Abstract
Protein S-nitrosation is a reversible post-translation modification critical for redox-sensitive cell signaling that is typically studied using the Biotin Switch method. This method and subsequent modifications usually require avidin binding or Western blot analysis to detect biotin labeled proteins. We describe here a modification of the Biotin Switch assay that eliminates the need for Western blot or avidin enrichment protocols and allows direct comparison of the S-nitrosation state proteins from two different samples in the same gel lane or on the same 2D gel. This S-FLOS method offers detection, identification and quantification of S-nitrosated proteins, with the potential for site-specific identification of nitrosation events.
Keywords: S-nitrosylation, Nitric oxide, Biotin switch, SNO, Proteomics
Protein S-nitrosation (also referred to as S-nitrosylation), a reversible post-translation modification of cysteines, affects many cell signaling pathways [1,2]. Emerging evidence suggests that dysregulation of this redox-sensitive modification is a marker of, or contributes to the pathophysiology of many disease processes including arthritis, pre-eclampsia, asthma, and stroke[3,4].
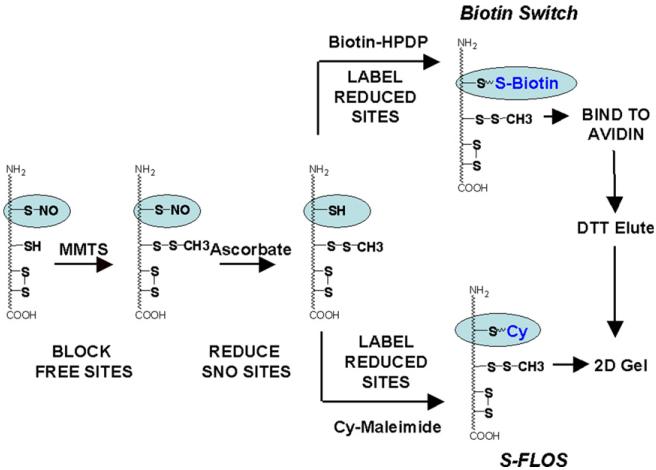
Rapid and global detection of biologically relevant nitrosated proteins would help to identify novel NO signaling pathways and molecular mechanisms of many redox-sensitive pathophysiologies[5,6]. The Biotin Switch assay [2] is one of several methods ([7] for recent review) that has been used to study S-nitrosation in a variety of proteomes [8-11]. It involves three steps aimed at replacing the cysteine linked nitrosothiol with a biotin tag at the S-nitrosation sites: (1) block free thiols, (2) selectively reduce S-nitrosated cysteines, and (3) biotinylate the newly released cysteine thiols (Fig. 1). The nitrosated proteins are then detected by Western blotting for the biotin label or enriched using streptavidin resins or anti-biotin antibodies for proteomic applications. S-nitrosation sites can be identified by analyzing biotinylated peptides from proteolytic digests that bind to an avidin column as in the SNOSID assay [6,12].
Fig. 1.
Comparison of the Biotin Switch and S-FLOS labeling schemes.
Thus, the Biotin Switch assay is a powerful method to identify and map protein S-nitrosation. However, this method is dependent on the efficiency and specificity of thiol blocking and ascorbate reduction, and requires secondary methods of detection via avidin binding or Western blot analysis. Moreover, quantifying relative changes in S-nitrosation between samples requires low background, a high signal to noise ratio and highly reproducible band or spot matching of avidin binding or antibody signals in different gel lanes or on separate 2D gels. We describe here a modification of the Biotin Switch assay; Selective Fluorescent Labeling Of S-nitrosothiols (S-FLOS, Fig. 1) which eliminates the need for Western blot or avidin enrichment protocols and allows direct comparison of the S-nitrosation state between two samples in the same gel lane or on the same 2D gel with a low background and a high signal to noise ratio. In addition, our method2 is compatible with in situ tissue staining (Fig. S2 in supplemental data), analysis of mitochondrial protein S-nitrosation [13], and identification of S-nitrosated protein by mass spectrometry with the potential to map S-nitrosation sites.
Experimental procedures
Protein extracts from mouse brains or cell cultures
NOS1 knockout (NOS1−/−) and wild-type C57BL/6J mice (9–11 weeks old) were purchased from The Jackson Laboratory (jax-mice.jax.org). All animals were treated according to NIH guidelines. Mice were anesthetized and perfused with normal saline to remove blood. Brains were dissected, snap frozen, and stored at −80 °C. Frozen brains (1 gm) in 5 ml of Lysis Buffer (50 mM Tris–HCl buffer, pH 7.5) containing protease inhibitors (Roche Diagnostics Corp., www.roche-applied-science.com) and 10 μM neocuproine (Sigma-Aldrich Co., www.sigmaaldrich.com) were homogenized (VWR PowerMax™ AHS 200, VWR International, LLC, www.vwrsp.com) for three rounds of 45 s homogenizing and 2 min on ice. Homogenates were centrifuged at 10,000 rpm at 4 °C for 5 min to recover supernatant. Mouse RAW264.7 cells were purchased from ATCC Global Bioresource Center (www.atcc.org) and cultured according to ATCC protocols. After removing culture medium, cells were lysed by scraping in Lysis Buffer. Protein concentrations of brain homogenates or cell lysates were determined using Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., www.bio-rad.com), adjusted to 1 mg/ml with Lysis Buffer and used immediately.
