Abstract
Background & objectives:
ADAM33 is a member of a family of genes that encode membrane-anchored proteins with a disintegrin and a metalloprotease domain, primarily expressed in lung fibroblasts and bronchial smooth muscle cells. ADAM33 has been identified as a risk factor for asthma and is known as a gene associated with airway remodelling. The present study was conducted with the aims to investigate the expression of ADAM33 protein in patients of asthma and non-asthmatic controls, and to assess if the expression of ADAM33 protein relates with severity of asthma.
Methods:
A total of 35 subjects, including 27 patients with asthma and eight non-asthmatic controls were included using Global Initiative for Asthma guidelines 2005. Bronchial biopsy tissues were collected and paraffin sections were made to store all study samples. Immunohistochemistry was performed using standardized protocol.
Results:
An increase in expression of ADAM33 protein was observed in the epithelium, smooth muscle and mesenchymal cells of asthma cases when compared to controls but there was no relationship with severity of asthma.
Interpretation & conclusions:
A higher expression of ADAM33 protein was seen in asthma patients compared to controls. Large prospective studies need to be done with adequate study design to confirm these preliminary finding.
Keywords: ADAM33 protein expression, asthma, bronchial biopsy, immunohistochemistry, severity of asthma
Asthma is one the most common chronic disorders with several overlapping phenotypes1, characterized by symptoms of intermittent airway obstruction2. In asthma, the airways show features of acute and chronic inflammation that are associated with airway remodelling which include thickening of the airway wall, subepithelial fibrosis, increased smooth muscle mass that may contribute to the development of airflow limitation by increasing airway resistance.
The ADAM gene family is a sub group of the zinc-dependent metalloproteinase having surface proteins with adhesion and protease activity3. ADAM33 is the member of the ADAM family and is most closely related to human ADAM12, 15, 19 and Xenopus ADAM13, all of which possess proteolytic activity4,5,6,7. ADAM33 represents a complex domain structure, also contains a transmembrane and cytoplasmic domain. The domain encodes signal sequence, pro, catalytic (metalloproteinase), disintegrin-like, cysteine-rich, and epidermal growth factor (EGF)3,4,8. This complex domain construction allows ADAM proteins to be implicated in diverse functions including cell proliferation, differentiation, migration, and embryogenesis via ecto-domain shedding of growth factors, ligands and receptors, membrane fusion, and cell adhesion via interactions with integrins and extracellular matrix proteins9,10,11,12,13. From a functional standpoint, these different domains interpret different functions of ADAM33, which include role in cell-cell and cell-matrix interactions14, cell migration12,15, cell-cell adhesion16 and signal transduction17. ADAM proteins are membrane anchored proteins and can function as a sheddases, and can “release a variety of cell-surface proteins, including growth factors, cytokines, cell adhesion molecules and receptors”3,14,18. Additionally, soluble form of ADAM33 promotes angiogenesis which has been described as the first biological function19.
Van Eerdewegh et al20 identified ADAM33 as an asthma susceptibility gene with the help of positional cloning method on an outbred Caucasian population. Linkage analysis and association studies of single nucleotide polymorphisms (SNPs) and haplotypes provide strong evidence for the involvement of several SNPs of ADAM33 for susceptibility to asthma and bronchial hyperresponsiveness (BHR)21. Many association studies have confirmed association of ADAM33 gene polymorphisms with asthma susceptibility and airway hyperreactivity, whereas other studies have shown absence of association21.
Studies on the expression of ADAM33 mRNA in human cells and tissues have revealed exclusive distribution of this protein in cells of mesenchymal origin, such as fibroblasts and smooth muscle cells, which indicates a possible role of ADAM33 in airway remodelling22,23,24. Studies show ADAM33 gene as a main challenge for asthma development and bronchial hyperreactivity. Therefore, the objectives of the present study were to investigate the expression of ADAM33 protein in patients of asthma and controls, and to assess if the expression of ADAM33 protein relates with severity of asthma.
Material & Methods
From August 2007 to December 2010 a case-control study was conducted in the Departments of Pulmonary Medicine, Pediatrics and Pathology of Chattrapati Shahuji Maharaj Medical University, a tertiary care, referral center at Lucknow, Uttar Pradesh, India. The study protocol was approved by the institutional ethics committee and written informed consent for participation was obtained from all the patients and controls.
