Abstract
Background. Transfusion associated bacterial infection has remained more frequent with a sever risk of morbidity and mortality. This study assessed the bacteriological safety of blood collected for transfusion. Method. A cross-sectional study was conducted at University of Gondar hospital blood bank from December 2011 to June 2012. Bacterial isolation, identification, and antimicrobial susceptibility tests were done as per the standard procedure. Chi-square test and P value were used to assess associations between risk factors and the bacterial isolation rate. Results. Twenty-one (15.33%) blood units were found contaminated with bacteria, and 95.24% contamination was due to external sources. The commonly isolated bacteria were Staphylococcus aureus, Coagulase negative Staphylococci, Escherichia coli, Klebsiella species, Streptococci species, Enterobacter species, and Citrobacter species. All of the bacteria isolated were 100% sensitive to Gentamicin, Chloramphenicol, Amoxicillin, and Doxycycline. Multiple antimicrobial resistances were observed in 66.7% of the isolates. Not using glove by phlebotomist, touching disinfected phlebotomy site and double puncture at the same hand or both hands of a donor were found to be risk factors for bacterial contamination. Conclusion. Bacterial contamination of blood to be transfused is a common problem in the hospital. So attention should be given to activities performed at the blood bank for safe transfusion practices.
1. Introduction
The development of modern blood transfusion medicine represents one of the greatest achievements of medicine in the 20th century with about 75 million blood units being collected and transfused yearly [1, 2]. Although millions of lives are saved by blood transfusion, limited access to transfusion and the provision of unsafe blood is putting millions of people at risk of transfusion transmissible infections (TTI) [1, 3, 4].
Due to stringent donor selection, improved mandatory tests, and close surveillance of new emerging infections, the risk of TTI in developed countries is very low [5], but blood safety remains an important public health concern in Africa where lack of availability and provision of unsafe blood adversely impacts morbidity and mortality in the region [6, 7].
Transfusion associated bacterial infection (TABI) has remained more frequent than viral infections and is associated with high mortality due to rapid occurrence of septic shock [8–11]. Contaminated blood units may contain a numbers of virulent bacteria as well as endotoxins that are considered to be fatal to the recipient [12, 13]. The potential sources of bacterial contamination of blood collected for transfusion are skin flora introduced at the time of phlebotomy and from bacteria in the donor's blood because of an underlying condition causing donor bacteremia [9, 14, 15].
In many African countries concern over TTIs usually focuses on viral risk, yet the risk of bacterial contamination, incurred during collection and processing, is 2500 times higher than in developed countries [4, 16]. Moreover countries in the region relay only on visual examination of the blood bag for evidence of hemolysis just prior to transfusion with its defect as a screening method to check for bacterial contamination [10, 17]. This makes the burden of TABI be more prevalent.
Studies from Africa including from Kenya [16], Nigeria [18], Ghana [19], and elsewhere in the world [13, 20–23] have indicated the magnitude of the problem, but, bacteriological safety of blood for transfusion is not well addressed in Ethiopia in general and in the study area in particular. So the aim of this study was to assess rate of bacterial contamination of blood collected for transfusion and to determine the antimicrobial susceptibility pattern of the isolated bacteria.
2. Materials and Methods
2.1. Study Area and Study Design
A cross-sectional study was conducted from December 2011 to June 2012 at University of Gondar hospital blood bank which is a teaching hospital that provides health service to over five million inhabitants in Northwest Ethiopia. According to the data obtained from the blood bank the average annual number of blood units collected is 2000 and majority of which are provided for surgical and gynecological cases.
2.2. Sample Size Determination and Sampling Procedure
The sample size was calculated based on assumption of prevalence of bacteria isolated from blood for transfusion in Kenya and Nigeria with each reporting 8.8% [16, 18]. With 5% margin of error and 95% confidence interval (alpha = 0.05), the actual sample size for the study was computed using one sample population proportion formula (Cochran's sample size formula) as indicated in the following:
| (1) |
Using systematic random sampling, a total of 137 blood donors and blood units were investigated.