GSNO transnitrosation of purified proteins or cell lysates
One milligram/milliliter of Creatine Kinase (0.25 mg/ml), Bovine Serum Albumin (0.25 mg/ml), Arginase I (0.25 mg/ml), and Lysozyme (0.25 mg/ml) (Sigma-Aldrich Co.) or 1 mg/ml RAW264.7 cell lysates in 100 μl of 50 mM Tris–HCl, pH 7.4, containing 1% SDS and 10 μM neocuproine were incubated with GSNO (3–100 μM) for 30 min at room temperature in the dark with constant agitation. For Fig. 2C, 20 mg/ml BSA solution in 500 μl of a 50 mM Tris–HCl (pH 7.5) buffer was treated with increasing doses of GSNO for 1 h at room temperature in the dark.
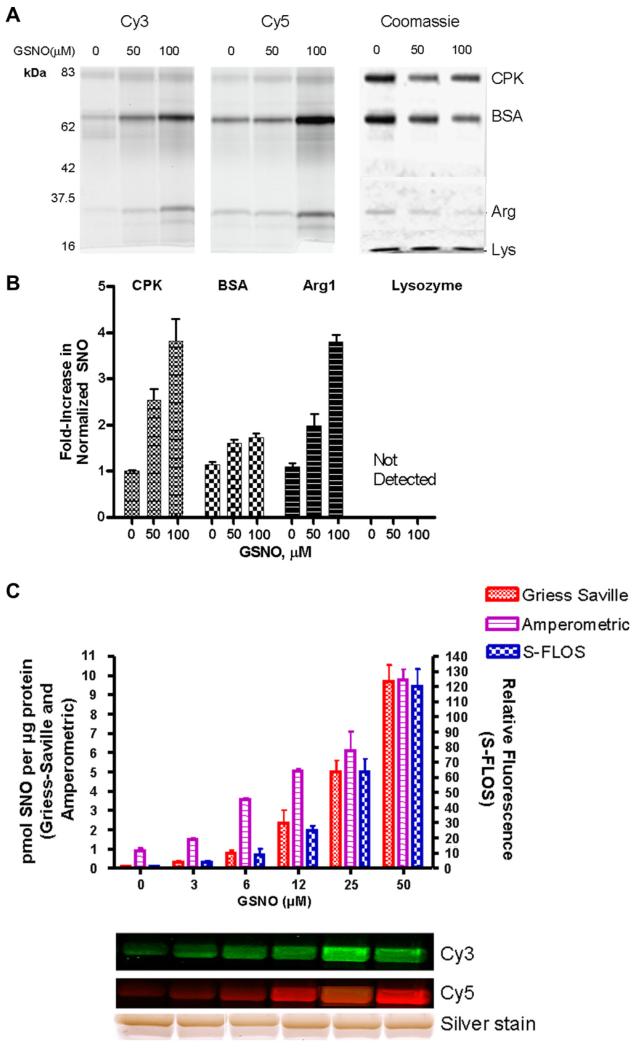
Fig. 2.
Detecting and quantifying S-nitrosation in purified proteins transnitrosated with GSNO using the S-FLOS assay. (A) A 100 μg mixture of Creatine Kinase (CPK), Bovine Serum Albumin (BSA), Arginase I (Arg), and Lysozyme (Lys) treated with 0, 50 and 100 μM GSNO was labeled with either Cy3 or Cy5 using the S-FLOS assay. Cy3 (12.5 μg) and Cy5 (12.5 μg) labeled proteins were mixed, separated by SDS–PAGE and visualized using a Typhoon fluorescence scanner (Cy3 left panel, Cy5 middle panel). After fluorescence imaging, the gel was stained for total protein using Coomassie blue (right panel) to determine protein recovery. (B) Relative changes in fluorescence from the four proteins in A were quantified using ImageQuant and normalized to protein content from densitometry of Coomassie blue stained bands. Histograms represent averages from three Cy3-and three Cy5-labeled samples. Error bars represent +/− standard deviations of three independent experiments. Fluorescence intensity for lysozyme was not detected, suggesting that lysozyme is not nitrosated under these conditions. (C) Two hundred microgram of BSA (1 mg/ml) was treated with different doses of GSNO (0–50 μM) for 30 min at 37 °C in the dark. BSA–SNO content for 100 μg of BSA at each GSNO dose was quantified using an amperometeric assay (6 replicates) and the remaining 100 μg BSA using the S-FLOS assay (6 replicates) as in A. Five microgram of Cy3 or Cy5-label BSA was analyzed by SDS–PAGE and imaged for fluorescence (middle panels) or total protein (silver stain, bottom panel). S-FLOS relative fluorescence intensities were calculated from fluorescence with GSNO per μg protein divided by fluorescence without GSNO per μg protein using ImageQuant and normalized to the protein load based on densitometry silver stain images. BSA–SNO content was also quantified using the Griess–Saville assay alone (9 replicates) from 1 mg BSA (20 mg/ml) treated with increasing doses of GSNO for 1 h at room temperature in the dark. Error bars represent +/− standard deviation.
Nitric oxide synthase stimulation in cell cultures
RAW264.7 cells were cultured for 0–48 h in the presence of 100 U/ml γ interferon (IFNγ, Roche Diagnostics Corp.) and 5 μg/ml bacterial lipopolysaccharide (LPS, Sigma-Aldrich Co.) before harvesting and lysis.
