Abstract
Systemic lupus erythematosus (SLE) is a multisystem complex autoimmune disease of uncertain etiology (OMIM 152700). Over recent years a genetic component to SLE susceptibility has been established1–3. Recent successes with association studies in SLE have identified genes including IRF5 (refs. 4,5) and FCGR3B6. Two tumor necrosis factor (TNF) superfamily members located within intervals showing genetic linkage with SLE are TNFSF4 (also known as OX40L; 1q25), which is expressed on activated antigen-presenting cells (APCs)7,8 and vascular endothelial cells9, and also its unique receptor, TNFRSF4 (also known as OX40; 1p36), which is primarily expressed on activated CD4+ T cells10. TNFSF4 produces a potent co-stimulatory signal for activated CD4+ T cells after engagement of TNFRSF4 (ref. 11). Using both a family-based and a case-control study design, we show that the upstream region of TNFSF4 contains a single risk haplotype for SLE, which is correlated with increased expression of both cell-surface TNFSF4 and the TNFSF4 transcript. We hypothesize that increased expression of TNFSF4 predisposes to SLE either by quantitatively augmenting T cell–APC interaction or by influencing the functional consequences of T cell activation via TNFRSF4.
We genotyped a total of 45 SNPs across TNFSF4 and 4 SNPs across TNFRSF4 in a collection of 472 UK nuclear families (Supplementary Table 1 online). After preliminary genotyping, we used 36 markers across TNFSF4 and 4 markers in TNFRSF4 for analysis, removing 9 markers because they failed quality control, as described in Methods (Supplementary Table 2 online).
We analyzed the variants across TNFRSF4 and found that the haplotype structure consisted of a single block across the entire gene, comprising three major haplotypes (Supplementary Fig. 1 online). Transmission-disequilibrium test (TDT) analysis of both individual SNPs and haplotypes across TNFRSF4 showed that none of the SNPs (Pu > 0.05) and none of the haplotypes reached significance (Pu > 0.05; data not shown).
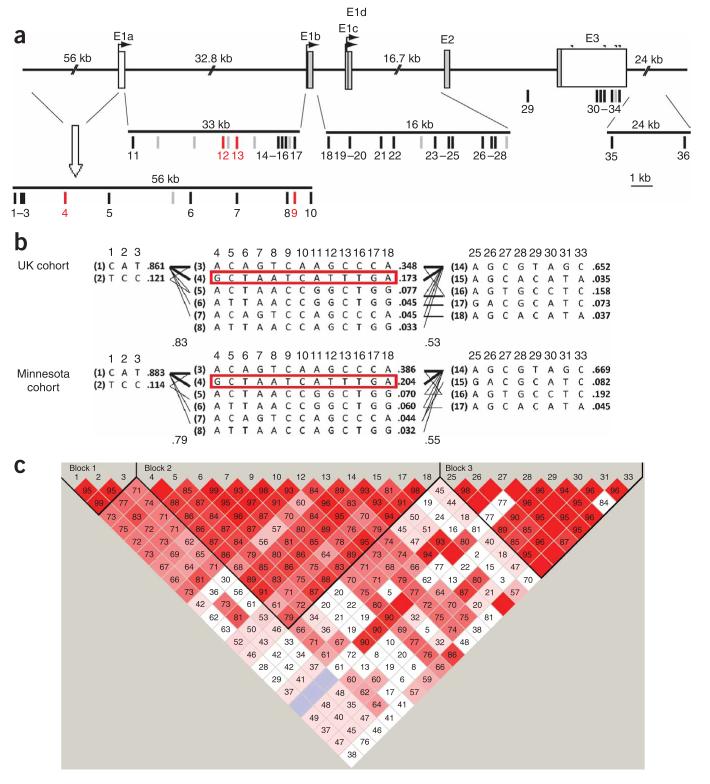
We defined the boundaries of the TNFSF4 haplotype blocks across a 182-kb region containing the entire 137-kb gene and its flanking sequence (Fig. 1). There is a bipartite pattern of linkage disequilibrium (LD), with strong LD across haplotype blocks 1 and 2 making up the 90-kb upstream region, as indicated by an interblock D′ score exceeding 0.83. There is some breakdown of LD at the start of intron 1, between haplotype blocks 2 and 3. Pairwise r2 values between any variant from the two upstream blocks and one from block 3 were below 0.30 (Supplementary Fig. 2 online). Both individual SNP and haplotype-TDT analyses (discussed in more detail below) showed strong associations. Taking the minimum of the single SNP P values and correcting for multiple testing with 100,000 permutations, we obtained a summary overall P value for the gene of 0.0020.
Figure 1.
Gene structure, haplotypic architecture and haplotype-TDT analysis in TNFSF4. (a) Human TNFSF4 consists of three exons. The translated exons are illustrated as gray boxes and the 5′ and 3′ UTRs as black boxes. The four potential poly(A) signals in the 3′ UTR are denoted by asterisks. In the diagram the SNPs used in the analysis have been recoded as numbers from 1 to 36: SNP 1 (rs10798267), SNP 2 (rs12118748), SNP 3 (rs4916318), SNP 4 (rs10912580), SNP 5 (rs844665), SNP 6 (rs844654), SNP 7 (rs844648), SNP 8 (rs2795288), SNP 9 (rs12039904), SNP 10 (rs844644), SNP 11 (rs844643), SNP 12 (rs2205960), SNP 13 (rs1234317), SNP 14 (rs3861953), SNP 15 (rs7535152), SNP 16 (rs1234315), SNP 17 (rs1234314), SNP 18 (rs3850641), SNP 25 (rs7518045), SNP 26 (rs6661173), SNP 27 (rs4113832), SNP 28 (rs7513384), SNP 29 (rs3861950), SNP 30 (171420977C<A), SNP 31 (rs7514229) and SNP 33 (rs3900307) as in Supplementary Table 2 online. The markers that failed quality control are place-marked in the sequence by gray lines. SNPs 4 (rs10912580), 9 (rs12039904), 12 (rs2205960) and 13 (rs1234317) are shown in red because they have overtransmitted minor alleles that tag overtransmitted haplotypes. (b) The haplotype block structure across TNFSF4 constructed from 413 European Caucasian parent-proband trios in the UK study cohort and in 262 US Minnesota parent-proband trios. There are three haplotype blocks across the gene. The haplotypes are numbered on the left of each haplotype in brackets from 1–13, with the haplotype frequencies shown to the right of each haplotype. Only haplotypes with a frequency of greater than 2.5% are shown. The SNP numbers across the top of the haplotypes correspond to those in the gene diagram. The red-boxed haplotypes are those showing overtransmission in both populations. (c) LD D′ prime chart from Haploview that summarizes the pattern of LD in the UK SLE families as a colored plot. Bright red represents regions of high pairwise D′, and white represents regions of low pairwise D′. The numbers in the boxes are the pairwise D′ values.