Characterization of phenotype: All subjects who underwent bronchoscopic examination from August 2007 to December 2010 were screened for suitability for inclusion in the study. Of the total 859 subjects, 27 subjects as asthma cases and eight as non-asthmatic controls satisfied the inclusion and exclusion criteria. Inclusion criteria for cases were (i) diagnosis of asthma according to the treating physician, (ii) symptoms, (iii) use of medications for asthma, (iv) reversible airflow limitation according to GINA (Global Initiative for Asthma) guidelines; FEV1 reversibility >12 per cent and 200 ml after a post-bronchodilator spirometry, (v) on any regular medication for asthma such as corticosteroids, β-2 agonist, methylxanthines, leukotriene modifiers and cromones, (vi) at least one urgent care visit in the previous 12 months, and (vii) at least 3 year records of medically diagnosed asthma. Additionally, all considered patients had positive family history of asthma. Asthma severity was classified according to GINA guidelines2.
The inclusion criteria for controls were: (i) no past or present diagnosis of asthma and other pulmonary diseases, (ii) no history of wheezing, shortness of breath, and other symptoms of allergic diseases such as nasal and skin symptoms, (iii) no use of medications for asthma, and (iv) absence of first-degree relatives with a history of asthma.
Spirometry: Pulmonary function test (PFT) was done using a spirometer (Spiro lab II, MIT II, Longfian Scitech Company, Korea) of all cases and controls before they underwent a bronchoscopic examination according to standard guidelines25. Post-bronchodilator reversibility testing was performed 15-30 min after 2 puffs of salbutamol. Controls underwent only a pre-bronchodilator study.
Bronchosopy: Flexible bronchoscope (Model- PENTAX FB-18V (South India Surgical Co. Ltd., New Delhi Bronchoscope from Japan) was used for performing bronchoscopy. Accessories for tissue sampling used were bronchial brushes [Bigger (Model-CS6015ST); Medium (Model-CSC3S) and Small (Model-CS6002SN)], aspiration needle and endobronchial biopsy forceps (Model-KW24159).
All bronchial biopsies were fixed in 10 per cent buffered formalin and embedded in paraffin. The whole tissue sample was blocked and 3-μm sections were taken for the immunohistochemistry analysis.
Immunohistochemistry: The sections were dewaxed in xylene and dehydrated through graded alcohol concentration and washed in tris buffer (pH 7.2). Endogenous peroxidase activity was blocked using 3 per cent hydrogen peroxide in methanol for 30 min; antigen retrieval was done with citrate buffer (pH 6.2) at 90°C for 90 min in a water bath. After washing in tris-buffer (pH 7.4), sections were incubated with the primary polyclonal ADAM33 antibody (Rb pAb –abcam ab39193-100) (Source: Allied Scientific Products, Kolkata) at a dilution range of 1:100 and left overnight at 4°C in a humid chamber. The next day, sections were brought to room temperature and further procedures were performed involving the following steps; washing in tris-buffer, incubation with secondary antibody directly for 15 min [MACH 4 Universal HRP Polymer Kit with DAB (Source: Biocare Medical, USA)] again washing in tris-buffer, 3’diaminobenzidine tetrahydrochloride was used to reveal the peroxidase complex. The sections were counterstained with hematoxylin and dehydrated, cleared, and mounted. Parallel control slides were run with all batches including positive and negative controls. Negative controls were those sections on which the primary antibodies were absent and positive controls were those sections on which standardization was done. All buffers and reagents used for the experiment were either from MERCK (Merck Limited, Mumbai, India) or SRL (SISCO Research Laboratories Pvt. Ltd., Mumbai, India).
Quantitative measurement of ADAM33 expression: Slides were examined and images were acquired by a ZEISS, Imager Z2 attached to a digital camera Axiocam HRc using the DELL PRECISION T3500 system. Scoring of epithelial staining (which refers to the percentage of the epithelium stained positively) was performed blindly. The scoring system was as follows: 0 = no staining, 1=1 to 12.5%, 2=12.5 to 25%, 3=25 to 37.5%, 4=37.5 to 50%, 5=50 to 62.5%, 6=62.5 to 75%, 7=75 to 87.5%, and 8=87.5 to 100 per cent. All positive stained epithelial cells were counted independent of intensity.
