.67–4.35 | 0.257 |
3.3. SNPs and associated cytokine levels
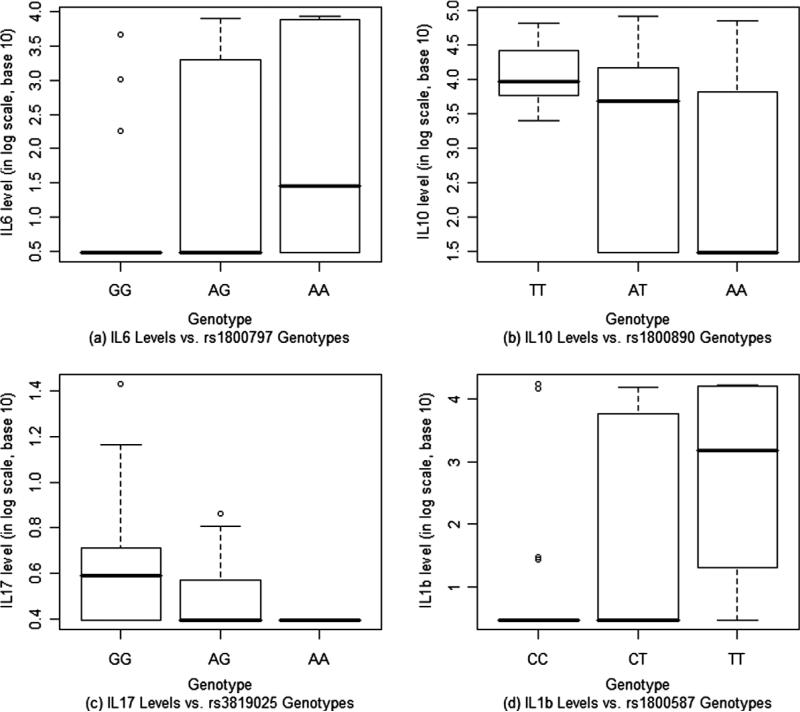
We analyzed the relationship between genotypes of the SNP and the associated cytokine levels by examining the effect of the SNP genotypes and disease status while taking into account the matching effect. This was done through the general linear model with both SNP genotypes and disease status being the factors of interest and the pairs of matched case and control representing the randomized blocks. The p-values from the ANOVA table for testing effects of the SNP genotypes and disease status on the cytokine levels are presented in Table 5. The mean cytokine levels (in log scale) for each of the three possible genotypes for all SNPs are also presented in Table 5, which show how the associated cytokine level changes across the three genotypes for each SNP. SNP rs3819025, in the exon of IL-17 gene, showed a significant association with its respective cytokine concentration while adjusting for the effect of disease status. Similarly, SNPs rs1800890 and rs1800797 are significantly associated with the cytokine IL-10 and IL-6, respectively. The IL-1α SNP (rs1800587) was marginally associated with IL-1 serum concentrations after adjusting for the effect of disease status. All four SNPs showed a monotonic increasing or decreasing relationship between the genotype and the cytokine serum levels (Fig. 1). We noticed that in Table 3 cytokine IL-6 level differs significantly between the case and control. However this association became marginally significant after taking into account the effect of the SNP (rs1800797) genotype, implying that the association between the cytokine IL-6 level and the disease may be partly explained by that between IL-6 and SNP rs1800797.
Table 5.
Association of cytokine with genotype and disease status.