S-FLOS assay
The entire assay, through scanning, was performed in the dark. Free cysteines in 1 mg/ml of purified proteins or protein extracts were blocked in 100 μl of 20 mM methyl methanethiosulfonate (MMTS) in 50 mM Tris–HCl, 10 μM neocuproine and 1% SDS at 50 °C for 1 h. Excess GSNO and MMTS were removed by precipitation with four volumes of cold acetone for 1 h. Proteins were redissolved in 100 μl Reducing Buffer (50 mM Tris–HCl pH 7.4, 4% CHAPS, 5 mM ascorbate) and incubated at room temperature for 1 h. The reduced proteins were buffer exchanged into 120 μl Labeling Buffer (50 mM Tris–HCl pH 7.0, 7 M Urea, 4% CHAPS) using protein desalting spin columns (Pierce, www.piercenet.com). To optimize Cy-dye labeling, Cy-dye was titrated from 4 to 40 pmol dye per μg protein. For the purified proteins, 5 μg of each protein sample was labeled with 10 pmol of Cy3 or Cy5-maleimide (Cy-Dye DIGE Fluor for Scarce Samples, GE Healthcare, www5.gelifesciences.com) at 37 °C for 30 min. For cell lysates or brain homogenates, protein concentration was determined after buffer exchange into Labeling Buffer using the 2D Quant kit (GE Healthcare). 12.5 μg of total protein was then labeled with 40 pmol of either Cy3 or Cy5-maleimide at 37 °C for 30 min. The ascorbate reduction and Cy-dye labeling steps were performed sequentially because ascorbate sometimes interfered with the Cy-dye labeling. The Cy3 and Cy5-labeled proteins were either resolved individually using SDS–PAGE or for direct comparison, mixed and resolved using either 1D or 2D gel electrophoresis. Fluorescence images were acquired on a Typhoon 9400 (GE Healthcare) scanning at 500 PMT with excitation/emission filters for Cy3 or Cy5. ImageQuant (version 5.2, GE Healthcare) was used to determine relative fluorescence intensities. Gels were post stained with colloidal coomassie blue (SimplyBlue SafeStain, Invitrogen Corp., catalog.invitrogen.com) or silver (Silver Stain Plus kit, Bio-Rad Laboratories, Inc.) and scanned to determine protein loads using densitometry (UMAX PowerScan III).
Biotin Switch assay
Nitrosated proteins were detected based on a modified Biotin Switch assay using the NitroGlo assay kit (Perkin-Elmer Inc., las.perkinelmer.com) according to the manufacturer’s protocol. Biotinylated proteins were enriched as previously described [15] by binding 50 μl of streptavidin-agarose beads (Pierce) at 4 °C overnight.
2D Gel electrophoresis
Cy3 and Cy5-labeled protein samples (12.5 μg of each) were mixed and buffer exchanged into 120 μl of Rehydration Buffer (8 M Urea, 4% CHAPS, 0.2% DTT, 0.0002% bromophenol blue) using protein desalting spin columns. Ampholytes (1.8 μl of pH 4–7 IPG buffer, GE Healthcare) were added, and the proteins were resolved on a 7 cm pH 4–7 IPG strip (GE Healthcare) followed by 4–12% gradient SDS–PAGE (NuPage, Invitrogen). All gels were run in the dark.
Protein identification
Silver stained protein spots excised from 2D gels were destained using 30 μl of 1:1 mixture of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate according to Gharahdaghi et al., 1999 [16] and digested with trypsin (sequencing grade, Promega Corp., www.promega.com) in 20 mM ammonium bicarbonate at 37 °C overnight as previously described [17]. Extracted peptides were fractionated on a 5–40% acetonitrile gradient in 0.1% formic acid over 25 min at 300 nl/min on a 75 μm × 100 mm column with a 8 μm emitter (New Objectives, Inc., www.newobjective.com) and packed with 5 μm, 120Å C18 beads (YMC ODS-AQ, Waters Corp., www.waters.com). Eluting peptides were analyzed by collision-induced dissociation (CID) using nanoLC tandem mass spectrometry analysis on a QSTAR/Pulsar (Applied Biosystems/MDX Sciex, home.appliedbiosystems.com) interfaced with an Eksigent 2D nano-LC system (www.eksigent.com). Survey scans were acquired from m/z 350–1200 with up to three precursors selected for MS/MS using a dynamic exclusion of 30 s. A rolling collision energy was used to promote fragmentation. Peptide sequences were identified by screening the fragmentation data against the NCBI non-redundant database using in-house Mascot server and Mascot Daemon as an interface. Peptides with scores higher than Mascot’s calculated 95% confidence probability threshold were considered reliable peptide sequences. Proteins with two or more reliable peptides were considered significant protein identifications.
Amperometric detection of S-nitrosothiols
Absolute levels of S-nitrosothiols were determined using an amperometric NO probe (ISO-NOP70L probe, World Precision Instruments, Inc., www.wpiinc.com) according to vendor’s protocol and based on Zhang et al., 2002 [18]. In brief, the ISO-NOP70L probe was polarized in 20 ml of 50 mM Tris–HCl buffer containing 10 mM Cu2+ at room temperature and calibrated by adding increasing concentrations of GSNO and SNAP. BSA (1 mg/ml) samples were treated with GSNO as described above. Excess GSNO was removed by acetone precipitation in four volumes of cold acetone for 1 h followed by desalting on protein desalting spin columns into 120 μl 50 mM Tris–HCl (pH 7.4). Protein concentration was determined with Bio-Rad Protein Assay in order to add 100 μg of BSA directly to the probe bath to determine the BSA–SNO levels. For untreated and low GSNO doses, 500–1000 μg BSA was required to detect signal.