Statistical analysis: Data were analyzed using SPSS 11.5 (SPSS Inc., Chicago, IL). Demographic characteristics of patients and controls were described as frequencies and percentages, whereas descriptive statistics of patients and controls were presented as mean and standard deviations for continuous measures. Categorical data were compared by Chi square (χ2) test and Fisher's exact test, and student's t test was applied for continuous variables. To find out association of epithelial staining, single (1-way) ANOVA was used to analyze differences in between the groups followed by Tukey post-hoc multiple comparison test.
Results
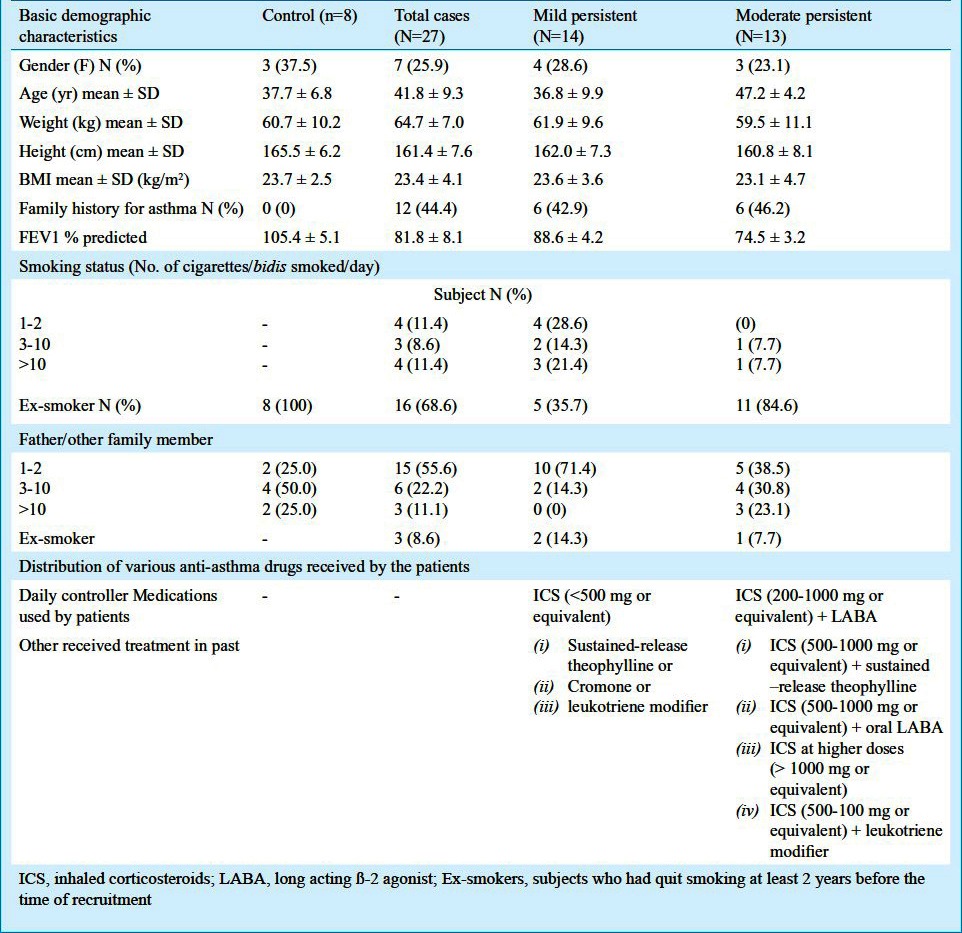
A total of eight non-asthmatic controls and 27 asthma patients were selected during the study period. GINA grading2 was used to classify asthma severity and the subjects with the following asthma severity were included: (i) mild persistent [n=14 (51.9%)], (ii) moderate persistent [n=13 (48.1%)]. Table I illustrates the demographic characteristics of the study population. Eleven patients (31.4%) were current smokers and the remaining 68.6 per cent were ex-smokers. All eight controls had stopped smoking; however, were ex-smokers. In controls, exposure to second hand smoke was 100 per cent.
Table I.
