SUMMARY
The transition of ductal carcinoma in situ (DCIS) to invasive carcinoma is a poorly understood key event in breast tumor progression. Here, we analyzed the role of myoepithelial cells and fibroblasts in the progression of in situ carcinomas using a model of human DCIS and primary breast tumors. Progression to invasion was promoted by fibroblasts and inhibited by normal myoepithelial cells. Molecular profiles of isolated luminal epithelial and myoepithelial cells identified an intricate interaction network involving TGFβ, Hedgehog, cell adhesion, and p63 required for myoepithelial cell differentiation, the elimination of which resulted in loss of myoepithelial cells and progression to invasion.
INTRODUCTION
The natural history of breast cancer involves progression through clinical and pathologic stages starting with abnormal epithelial proliferation, progressing into in situ and invasive carcinomas, and culminating in metastatic disease (Burstein et al., 2004). DCIS is thought to be a precursor of invasive ductal carcinoma based on molecular, epidemiological, and pathological studies (Burstein et al., 2004). Surgical margins and histologic features have been associated with increased risk of subsequent tumor events, but none of these predicted the risk of invasive recurrence (Fisher et al., 1999; Gauthier et al., 2007). Comprehensive molecular profiling studies comparing DCIS and invasive ductal carcinomas have failed to identify tumor-stage-specific signatures (Chin et al., 2004; Ma et al., 2003; Porter et al., 2003; Yao et al., 2006). However, these studies have focused mainly on the tumor epithelial cells, and the role of the microenvironment in tumor progression has not been explored.
Epithelial-mesenchymal interactions are important for normal mammary gland development and for breast tumorigenesis (Howlett and Bissell, 1993). In vivo and in vitro studies have demonstrated that ECM (extracellular matrix) molecules and cells composing the microenvironment modulate tissue-specificity of the normal breast as well as the growth, survival, polarity, and invasive behavior of breast cancer cells (Bissell et al., 2005; Weinberg and Mihich, 2006). In the normal mammary gland, a layer of myoepithelial cells that produces and is in contact with the basement membrane surrounds luminal epithelial cells, which in turn line the ducts and the alveoli. In addition to playing a role in expelling milk from the ducts during lactation via their contractile function, myoepithelial cells increasingly are recognized as important regulators of normal mammary gland development and function due to their effect on luminal epithelial cell polarity, branching, and differentiation (Bissell et al., 2005). Myoepithelial cells also have been labeled “natural tumor suppressors” due to their negative effects on tumor cell growth, invasion, and angiogenesis achieved via secretion of protease inhibitors and downregulation of MMP (matrix metalloprotease) levels (Barsky and Karlin, 2005; Polyak and Hu, 2005). These conclusions have been largely based on coculture assays; the role of myoepithelial cells in tumorigenesis and the basis of their disappearance during invasive progression have not been explored. The tumor-suppressive function of myoepithelial cells progressively gets lost during the in situ to invasive carcinoma transition. Indeed, the diagnostic criterion that distinguishes invasive from in situ carcinomas is the disappearance of the organized myoepithelial cell layer and the basement membrane (Lerwill, 2004).