To seek replication of the UK association in TNFSF4, we genotyped selected variants in an independent collection of 263 Minnesota SLE parental-affected trios. Parental allele frequencies were similar in the UK and Minnesota populations (Supplementary Table 3 online), as was the pattern of LD across TNFSF4 between the two populations (Fig. 1b). The overall P value for the gene in the Minnesota samples was 0.0057.
Haplotype-TDT analysis within the upstream region confirmed that there is a single overtransmitted GCTAATCATTTGA haplotype (4) in both the UK (Pu = 0.0104) and Minnesota datasets (Pu = 3.00 × 10−4; Table 1). This overtransmitted haplotype was tagged by multiple overtransmitted rare alleles (SNPs rs10912580, rs12039904, rs2205960 and rs1234317). Both populations also contain an undertransmitted ACAGTCAAGCCC haplotype (3) in block 2, which was tagged by the rare A allele of rs844644. The undertransmitted effect was stronger in the UK population than in the Minnesota population, but the reverse was true for the risk haplotype.
Table 1. Haplotype-TDT analysis across the promoter of TNFSF4 in UK and Minnesota SLE families.
| UK (n = 416 families) |
Minnesota (n = 262 families) |
UK-Minnesota (n = 778 families) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype number | Constituent SNPs | Haplotype | χ 2 | P u | T/U | χ 2 | P u | T/U | χ 2 | P u | T/U | P p |
| 3 | 4-5-6-7-8-9-10-11-12-13-16-17-18 | ACAGTCAAGCCC | 9.054 | 0.003 | 0.70 | 4.22 | 0.04 | 0.76 | 11.9 | 6.00 × 10−4 | 0.73 | <1 × 10−5 |
| 4 | 4-5-6-7-8-9-10-11-12-13-16-17-18 | GCTAATCATTTG | 6.56 | 0.0104 | 1.46 | 13.0 | 3.00 × 10−4 | 1.79 | 18.3 | 1.86 × 10−5 | 1.61 | <1 × 10−5 |
The table shows the result of haplotype-TDT analysis in 416 UK SLE trios and in 262 US Minnesota families, using GENEHUNTER. For each haplotype, the base composition is given. Only the haplotypes having an uncorrected P value (Pu) of < 0.05, with 1 degree of freedom, in either the UK or Minnesota families are shown. The value of T/U represents a normalized T:U ratio. For the joint UK-Minnesota dataset, the P value after permutation analysis with 100,000 permutations (Pp) is also given.
Table 2 indicates that seven SNPs (rs10912580, rs844654, rs844648, rs2795288, rs12039904, rs1234315 and rs1234314) showed independent association in both the UK and Minnesota parental-affected trios (Pu < 0.05). There is some apparent heterogeneity in the pattern of association in the two populations, as several variants (rs844654, rs844648, rs2795288 and rs844644) showed association in the UK population but weaker signals in the Minnesota samples, and both rs2205960 and rs1234317 showed a stronger pattern of association in the Minnesota samples than in the UK families. However, we believe that these differences are due to chance: testing for heterogeneity in transmission ratios showed no significant differences (P > 0.1) after analysis with Pearson’s χ2 to compare the T:U ratio (the ratio of informative families showing transmission of the quoted minor allele to families not showing transmission; Supplementary Table 4 online). It therefore seemed reasonable to carry out a joint analysis of the two datasets; this showed multiple associated individual SNPs in the upstream region (Table 2) and associations from the upstream haplotypes (3 and 4), which remained significant after permutation analysis (Pp < 1 × 10−5; Table 1).
Table 2. TDT analysis of single SNPs across TNFSF4.
| UK (n = 416 families) |
Minnesota (n = 262 families) |
UK-Minnesota (n = 778 families) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | SNP analyzed |
MA | T:U | χ 2 | P u | T/U | T:U | χ 2 | P u | T/U | T:U | χ 2 | P u | T/U | P p |
| rs10798267 | 1 | T | 68:76 | 0.444 | 0.505 | 0.895 | 49:53 | 0.157 | 0.692 | 0.925 | 117:129 | 0.585 | 0.444 | 0.907 | 0.999 |
| rs12118748 | 2 | C | 72:77 | 0.168 | 0.682 | 0.935 | 48:51 | 0.091 | 0.763 | 0.941 | 120:128 | 0.258 | 0.612 | 0.938 | 1.000 |
| rs4916318 | 3 | C | 74:79 | 0.163 | 0.686 | 0.937 | 49:53 | 0.157 | 0.692 | 0.925 | 123:132 | 0.312 | 0.573 | 0.932 | 1.000 |
| rs10912580 | 4 | G | 142:95 | 9.32 | 2.00 × 10−3 | 1.49 | 134:90 | 8.64 | 3.28 × 10−3 | 1.49 | 276:185 | 18.0 | 2.25 × 10−5 | 1.49 | 4.00 × 10−4 |
| rs844665 | 5 | T | 62:46 | 2.37 | 0.123 | 1.35 | 33:40 | 0.671 | 0.413 | 0.825 | 95:86 | 0.448 | 0.504 | 1.11 | 0.999 |