| Cytokine | SNP | Cytokine levels for different genotypes |
p-Value (effect on cytokine) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Mean | Genotype | Mean | Genotype | Mean | Genotype | Disease status | ||
| IL-6 | rs1800797 | G | 0.872 | AG | 1.737 | A | 2.005 | 0.0398 | 0.0781 |
| IL-1α | rs1800587 | C | 1.000 | CT | 1.493 | T | 2.756 | 0.0638 | 0.0139 |
| IL-1β | rs1143634 | T | 2.151 | CT | 1.914 | C | 1.298 | 0.2441 | 0.0197 |
| IL-1β | rs1143627 | A | 1.075 | AG | 1.692 | G | 1.294 | 0.4550 | 0.0051 |
| IL-1β | rs16944 | A | 1.294 | AG | 1.750 | G | 1.075 | 0.3893 | 0.0036 |
| IL-10 | rs1800896 | G | 3.657 | AG | 3.022 | A | 2.496 | 0.1644 | 0.3623 |
| IL-10 | rs1800890 | T | 4.075 | AT | 3.102 | A | 2.377 | 0.0222 | 0.3632 |
| IL-10 | rs1800872 | C | 2.970 | AC | 2.800 | A | 2.926 | 0.9087 | 0.1905 |
| IL-10 | rs1800871 | C | 2.995 | CT | 2.640 | T | 2.926 | 0.6571 | 0.1420 |
| IL-2 | rs2069762 | T | 2.401 | GT | 1.705 | G | 1.949 | 0.4029 | 0.1620 |
| IL-13 | rs20541 | C | 4.028 | CT | 4.760 | T | 4.471 | 0.2604 | 0.2036 |
| IL-13 | rs1800925 | T | 5.045 | CT | 4.426 | C | 4.285 | 0.5398 | 0.4403 |
| IL-4 | rs2243248 | T | 2.502 | GT | 2.482 | G | 2.805 | 0.9709 | 0.1453 |
| IL-4 | rs2243250 | T | 2.008 | CT | 3.086 | C | 2.186 | 0.1286 | 0.0745 |
| IFN-γ | rs2069705 | C | 3.257 | CT | 2.439 | T | 3.016 | 0.2776 | 0.2080 |
| IL-5 | rs2069812 | C | 1.839 | CT | 1.847 | T | 1.383 | 0.5136 | 0.0057 |
| TNF-α | rs361525 | G | 1.308 | AG | 0.602 | A | 0.3897 | 0.0086 | |
| TNF-α | rs1800629 | G | 1.413 | AG | 0.879 | A | 0.1412 | 0.0014 | |
| TNF-β | rs909253 | C | 1.180 | CT | 1.139 | T | 1.549 | 0.6004 | 0.0079 |
| IL-17 | rs3819025 | G | 0.584 | AG | 0.492 | A | 0.394 | 0.0048 | 0.4463 |
| IL-12 | rs2243113 | G | AG | 1.280 | A | 1.216 | 0.7108 | 0.1053 | |
| IL-8 | rs2227306 | T | 1.848 | CT | 1.992 | C | 1.985 | 0.8105 | 0.0989 |
| IL-8 | rs4073 | T | 1.957 | AT | 2.059 | A | 1.792 | 0.4522 | 0.0680 |
Fig. 1.
Boxplots for cytokine levels of SNP genotypes.
4. Discussion
Three of the five significant cytokines, specifically IL-1β, IL-2 and TNF-α are critical cytokines representing results of the immune system's tumor surveillance activities. Interestingly, both antigen presenter cells related cytokines (IL-1β, TNF-α) and Th1 type T cells activity (IL-2) as strongly associated with the disease status among our breast cancer participants. Our results are in accordance with MD Anderson Cancer Center breast cancer patients’ results [20] in which inflammatory cytokines were also detected at elevated levels at breast cancer diagnosis. However, it is essential to mention that other immune disorders and conditions can also influence and increase pro-inflammatory cytokine levels in the serum, therefore those should be carefully recorded and monitored in similar studies. From Th2 type cytokines, IL-5 serum concentration was also significantly increased among breast cancer patients compared to disease free controls. A previous study reported eosinophils as members of the tumor infiltrating lymphocytes [21]. Based on in vitro experiments, eosinophil granulocytes, pretreated with IL-5 before interaction with the tumor or endothelial cells, bound aggressively to the endothelial cells, thereby preventing tumor attachment. Our findings raise the possibility that the detected increased IL-5 level may represent a molecular signal for eosinophil granulocytes attracting them to the cancer cells. It is also raises the possibility that eosinophil granulocytes’ potential for future therapeutic investigations.
In addition, we have found increased serum level of IL-6 cytokine among our breast cancer patients. This is in accordance with previous works indicating IL-6 involvement in metastatic potential of breast cancer [12]. This cytokine has been linked repeatedly to inflammatory response in liver and pancreatic carcinomas [22,23]. The detected serum cytokine results, especially in the case of IL-2 and TNF-α may direct our research interest to further evaluate their role among New Mexican breast cancer patients.
Based upon our results, the presence of the INF-γ (rs206970) variant (CC vs. CT and TT) of the SNPs chosen for this study shows significant association with breast cancer. Each SNP was in Hardy–Weinberg equilibrium and confirmed the frequency distribution of these SNPs published. Furthermore, we compared our population-based frequency results to Non-Hispanic White HapMap frequency data. The genotype frequencies we observed in the New Mexican Non-Hispanic Whites genotype frequencies were very close to the overall Caucasian data (Table 2). Given that no complete or reliable data on cytokine gene variant frequencies exists for Hispanics, we conducted separate analysis for Hispanic cases and controls (data not shown) as well. Because only 17.5% of our cases and 20.0% of the controls reported Hispanic ethnicity (Table 1), we did not expect to find large significant allelic differences when compared to Non-Hispanic Whites. However, there were a few SNPs that appear to be marginally different among cases (rs1800890, rs2243113, rs2243248, rs4073) and should be evaluated further.