To explore the involvement of the microenvironment in tumor progression, we previously analyzed the gene expression and DNA methylation profiles of different cell types from normal breast tissue, DCIS, and invasive breast carcinomas and observed dramatic changes in all cell types during tumor progression (Allinen et al., 2004; Hu et al., 2005). Importantly, myoepithelial cells associated with DCIS were not phenotypically normal; they had lost some of their differentiation markers and had upregulated genes promoting angiogenesis and invasion. While the physiological relevance of these molecular changes was unknown, based on our data we hypothesized that abnormal DCIS-associated myoepithelial cells, together with different stromal cells, degrade the basement membrane resulting in the progression of in situ carcinomas to invasive tumors. Testing this hypothesis required an experimental model of DCIS that faithfully reproduced human disease, since analysis of human tissue allows only correlative studies. The MCF10A series is one of the few human models of breast tumor progression (Miller, 2000), although it is likely to reflect only a subset of breast tumors with basal-like features. A derivative of MCF10A cells is the MCF10ADCIS.com cell line (subsequently referred to as MCFDCIS) (Miller et al., 2000), which reproducibly forms comedo DCIS-like lesions that spontaneously progress to invasive tumors. This MCFDCIS xenograft model highly resembles human disease with respect to histopathology and natural history. Furthermore, we here show that the gene expression profiles of epithelial and myoepithelial cells purified from MCFDCIS xenografts are also highly similar to those from primary human DCIS tumors. We used this model to explore the relative importance of myoepithelial cells and stromal fibroblasts in the in situ to invasive breast carcinoma transition. In this study, we demonstrated that normal myoepithelial cells suppress, but fibroblasts enhance, tumor growth and invasive progression in the absence of detectable genomic alterations in the tumor epithelial cells. We identified an intricate network involving TGFβ, Hedgehog (Hh), cell adhesion, and p63 that appears to play an essential role in myoepithelial cell differentiation, as elimination of key mediators of these pathways led to the loss of myoepithelial cells and progression to invasion. The gene expression profiles of epithelial and myoepithelial cells from primary human breast tissue correlated with these findings and supported the hypothesis that in situ to invasive carcinoma transition is regulated by loss of normal myoepithelial cell function.
RESULTS
Characterization of the MCFDCIS Cells and Their Xenografts
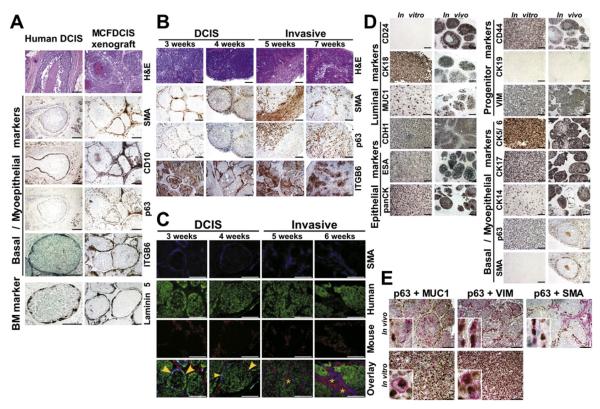
To explore whether MCFDCIS provided a good model for primary human DCIS, xenografts were analyzed for histology and molecular markers. We chose subcutaneous instead of orthotopic injections because the progression and phenotype of the tumors were different in the mammary fat pad presumably due to the local microenvironment (Figures S1A and S1B). Subcutaneous xenografts were similar to human high-grade comedo DCIS. The duct-like structures were surrounded by basement membrane positive for laminin 5 and contained a layer of cells positive for myoepithelial markers (smooth muscle actin-SMA, CD10, and p63) (Figure 1A). Analysis of the tumors at different time points (3–8 weeks) after injection revealed a progression from DCIS to invasive histology (Figure 1B). In DCIS, both SMA and p63 were expressed in the myoepithelial cell layer, whereas in invasive tumors, SMA-positive cells were stromal myofibroblasts, and a subset of tumor epithelial cells was p63 positive.
Figure 1. Properties of MCFDCIS Cells and Derived Xenografts.
(A) Comparison of MCFDCIS xenografts and human high-grade comedo DCIS analyzed by hematoxylin-eosin staining (H&E) to depict histology and immunohistochemistry for the expression of SMA, CD10, p63, ITGB6, and laminin 5.
(B) Progression of the MCFDCIS xenografts. Histology of the tumors (H&E) and expression of SMA, p63, and ITGB6 were analyzed at the indicated time points after injection.
(C) iFISH analysis of a time course experiment. Immunofluorescence using SMA antibody (blue) identifies myoepithelial cells or myofibroblasts. Fluorescently labeled human (green) and mouse (red) Cot1 DNA were used as probes for FISH. Yellow arrows and stars indicate human myoepithelial cells and mouse myofibroblasts, respectively.