| rs844654 | 6 | T | 231:151 | 16.8 | 4.26 × 10−5 | 1.53 | 146:110 | 5.06 | 0.024 | 1.33 | 377:261 | 21.1 | 4.38 × 10−6 | 1.44 | 1.00 × 10−4 |
| rs844648 | 7 | A | 217:139 | 17.1 | 3.57 × 10−5 | 1.56 | 152:110 | 6.73 | 0.009 | 1.38 | 369:249 | 23.3 | 1.39 × 10−6 | 1.48 | 1.00 × 10−5 |
| rs2795288 | 8 | A | 147:104 | 7.37 | 7.00 × 10−3 | 1.41 | 133:98 | 5.30 | 0.0212 | 1.36 | 280:202 | 12.6 | 3.81 × 10−4 | 1.39 | 5.80 × 10−3 |
| rs12039904 | 9 | T | 164:105 | 13.0 | 3.04 × 10−4 | 1.56 | 123:73 | 12.37 | 3.55 × 10−4 | 1.69 | 287:178 | 25.5 | 4.31 × 10−7 | 1.61 | 1.00 × 10−5 |
| rs844644 | 10 | A | 149:210 | 10.4 | 1.28 × 10−3 | 0.710 | 93:115 | 2.33 | 0.127 | 0.809 | 242:325 | 12.2 | 4.91 × 10−4 | 0.745 | 7.10 × 10−3 |
| rs844643 | 11 | G | 63:50 | 1.50 | 0.221 | 1.26 | 55:68 | 1.37 | 0.241 | 0.809 | 118:118 | 0.258 | 0.612 | 0.938 | 1.000 |
| rs2205960 | 12 | T | 128:109 | 1.52 | 0.217 | 1.17 | 111:78 | 5.76 | 0.0164 | 1.42 | 239:187 | 6.35 | 0.012 | 1.28 | 0.191 |
| rs1234317 | 13 | T | 123:106 | 1.26 | 0.261 | 1.16 | 121:82 | 7.49 | 6.12 × 10−3 | 1.48 | 244:188 | 7.26 | 7.05 × 10−3 | 1.33 | 0.078 |
| rs3861953 | 14 | T | 78:55 | 3.98 | 0.0460 | 1.42 | – | – | – | – | – | – | – | – | |
| rs7535152 | 15 | G | 81:63 | 2.25 | 0.130 | 1.29 | – | – | – | – | – | – | – | – | |
| rs1234315 | 16 | T | 147:87 | 15.4 | 8.80 × 10−5 | 1.69 | 152:109 | 7.08 | 7.80 × 10−3 | 1.39 | 299:196 | 21.4 | 3.67 × 10−6 | 1.53 | 8.00 × 10−5 |
| rs1234314 | 17 | G | 161:117 | 6.96 | 8.00 × 10−3 | 1.38 | 153:102 | 10.2 | 1.40 × 10−3 | 1.50 | 314:219 | 16.9 | 3.87 × 10−5 | 1.39 | 7.00 × 10−4 |
| rs3850641 | 18 | G | 103:79 | 3.17 | 0.0750 | 1.30 | 48:68 | 3.45 | 0.0630 | 0.706 | 151:147 | 0.0540 | 0.817 | 1.03 | 0.993 |
| 171440443C<T | 19 | C | 99:98 | 5.00 × 10−3 | 0.940 | 1.01 | – | – | – | – | – | – | – | – | |
| 171440442T<A | 20 | T | 62:63 | 8.00 × 10−3 | 0.930 | 0.980 | – | – | – | – | – | – | – | – | |
| 171438300G<C | 21 | G | 167:143 | 1.86 | 0.170 | 1.17 | – | – | – | – | – | – | – | – | |
| 171435020A<G | 22 | A | 138:114 | 2.29 | 0.130 | 1.21 | – | – | – | – | – | – | – | – | |
| rs4916313 | 23 | C | 136:133 | 0.0300 | 0.860 | 1.02 | – | – | – | – | – | – | – | – | |
| rs7518129 | 24 | G | 117:107 | 0.450 | 0.500 | 1.09 | – | – | – | – | – | – | – | – | |
| rs7518045 | 25 | G | 65:39 | 6.50 | 0.011 | 1.67 | 29:41 | 2.08 | 0.152 | 0.707 | 94:80 | 1.13 | 0.289 | 1.18 | 0.797 |
| rs6661173 | 26 | A | 68:39 | 7.86 | 5.00 × 10−3 | 1.74 | 36:47 | 1.46 | 0.227 | 0.766 | 104:86 | 1.71 | 0.192 | 1.21 | 0.740 |
| rs4113832 | 27 | T | 83:80 | 0.0600 | 0.814 | 1.04 | 68:93 | 3.88 | 0.0490 | 0.731 | 151:173 | 1.49 | 0.222 | 0.873 | 0.992 |
| rs7513384 | 28 | A | 15:38 | 9.98 | 2.00 × 10−3 | 0.394 | 16:17 | 0.0300 | 0.861 | 0.941 | 31:55 | 6.70 | 0.0100 | 0.395 | 0.149 |
| rs3861950 | 29 | C | 114:115 | 4.00 × 10−3 | 0.947 | 0.991 | 84:121 | 6.68 | 0.0100 | 0.694 | 198:236 | 3.33 | 0.068 | 0.839 | 0.833 |
| 171424387A<G | 30 | C | 79:106 | 3.94 | 0.0470 | 0.745 | 62:87 | 4.20 | 0.0410 | 0.713 | 141:193 | 8.10 | 4.00 × 10−3 | 0.731 | 0.522 |
| rs7514229 | 31 | T | 110:120 | 0.435 | 0.510 | 0.917 | 78:117 | 7.80 | 5.00 × 10−3 | 0.667 | 188:237 | 5.65 | 0.0170 | 0.793 | 0.648 |
| 171420860G<A | 32 | G | 64:42 | 3.85 | 0.0500 | 1.48 | – | – | – | – | – | – | – | – | |
| rs3900307 | 33 | A | 16:45 | 13.8 | 2.00 × 10−4 | 0.356 | 23:22 | 0.0220 | 0.882 | 1.05 | 39:67 | 7.40 | 6.00 × 10−3 | 0.582 | 0.109 |
| rs16845536 | 34 | T | 66:41 | 5.84 | 0.0160 | 1.61 | 41:30 | 1.70 | 0.192 | 1.37 | 107:71 | 7.28 | 7.00 × 10−3 | 1.51 | 0.342 |
| 171417779T<G | 35 | T | 73:86 | 1.06 | 0.300 | 0.850 | – | – | – | – | – | – | – | – | |
| rs1234310 | 36 | G | 125:134 | 0.310 | 0.580 | 0.930 | – | – | – | – | – | – | – | – | |
TDT analysis by GENEHUNTER for each SNP tested in the UK SLE families and in the Minnesota dataset. A separate analysis was carried out for the UK and Minnesota families and also for joint UK-Minnesota samples. The column marked “SNP analyzed” gives the order of analyzed SNPs shown in Figure 1. For each variant, the uncorrected P value is denoted “Pu” and is quoted with one degree of freedom. Permuted P values (Pp) after 100,000 permutations are shown for the UK-Minnesota joint dataset. The column marked “T:U” is the ratio of informative families showing transmission of the quoted minor allele to those families not showing transmission. The column marked T/U represents the normalized transmission/untransmission ratio.