2.3. Data Collection and Laboratory Procedure
2.3.1. Sociodemographic Characteristics and Other Factors
The sociodemographic characteristics of blood donors (age, sex, occupation, and others) and other factors like type of blood donors, use of glove by phlebotomist during blood collection, and double puncture at the same hand or both hands of a donor were collected using interview administered structured questionnaire and direct observation of bleeding procedures.
2.3.2. Sample Collection
Five milliliter of venous blood was collected directly from the donors and another 5 mL of blood was taken from the blood units ready for transfusion. Blood units collected from the sampled donor were mixed and kept inverted down for 30 to 45 minutes. This allows some sediments of blood (to maximize sensitivity of bacterial isolation) to slip out of the blood bag into the part of the septum at which the sample will be collected. After the period of sedimentation the septum near to the bag was clipped in order to block blood leakage through the septum that will be punctured. Finally 5 mL of blood was drawn from the part of septum already prepared for this purpose by sterile syringe with needle after disinfection with tincture of iodine [24].
2.3.3. Bacterial Isolation and Identification
Blood samples were inoculated in duplicate on to 45 mL of brain heart infusion (BHI) medium, incubated at 37°C, and observed daily after 48 hours of incubation for 5 to 7 consecutive days for presence of turbidity, hemolysis, and color changes which are evidence of microbial growth. Whenever visible sign of growth appears, small amount of the cultures was subcultured on to blood agar plate (BAP) and MacConkey (MAC) agar and incubated for 24 to 48 hours at 37°C. Pure colonies were examined by Gram's staining and further identification was made using different biochemical tests including catalase and coagulase tests for Gram-positive bacteria and hydrogen sulphide production (H2S), indole test, citrate utilization, lysine decarboxylase (LDC) test, gas production, and carbohydrate metabolism for Gram-negative bacteria [24–26].
2.3.4. Antimicrobial Susceptibility Test
Isolated bacteria were tested for their susceptibility pattern according to Kibry-Bauer disk diffusion method on Muller-Hinton agar [26] using a panel of 14 antimicrobials including Ampicillin (10 μg), Gentamicin (10 μg), Tetracycline (30 μg), Ciprofloxacin (5 μg), Chloramphenicol (30 μg), Trimethoprim-sulfamethoxazole (25 μg), Ceftriaxone (30 μg), Amoxicillin (30 μg), Penicillin G (10 units), Methicillin (5 μg), Vancomycin (30 μg), Clindamycin (2 μg), Erythromycin (15 μg), and Doxycycline (30 μg). Pure colonies of the test organism were taken using a sterile wire loop and emulsified in 3-4 mL of sterile nutrient broth. Bacterial suspensions were compared with 0.5 McFarland standard. Then a sterile cotton swab was dipped into the suspension and bacteria were inoculated onto the Muller-Hinton agar. The discs were placed on to the surface of inoculated media by using disc dispenser and incubated for 24 hr at 37°C. Results were read and recorded by measuring inhibition zone diameters to the nearest millimeter and interpreted after comparing with the standards, and isolates were classified as susceptible, intermediate, or resistant to the tested antibiotics [26].
2.4. Data Quality Control
All culture media were prepared following the manufacturer's instructions. Each batch of the prepared media was checked for sterility by incubating a sample medium at 37°C for 24 hr. Known bacterial species were inoculated and incubated at 37°C for 24 hr for the performance check [26]. E. coli ATCC25922 and S. aureus ATCC25923 sensitive to all antimicrobial agents were used as control strains.
2.5. Data Analysis
Statistical analysis was done using SPSS version 16.00 statistical software. Frequency and percentage were employed to summarize the results and presented in tables and graphs. Chi-square (χ 2) and P value were used to determine the association and strength of risk factors with the bacterial isolation rate from the collected blood for transfusion. A P value of less than 0.05 was considered as statistically significant.
2.6. Ethical Consideration
Ethical approval of the research was obtained from Ethical Review Committee of School of Biomedical and Laboratory Sciences College of Medicine and Health Sciences, and official letter was directed to University of Gondar hospital blood bank. Informed consent was obtained from the blood donors. Donor with bacteremia was contacted by the address registered on the donor's card and advised to communicate with clinicians.