Furthermore, the presence of IL-1β C-31T variant was less frequent among cases than controls, however this finding did not reached statistical significance. This was in accordance with previous works on risk reduction among breast cancer cases in Japan as well and our reduced odds ratio is in the comparable range to their findings [24]. Other cytokine variants (rs16944, rs2069762, rs1800797, rs2227306, rs4073, rs1982073, rs1800469, rs361525, rs1800629) summarized in Table 4 were also less frequently identified among our New Mexican breast cancer patients, therefore, the corresponding odds ratios showed decreased risk estimates. However, we will evaluate their probable role in more details and a larger sample size as these results did not reach statistical significance.
We also explored the associations between cytokine polymorphisms and serum cytokine concentrations (Table 3). Our ultimate goal is to uncover associations between genotypes and corresponding phenotypes of important immune system modifiers represented in cytokine pathways in breast cancer patients that could serve as useful biomarkers for tailoring therapy and/or predict outcomes from the disease.
We assessed the role of serum cytokine concentrations and the SNPs in relationship to breast cancer in this group of women. Statistical analysis results show that T variant of the IL-10 SNP (rs1800890) was associated with increased serum cytokine concentration. Similarly, the presence of IL-17 (rs3819025) gene G allelic variant was detected more frequently among those who had increased serum IL-17 levels.
Based on our analysis, the presence of the A variant of IL-6 SNP was significantly associated with increased serum cytokine production and that was also related to the disease status as well, even though this association was not statistically significant (Fig. 1). IL-1β serum concentrations were detected at an increased level among cases compared to controls; however, based on our results, we could not pinpoint which allelic variant (CC or CT) contributed to greater serum concentrations. Likewise, tumor necrosis factor, IL-2 and IL-5 serum levels were elevated among breast cancer patients compared to controls; however none of the studied SNPs can be detected as significant contributors to the serum cytokine production.
The presence of increased serum levels of pro-inflammatory cytokines at diagnosis might be indicative to certain genetic susceptibility markers; however, the existence of other inflammatory conditions should be carefully recorded and considered as well. Our study population did not allow us to examine potential linkage associations especially on chromosome 6 locations.
These findings might guide us to further understand the complexity of the presence of SNPs in cytokine genes. The variability in those genes and the corresponding cytokine serum concentrations among controls and breast cancer patients is an important area of research in tumor immunology.
We also recognize that ethnicity is an important contributor of allelic frequency variations along the entire genome. Ancestry associated examinations of certain single nucleotide polymorphisms are imperative in further molecular epidemiological studies. This is certainly true for studying New Mexicans as our Hispanic population has a high proportion of Spanish Jewish (Sephardic, Convertos) heritage [25–27], potentially putting them at higher risk for developing breast and other cancers due to the higher prevalence of inherited and familial breast cancer syndromes among this ethnic population.
We plan to include detailed ancestry evaluations using intronic SNPs in our next breast cancer and in other cancer studies in New Mexico similarly as in other breast cancer study among Latinas [28]. Based on our experience, the self-identified ethnicity is a strong predictor of ethnic background among New Mexicans. However, we believe that accurate identification of the presence of protective genetic factors potentially linked with admixture and ancestry are promising in cancer research and molecular epidemiology. Therefore, our future goal is to explore these factors among New Mexicans while working further on revealing the role of cytokines and the immune system in human cancers.
References
- 1.Ries LAG, Melbert D, Krapcho M, Stinchcomb DG, Howlader N, Horner MJ, et al., editors. SEER cancer statistics review, 1975–2006. National Cancer Institute; Bethesda, MD: 2009. Available from: http://seer.cancer.gov/csr/1975_2006/, based on August 2009 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Roitt I, editor. Immunology. C.V. Mosby; New York: 2006. [Google Scholar]
- 3.Cooke A, Owen M, Trowsdale J, Champion B. Advanced immunology. Mosby; London: 1996. chapter 10.1–10.13. [Google Scholar]