Nitrite detection in cell culture media (Griess assay)
Nitrite concentrations accumulating in cell culture media or released from BSA using 4 mM Hg2+ was determined using the Nitrite/Nitrate assay kit (NOS Assay Kit, Colorimetric, Calbiochem, www.emdbiosciences.com/html/CBC/home.html) following manufacturer’s instructions and is based on Nims et al., 1996 [19]. Briefly, cell cultures and BSA samples were treated as described above. Proteins were recovered by cold acetone precipitation followed by protein desalting columns into 50 mM Tris–HCl (pH 7.5) buffer. Volumes were made up to 500 μl in each sample. For each sample, protein concentrations were measured by Bio-Rad Protein Assay and SNO content was measured by the NOS Assay Kit calibrated with GSNO standards.
Results
The S-FLOS assay requires the same blocking and reduction steps as the Biotin Switch assay (Fig. 1). However, instead of HPDP–Biotin, S-FLOS uses maleimide conjugated Cy-dyes under conditions optimized for selectively labeling only reduced cysteine thiols, leaving disulfides intact. These Cy-dyes have been successfully used to detect free thiols in protein samples [20,21], but have not been used to detect or quantify S-nitrosated proteins. Two Cymaleimide dyes (Cy3 and Cy5) are commercially available from GE Healthcare and allow direct comparison of two different protein samples on the same electrophoresis gel. Relative differences between the label proteins can then be quantified using fluorescent imaging technology. Other maleimide linked dyes such as Alexa Fluors, Texas red, and BODIPY can also be used for SDS–PAGE and in situ staining applications; however, unlike Cy-dyes, these dyes are not charge balanced and, thus, are unsuitable for 2D gels.
To determine if the S-FLOS method can detect changes S-nitrosation, a mixture of four proteins were exogenously nitrosated by direct S-transnitrosation using S-nitrosoglutathione (GSNO) (Fig 2A). Hundred micrograms of a mixture of Creatine Kinase (CPK), Bovine Serum Albumin (BSA), Arginase I, and Lysozyme was incubated with increasing doses of GSNO. CPK [22] and BSA [23] are known targets of nitrosation; and Arg 1 has been identified recently as a target for S-nitrosation in this laboratory [24]. Lysozyme served as a negative control for the S-FLOS assay. The GSNO treatment was followed by labeling ‘nitrosated’ proteins with either Cy3 or Cy5 using the S-FLOS assay (Fig 2A). The fluorescence intensities were calculated using ImageQuant software, and normalized to protein levels (Fig 2B). Although reproducible losses in protein recovery occurred during the acetone precipitation step in the presence of GSNO (Fig 2A right panel), fluorescence intensities clearly increased in samples treated with GSNO (Fig. 2A, left and middle panels). Relative increases in fluorescence intensities were similar for either dye as demonstrated by small error bars in Fig 2B. GSNO can also glutathionylate proteins such as creatine kinase [25], ([26] for review). The conditions driving nitrosation versus glutathionylation are not understood. BSA, however, is only S-nitrosated by GSNO [25]. Following treatment with GSNO, both the amperometric probe and the Griess-Saville method independently measured an increase in the S-nitrosothiol content of BSA (Figure 2C). Therefore, S-FLOS can detect and quantify relative changes in exogenously S-nitrosated proteins. It has been reported that it is difficult to reduce S-nitrosated albumin with ascorbate [7]. However, the S-FLOS signal from BSA increased with increasing transnitrosation (Fig. 2), but was absent when the ascorbate reduction step was omitted (data not shown).
The S-FLOS assay also detected changes in S-nitrosation in RAW264.7 cells protein extracts (Fig. 3). The S-FLOS signal markedly increased in protein extracts treated with GSNO (Fig 3A, GSNO + lanes) as compared to the untreated extracts (Fig 3A GSNO − lanes). The source of background S-FLOS signal in the untreated extracts was not determined. However, pretreating proteins with ascorbate before the S-FLOS assay led to a nearly complete loss of the S-FLOS signal (Fig 3A, Asc + lanes).
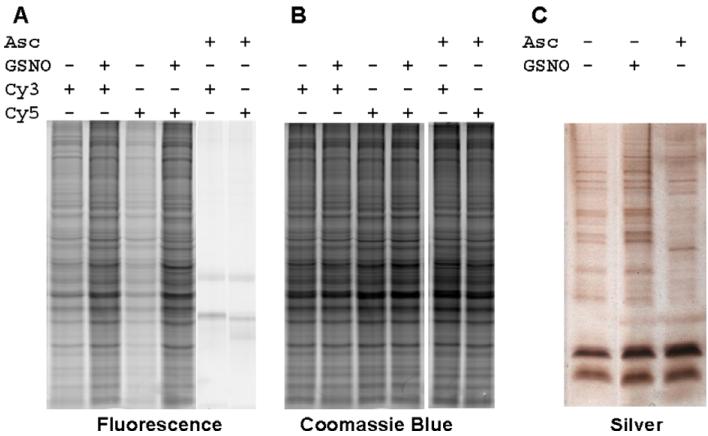
Fig. 3.