We sought to further corroborate the association signals from the family-based cohorts in an independent case-control collection of 424 unrelated UK SLE cases with 642 control samples from the British 1958 Birth Control Cohort. All the cases used were separate from those in the family-based TDT analysis (Table 2). The variants chosen for this subsequent analysis included haplotype-tagging SNPs and those showing association in the UK SLE trios (Table 2). The results of this case-control analysis (Table 3) replicated the association of variants upstream of TNFSF4 with SLE. There are significant associations from four SNPs, including two haplotype-tagging variants, one of which tags the overtransmitted haplotype (rs2205960 Pu = 7.00 × 10−3, odds ratio (OR) = 1.28, 95% confidence interval (CI) = 1.07–1.53) and the other of which tags the undertransmitted haplotype (rs844644 Pu = 6.83 × 10−5, OR = 0.710, 95% CI = 0.597–0.840).
Table 3. Case-control analysis.
| Allelic association test |
||||||
|---|---|---|---|---|---|---|
| SNP | SNP analyzed | MA | MAa | MAu | χ 2 | P u |
| rs10798267 | 1 | T | 0.106 | 0.08 | 4.71 | 0.03 |
| rs10912580 | 4 | G | 0.287 | 0.25 | 3.86 | 0.05 |
| rs844665 | 5 | T | 0.115 | 0.0762 | 8.71 | 3.2 × 10−3 |
| rs844654 | 6 | T | 0.493 | 0.444 | 5.36 | 0.02 |
| rs844648 | 7 | A | 0.485 | 0.439 | 4.63 | 0.03 |
| rs2795288 | 8 | A | 0.471 | 0.416 | 3.34 | 0.07 |
| rs12039904 | 9 | T | 0.264 | 0.243 | 1.27 | 0.26 |
| rs844644 | 10 | A | 0.379 | 0.463 | 15.9 | 6.8 × 10−5 |
| rs2205960 | 12 | T | 0.276 | 0.233 | 7.38 | 7.0 × 10−3 |
| rs1234317 | 13 | T | 0.299 | 0.255 | 4.44 | 0.04 |
| rs3861953 | 14 | T | 0.0858 | 0.0726 | 1.20 | 0.27 |
| rs1234315 | 16 | T | 0.438 | 0.480 | 2.28 | 0.13 |
| rs1234314 | 17 | G | 0.456 | 0.429 | 1.76 | 0.18 |
| 171420860G<A | 32 | T | 0.360 | 0.310 | 7.43 | 7.0 × 10−3 |
| 171417779T<G | 35 | T | 0.100 | 0.070 | 6.08 | 0.01 |
An independent case-control analysis across TNFSF4 in 424 UK unrelated cases and 642 control samples taken from the 1958 British Birth Control Cohort, using PLINK. The column marked “SNP analyzed” gives the order of analyzed SNPs shown in Figure 1. The column marked “MA” gives the identity of the minor allele, with the frequency of this minor allele given in affected (MAa) and unaffected individuals (MAu). In the allelic association analysis, the uncorrected P value (Pu) is quoted for each marker.
To maximize the information available from both populations, we combined the P values in the parental-affected trios from both populations (n = 778) with the UK case-control cohort (424 cases, 642 controls) by using Fisher’s test (Supplementary Table 5 online). There was evidence of a strong association from variants across the entire upstream region of TNFSF4. The two variants with the strongest associations were the overtransmitted haplotype-tagging variant, rs12039904 (P = 1.91 × 10−6), and rs844644, which tags the under-transmitted haplotype (P = 6.08 × 10−7).
To further investigate the relationship between genetic variants in TNFSF4 and SLE, we conducted a genotype association analysis using 840 cases (taken from the UK trios and unrelated cases) and 642 unrelated controls (Table 4). A recessive model was least favored by the data. In an attempt to dissect out the causative alleles in the upstream region, we used conditional logistic analysis to determine which variants made the strongest contribution to the association, using SNPs that were either significantly associated in the joint UK-Minnesota trios after permutation analysis or that were haplotype tagging (Table 1 and Fig. 1b). The results of this conditional analysis showed that the effect of each individual SNP disappeared when it was estimated conditional on the haplotype background and supports the hypothesis that no single genotyped variant is responsible for the association in the upstream region of TNFSF4 (Supplementary Table 6 online). The exception is rs844644, which showed a moderate P value (P = 0.015) by conditional analysis. This could be because rs844644 was the only variant tested without a unique minor allele tagging the overtransmitted haplotype, but instead has a rare A allele tagging the undertransmitted haplotype.
Table 4. Modeling the pattern of inheritance in UK total cases and controls.