Demographic profile of cases and controls
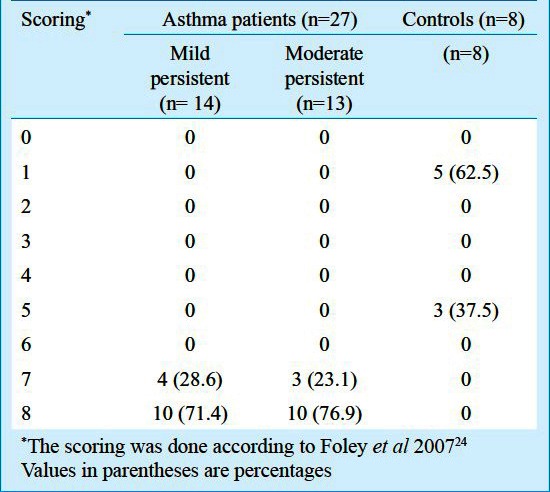
Higher expression of ADAM33 was identified in the epithelium, subepithelium, submucosal inflammatory cells, and smooth muscles of the subjects with asthma as compared with controls (Figure). The epithelium stained positively for ADAM33 protein in subjects with asthma and the staining was between 75-100 per cent. The epithelium of control biopsies stained positive weakly (staining between 0-62.5%). The scores for epithelial staining of ADAM33 are shown in Table II. No significant difference was found between mild persistent and moderate persistent subgroups of asthma. However, both mild and moderate persistent subgroups were found to be significantly associated with highly positive epithelium cells compared with control (P<0.001 and 0.001, respectively).
Fig.
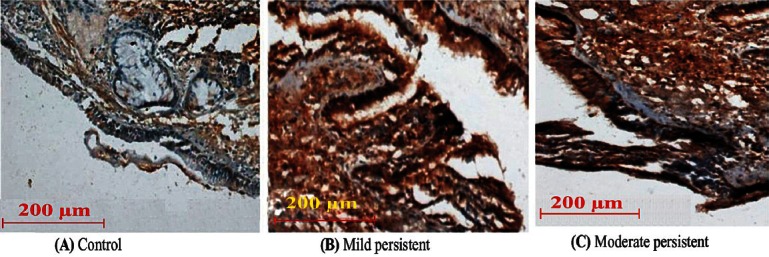
Representative examples of immunohistochemistry of bronchial biopsies showing staining for ADAM33 in a subject; (A) Control, (B) Mild persistent, (C) Moderate persistent. Magnification X 200.
Table II.
Epithelium scoring of subjects
Discussion
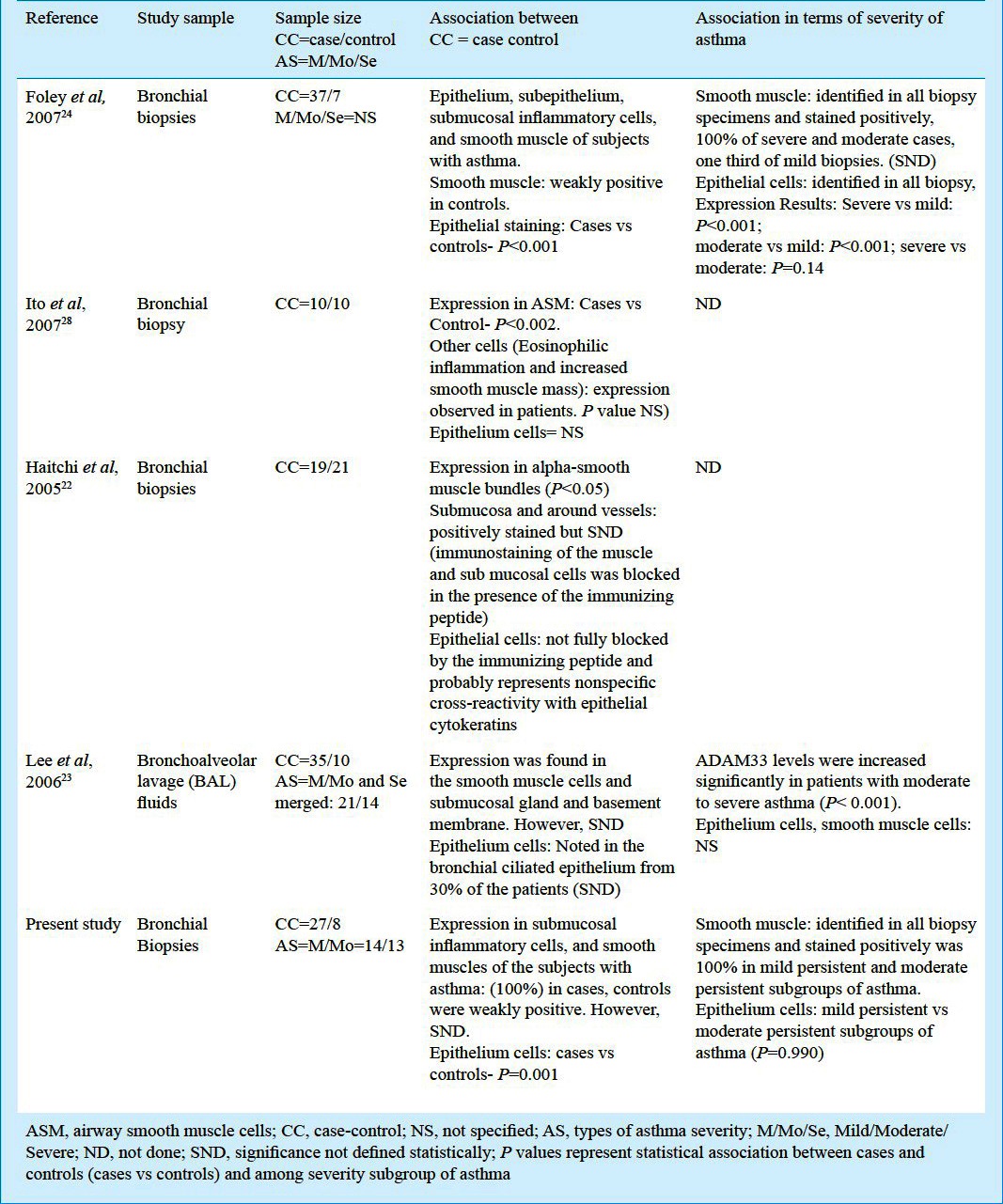
In the current study, bronchial biopsies were taken from 35 subjects (27 asthma cases and 8 non-asthmatic controls). An increase in expression of ADAM33 protein was seen in the epithelium, smooth muscle and mesenchymal cells of cases when compared to controls but there was no relationship with severity of asthma. Controls also showed weak positivity of expression of ADAM33 protein in epithelium cells. The reason may be that all controls had past history of smoking and had exposure to second hand smoke, and earlier studies have confirmed that smoking is responsible for airway damage26,27. Table III shows expression of ADAM33 in patients of asthma compared with controls and relation with severity of asthma in earlier studies along with the present results.
Table III.
Expression of ADAM33 in patients of asthma compared with controls and protein relation with severity of asthma in other previous published studies and with our results