Comparison of the Biotin Switch and S-FLOS assays in RAW264.7 cells. (A) Proteins (25 μg) from lysates of RAW264.7 cells treated with (lanes 2 and 4) or without (lanes 1, 3, 5 and 6) 100 μM GSNO were labeled with Cy3 or Cy5 using S-FLOS, separated by SDS–PAGE and imaged for fluorescence. A clear increase in fluorescence is observed in treated samples (lanes 2 and 4). Proteins in lanes 5 and 6 were pretreated with ascorbate (Asc +) prior to the S-FLOS assay and completely blocked the fluorescence labeling. Lanes 5 and 6 were scanned at a higher PMT (650 versus 500 for lanes 1–4) for increased fluorescence detection. (B) Coomassie blue staining for total protein of gel in Fig. 3A showing equal protein load all lanes. (C) For comparison with S-FLOS analysis, the Biotin Switch assay was performed on the same samples in Fig. 3A according to Jaffrey and Snyder, 2001 [15]. After HPDP–biotinylation, nitrosated proteins were enriched using streptavidin coated agarose, resuspended in Laemmli sample buffer, resolved by SDS–PAGE, and silver stained for total protein. Differences in GSNO treated (lane 2) versus untreated (lane 1) proteins were similar to S-FLOS results. However, pre-treatment with ascorbate (lane 3) prior to the Biotin Switch assay did not eliminate proteins binding to streptavidin.
The same experiment was performed using HPDP–biotin in the Biotin Switch assay. Biotinylated proteins isolated by binding to avidin-linked beads were analyzed on silver stained gels (Fig. 3C). The intensity of many of the protein bands increased after treating with GSNO. The Biotin Switch assay, however, has a high background noise since many protein bands were still present after pretreating proteins with ascorbate before the Biotin Switch assay (Fig 3C, Asc + lane). These ‘background’ proteins were likely non-specific binding proteins or endogenously biotinylated.
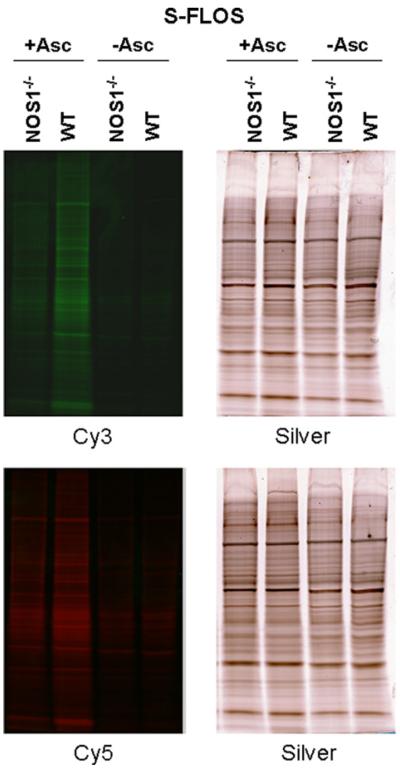
Brain homogenates of NOS1−/− mice show reduced S-FLOS signal as compared to wild-type brain homogenates (Fig. 4). Brain homogenates of wild-type and NOS1−/− mice were subjected to S-FLOS and resolved using SDS–PAGE. Differences in signal intensity between the wild-type and NOS1 knockout mice were clearly distinguished. The S-FLOS signal intensity was dependent on the ascorbate reduction step and results were similar with either Cy-dye. NOS1−/− samples were not devoid of signal presumably because of the presence of other NOS isoforms, particularly NOS3.
Fig. 4.
Comparison of S-FLOS labeling in brain homogenates from wild-type and NOS1 knockout mice. Protein extracts from wild-type (WT) and NOS1 knockout (NOS1−/−) mouse brains were labeled with either Cy3 (top panels) or Cy5 (bottom panels) with (+Asc) or without (−Asc) using the ascorbate reduction step in the S-FLOS assay. Labeled proteins (12.5 μg per lane) were separated by SDS–PAGE and imaged for fluorescence (left panels) or silver stained for total protein (right panels). Both Cy3 and Cy5 fluorescence are lower in NOS1−/− compared to WT samples and have very low signal in the absence of the ascorbate reduction step. Protein loads were equal in all lanes (right panels).
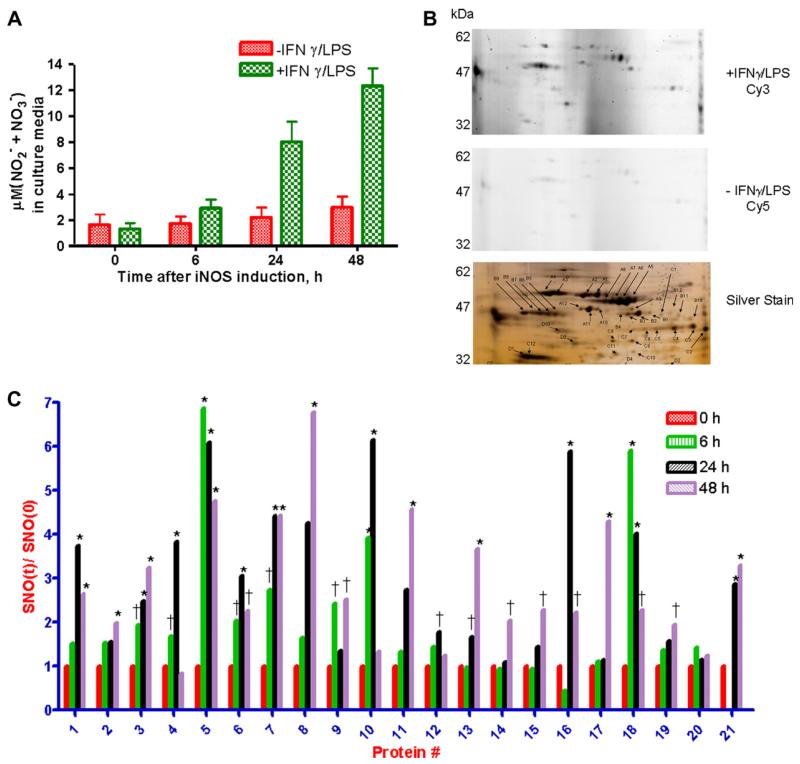
Proteins from RAW264.7 cells stimulated with 100 U/ml IFNγ and 5 μg/ml LPS to induce NOS2-dependent NO production, and thereby, S-nitrosation [8] were assayed for changes in the S-FLOS signal using 2D SDS–PAGE (Fig. 5). Proteins from three biological replicates were extracted at four time points over 48 h. Untreated RAW264.7 cells, harvested at the same time intervals as the treated samples, served as controls. Nitric oxide production was confirmed in stimulated cells by quantifying the accumulation of NO2− and NO3− in culture media using the Griess assay (Fig 5A). The lysates were subjected to S-FLOS. At each time point, stimulated samples were labeled with Cy3-maleimide; the unstimulated controls were labeled with Cy5-maleimide; Cy3 and Cy5-labeled proteins were mixed and resolved on a single 2D gel. Comparing the Cy3 and Cy5 fluorescent images after 24 h stimulation shows an increase in S-FLOS signal correlating with an increase in NO production in IFNγ/LPS stimulated RAW264.7 cells (Fig 5B, top and middle panels). Comparing the Cy3 image (Fig 5B top panel) and the silver stain image (Fig 5B, bottom panel) highlights the selectivity, and sensitivity of the S-FLOS method. Various high abundance proteins such as spots B2, C12, and D1 have no fluorescence signal whereas several low-abundance proteins (e.g. spot A9) have the fluorescent tag. Dye switching showed the same results (Fig. S1 in supplemental data).