(D and E) Single (D) and dual (E) immunohistochemical analyses of the indicated markers in MCFDCIS cells (in vitro) or xenografts (in vivo). Insets show colocalization of the indicated markers in a single cell and the mutually exclusive expression of p63 and MUC1 in xenografts.
Scale bars correspond to 100 μm in all panels.
We tested several other human breast cancer cell lines in xenograft assays; however, none of these formed tumors with DCIS histology (Figure S1C). We reasoned that the unique characteristic of MCFDCIS cells may be due to their proposed bipotential progenitor properties (Santner et al., 2001). To investigate this, we performed immunofluorescence in situ hybridization (iFISH) analysis of MCFDCIS xenografts to test whether both epithelial and myoepithelial cells were derived from MCFDCIS cells and if the myoepithelial cell layer disappeared as tumors became invasive. Samples collected at different time points after injection were hybridized with fluorescently labeled human and mouse cot-1 DNA as probes for FISH and stained with pancytokeratin (panCK) and SMA antibodies for epithelial and myoepithelial cells, respectively. As anticipated, panCK+ tumor epithelial cells were indeed of human origin in all tumors (Figure S2A). SMA+ myoepithelial cells were also of human origin (Figure 1C), confirming that MCFDCIS cells are bipotential progenitors. In contrast, SMA+ myofibroblasts in the stroma were of murine origin in all xenografts. iFISH analysis also revealed the gradual disappearance of SMA+ myoepithelial cells coinciding with the progression of DCIS to invasive tumors (Figure 1C). Since other MCF10A derivatives were also proposed to be progenitors (Santner et al., 2001), we tested these in xenograft assays as well, but they were either nontumorigenic (MCF10AT cells) or formed invasive tumors (MCF10CA cells) (data not shown). Thus, of all the cells tested, the MCFDCIS cell line was the only human breast cancer cell line that formed DCIS xenografts in our hands.
To dissect the progenitor properties of MCFDCIS cells, we analyzed the expression of known basal/progenitor and differentiated luminal and myoepithelial cell markers (Table S1) in cultured cells and xenografts. Immunohistochemistry and FACS analyses showed that MCFDCIS cells in 2D culture were uniformly positive for panCK, CK18, ESA, CDH1, CD44, CK17, CK5/6, p63, VIM, ITGA6, and ITGB6; partially positive for MUC1 and CK14; and negative for CD24, SMA, CK19, and CD10 (Figure 1D and Figure S2B). In xenografts, most tumor epithelial cells were positive for panCK, ESA, CD44, CK17, CDH1, and VIM. In DCIS, p63 and SMA expression was limited to the myoepithelial cells that also showed decreased or no expression of MUC1, CD24, and CK18, while CK5/6 and CK14 demonstrated heterogeneous staining patterns with all cells positive in some areas and only the myoepithelial cells positive in others (Figure 1D). Doublestaining confirmed the colocalization of multiple markers characteristic of a particular cellular phenotype within one cell and the mutually exclusive expression of p63 and MUC1 in xenografts following differentiation (Figure 1E). Thus, based on our analyses, in culture, MCFDCIS cells had progenitor characteristics, and in xenografts, they differentiated into luminal and myoepithelial cells.
To determine if all or only a subpopulation of MCFDCIS cells had progenitor properties, we sorted the cells into MUC1+ and MUC1− fractions and injected them into mice (Figure S2C). MUC1 expression has been used for identification of cells with progenitor properties (Gudjonsson et al., 2002b; Stingl et al., 2005), and this was the only cell surface marker that we found heterogeneously expressed in MCFDCIS cells in culture. Both MUC1+ and MUC1− cells gave rise to DCIS tumors (Figure S2C), suggesting that both populations have bipotential progenitor properties. We also derived single-cell clones of MCFDCIS cells with different CK14 and MUC1 expression levels (Figures S2D and S2E). Four of these independent clones were injected into mice and resulting xenografts from all four had DCIS histology and showed the same CK14, MUC1, SMA, and p63 expression patterns (Figure S2F). Thus, using this approach, we found that all or a significant fraction of MCFDCIS cells may have bipotential progenitor properties, and the expression patterns of some luminal (MUC1 and CD24) and myoepithelial (CK14, p63, and SMA) markers were discordant in 2D culture and in xenografts, suggesting that differentiation only occurs in vivo.