Bronchial hyperresponsiveness (BHR) is the marker that distinguishes asthma from other diseases of the airways. It has also been suggested that the increase in adventitial thickness could cause an uncoupling of the airway smooth muscle from the load provided by surrounding parenchymal recoil to cause embroidered airway narrowing. These alterations in smooth muscle mass relative to disease severity are thought to be the most likely explanations for BHR and the unnecessary airway narrowing observed in the established disease27. The role of the epithelium in airway remodelling has previously been suggested28. Earlier studies have confirmed ADAM33 gene as a newly identified protease involved in airway remodelling30,31.
Expression of ADAM33 was first observed in cells of mesenchymal origin, such as fibroblasts and smooth muscle cells, indicating a possible role in airway remodelling20. Another study demonstrated the existence of multiple novel isoforms of the ADAM33 in human airway fibroblasts32 and gene to be selectively expressed by mesenchymal cells18.
Similar to our study, Foley et al24 found increased expression of ADAM33 epithelial staining in more than 80 per cent of biopsies from subjects with asthma with weak staining in control epithelium. Unlike our results, they observed a statistically significant increase in the epithelial staining as asthma severity progressed from mild to severe24. One possible reason for this difference may be due to the fact that in our study patients in intermittent and severe persistent subgroups of asthma could not be included. Cases of asthma recruited by us were somewhat homogeneous as far as clinical severity of disease was concerned and in accordance to GINA guidelines2.
Another study reported no difference in ADAM33 mRNA expression between bronchial biopsies from subjects with asthma and control as well as reported weak immunoreactivity in bronchial epithelial cells, attributing this to nonspecific cross-reactivity with epithelial cytokeratins22. Lee et al23 have previously reported a significant negative correlation between protein expression of ADAM33 in bronchoalveolar lavage samples and FEV1% predicted in asthma and observed positive epithelial staining expression of ADAM33 protein, though at a frequency of less than 50 per cent in the subjects with asthma. Ito et al28 also showed association of ADAM33 protein expression in airway smooth muscle cells, but not defined positivity of epithelium cell with cases of asthma and controls. Dijkstra et al30 assessed the expression of various ADAMs (ADAM8, ADAM9, ADAM10, ADAM17, ADAM19 and ADAM33) in human lung tissue and found that all were usually distributed over the bronchial epithelium.