Fig. 5.
Relative quantification of NOS2-dependant nitrosation in IFNγ/LPS stimulated RAW264.7 cells using S-FLOS. (A) Using the Griess method, NO production was measured in culture media at 0, 6, 24 and 48 h from RAW264.7 cells incubated with or without 100 U/ml IFNγ/5 μg/ml LPS to induce endogenous NOS2-dependent NO production. Error bars represent +/− standard deviation for three biological replicates; (B) Protein (12.5 μg) from stimulated or unstimulated RAW264.7cells at the 24 h time point in A were labeled Cy3 or Cy5, respectively, using the S-FLOS assay. Cy3 and Cy5-labeled proteins were mixed, separated on a 2D SDS–PAGE gel, imaged for fluorescence (Cy3, top and Cy5, middle panels) and silver stained for total protein (bottom panel). Spots picked for identification by mass spectrometry are shown on silver stain image. (C) RAW264.7 cells treated with IFNγ/LPS were harvested at 0, 6, 24 and 48 h. As in B, at each time point protein extracts were subjected to S-FLOS, using Cy3 for stimulated cells and Cy5 for unstimulated controls and resolved on 2D gels. The relative changes in fluorescence in 21 proteins from stimulated cells, normalized to unstimulated controls, are shown. The data are averages of three independent experiments. Error bars are omitted for clarity (*p < 0.01, †p < 0.05).
The relative changes in fluorescence intensities of 21 spots were measured over the 48 h stimulation (Fig. 5C). Spots had different time dependent increases in fluorescent intensity, with the S-FLOS signal either peaking at different time points (e.g. proteins 5, 10 and 11) or not changing (e.g. protein 20). The subset of proteins associated with increased Cy fluorescence, excised and identified by tandem mass spectrometry, are presented in Table 1. Five are known targets for S-nitrosation. Two proteins are novel targets for S-nitrosation and also contain the loose consensus sequence for S-nitrosation ((K/R/H/D/E) C (D/E)) [27]. The two proteins that did not have Cy-fluorescence also do not have this consensus sequence and are not reported to be S-nitrosated in the literature.
Table 1.
Proteins identified from Fig. 5B with increased S-FLOS signal are known targets for S-nitrosation or contain the proposed consensus sequence
| Spot | Protein identification | Reference for nitrosation | S-FLOS signal |
|---|---|---|---|
| Known targets | |||
| A5/A6/A7 | Bovine serum albumin* | Rafikova et al. (2002) PNAS 99:5913 | Yes |
| B2 | Protein disulfide isomerase | Uehara et al. (2006) Nature 441:513 | Yes |
| B5–B7 | Cysteine proteinase inhibitor* | Salvati et al. (2001) Biochim. Biophys. Acta 1545:357 | Yes |
| 78-kDa Glucose-regulated protein (GRP78) | Moon et al. (2006) Hepatology 44:1218 | Yes | |
| C3/C4/C5 | Enolase1 | Gao et al. (2005) Nitric Oxide 12:121 | Yes |
| Novel targets | |||
| A2/A3 | Heat shock protein-70 | No reference contains “consensus” sequence | Yes |
| A4 | Heat shock 70kDa protein 5 | No reference contains “consensus” sequence | Yes |
| A11 | Heat shock protein-65 | No reference contains “consensus” sequence | Yes |
| Negative controls | |||
| D1/C12 | Nucleolar phospho-protein | No reference | |
| No consensus sequence | No | ||
| D2 | Aldolase A | No reference | |
| No consensus sequence | No |
A subset of proteins in spots excised from the gel shown in Fig. 5B (bottom panel) where digested with trypsin in gel. Extracted peptides were analyzed by tandem mass spectrometry. Proteins were identified using MASCOT and are based on two or more peptide sequences with scores above the 95% confidence (see supplemental data). S-FLOS signal is based on Cy3 fluorescence in Fig. 5B.