| Cases (n = 840) |
Control (n = 642) |
P value for model of inheritance |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | SNP analyzed | Minor, ‘a’ | Major, ‘A’ | ‘AA’ | ‘Aa’ | ‘aa’ | ‘AA’ | ‘Aa’ | ‘aa’ | Additive | Recessive | Dominant |
| rs10798267 | 1 | T | C | 11 | 137 | 557 | 12 | 75 | 554 | 5.1 × 10−4 | 0.66 | 3.4 × 10−4 |
| rs10912580 | 4 | G | A | 63 | 286 | 353 | 50 | 241 | 400 | 0.017 | 0.24 | 4.4 × 10−3 |
| rs844665 | 5 | T | C | 11 | 134 | 549 | 5 | 97 | 600 | 5.5 × 10−3 | 0.13 | 1.8 × 10−3 |
| rs844654 | 6 | T | A | 207 | 402 | 182 | 186 | 437 | 289 | 1.0 × 10−4 | 4.8 × 10−3 | 6.5 × 10−5 |
| rs844648 | 7 | A | G | 204 | 396 | 186 | 180 | 435 | 290 | 1.4 × 10−4 | 3.0 × 10−3 | 1.3 × 10−4 |
| rs2795288 | 8 | A | T | 120 | 249 | 115 | 138 | 297 | 254 | 1.2 × 10−5 | 0.052 | 2.0 × 10−6 |
| rs12039904 | 9 | T | C | 68 | 309 | 407 | 57 | 330 | 526 | 0.029 | 0.056 | 0.019 |
| rs844644 | 10 | A | C | 119 | 371 | 304 | 197 | 452 | 264 | 1.8 ×10−5 | 4.7 × 10−4 | 4.2 × 10−5 |
| rs2205960 | 12 | T | G | 59 | 277 | 389 | 48 | 327 | 534 | 0.024 | 0.020 | 0.039 |
| rs1234317 | 13 | T | C | 60 | 266 | 328 | 48 | 261 | 392 | 0.066 | 0.11 | 0.034 |
| rs3861953 | 14 | T | C | 9 | 137 | 628 | 4 | 93 | 599 | 0.032 | 0.23 | 0.011 |
| rs1234315 | 16 | T | C | 99 | 227 | 159 | 209 | 440 | 245 | 0.094 | 0.21 | 0.036 |
| rs1234314 | 17 | G | C | 162 | 336 | 186 | 321 | 888 | 573 | 2.3 × 10−3 | 1.5 × 10−3 | 0.017 |
The mode of inheritance was modeled in the 840 cases taken from 416 UK SLE trios, together with the 424 unrelated UK cases and the 642 controls taken from the 1958 British Birth Control Cohort. The column marked “SNP analyzed” gives the order of analyzed SNPs shown in Figure 1. The identity of the minor allele ‘a’ and the major allele ‘A’ are given. The values in the columns marked “cases” and “controls” represent the numbers of samples corresponding to each class of genotype, “AA”, “Aa” and “aa” represent a given variant. The P values quoted show the strength of agreement with either an additive, recessive or dominant model of inheritance, as determined by PLINK.
We carried out subphenotype analysis in the UK SLE families using all the SNPs tagging the overtransmitted upstream haplotypes. We analyzed three subtypes of SLE: antiphospholipid syndrome, Sjøgren’s syndrome and lupus nephritis. All the two-sample t-tests for all possible SNP-phenotype combinations exceeded 0.2, indicating that the disease susceptibility variants in TNFSF4 did not influence the lupus phenotype toward any of the three subtypes tested.
The two variants closest to the start of exons in the upstream region of TNFSF4 are rs12039904 (321 bp away from exon 1a) and rs1234314 (931 bp away from exon 1b). Neither variant falls within a transcription factor binding site. However, the T allele of rs1234317, which uniquely tags the overtransmitted haplotype, is predicted by the Genomatix SNP analysis web tool to destroy the DNA binding site for the transcriptional repressor E4BP4, a transcription factor with a role in the survival of early B cell progenitors12.
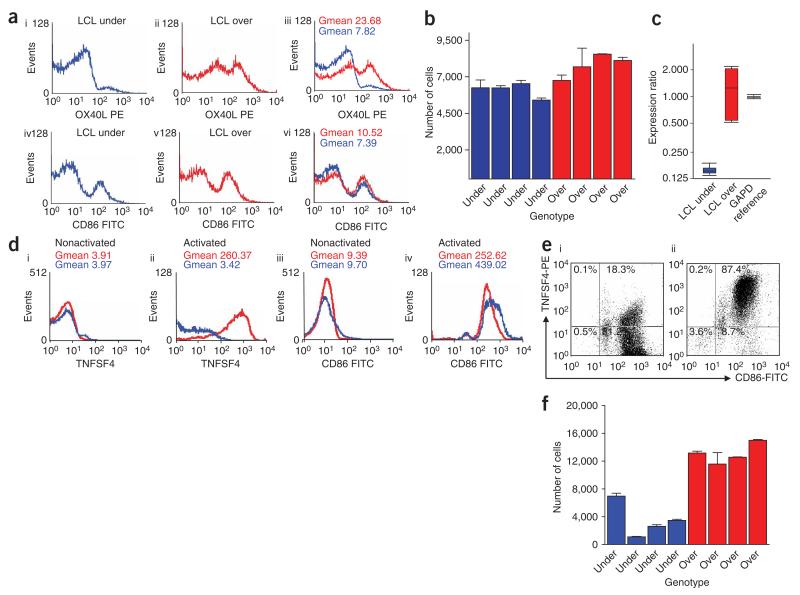
To better understand how TNFSF4 may act as a disease susceptibility gene in SLE, we asked whether the upstream haplotypes influenced TNFSF4 expression in lymphoblastoid cell lines (LCL cells) and in peripheral blood lymphocytes (PBLs) from our UK SLE collection. We genotyped more than 1,000 LCL cells for rs2205960, rs1234314 and rs7514229. The LCL cells selected were homozygous for either the overtransmitted haplotype 4 (LCL over) or the undertransmitted haplotype 3 (LCL under). Further resequencing for SNPs rs10912580, rs12039904, rs1234317 and rs844644 confirmed homozygosity for the haplotype-tagging SNPs in these cell lines. After activation, the LCL cells were shown to upregulate the cell-surface expression of CD86 and TNFSF4 (Fig. 2a), with significantly more TNFSF4-positive cells in the LCL-over cells than in the LCL-under cells (P = 0.02; Fig. 2b). Quantification of TNFSF4 mRNA levels by qRT-PCR showed that the LCL-over cells had a 6.7-fold (P 0.008) increase in the level of TNFSF4 transcript compared to the LCL-under cells (Fig. 2c). Cell-surface TNFSF4 was also upregulated after activation of PBLs (Fig. 2d), and PBL-over cells had a greater proportion of cells with higher TNFSF4 expression compared with PBL-under cells (Fig. 2e). This difference in expression was consistent in eight individuals (P = 0.02; Fig. 2f).
Figure 2.