Similarity of the MCFDCIS Model to Human DCIS
To ensure that results obtained using this model are relevant to human disease, it was essential to establish if the MCFDCIS xenografts reproduce human breast tumors. To compare genetic alterations in cells from the MCF10A-series and MCFDCIS-derived xenografts to those found in primary human breast tumors, we performed SNP (single nucleotide polymorphism) array analyses. All MCF10A-derived cells had copy number gain at chromosome 8q24 and homozygous deletion of CDKN2A (Figures S3A and S3B). Both of these genetic alterations are known to occur in human breast carcinomas (Cairns et al., 1995; Yokota et al., 1999), supporting the similarity between the MCFDCIS model and human disease.
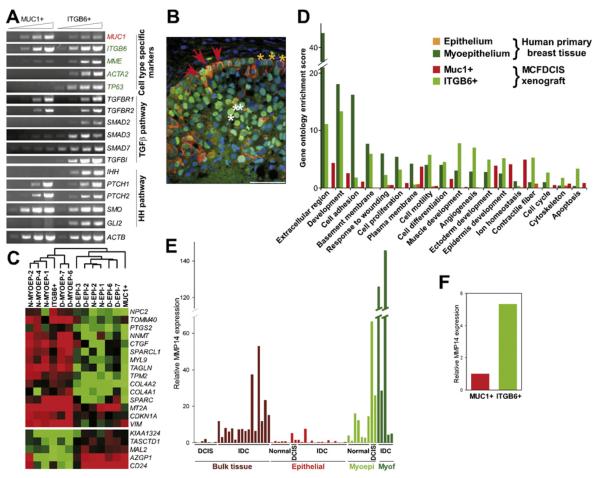
To define the similarity of MCFDCIS xenografts to human DCIS at the cellular level, we purified luminal and myoepithelial cells using MUC1 and integrin β6 (ITGB6) as cell surface markers, respectively. ITGB6 has been identified as a gene upregulated by p63 (Carroll et al., 2006) and TGFβ1 (Zambruno et al., 1995). In MCFDCIS-derived DCIS xenografts and in human DCIS, ITGB6 was specifically expressed in myoepithelial cells (Figure 1A), making it an ideal cell surface marker for their purification. The purity of the MUC1+ and ITGB6+ cells was confirmed by semiquantitative RT-PCR analysis of cell type-specific markers. Myoepithelial cell markers CD10 (MME), SMA (ACTA2), and p63 (TP63) were present only in ITGB6+ cells (Figure 2A), consistent with their myoepithelial phenotype. Although the cells were separated based on MUC1 and ITGB6 cell-surface markers, the expression of MUC1 and ITGB6 mRNAs were not completely mutually exclusive in the two cell populations.
Figure 2. Similarity of the MCFDCIS Model to Human DCIS.
(A) Semiquantitative RT-PCR analysis of the expression of the indicated genes in MUC1+ and ITGB6+ cells purified from DCIS xenografts.
(B) Immunofluorescence analysis of CK5 (orange) and p53 (green) expression in human DCIS. Red arrows mark double-positive abnormal myoepithelial cells in the basal layer, whereas yellow and white arrows mark CK5+ normal myoepithelial and p53+ tumor epithelial cells, respectively. Scale bar corresponds to 50 μm.
(C) Dendogram depicting cluster analysis of SAGE libraries to delineate similarities of MUC1+ and ITGB6+ cells to human breast epithelial and myoepithelial cells, respectively.
(D) Gene ontology categories enriched in ITGB6+ and MUC1+ cells from MCFDCIS xenografts and epithelial and myoepithelial cells from primary human breast tissue. Scores > 1.3 correspond to p < 0.05.