Proteins in spots A5/A6/A7 and B5/B6/B7 series are contaminants from growing cells in DMEM containing FBS. These proteins will be nitrosated as the cells release NO upon stimulation, and were detected by the S-FLOS assay.
Discussion
Proteins known to be modulated by S-nitrosation include structural proteins, ion-channels, and enzymes [1] which can be activated (e.g. ryanodine receptors [28], COX-2 [29], HIF-1 [30], arginase 1 [24]) or inhibited (creatine kinase, eNOS [31], caspases [1]) by this reversible redox-sensitive modification of specific cysteines. Identifying proteins whose activity/function is affected by changes in S-nitrosation is key to understanding the basic cellular mechanisms and the pathophysiology of many diseases. The Biotin Switch assay and some of its variations has played a significant role [8-11] in identifying S-nitrosated proteins and their S-nitrosation sites. Other methods for determining total free thiols in protein extracts using maleimide-linked Cy-dyes have recently been described [32]. Our S-FLOS method is a new modification2 of the Biotin Switch assay, using Cy-maleimide dyes in place of HPDP–Biotin. These dyes selectively form adducts with protein thiols at pH 7, which are not reduced by common reducing agents used in gel electrophoresis (e.g. DTT and TCEP) as is HPDP–Biotin. Detecting the fluorescent labeled proteins does not require Western blot analysis or an avidin enrichment step as with the Biotin Switch assay and this two Cy-dye system allows direct comparison the fluorescent intensities of proteins from two different samples on the same 2D gel.
Using any version of the Biotin Switch method to detect S-nitrosation in proteomic studies, however, raises concerns over the selectivity and sensitivity of the thiol blocking and S-nitrosothiol reducing agents, as well as the efficiency of blocking a large total protein thiol signal to detect a small S-nitrosation content [7]. Although the possibility that the S-FLOS assay also detects disulfides and relatively rare sulfenic acid modifications of thiols [33] is not completely ruled, our results demonstrate that the S-FLOS method can detect changes in S-nitrosation. The S-FLOS signal increased under the same conditions and with the same time dependency as increases in total protein S-nitrosothiol content as measured by the amperometric probe or the Griess–Saville method. Proteins having an increase S-FLOS signal under these conditions and identified by mass spectrometry where previously shown to be S-nitrosated or have the loose consensus sequence for S-nitrosation ((K/R/H/D/E)C(D/E)) [27]. Proteins identified without an S-FLOS signal do not have this sequence and are not reported to be S-nitrosated. The S-FLOS signal also is reduced in NOS1 deficient mice, indicating detection of S-nitrosation sites.
The S-FLOS conditions for thiol blocking and S-nitrosothiol reduction was selective and efficient as demonstrated by the virtually complete loss of the S-FLOS signal in samples pretreated with reducing agents before performing the S-FLOS assay or in omitting the Asc reduction of S-nitrosothiols step in the S-FLOS assay. Although an ascorbate dose–response curve for the effect of Asc reduction in the S-FLOS method was not performed. The Asc reduction step was required to obtain an S-FLOS signal. S-nitrosothiol reduction before MMTS blocking also completely blocked the S-FLOS signal showing the MMTS blocking of free thiols was efficient and not removed by the Asc reduction step.
The S-FLOS assay data has low background noise. Clear increases in S-FLOS signals correlated with exogenous GSNO transnitrosation or NOS2 dependent nitrosation and with endogenous nitrosation by constitutive NOS (WT vs NOS1 deficient mice). The S-FLOS assay, therefore, has the necessary and sufficient signal to noise to detect relative changes in S-nitrosothiol content of individual proteins under conditions that affect the total protein S-nitrosation state.
In addition to MMTS blocking and ascorbate reduction concerns, it is crucial to strictly control pH between 7.0 and 7.5 during the Cy-dye labeling step in the S-FLOS assay. Higher pH’s can lead to non-specific Cy-maleimide labeling of lysines, independent of blocking and reduction, and lead to erroneous detection of protein nitrosothiols. Furthermore, for purified proteins, the Cy-dye must be titrated from 4 to 40 pmol of dye per μg protein to optimize Cy-dye labeling for each protein. The optimal amount of Cy-dye is likely a function of the number and ratio of nitrosated cysteines to total cysteines in the purified protein. For protein mixtures, 40 pmol per μg protein serves as a suitable proportion for labeling.
As with any difference gel electrophoresis (DIGE) analysis, dye switching is required to eliminate potential dye specific effects [34]. Although absolute fluorescence intensities may differ slightly between dyes, relative intensities of between samples for each dye were the same. For comparison across dyes, the fluorescence intensities should be normalized to the relative contribution of any one band or spot’s fluorescence to the sample’s total protein fluorescence for each dye. This normalization will distinguish between true changes in fluorescence of a spot or band from merely a general increase in fluorescence labeling. Supplemental data Fig. S1 is a biological replicate and dye swap (Cy5 stimulated, Cy3 unstimulated) of the experiment in Fig 5B. Both show similar backgrounds for the S-FLOS fluorescence signal in protein samples from the stimulated cells.
The S-FLOS assay, therefore, has the low background, high sensitivity and high signal to noise to detect relative changes in S-nitrosothiol content of individual proteins under conditions that affect the total protein S-nitrosation state. It provides direct relative quantification in 2D electrophoresis applications, eliminating streptavidin affinity purification steps or Western blot analysis. While S-FLOS can detect changes in S-nitrosation, it currently does not distinguish between changes due to protein expression versus changes in the number of modified cysteines. To distinguish between these two possibilities, a second method, such as a traditional DIGE analysis [35], must be performed in parallel to the S-FLOS assay to quantify relative changes in protein expression. However, using S-FLOS to detect and identify proteins showing quantitative differences in S-nitrosation between two different samples on a single 2D gel will provide quantitative, spatial and temporal information on changes in S-nitrosation useful to investigating the role of S-nitrosation in normal cellular signaling [36] as well as in the pathophysiology of diseases associated with NO dysregulation.