3. Results
3.1. Sociodemographic and Related Characteristics of Blood Donors
A total of 137 blood donors and blood units collected from them were included in this study. The mean age of the study participants was 30.3 years with a standard deviation of 10.1, and the majority, 59.1%, of them were under the age group of 22−35 years. Males comprise about 81% of the study participants. Large number of the donors, 72.3%, 51.1%, 75.2%, and 44.5%, were urban residents, married, Christians by religion, at level of college/university in education, respectively. About 77% of the donors were family replacement donors, and 72.3% of the donors have no history of previous donation (Table 1).
Table 1.
Sociodemographic variables of blood donors at University of Gondar hospital blood bank, Northwest Ethiopia, 2012.
| Variables | Frequency (n) | Percent (%) |
|---|---|---|
| Age | ||
| Under 21 | 19 | 13.9 |
| 22–35 | 81 | 59.1 |
| 36–49 | 27 | 19.7 |
| Above 50 | 10 | 7.3 |
| Sex | ||
| Male | 111 | 81.0 |
| Female | 26 | 19.0 |
| Place of residence | ||
| Rural | 38 | 27.7 |
| Urban | 99 | 72.3 |
| Secondary school | 26 | 19.0 |
| College/university | 61 | 44.5 |
| Type of donor | ||
| Volunteer | 27 | 19.7 |
| Replacement | 106 | 77.4 |
| Paid | 4 | 2.9 |
| Number of previous donation | ||
| 0 times | 99 | 72.3 |
| Once | 16 | 11.7 |
| Twice | 3 | 2.2 |
| Three times | 2 | 1.5 |
| More than three times | 17 | 12.4 |
3.2. Rate of Bacterial Isolation
Over the study period, 21 blood units, out of 137, were found to be contaminated with bacteria making the bacterial isolation rate of 15.3%. Only in a single instant bacteria were isolated from the donor as well as from the respective blood unit, representing 1/21 (4.8%) of contaminated blood units. The majority, 66.7% (14/21), of the organisms isolated were Gram-positive mainly S. aureus (42.9%), Coagulase negative Staphylococci species (19.05%), and Streptococci species (4.8%). Gram-negative bacteria isolated include E. coli (14.2%), Klebsiella species (9.52%), Enterobacter species (4.8%), and Citrobacter species (4.8%) (Figure 1).
Figure 1.
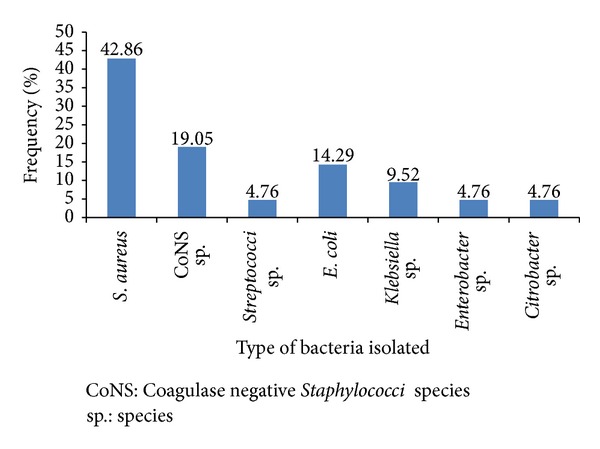
Type and frequency of bacteria isolated from blood units collected for transfusion at University of Gondar hospital blood bank, Northwest Ethiopia, 2012.
3.3. Phlebotomy Procedures and Association with Rate of Bacterial Contamination
During collection of blood for transfusion only 70% alcohol was used as a disinfectant, and about 70.8% (97/137) of sampled blood units were collected without using glove by the phlebotomist. Moreover 10.2% (14/137) of sampled blood units were collected from the donors whose disinfected phlebotomy site had been touched by the hand of blood collectors and 7.3% (10/137) had been collected by double puncture at the same hand or both hands of a donor. When these activities were tested for association with rate of bacterial contamination not using glove by phlebotomist during blood collection, touching disinfected phlebotomy site and double puncture at the same hand or both hands of a donor have shown statistically significant association with bacterial contamination (Table 2).
Table 2.