(E) qPCR analysis of MMP14 in bulk DCIS and invasive ductal carcinoma (IDC) and in epithelial and myoepithelial cells, and myofibroblasts from normal human breast tissue, in situ carcinomas, and invasive carcinomas.
(F) qPCR analysis of MMP14 in MUC1+ and ITGB6+ cells.
SNP array analysis of the isolated cells confirmed that both MUC1+ and ITGB6+ cells are genetically abnormal and have the same copy number changes as parental MCFDCIS cells (Figures S3D and S3E). Our previous study in human tumors found that DCIS-associated myoepithelial cells do not have clonal somatic copy number alterations (Allinen et al., 2004). This seeming discrepancy could be due to the fact that MCFDCIS cells are bipotential tumor-initiating cells, whereas the majority of myoepithelial cells in DCIS represent cells of normal ducts/alveoli into which tumors spread (Figure S4). A prediction of this hypothesis is that in basal-like DCIS (which the MCFDCIS model represents) rare genetically abnormal myoepithelial cells can be found in the area of tumor initiation. To test this hypothesis, we performed combined immunofluorescence staining for p53, a gene frequently mutated and overexpressed in basal-like tumors, and CK5, a myoepithelial cell marker, on human basal-like DCIS. As depicted in Figure 2B, we observed occasional p53+/CK5+ cells in the basal layer of some human DCIS potentially identifying genetically abnormal bipotential progenitors and myoepithelial cells. Analysis of 169 DCIS samples revealed CK5+/p53+ positive cells in 8% of cases and enrichment in basal-like tumors. These findings strengthen the similarity of the MCFDCIS model to human basal-like DCIS and imply that findings obtained in this model are likely to be relevant to human disease.
Next we analyzed the comprehensive gene expression profiles of MUC1+ and ITGB6+ cells using SAGE. SAGE data further supported the myoepithelial and luminal characteristics of ITGB6+ and MUC1+ cells, respectively, since several known markers of these cell types were almost mutually exclusively expressed in the respective SAGE libraries (Table 1 and Table S5). Furthermore, genes differentially expressed between ITGB6+ and MUC1+ cells were also differentially expressed between myoepithelial and epithelial cells from human breast tissue (Table 1 and Table S5). Hierarchical cluster analysis of SAGE libraries also demonstrated the myoepithelial and luminal features of ITGB6+ and MUC1+ cells, respectively (Figure 2C).
Table 1.
Similarity of MUC1+ and ITGB6+ Cells to Epithelial and Myoepithelial Cells, Respectively, from Primary Human Tissue
| ITGB6+ | MUC1+ | N-MYOEP-1 | N-MYOEP-2 | N-MYOEP-4 | D-MYOEP-6 | D-MYOEP-7 | N-EPI-1 | N-EPI-2 | D-EPI-2 | D-EPI-3 | D-EPI-6 | D-EPI-7 | I-EPI-7 | I-EPI-8 | I-EPI-9 | Unigene ID No. | Gene Symbol | Gene Description |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes High in ITGB6+ and MYOEP | ||||||||||||||||||