Our study had the following limitations: (i) sample size determination was not performed, (ii) controls were only eight which may not be sufficient to obtain robust conclusions, (iii) due to the difficulty in obtaining healthy controls for the study only controls who were not healthy, were smokers and suffered from lung cancer, were included, (iv) due to low sample size further sub-group analysis to evaluate the impact of medications such as ICS (inhaled corticosteroids) on ADAM33 expression could not be performed, (v) post-bronchodilator study could not performed in controls and therefore, the exclusion of diagnosis of asthma in this group could not be confirmed, (vi) a significant proportion of subjects was current smokers and the rest were ex-smokers whereas all controls were ex-smokers. Since smoking is known to be associated with increased ADAM33 expression, the effect of smoking and asthma in the increased expression of ADAM33 could not be differentiated.
It is already known that epithelium cells play an important role in airway remodelling28. Significantly unregulated expression of ADAM33 protein in bronchial epithelium cells has also been reported earlier23,24,31 as observed in the present study also.
In conclusion, a higher expression of ADAM33 protein was observed in bronchial epithelium of asthma patients compared to controls. This observation and its clinical significance in the Indian population need to be confirmed in larger, prospective studies.
Acknowledgment
Authors acknowledge the financial support received from Department of Biotechnology, Government of India, New Delhi (BT/PR7115/Med/14/955/2006) for carrying out this work. Authors also thank all the patients and controls for providing samples for ADAM33 protein analysis.
References
- 1.Cookson WO, Moffatt MF. Genetics of asthma and allergic disease. Hum Mol Genet. 2000;9:2359–64. doi: 10.1093/hmg/9.16.2359. [DOI] [PubMed] [Google Scholar]
- 2.Global Initiative for Asthma (Gina), National Heart, Lung and Blood Institute, US Department of Health and Human Services: National Institute of Health (NIH) Publication No 02-3659 (2005) Available from: http://www.ginasthma.org/Guidelines/guidelines-resources.html .
- 3.Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–7. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- 4.Yoshinaka T, Nishii K, Yamada K, Sawada H, Nishiwaki E, Smith K, et al. Identification and characterization of novel mouse and human ADAM33s with potential metalloprotease activity. Gene. 2002;282:227–36. doi: 10.1016/s0378-1119(01)00818-6. [DOI] [PubMed] [Google Scholar]
- 5.Gunn TM, Azarani A, Kim PH, Hyman RW, Davis RW, Barsh GS. Identification and preliminary characterization of mouse Adam33. BMC Genet. 2002;3:2. doi: 10.1186/1471-2156-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawaguchi N, Xu X, Tajima R, Kronqvist P, Sundberg C, Loechel F, et al. ADAM 12 protease induces adipogenesis in transgenic mice. Am J Pathol. 2002;160:1895–903. doi: 10.1016/S0002-9440(10)61136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirakabe K, Wakatsuki S, Kurisaki T, Fujisawa SA. Roles of meltrin beta /ADAM19 in the processing of neuregulin. J Biol Chem. 2001;276:9352–8. doi: 10.1074/jbc.M007913200. [DOI] [PubMed] [Google Scholar]
- 8.Wolfsberg TG, Straight PD, Gerena RL, Huovila AP, Primakoff P, Myles DG, et al. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev Biol. 1995;169:378–83. doi: 10.1006/dbio.1995.1152. [DOI] [PubMed] [Google Scholar]
- 9.Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–30. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- 10.Eto K, Puzon-McLaughlin W, Sheppard D, Sehara-Fujisawa A, Zhang XP, Takada Y. RGD-independent binding of integrin alpha9beta1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J Biol Chem. 2000;275:34922–30. doi: 10.1074/jbc.M001953200. [DOI] [PubMed] [Google Scholar]
- 11.Gilpin BJ, Loechel F, Mattei MG, Engvall E, Albrechtsen R, Wewer UM. A novel, secreted form of human ADAM 12 (meltrin alpha) provokes myogenesis in vivo. J Biol Chem. 1998;273:157–66. doi: 10.1074/jbc.273.1.157. [DOI] [PubMed] [Google Scholar]