Procedural activities performed during the collection for transfusion and their association with rate of bacterial contamination.
| Activities | Culture results | χ 2 (P value) | |
|---|---|---|---|
| Positive | Negative | ||
| Glove use | |||
| Yes | 2 | 38 | 4.64 (0.031) |
| No | 19 | 78 | |
| Touching disinfected phlebotomy site | |||
| Yes | 11 | 3 | 48.05 (0.000) |
| No | 10 | 113 | |
| Double puncture | |||
| Yes | 9 | 1 | 46.34 (0.000) |
| No | 12 | 115 | |
| Use of tincture of iodine for disinfection | |||
| Yes | |||
| No | 21 | 116 | |
3.4. Antimicrobial Susceptibility Test Result
The bacterial isolates showed diverse susceptibility patterns to the antibiotics tested. All the bacteria isolated were 100% sensitive to Gentamicin, Chloramphenicol, Amoxicillin, and Doxycycline. All Gram-positive isolates were 100% sensitive to Vancomycin, Ciprofloxacin, and all Gram-negative isolates were 100% sensitive to Ceftriaxone and Erythromycin. However, all Gram-negative bacteria were 100% resistant to Penicillin G (Table 3). Multiple antimicrobial resistances were observed in 66.7% (14/21) of the isolated bacteria.
Table 3.
Antimicrobial susceptibility pattern bacteria isolated from blood units collected for transfusion.
| Drugs tested | Species isolated n (%) | ||||||
|---|---|---|---|---|---|---|---|
| S. aureus | CoNS | Streptococci | E. coli | Klebsiella | Enterobacter | Citrobacter | |
| AMP | |||||||
| S | 5 (55.6) | 2 (50) | 1 (100) | 2 (66.7) | 2 (100) | 0 (0) | 1 (100) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| R | 4 (44.4) | 2 (50) | 0 (0) | 1 (33.3) | 0 (0) | 0 (0) | 0 (0) |
| CN | |||||||
| S | 9 (100) | 4 (100) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 1 (100) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| MET | |||||||
| S | 8 (88.9) | 4 (100) | 1 (100) | NA | NA | NA | NA |
| I | 1 (11.1) | 0 (0) | 0 (0) | NA | NA | NA | NA |
| R | 0 (0) | 0 (0) | 0 (0) | NA | NA | NA | NA |
| TE | |||||||
| S | 7 (77.8) | 4 (100) | 1 (100) | 2 (66.7) | 1 (50) | 1 (100) | 0 (0) |
| I | 0 (0) | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) | 0 (0) | 1 (100) |
| R | 2 (22.2) | 0 (0) | 0 (0) | 0 (0) | 1 (50) | 0 (0) | 0 (0) |
| VA | |||||||
| S | 9 (100) | 4 (100) | 1 (100) | NA | NA | NA | NA |
| I | 0 (0) | 0 (0) | 0 (0) | NA | NA | NA | NA |
| R | 0 (0) | 0 (0) | 0 (0) | NA | NA | NA | NA |
| P | |||||||
| S | 4 (44.4) | 2 (50) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| R | 5 (55.6) | 2 (50) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 1 (100) |
| CIP | |||||||
| S | 9 (100) | 4 (100) | 1 (100) | 0 (0) | 1 (50) | 0 (0) | 0 (0) |
| I | 0 (0) | 0 (0) | 0 (0) | 2 (66.7) | 0 (0) | 0 (0) | 0 (0) |
| R | 0 (0) | 0 (0) | 0 (0) | 1 (33.3) | 1 (50) | 1 (100) | 1 (100) |
| C | |||||||
| S | 9 (100) | 4 (100) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 1 (100) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| SXT | |||||||
| S | 8 (88.9) | 4 (100) | 1 (100) | 3 (100) | 1 (50) | 0 (0) | 1 (100) |
| I | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) | 1 (50) | 1 (100) | 0 (0) |
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| DA | |||||||
| S | 7 (77.8) | 4 (100) | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) |
| I | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| R | 2 (22.2) | 0 (0) | 0 (0) | 3 (100) | 1 (50) | 1 (100) | 0 (0) |
| CRO | |||||||
| S | 9 (100) | 2 (50) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 0 (0) |
| I | 0 (0) | 2 (50) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| AMC | |||||||
| S | 9 (100) | 4 (100) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 1 (100) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| E | |||||||
| S | 7 (77.8) | 4 (100) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 1 (100) |
| I | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| R | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| DO | |||||||
| S | 9 (100) | 4 (100) | 1 (100) | 3 (100) | 2 (100) | 1 (100) | 1 (100) |
| I | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| R | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
AMP: Ampicillin; CN: Gentamicin; TE: Tetracycline; CIP: Ciprofloxacin; C: Chloramphenicol; SXT: Trimethoprim-sulfamethoxazole; CRO: Ceftriaxone; AMC: Amoxicillin; P: Penicillin G; MET: Methicillin; VA: Vancomycin; DA: Clindamycin; E: Erythromycin; DO: Doxycycline; NA: not applicable; CoNS: Coagulase negative Staphylococci species; S: sensitive; I: intermediate; R: resistance.