SLE susceptibility alleles are associated with an increase in TNFSF4 expression. (a) Histograms of CD40L anti-IgD-activated EBV-LCL cells showing expression of TNFSF4 ((i), (ii) and (iii)) and CD86 ((iv), (v) and (vi)) for representative samples that were homozygous for the either the undertransmitted susceptibility haplotype (LCL under, (i) and (iv); shown in blue) and for the overtransmitted haplotype (LCLover, (ii) and (v); shown in red), respectively. The geometric mean (Gmean) values of the fluorescent intensity are shown for each marker. The data presented are from one of two independent experiments, with similar results on the same cell lines. (b) The bar chart shows the numbers of CD40L and anti-IgD-stimulated cells in eight different homozygous cell lines (four LCL under and four LCL over). Each bar represents the mean of two independent replicates. (c) By carrying out quantification by qRT-PCR of TNFSF4 mRNA in two LCL-under and three LCL-over cell lines and pairwise analysis by REST, we generated a whisker box-plot representation of the variation between TNFSF4 transcript levels. LCL-over samples, which carry SLE-susceptibility alleles, were associated with a 6.7-fold increase in RNA expression (P = 0.008). TNFSF4 expression was normalized to GAPD mRNA expression in the same sample. The bars indicate the relative TNFSF4 expression in three independent replicates for each sample, ± s.d. (d) Overlaid histograms showing TNFSF4 expression (i and ii) and CD86 expression (iii and iv) in CD40L/anti-IgD–stimulated and unstimulated PBLs. These PBLs were taken from UK SLE probands that were homozygous for the undertransmitted haplotype (shown in blue) and for the overtransmitted haplotype (shown in red). The Gmean values presented are shown for the cell-surface markers in two individuals. (e) Representative FACS plots of stimulated PBLs taken from probands expressing (i) the undertransmitted haplotype and (ii) the overtransmitted haplotype. The percentage of CD86+TNFSF4+ cells within the activated cell population is indicated in the upper left quadrant. Cells were designated TNFSF4-positive if TNFSF4 expression fell within background staining compared to a mouse IgG1 negative control mAb (MOPC 31C, Ancell). (f) The numbers of TNFSF4-positive PBLs taken from eight SLE-affected probands (four homozygotes for each of the over- and undertransmitted haplotypes), 48 h after CD40L- anti-IgD stimulation. Each bar represents the mean of two independent replicates.
Using both a family-based and case-control study design, we have shown that the upstream region of TNFSF4 contains a single risk haplotype for SLE, which carries a series of haplotype-tagging variants. There is a similar T:U for TNFSF4 (T:U = 1.61) and IRF5 (T:U = 1.92)5, which underlines the importance of TNFSF4 as a lupus susceptibility gene. This overtransmitted haplotype is correlated with increased expression of both cell-surface TNFSF4 and TNFSF4 transcript. We hypothesize that variation in the upstream region of the gene will increase the expression of TNFSF4, and, through TNFRSF4, increase co-stimulation for CD4+ T cells and/or further activate the APCs13 that express TNFSF4. This increased expression of TNFSF4 may act by destabilizing peripheral tolerance through inhibiting the generation of IL-10–producing CD4+ type 1 regulatory T cells14. Notably, TNFSF4 has also been associated with susceptibility to atherosclerosis15, and individuals with SLE are prone to accelerated arterial disease. The role of TNFSF4 in the pathogenesis of SLE highlights the importance of the role of the T cell–APC interaction in this disease, a conclusion supported by the genetic influence, albeit a modest one, arising from the CTLA4-ICOS locus16.
METHODS
UK family collection
A large collection of SLE-affected nuclear families was established in the laboratory. A diagnosis of SLE was established by telephone interview, health questionnaire and details from clinical notes. All probands conformed to the American College of Rheumatology criteria for SLE17, and written consent was obtained from participants, including unaffected relatives. Ethical approval was obtained from the Multi-Centre Research Ethics Committee (MREC). The composition of the UK family cohort consisted of 416 European Caucasian parental-affected trios (Table 1). There was a separate collection of 424 independent UK SLE cases and 642 controls taken from the British 1958 Birth Control Cohort (see URLs section below).
Non-UK family collection for corroborative genotyping
We corroborated the UK association study using a total of 263 US parental-affected trios from the Minnesota SLE collection. These studies were approved by the Human Subject Institutional Review Boards at the University of Minnesota, and informed consent was obtained from all subjects.
Preparation of genomic DNA and cDNA
We isolated genomic DNA from anticoagulated whole blood by a standard phenol-chloroform extraction. Lymphocytes were separated from anticoagulated whole blood by centrifugation through Histopaque-1077 (Sigma-Aldrich) in Accuspin tubes (Sigma-Aldrich) using a method based on one previously described18.
SNP identification
The majority of the polymorphisms across TNFSF4 were identified from the public database, dbSNP. However, eight variants in TNFSF4 and all four variants in TNFRSF4 were initially identified by resequencing, in a cohort of 60 SLE probands from our total family collection. The details of all sequencing primers are available on request.
After initial genotyping in 416 UK trios, we excluded markers from the analysis if their genotyping frequency was low (<85%), the number of mendelian errors identified using pedCHECK was greater than 5%, or the Hardy-Weinberg equilibrium P value in the parental samples was less than P = 0.05.
Genotyping
The predominant genotyping method in the UK cohort was MALDI-TOF mass spectrometry (Sequenom)19. However, we typed TNFSF4 SNPs 171440443C<T, 171440442T<A and 171420977C<A using pyrosequencing methodology. PCR-RFLP was used to type TNFSF4 SNPs rs3861950, rs7514229 and 171417779T<G and TNFRSF4 SNPs 19FRG<A, 6FRC<G and 9FRA<C. We used the restriction enzymes HpaI (rs3861950), DraI (rs7514229) and AciI (417779T<G) for the polymorphisms in TNFSF4 and AluI (19FRG<A), PflmI (6FRC) and Alw26I (11FR3A<G) for those in TNFRSF4. All PCRs were carried out on DNA Engine Tetrads (MJ Research Inc.), in either 96-well or 384-well format. Details of methodology and primer sequences are available on request. In the Minnesota cohort, the variants were typed using the Sequenom platform and are listed in Supplementary Table 2.
Statistical analysis
All sample genotype and phenotype data was managed by, and analysis files generated with, BC/GENE and BC/CLIN software (Biocomputing Platforms Ltd.). For population controls, we counted alleles in parental samples for each SNP genotyped across TNFSF4 and TNFRSF4. We calculated these allele frequencies for each allele as a fraction of the total alleles for each SNP. A comparison of the parental allele frequencies between the UK and Minnesota collections was made from χ2 analysis using a 3 × 2 contingency table with a P = 0.05 level of significance.
For SNPs in each gene, we generated haplotype patterns using the algorithm described in Haploview20. This program constructs haplotypes based on the D′ measure of linkage disequilibrium (LD)21, together with a LOD score as measure of significance and 95% confidence intervals to state the accuracy of the P value. The pairwise linkage disequilibrium (LD) for SNPs across each gene was confirmed by the r2 values22,23. Only markers having a minor allele frequency greater than 5% were included in the haplotype constructions, and haplotypes with a frequency greater than 2.5% were included in the LD diagrams. The haplotype block definitions were based on confidence limits for strong LD of 0.85 (upper) and 0.70 (lower), upper confidence interval maximum for strong recombination of 0.85, and at least 80% of strong LD in informative comparisons20.