| 63 | 7 | 59 | 17 | 11 | 597 | 173 | 2 | 0 | 2 | 7 | 25 | 7 | 10 | 59 | 17 | 111779 | SPARC | Secreted protein, acidic, cysteine-rich |
| 90 | 4 | 112 | 136 | 263 | 260 | 204 | 53 | 14 | 8 | 17 | 56 | 64 | 14 | 8 | 5 | 534330 | MT2A | Metallothionein 2A |
| 54 | 3 | 91 | 33 | 54 | 12 | 10 | 33 | 20 | 3 | 0 | 3 | 1 | 5 | 0 | 0 | 591484 | LAMC2 | Laminin, gamma 2 |
| 24 | 1 | 114 | 45 | 149 | 36 | 58 | 0 | 6 | 5 | 0 | 8 | 4 | 3 | 15 | 3 | 632099 | TAGLN | Transgelin |
| 24 | 1 | 14 | 41 | 39 | 16 | 17 | 2 | 0 | 0 | 3 | 3 | 1 | 5 | 0 | 0 | 300772 | TPM2 | Tropomyosin 2 (beta) |
| 20 | 1 | 9 | 20 | 7 | 6 | 15 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 508716 | COL4A2 | Collagen, type IV, alpha 2 |
| 13 | 0 | 45 | 31 | 13 | 26 | 38 | 2 | 2 | 6 | 3 | 5 | 3 | 5 | 8 | 20 | 504687 | MYL9 | Myosin, light polypeptide 9, regulatory |
| 121 | 13 | 7 | 48 | 26 | 91 | 56 | 4 | 2 | 3 | 16 | 4 | 2 | 7 | 23 | 5 | 110675 | TOMM40 | Translocase of outer mitochondrial membrane 40 homolog |
| 12 | 0 | 16 | 5 | 5 | 20 | 31 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 590970 | AXL | AXL receptor tyrosine kinase |
| 12 | 1 | 23 | 42 | 17 | 59 | 60 | 18 | 0 | 0 | 9 | 5 | 2 | 3 | 11 | 0 | 503911 | NNMT | Nicotinamide N-methyltransferase |
| 23 | 3 | 58 | 53 | 28 | 106 | 162 | 0 | 0 | 8 | 21 | 10 | 7 | 4 | 17 | 23 | 591346 | CTGF | Connective tissue growth factor |
| 9 | 0 | 26 | 39 | 18 | 96 | 37 | 0 | 6 | 2 | 3 | 8 | 7 | 7 | 8 | 2 | 62886 | SPARCL1 | SPARC-like 1 |
| 8 | 0 | 12 | 6 | 0 | 31 | 17 | 0 | 2 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 17441 | COL4A1 | Collagen, type IV, alpha 1 |
| 8 | 0 | 12 | 8 | 24 | 5 | 23 | 6 | 4 | 3 | 3 | 23 | 9 | 0 | 3 | 2 | 474833 | CSNK1E | Casein kinase 1, epsilon |
| 8 | 0 | 10 | 16 | 3 | 14 | 4 | 0 | 0 | 0 | 3 | 0 | 1 | 1 | 3 | 0 | 111779 | SPARC | Secreted protein, acidic, cysteine-rich |
| 24 | 6 | 37 | 47 | 29 | 76 | 13 | 18 | 8 | 14 | 5 | 5 | 8 | 16 | 2 | 26 | 370771 | CDKN1A | Cyclin-dependent kinase inhibitor 1A |
| 42 | 10 | 87 | 34 | 18 | 11 | 6 | 16 | 16 | 5 | 5 | 4 | 10 | 5 | 6 | 5 | 133892 | TPM1 | Tropomyosin 1 (alpha) |
| 17 | 4 | 3 | 14 | 7 | 5 | 8 | 0 | 2 | 2 | 2 | 4 | 0 | 1 | 5 | 0 | 298654 | DUSP6 | Dual specificity phosphatase 6 |
| 82 | 22 | 66 | 122 | 159 | 44 | 29 | 16 | 26 | 5 | 3 | 4 | 1 | 1 | 3 | 2 | 433845 | KRT5 | Keratin 5 |
| 107 | 37 | 38 | 36 | 101 | 102 | 121 | 4 | 8 | 3 | 10 | 15 | 6 | 7 | 20 | 15 | 533317 | VIM | Vimentin |
| 28 | 10 | 19 | 14 | 9 | 31 | 23 | 2 | 8 | 3 | 3 | 8 | 9 | 34 | 0 | 5 | 223678 | JMJD3 | Jumonji domain containing 3 |
| Genes High in MUC1+ and EPI | ||||||||||||||||||