4. Discussion
The present study showed 15.33% bacterial contamination rate of whole blood collected for transfusion at University of Gondar hospital blood bank which is strikingly higher than the rates detected in United kingdom (0.19%), Canada (0.2 to 0.4%), and Japan (6.3%) [12, 21, 27]. The lower prevalence in other countries may be due to close surveillance of emerging infections and the meticulous care of blood collection procedure with stringent donor selection and presence of efficient infection prevention controls protocol, which are too poor in developing countries [1, 28, 29]. The current result is also higher than rates reported from African countries including Ghana (9%), Nigeria (8.8%), and Kenya (8.8%) [16, 18, 19]. The higher prevalence in our study may be due to the unusual practice of glove use and touching the site of phlebotomy after disinfection as well as double puncture which are observed in this study area even though such factors are not well reported from other related studies, but this rate is relatively lower than reports from Ghana where contamination rate was 17.5% [30].
The most frequently isolated bacteria were S. aureus followed by Coagulase negative Staphylococci, E. coli and Klebsiella species. Similar findings were reported elsewhere [14, 16, 18, 22, 27, 30, 31].
The potential source of bacterial contamination of blood collected for transfusion is either bacteria in the donor's blood because of an underlying condition causing donor bacteremia or external contaminants introduced at the time of phlebotomy [9, 14, 15]. The source of bacterial contaminant of sampled blood units accounted by the donor bacteremia was only 4.76%. However, 95.21% of contaminations were contributed by contaminants introduced at the time of phlebotomy. Different reports indicated that proper blood donor skin disinfection has long been recognized as a definite way to reduce blood contamination [27, 32]. This is strongly supported by this study as glove use, touching the disinfected phlebotomy site, and double puncture have shown statistically significant association with the bacteriological culture positivity of blood units. In addition the isolates obtained in our study were mostly skin associated organisms, and total coli form groups, which are often considered contaminants related to procedure during blood collection rather than donor bacteremia.
The antimicrobial resistance pattern observed in this study was in agreement with reports from Ghana, where most of the isolated organisms showed to be susceptible to Gentamicin and Erythromycin while they were resistant to Penicillin G and Tetracycline. The rate of multiple antimicrobial resistances observed was also in agreement with these studies [19, 30].
The resistance rates in the organisms isolated highlight the growing problem of antimicrobial resistance. The risks of transfusing contaminated donor blood are high, and transfusing blood with drug resistant strains of bacteria may worsen the difficulty of the already sick and the immunocompromised individuals as these organisms are capable of causing serious risk of fatality when transfused to patients [32].
5. Conclusion
Knowledge of the prevalence of bacterial contamination of blood for transfusion and the sources or causes of contamination in different parts of the world, particularly in Africa, is important for the planning of preventive measures at blood transfusion centers and the reduction of TABI.
From this study it can be concluded that bacterial contamination of donated blood is highly prevalent in the study area which indicates a potential risk of health care associated infection to patients. This has been approved by study conducted in USA [33]. Moreover high resistance patterns observed for single and multiple antimicrobials are also a great concern that needs urgent attention. Therefore, this study calls a need for supervision and corrective actions for mistakes that are made during blood collection, for introduction of policies for safe transfusion practices, and further research to clarify the extent and nature of the problem.
Conflict of Interests
The authors announce that they have no any competing interests.
Acknowledgments
The authors' gratefulness is also extended to the Amhara Regional Health Bureau for financial support, University of Gondar Hospital laboratory for materials and reagents support, and the blood donors for their voluntary participation in the study.