Supplementary Material
Acknowledgments
Funding was provided by a Grant from the National Heart, Lung, and Blood Institute, contract N01-HV-28180 (to R.N.C) and by NIH Grant R01 AG021523 (to D.E.B).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.niox.2008.07.007.
Footnotes
A similar approach Sun et al., 2007 [14] was published while this paper was under review.
References
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 3.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol. Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 5.Derakhshan B, Wille PC, Gross SS. Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat. Protoc. 2007;2:1685–1691. doi: 10.1038/nprot.2007.210. [DOI] [PubMed] [Google Scholar]
- 6.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. Proteomic methods for analysis of S-nitrosation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;851:152–159. doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao C, Guo H, Wei J, Mi Z, Wai PY, Kuo PC. Identification of S-nitrosylated proteins in endotoxin-stimulated RAW264.7 murine macrophages. Nitric Oxide. 2005;12:121–126. doi: 10.1016/j.niox.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Kuncewicz T, Sheta EA, Goldknopf IL, Kone BC. Proteomic analysis of S-nitrosylated proteins in mesangial cells. Mol. Cell Proteomics. 2003;2:156–163. doi: 10.1074/mcp.M300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005;137:921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Ruiz A, Lamas S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch. Biochem. Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Lu XM, Lu M, Tompkins RG, Fischman AJ. Site-specific detection of S-nitrosylated PKB α/Akt1 from rat soleus muscle using CapLC-Q-TOF(micro) mass spectrometry. J. Mass Spectrom. 2005;40:1140–1148. doi: 10.1002/jms.885. [DOI] [PubMed] [Google Scholar]
- 13.Varma VA, Cerjan CM, Abbott KL, Hunter SB. Non-isotopic in situ hybridization method for mitochondria in oncocytes. J. Histochem. Cytochem. 1994;42:273–276. doi: 10.1177/42.2.8288868. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ. Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 15.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2000:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 16.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Kislyak Y, Lin J, Dickson A, Cardosa L, Broderick M, Fein H. Nanometer size electrode for nitric oxide and S-nitrosothiols measurement. Electrochem. Commun. 2002;4:11–16. [Google Scholar]
- 19.Nims RW, Cook JC, Krishna MC, Christodoulou D, Poore CM, Miles AM, Grisham MB, Wink DA. Colorimetric assays for nitric oxide and nitrogen oxide species formed from nitric oxide stock solutions and donor compounds. Methods Enzymol. 1996;268:93–105. doi: 10.1016/s0076-6879(96)68012-4. [DOI] [PubMed] [Google Scholar]
- 20.Maeda K, Finnie C, Svensson B. Cy5-maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. Biochem. J. 2004;378:497–507. doi: 10.1042/BJ20031634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Loscalzo J. S-nitrosoprotein formation and localization in endothelial cells. Proc. Natl. Acad. Sci. USA. 2005;102:117–122. doi: 10.1073/pnas.0405989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolosker H, Panizzutti R, Engelender S. Inhibition of creatine kinase by S-nitrosoglutathione. FEBS Lett. 1996;392:274–276. doi: 10.1016/0014-5793(96)00829-0. [DOI] [PubMed] [Google Scholar]
- 23.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santhanam L, Lim HK, Lim HK, Miriel V, Brown T, Patel M, Balanson S, Ryoo S, Anderson M, Irani K, Khanday F, Di Costanzo L, Nyhan D, Hare JM, Christianson DW, Rivers R, Shoukas A, Berkowitz DE. Inducible NO synthase dependent S-nitrosylation and activation of arginase1 contribute to age-related endothelial dysfunction. Circ. Res. 2007;101:692–702. doi: 10.1161/CIRCRESAHA.107.157727. [DOI] [PubMed] [Google Scholar]
- 25.Giustarini D, Milzani A, Aldini G, Carini M, Rossi R, Dalle-Donne I. S-nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid. Redox. Signal. 2005;7:930–939. doi: 10.1089/ars.2005.7.930. [DOI] [PubMed] [Google Scholar]
- 26.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 28.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc. Natl. Acad. Sci. USA. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 30.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of HIF-1α stability through S-nitrosylation. Mol. Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravi K, Brennan LA, Levic S, Ross PA, Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc. Natl. Acad. Sci. USA. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurd TR, Prime TA, Harbour ME, Lilley KS, Murphy MP. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J. Biol. Chem. 2007;282:22040–22051. doi: 10.1074/jbc.M703591200. [DOI] [PubMed] [Google Scholar]
- 33.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 34.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann J, Dimmeler S, Haendeler J. Shear stress increases the amount of S-nitrosylated molecules in endothelial cells: important role for signal transduction. FEBS Lett. 2003;551:153–158. doi: 10.1016/s0014-5793(03)00917-7. [DOI] [PubMed] [Google Scholar]
- 37.Ckless K, Reynaert NL, Taatjes DJ, Lounsbury KM, van der Vliet A, Janssen-Heininger Y. In situ detection and visualization of S-nitrosylated proteins following chemical derivatization: identification of Ran GTPase as a target for S-nitrosylation. Nitric Oxide. 2004;11:216–227. doi: 10.1016/j.niox.2004.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.