| 47 | 99 | 2 | 23 | 11 | 7 | 4 | 39 | 53 | 139 | 14 | 22 | 76 | 33 | 56 | 21 | 301350 | FXYD3 | FXYD domain containing ion transport regulator 3 |
| 24 | 55 | 2 | 78 | 5 | 0 | 17 | 47 | 154 | 106 | 35 | 118 | 1090 | 347 | 51 | 120 | 520943 | NCF1 | Neutrophil cytosolic factor 1 |
| 16 | 37 | 3 | 5 | 4 | 2 | 0 | 2 | 12 | 14 | 17 | 0 | 9 | 7 | 5 | 5 | 346868 | EBNA1BP2 | EBNA1 binding protein 2 |
| 11 | 32 | 3 | 22 | 12 | 2 | 0 | 31 | 26 | 88 | 7 | 19 | 43 | 16 | 12 | 8 | 464210 | SYNGR2 | Synaptogyrin 2 |
| 7 | 21 | 0 | 2 | 0 | 0 | 0 | 2 | 6 | 36 | 0 | 12 | 15 | 5 | 21 | 5 | 485158 | SPDEF | SAM pointed domain containing ets transcription factor |
| 7 | 21 | 0 | 9 | 5 | 0 | 0 | 33 | 10 | 18 | 5 | 7 | 10 | 16 | 3 | 9 | 194385 | STAP2 | Signal-transducing adaptor protein-2 |
| 13 | 59 | 0 | 42 | 4 | 1 | 2 | 62 | 168 | 228 | 5 | 167 | 105 | 68 | 11 | 174 | 375108 | CD24 | CD24 molecule |
| 0 | 7 | 0 | 5 | 0 | 0 | 0 | 4 | 6 | 12 | 2 | 10 | 19 | 7 | 14 | 3 | 642705 | KIAA1324 | KIAA1324 |
| 0 | 9 | 2 | 5 | 5 | 0 | 4 | 6 | 18 | 11 | 7 | 16 | 19 | 1 | 3 | 11 | 542050 | TACSTD1 | Tumor-associated calcium signal transducer 1 |
| 0 | 9 | 37 | 20 | 14 | 16 | 10 | 88 | 51 | 207 | 33 | 30 | 109 | 68 | 47 | 27 | 437638 | XBP1 | X-box binding protein 1 |
| 1 | 10 | 5 | 2 | 0 | 1 | 4 | 6 | 18 | 8 | 2 | 4 | 10 | 4 | 11 | 18 | 130413 | TM9SF2 | Transmembrane 9 superfamily member 2 |
| 30 | 275 | 0 | 2 | 1 | 0 | 0 | 12 | 14 | 23 | 17 | 1 | 1 | 3 | 60 | 8 | 46452 | SCGB2A2 | Secretoglobin, family 2A, member 2 |
| 0 | 21 | 3 | 3 | 4 | 2 | 0 | 10 | 6 | 27 | 5 | 15 | 15 | 10 | 6 | 14 | 201083 | MAL2 | Mal, T-cell differentiation protein 2 |
| 1 | 61 | 0 | 2 | 0 | 0 | 0 | 8 | 4 | 2 | 2 | 3 | 6 | 10 | 12 | 2 | 105887 | LOC124220 | Similar to common salivary protein 1 |
| Genes High in ITGB6+ and DCIS-Associated MYOEP | ||||||||||||||||||
| 15 | 1 | 5 | 8 | 5 | 55 | 77 | 14 | 22 | 45 | 0 | 11 | 4 | 3 | 3 | 12 | 448588 | NGFRAP1 | Nerve growth factor receptor associated protein 1 |
| 13 | 0 | 0 | 2 | 0 | 15 | 48 | 0 | 0 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 821 | BGN | Biglycan |
| 9 | 0 | 3 | 5 | 1 | 45 | 58 | 2 | 2 | 6 | 9 | 29 | 8 | 15 | 8 | 20 | 522584 | TMSB4X | Thymosin, beta 4, X-linked |
| 40 | 9 | 2 | 2 | 1 | 21 | 19 | 0 | 0 | 2 | 5 | 1 | 0 | 0 | 6 | 0 | 7835 | MRC2 | Man nose receptor, C type 2 |
| 17 | 4 | 0 | 6 | 3 | 69 | 27 | 0 | 4 | 0 | 0 | 4 | 3 | 4 | 8 | 5 | 529053 | C3 | Complement component 3 |
| 42 | 13 | 0 | 5 | 4 | 20 | 79 | 6 | 0 | 6 | 2 | 7 | 22 | 8 | 5 | 18 | 289019 | LTBP3 | Latent transforming growth factor beta binding protein 3 |
Detailed description of SAGE libraries and their analysis are included in the Supplemental Data. Normalized tag counts, Unigene ID, gene symbol, and gene description are listed.