References
- 1.Vamvakas EC, Blajchman MA. Transfusion-related mortality: the ongoing risks of allogeneic blood transfusion and the available strategies for their prevention. Blood. 2009;113(15):3406–3417. doi: 10.1182/blood-2008-10-167643. [DOI] [PubMed] [Google Scholar]
- 2.Jani P, Howard A. Blood transfusion in surgery in Africa. East and Central African Journal of Surgery. 2011;16:2073–9990. [Google Scholar]
- 3.Diro E, Alemu S, Yohannes A. Blood safety & prevalence of transfussion transmissible viral infections among donors at the Red Cross Blood Bank in Gondar University Hospital. Ethiopian Medical Journal. 2008;46(1):7–13. [PubMed] [Google Scholar]
- 4.Bloch EM, Vermeulen M, Murphy E. Blood transfusion safety in Africa: a literature review of infectious disease and organizational challenges. Transfusion Medicine Reviews. 2012;26(2):164–180. doi: 10.1016/j.tmrv.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendel S. Rational testing for transmissible diseases. ISBT Science Series. 2007;2(1):19–24. [Google Scholar]
- 6.Tapko J, Mainuka P, Diarra-Nama AJ. Status of Blood Safety in the WHO African Region: Report of the 2006 Survey. http://www.afro.who.int/en/clusters-a-programmes/hss/blood-safety-laboratories-a-health-technology/htl-publications.html.
- 7.Tagny CT, Mbanya D, Tapko J-B, Lefrère J-J. Blood safety in Sub-Saharan Africa: a multi-factorial problem. Transfusion. 2008;48(6):1256–1261. doi: 10.1111/j.1537-2995.2008.01697.x. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb T. Hazards of bacterial contamination of blood products. Anaesthesia and Intensive Care. 1993;21(1):20–23. doi: 10.1177/0310057X9302100108. [DOI] [PubMed] [Google Scholar]
- 9.Kuehnert MJ, Roth VR, Haley NR, et al. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion. 2001;41(12):1493–1499. doi: 10.1046/j.1537-2995.2001.41121493.x. [DOI] [PubMed] [Google Scholar]
- 10.Ribault S, Faucon A, Grave L, Nannini P, Faure IB. Detection of bacteria in red blood cell concentrates by the scansystem method. Journal of Clinical Microbiology. 2005;43(5):2251–2255. doi: 10.1128/JCM.43.5.2251-2255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alter HJ, Klein HG. The hazards of blood transfusion in historical perspective. Blood. 2008;112(7):2617–2626. doi: 10.1182/blood-2008-07-077370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreier J, Störmer M, Kleesiek K. Real-time polymerase chain reaction in transfusion medicine: applications for detection of bacterial contamination in blood products. Transfusion Medicine Reviews. 2007;21(3):237–254. doi: 10.1016/j.tmrv.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Vasconcelos E, Seghatchian J. Bacterial contamination in blood components and preventative strategies: an overview. Transfusion and Apheresis Science. 2004;31(2):155–163. doi: 10.1016/j.transci.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Sugai Y, Sugai K, Fuse A. Current status of bacterial contamination of autologous blood for transfusion. Transfusion and Apheresis Science. 2001;24(3):255–259. doi: 10.1016/s1473-0502(01)00067-2. [DOI] [PubMed] [Google Scholar]
- 15.Kleinman S, Chan P, Robillard P. Risks associated with transfusion of cellular blood components in Canada. Transfusion Medicine Reviews. 2003;17(2):120–162. doi: 10.1053/tmrv.2003.50009. [DOI] [PubMed] [Google Scholar]
- 16.Hassall O, Maitland K, Pole L, et al. Bacterial contamination of pediatric whole blood transfusions in a Kenyan hospital. Transfusion. 2009;49(12):2594–2598. doi: 10.1111/j.1537-2995.2009.02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janatpour KA, Paglieroni TG, Crocker VL, DuBois DJ, Holland PV. Visual assessment of hemolysis in red blood cell units and segments can be deceptive. Transfusion. 2004;44(7):984–989. doi: 10.1111/j.1537-2995.2004.03315.x. [DOI] [PubMed] [Google Scholar]