- 4.Hofmann SR, Ettinger R, Zhou YJ, Gadina M, Lipsky P, Siegel R, et al. Cytokines and their role in lymphoid development, differentiation and homeostasis. Curr Opin Allergy Clin Immunol. 2002;2:495–506. doi: 10.1097/00130832-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Harrington L, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;8:349–56. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 6.McKenzie BS, Kastelein RA, Cua DJ. Understanding the IL-23–IL-17 immune pathway. Trends Immunol. 2006;27:16–22. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Gallon J, Costes A, Sanche-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 8.Nedergaard Ladekarl M, Thomsen HF, Nyengaard JR, Nielsen K. Low density of CD3+, CD4+ and CD8+ cells is associated with increased risk of relapse in squamous cell cervical cancer. Br J Cancer. 2007;97:1135–8. doi: 10.1038/sj.bjc.6604001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucey DR, Clereci M, Shearer GM. Type 1 and type 2 cytokine dysregulation in human infectious, neoplastic, and inflammatory diseases. Clin Microbiol Rev. 1996;9:532–62. doi: 10.1128/cmr.9.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keen LJ. The extent and analysis of cytokine and cytokine receptor gene polymorphism. Transpl Immunol. 2002;10:143–6. doi: 10.1016/s0966-3274(02)00061-8. [DOI] [PubMed] [Google Scholar]
- 11.Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, Shen M, et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107(10):4101–8. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMichele A, Martin AM, Mick R, Gor P, Wray L, Klein-Cabral M, et al. Interleukin-6-174 G → C polymorphism is associated with improved outcome in high-risk breast cancer. Cancer Res. 2003;63:8051–6. [PubMed] [Google Scholar]
- 13.Saha A, Gupta V, Bairwa NK, Malhotra D, Bamezai R. Transforming growth factor-β1 genotype in sporadic breast cancer patients from India: status of enhancer, promoter, 5′-untranslated-region and exon-1 polymorphisms. Eur J Immunogenet. 2004;31:37–42. doi: 10.1111/j.1365-2370.2004.00442.x. [DOI] [PubMed] [Google Scholar]
- 14.Jin Q, Hemminki K, Grzybowska E, Klaes R, Soderberg M, Zientek H, et al. Polymorphisms and haplotype structures in genes for transforming growth factor β1 and its receptors in familial and unselected breast cancers. Int J Cancer. 2004;112:94–9. doi: 10.1002/ijc.20370. [DOI] [PubMed] [Google Scholar]
- 15.Caras I, Grigorescu A, Stavaru C, Radu DL, Mogos I, Szegli G, et al. Evidence for immune defects in breast and lung cancer patients. Cancer Immunol Immunother. 2004;53:1146–52. doi: 10.1007/s00262-004-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafountaine N, Roland S, et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol. 1998;113:401–6. doi: 10.1046/j.1365-2249.1998.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouma G, Crusius JB, Oudkerk PM, Kolkman JJ, von Blomberg BM, Kostense PJ, et al. Secretion of tumour necrosis factor-β and lymphotoxin-α in relation to polymorphisms in the TNF genes and HLA-DR alleles. Relevance for inflammatory bowel disease. Scand J Immunol. 1996;43:456–63. doi: 10.1046/j.1365-3083.1996.d01-65.x. [DOI] [PubMed] [Google Scholar]
- 18.Dixon WJ, Massey FJ. Introduction to statistical analysis. 4th ed. McGraw-Hill; 1983. [Google Scholar]
- 19.ÓBrien RG, Muller KE. Applied analysis of variance in behavioral science. Marcel Dekker; New York: 1983. pp. 279–344. [Google Scholar]
- 20.Pusztai L, Mendoza TR, Reuben JM, Martinez MM, Willey JS, Lara J, et al. Changes in plasma levels of inflammatory cytokines in response to paclitaxel chemotherapy. Cytokine. 2004;25(3):94–102. doi: 10.1016/j.cyto.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Furbert-Harris PM, Hunter KA, Vaughn TR, Parish-Gause D, Laniyan I, Harris D, et al. Eosinophils in a tri-cell multicellular tumor spheroid (MTS)/endothelium complex. Cell Mol Biol (Noisy-le-grand) 2003;49(7):1081–8. [PubMed] [Google Scholar]
- 22.Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links. Ann NY Acad Sci. 2009;1155:206–21. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 23.Gumbs AA. Obesity, pancreatitis and pancreatic cancer. Obes Surg. 2008;18(9):1183–9. doi: 10.1007/s11695-008-9599-3. [DOI] [PubMed] [Google Scholar]
- 24.Ito LS, Iwata H, Hamajima N, Saito T, Matsuo K, Mizutani M, et al. Significant reduction in breast cancer risk for Japanese women with interleukin 1B-31 CT/TT relative to CC genotype. Jpn J Clin Oncol. 2002;32(10):398–402. doi: 10.1093/jjco/hyf081. [DOI] [PubMed] [Google Scholar]
- 25.Hordes SM. The sephardic legacy in New Mexico: a history of the “crypto-Jews”. J West. 1996;35:82–90. [Google Scholar]
- 26.Wiilermet CM, Edgar HJ. Dental morphology and ancestry in Albuquerque, New Mexico. Homo. 2009;60(3):207–24. doi: 10.1016/j.jchb.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Bordenave K, Griffith J, Hordes SM, Williams TM, Padilla RS. The historical and geomedical immunogenetics of pemphigus among the descendants of Sephardic Jews in New Mexico. Arch Dermatol. 2001;137(6):825–6. [PubMed] [Google Scholar]
- 28.Ziv E, John EM, Choudhry S, Kho J, Lorizio W, Perez-Stable EJ, et al. Genetic ancestry and risk factors for breast cancer among Latinas in the San Francisco Bay Area. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1878–85. doi: 10.1158/1055-9965.EPI-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]