- 12.Martin J, Eynstone LV, Davies M, Williams JD, Steadman R. The role of ADAM 15 in glomerular mesangial cell migration. J Biol Chem. 2002;277:33683–9. doi: 10.1074/jbc.M200988200. [DOI] [PubMed] [Google Scholar]
- 13.Yuan R, Primakoff P, Myles DG. A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm-egg plasma membrane adhesion and fusion. J Cell Biol. 1997;137:105–12. doi: 10.1083/jcb.137.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 15.McFarlane S. Metalloproteases: carving out a role in axon guidance. Neuron. 2003;37:559–62. doi: 10.1016/s0896-6273(03)00089-8. [DOI] [PubMed] [Google Scholar]
- 16.Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. 2003;102:1186–95. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- 17.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 18.Umland SP, Garlisi CG, Shah H, Wan Y, Zou J, Devito KE, et al. Human ADAM33 messenger RNA expression profile and post-transcriptional regulation. Am J Respir Cell Mol Biol. 2003;29:571–82. doi: 10.1165/rcmb.2003-0028OC. [DOI] [PubMed] [Google Scholar]
- 19.Puxeddu I, Pang YY, Harvey A, Haitchi HM, Nicholas B, Yoshisue H, et al. The soluble form of a disintegrin and metalloprotease 33 promotes angiogenesis: implications for airway remodeling in asthma. J Allergy Clin Immunol. 2008;121:1400–6. doi: 10.1016/j.jaci.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2001;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 21.Sharma N, Tripathi P, Awasthi S. Role of ADAM33 gene and associated single nucleotide polymorphisms in asthma. Allergy Rhinol (Providence) 2011;2:e63–70. doi: 10.2500/ar.2011.2.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haitchi HM, Powell RM, Shaw TJ, Howarth PH, Wilson SJ, Wilson DI, et al. ADAM33 expression in asthmatic airways and human embryonic lungs. Am J Respir Crit Care Med. 2005;171:958–65. doi: 10.1164/rccm.200409-1251OC. [DOI] [PubMed] [Google Scholar]
- 23.Lee JY, Park SW, Chang HK, Kim HY, Rhim T, Lee JH, et al. A disintegrin and metalloproteinase 33 protein in patients with asthma: Relevance to airflow limitation. Am J Respir Crit Care Med. 2006;173:729–35. doi: 10.1164/rccm.200409-1175OC. [DOI] [PubMed] [Google Scholar]
- 24.Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, et al. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–71. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 25.Lambert RK, Wiggs BR, Kuwano K, Hogg JC, Paré PD. Functional significance of increased airway smooth muscle in asthma and COPD. J Appl Physiol. 1993;74:2771–81. doi: 10.1152/jappl.1993.74.6.2771. [DOI] [PubMed] [Google Scholar]
- 26.Miller MR, Crapo R, Hankinson J, Brusasco V, Burgos F, Casaburi R, et al. ATS/ERS Task Force. General considerations for lung function testing. Eur Respir J. 2005;26:153–61. doi: 10.1183/09031936.05.00034505. [DOI] [PubMed] [Google Scholar]
- 27.Davies DE. The role of the epithelium in airway remodeling in asthma. Proc Am Thorac Soc. 2009;6:678–82. doi: 10.1513/pats.200907-067DP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito I, Laporte JD, Fiset PO, Asai K, Yamauchi Y, Martin JG, et al. Downregulation of a disintegrin and metalloproteinase 33 by IFN-gamma in human airway smooth muscle cells. J Allergy Clin Immunol. 2007;119:89–97. doi: 10.1016/j.jaci.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Powell RM, Wicks J, Holloway JW, Holgate ST, Davies DE. The splicing and fate of ADAM33 transcripts in primary human airways fibroblasts. Am J Respir Cell Mol Biol. 2004;31:13–21. doi: 10.1165/rcmb.2003-0330OC. [DOI] [PubMed] [Google Scholar]
- 30.Dijkstra A, Postma DS, Noordhoek JA, Lodewijk ME, Kauffman HF, ten Hacken NH, et al. Expression of ADAMs (“a disintegrin and metalloprotease”) in the human lung. Virchows Arch. 2009;454:441–9. doi: 10.1007/s00428-009-0748-4. [DOI] [PubMed] [Google Scholar]
- 31.Holgate ST, Davies DE, Powell RM, Holloway JW. ADAM33: a newly identified protease involved in airway remodelling. Pulm Pharmacol Ther. 2006;19:3–11. doi: 10.1016/j.pupt.2005.02.008. [DOI] [PubMed] [Google Scholar]