- 18.Bolarinwa RA, Aboderin OA, Odetoyin BW, Adegunloye AB. Bacterial contamination of blood and blood components in a tertiary hospital setting in Nigeria. International Journal of Infection Control. 2011;7:1–11. [Google Scholar]
- 19.Adjei AA, Kuma GK, Tettey Y, et al. Bacterial contamination of blood and blood components in three major blood transfusion centers, Accra, Ghana. Japanese Journal of Infectious Diseases. 2009;62(4):265–269. [PubMed] [Google Scholar]
- 20.Asher D, Atterbury LJ, Chapman C, et al. Serious Hazards of Transfusion. SHOT Annual Report 2000-2001. 2002, http://www.shotuk.org/wp-content/uploads/2010/03/SHOT-Report-00-01.pdf. [Google Scholar]
- 21.Seghatchian J. Bacterial contamination of blood components. Transfusion and Apheresis Science. 2001;25(2):147–150. doi: 10.1016/s1473-0502(01)00096-9. [DOI] [PubMed] [Google Scholar]
- 22.Wagner SJ. Transfusion-transmitted bacterial infection: risks, sources and interventions. Vox Sanguinis. 2004;86(3):157–163. doi: 10.1111/j.0042-9007.2004.00410.x. [DOI] [PubMed] [Google Scholar]
- 23.Kleinman S, Chan P, Robillard P. Risks associated with transfusion of cellular blood components in Canada. Transfusion Medicine Reviews. 2003;17(2):120–162. doi: 10.1053/tmrv.2003.50009. [DOI] [PubMed] [Google Scholar]
- 24.Blood Culture Bottle for Use in the Culture of Microorganisms. A Qualitative Test for the Detection of Microorganisms in Blood. Guidelines of Laboratory Procedure. 1989, http://www.bd.com/ds/technicalCenter/clsi/clsi-septicheck.pdf. [Google Scholar]
- 25.Vandepitte J, Verhaegen J, Engbaek K, Rohner P, Piot P, Heuck CC. Basic Laboratory Procedures in Clinical Bacteriologyed. 2nd edition. Geneva, Switzerland: World Health Organization; 2003. [Google Scholar]
- 26.Cheesbrough M. District Laboratory Practice in Tropical Countries. 2nd edition. Cambridge, UK: Cambridge University Press; 2006. [Google Scholar]
- 27.Brecher ME, Hay SN. Bacterial contamination of blood components. Clinical Microbiology Reviews. 2005;18(1):195–204. doi: 10.1128/CMR.18.1.195-204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodd R, Roth WK, Ashford P, Dax EM, Vyas G. Transfusion medicine and safety. Biologicals. 2009;37(2):62–70. doi: 10.1016/j.biologicals.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Liumbruno GM, Catalano L, Piccinini V, Pupella S, Grazzini G. Reduction of the risk of bacterial contamination of blood components through diversion of the first part of the donation of blood and blood components. Blood Transfusion. 2009;7(2):86–93. doi: 10.2450/2008.0026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opoku-Okrah C, Feglo P, Amidu N, Dakorah MP. Bacterial contamination of donor blood at the Tamale Teaching Hospital, Ghana. African Health Sciences. 2009;9(1):13–18. [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs MR, Palavecino E, Yomtovian R. Don’t bug me: the problem of bacterial contamination of blood components—challenges and solutions. Transfusion. 2001;41(11):1331–1334. doi: 10.1046/j.1537-2995.2001.41111331.x. [DOI] [PubMed] [Google Scholar]
- 32.Perez P, Salmi LR, Folléa G, et al. Determinants of transfusion-associated bacterial contamination: results of the French BACTHEM Case-Control Study. Transfusion. 2001;41(7):862–872. doi: 10.1046/j.1537-2995.2001.41070862.x. [DOI] [PubMed] [Google Scholar]
- 33.Martínez F, Tarrand J, Lichtiger B. Impact on patient outcome following transfusion of bacterially contaminated platelets: the M.D. Anderson Cancer Center experience. American Journal of Clinical Pathology. 2010;134(2):207–212. doi: 10.1309/AJCPS9YV7YTOMRRH. [DOI] [PubMed] [Google Scholar]