We used several statistical tools to investigate different aspects of the genotype data. We tested association of alleles to SLE for both individual and multiple SNPs by the transmission disequilibrium test (TDT), which compares the observed and expected transmission of alleles from heterozygous parents to affected offspring. This analysis was carried out in parental-proband trios using GENEHUNTER 2.1r3beta24,25. We used PLINK v0.99o to calculate the overall P values for the gene in both the UK and Minnesota populations using 100,000 permutations, to perform the case-control association tests for single SNPs, to model the genotypic associations, and to run the Breslaw-Day test of heterogeneity in odds ratios between the UK and Minnesota populations.
We carried out haplotype-TDT analysis using Haploview. We calculated a total association for TNFSF4 by combining the P values from the UK and Minnesota parental-affected trios and the UK case-control collection using Fisher’s method26. To determine the variants on a haplotype making the strongest contribution to the haplotype association, we carried out conditional logistic regression using WHAP in samples from the UK and Minnesota parental-affected trios. The variants from haplotype block 2 selected for this analysis either were significantly associated after permutation analysis in the trios or were haplotype tagging (Table 4). This analysis tested whether there was still an effect from the haplotype block 2 after conditioning on each constituent variant in turn.
We used the Mann-Whitney test to compare the differences between the numbers of TNFSF4-positive cells carrying the overtransmitted upstream haplotype and those carrying the TNFSF4-positive undertransmitted upstream haplotypes. We carried out separate analyses for the Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (LCL cells) and peripheral blood lymphocytes (PBLs).
SLE phenotype analysis
In the UK SLE families, 45% of the cases had one or more phenotypes characteristic of antiphospholipid syndrome (APL), 46% probands had one or more of the phenotypes characteristic of Sjøgren’s syndrome and 23% of probands had renal disease. The major phenotypes of these syndromes are the presence of thrombosis, previous miscarriage and either IgG and/or IgM autoantibodies to cardiolipin IgG for antiphospholipid syndrome and the evidence of sicca symptoms and autoantibodies to Ro and/or La for Sjøgren’s syndrome. We used a calculated variable to classify the UK SLE probands into the APL or Sjøgren’s syndrome groupings if they had one or more of these major SLE phenotypes.
To investigate whether the transmission bias was greater in individuals with a given subphenotype, we summed score contributions from TRANSMIT analysis27 over the families. If the genetic effect is the same in each family, then these score contributions should be exchangeable; in particular, they should have the same mean. A two-sample t-test was used to compare the mean contribution in families where the proband had a certain phenotype with the test statistic in families where the proband did not have this phenotype.
Selection of samples for expression analysis
We carried out expression analysis for the upstream haplotypes, but not individual variants, on LCL cells (see Acknowledgments) and PBLs taken from our UK SLE cohort. Both the PBLs and the LCL cells were genotyped with SNPs rs2205960, rs1234314 and rs7514229. We determined the phase of the upstream haplotypes for SNPs rs2205960 and rs1234314 using PHASE version 2 (ref. 28). To control for potential variation in expression at the 3′ end of TNFSF4, samples were included in the expression study only if they were also homozygous for SNP rs7514229 in the 3′ UTR of the gene. The PBLs had been fully typed for all the haplotype-tagging SNPs in the upstream region, but we also resequenced the homozygous cell lines for the haplotype-tagging SNPs, rs10912580, rs12039904, rs1234317 and rs844644 (which tags the undertransmitted haplotype), to confirm homozygosity. Therefore, both the PBLs and LCL cells used in the expression studies were homozygous for all the tagging SNPs carried by the overtransmitted upstream haplotype 4 or those which were homozygous for the rs844644 carried by the undertransmitted upstream haplotype 3 (LCL under).
In vitro activation of PBLs and LCL cells
Peripheral blood lymphocytes (PBLs) and EBV-transformed cell lines (LCL cells) were suspended in complete RPMI medium, which comprised RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 10,000 U/ml penicillin (Invitrogen), 10,000 μg/ml streptomycin (Invitrogen), 200 mM L-glutamine supplement (Invitrogen), and grown in suspension at a concentration of 3 × 106 cells/ml. LCL cells were activated for 48 h in complete RPMI medium at 3 × 106 cells/ml, supplemented with 10 ng/ml rCD40L (Axxora), 20 ng/ml CD40L enhancer (Axxora) and 2 μg/ml goat anti–human α-anti-IgD polyclonal antibody (Serotec). We froze 2 × 106/ml activated cells in Trizol for RNA extraction and immediately resuspended 0.5 × 106 cells/ml in FACS staining buffer supplemented with fluor-conjugated antibodies for FACS analysis.
FACS analysis
PBLs and LCL cells were stained in FACS staining buffer using FITC-conjugated monoclonal antibody to human CD86 (MCA1118F, Serotec) as a marker of B cell activation, in combination with phycoerythrin-conjugated monoclonal antibody to human TNFSF4 (ANC10G1). Cells were size-gated and analyzed for expression of CD86 and TNFSF4. The cells were designated TNFSF4-positive if TNFSF4 expression fell within the background staining, compared to a mouse IgG1 negative control monoclonal antibody (MOPC 31C, Ancell). All analyses of FACS data was carried out on the FACScalibur cell sorter, using Cellquest software (Becton Dickinson), and cell plots were generated using WinMDI software (J. Trotter, The Scripps Institute).
TNFSF4 total RNA quantitative RT-PCR (qRT-PCR)
PBLs and LCL cells that were homozygous for either the over- or the undertransmitted upstream haplotypes were selected for qRT-PCR. We isolated total RNA from 1 × 106 CD40L + anti IgD treated or untreated cells using Trizol (Life Technologies), according to the manufacturer’s instructions. We synthesized first strand cDNA from 500 ng of RNA template using murine Moloney reverse transcriptase and oligo(dT)18 primers, which selectively anneal to the poly(A) tail of mRNA (RevertAid, Fermentas). We quantified the RNA using the Nanodrop ND-1000 Spectrophometer (Labtech International) and checked its quality on an Agilent 2100 Bioanalyser (Agilent). The primers used for qRT-PCR across TNFSF4 spanned exons E2 and E3 (primer sequences available on request).
We quantified the level of TNFSF4 transcript in each sample using qRT-PCR. Standard thermal cycling conditions were used as described in the manufacturer’s instructions for the ABsolute SYBR green ROX mix (AB Gene) on the AB 7500 Real Time PCR System (Applied Biosystems 7500 Fast System with version 1.3.0 software). Direct detection of PCR product was monitored by measuring the threshold cycle (CT) at which an exponential increase in fluorescence was caused by SYBR green dye intercalation. We carried out ABI7500 melting point analysis to generate a single TNFSF4, GAPD, HPRT1 or NONO- specific melting temperature curve. A mean CT value was calculated for each sample from three replicates. These mean CT values were used to calculate the relative expression levels of TNFSF4 compared to the three housekeeping genes human GAPD, HPRT1 and NONO transcripts, used as endogenous references. Amplification efficiency was determined from trireplicated, serially diluted aliquots of GAPD, HPRT1 and NONO amplified alongside each TNFSF4 test sample. Statistical significance after normalization was tested by a pairwise randomization test using the relative expression software tool, REST (Corbett Life Science).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Wellcome Trust through a Senior Fellowship awarded to T.J.V. We acknowledge the work of P. Spencer in recruiting individuals and families into the study, and we would like to thank our clinical colleagues for helping us recruit study participants. Our thanks and appreciation is extended to all the study participants and their relatives for generously donating blood samples, and to all the general practitioners and practice nurses for collecting them. J.D.R. is funded by grants from the National Institutes of Allergy and Infectious Diseases (AI065687; AI067152) and from the National Institute of Diabetes and Digestive and Kidney Diseases (DK064869; DK062432). We appreciate the contribution to the genotyping from the Broad Institute Center for Genotyping and Analysis, which is supported by grant U54 RR020278-01 from the National Center for Research Resources. We gratefully acknowledge the work of D. Smyth at the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory for genotyping five variants from the diabetes collection. We also thank H. Stevens and the Diabetes and Inflammation Laboratory DNA team for sending EBV-transformed lymphoblastoid cell lines. We acknowledge use of DNA from the British 1958 Birth Cohort collection (D. Strachan, S. Ring, W. McArdle and M. Pembrey), funded by the Medical Research Council grant G0000934 and Wellcome Trust grant 068545/Z/02. We thank J. Todd for LCL lymphocytes and for critical reading of this manuscript.
Footnotes
URLs. British 1958 Birth Control Cohort, http://www.b58cgene.sgul.ac.uk/; PLINK v0.99o, http://pngu.mgh.harvard.edu/~purcell/plink/; WHAP http://pngu.mgh.harvard.edu/purcell//whap/.
GenBank accession codes. Human TNFSF4, AL022310.1.
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Vyse TJ, Todd JA. Genetic analysis of autoimmune disease. Cell. 1996;85:311–318. doi: 10.1016/s0092-8674(00)81110-1. [DOI] [PubMed] [Google Scholar]
- 2.Deapen D, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 3.Forabosco P, et al. Meta-analysis of genome-wide linkage studies of systemic lupus erythematosus. Genes Immun. 2006;7:609–614. doi: 10.1038/sj.gene.6364338. [DOI] [PubMed] [Google Scholar]
- 4.Graham RR, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 5.Cunninghame Graham DS, et al. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum. Mol. Genet. 2007;16:579–591. doi: 10.1093/hmg/ddl469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohshima Y, et al. Expression and function of OX40 ligand on human dendritic cells. J. Immunol. 1997;159:3838–3848. [PubMed] [Google Scholar]
- 8.Godfrey WR, Fagnoni FF, Harara MA, Buck D, Engleman EG. Identification of a human OX-40 ligand, a costimulator of CD4+ T cells with homology to tumor necrosis factor. J. Exp. Med. 1994;180:757–762. doi: 10.1084/jem.180.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imura A, et al. The human OX40/gp34 system directly mediates adhesion of activated T cells to vascular endothelial cells. J. Exp. Med. 1996;183:2185–2195. doi: 10.1084/jem.183.5.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latza U, et al. The human OX40 homolog: cDNA structure, expression and chromosomal assignment of the ACT35 antigen. Eur. J. Immunol. 1994;24:677–683. doi: 10.1002/eji.1830240329. [DOI] [PubMed] [Google Scholar]
- 11.Gramaglia I, Weinberg AD, Lemon M, Croft M. Ox-40 ligand: a potent costimulatory molecule for sustaining primary CD4 T cell responses. J. Immunol. 1998;161:6510–6517. [PubMed] [Google Scholar]
- 12.Ikushima S, et al. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuber E, Neurath M, Calderhead D, Fell HP, Strober W. Cross-linking of OX40 ligand, a member of the TNF/NGF cytokine family, induces proliferation and differentiation in murine splenic B cells. Immunity. 1995;2:507–521. doi: 10.1016/1074-7613(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, et al. OX40 ligand shuts down IL-10-producing regulatory T cells. Proc. Natl. Acad. Sci. USA. 2006;103:13138–13143. doi: 10.1073/pnas.0603107103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 2005;37:365–372. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- 16.Graham DS, Wong AK, McHugh NJ, Whittaker JC, Vyse TJ. Evidence for unique association signals in SLE at the CD28-CTLA4-ICOS locus in a family-based study. Hum. Mol. Genet. 2006;15:3195–3205. doi: 10.1093/hmg/ddl395. [DOI] [PubMed] [Google Scholar]
- 17.Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 18.Boyum A. Separation of leukocytes from blood and bone marrow. Introduction. Scand. J. Clin. Lab. Invest. Suppl. 1968;97:7. [PubMed] [Google Scholar]
- 19.Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 20.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Lewontin RC. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill WG, Weir BS. Maximum-likelihood estimation of gene location by linkage disequilibrium. Am. J. Hum. Genet. 1994;54:705–714. [PMC free article] [PubMed] [Google Scholar]
- 23.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 24.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 25.Kruglyak L, Lander ES. Faster multipoint linkage analysis using Fourier transforms. J. Comput. Biol. 1998;5:1–7. doi: 10.1089/cmb.1998.5.1. [DOI] [PubMed] [Google Scholar]
- 26.Fisher RA. Statistical Methods for Research Workers. Hafner; New York: 1954. [Google Scholar]
- 27.Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am. J. Hum. Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.