N, normal; D, DCIS; I, invasive ductal carcinoma.
Genes related to TGFβ1 signaling are italicized.
Functional Analysis of Cell Type-Specific Gene Expression Patterns
Functional annotation of the genes differentially expressed between ITGB6+ and MUC1+ cells from xenografts and between myoepithelial and epithelial cells from human breast tissue revealed statistically significant enrichment of genes involved in ECM and basement membrane and development in ITGB6+ and myoepithelial cells (Figure 2D). The functional similarity between MUC1+ and epithelial cells was more limited, reflecting the higher diversity of luminal cells. We also performed network analysis of SAGE data using MetaCore as previously described (Shipitsin et al., 2007). These analyses strongly suggested that myoepithelial and ITGB6+ cells from human breast tissue and xenografts, respectively, have very similar phenotypes likely regulated by TGFβ and VEGFA, and their main functions involve ECM, cytoskeleton, and membrane remodeling and lipoprotein metabolism (Figure S5). Importantly, genes overexpressed in myoepithelial cells were also overexpressed in ITGB6+ cells and vice versa (Table S3). Both pair-wise comparisons (ITGB6+/MUC1+ and myoepithelial/epithelial cells) showed remarkably close enrichment pattern in GO processes, canonical pathway maps, and disease biomarkers, which is highly nonrandom, as MetaCore contains 600 pathway maps, over 2000 GO processes, and >500 disease categories. Common maps included cell adhesion, ECM remodeling, keratin filaments, TGFβ and Wnt signaling, and niacin/HDL metabolism; common GO processes included cell adhesion and actin cytoskeleton reorganization, whereas the most affected disease biomarkers included breast, endocrine gland, and urogenital neoplasms, and epithelial carcinoma (Table S4).
Both the TGFβ1 and Hh signaling pathways have been implicated in the regulation of progenitor cell function and myoepithelial differentiation, and SAGE and pathway analyses indicated their activation in ITGB6+ and in human myoepithelial cells (Figure S5; Table 1; Table S3). To dissect the possible mechanism of their cell type-specific activation, we analyzed the expression of their signaling components by semiquantitative RT-PCR in MUC1+ and ITGB6+ cells. TGFBR1 and TGFBR2 (receptors for TGFβ1) and PTCH1, PTCH2, and SMO (receptors for Hh) were equally present in both cell types, whereas SMAD2, SMAD3, and GLI2 (transcription factors); IHH (a ligand for PTCH); and TGFβ1 transcriptional targets SMAD7 and TGFBI were expressed more abundantly in ITGB6+ cells (Figure 2A). These data suggest that specific activation of TGFβ1 targets in ITGB6+ cells may be due to the restricted expression of SMAD2 and SMAD3 in these cells, a finding that was confirmed also by immunohistochemical analysis (data not shown). Hh signaling and GLI2 expression may be upregulated in ITGB6+ cells due to the high expression of IHH (Figure 2A) and BGN (Table 1).
The Importance of the Basement Membrane in Progression to Invasion
The gene expression profiles of ITGB6+ and myoepithelial cells from primary human breast tissue suggested that one of their main functions is the synthesis and maintenance of the basement membrane (BM). BM degradation is a hallmark of malignancy and the definition of invasive progression, yet the underlying molecular mechanism is undefined. Several MMPs have been implicated to play a role in this process, but a recent study suggested that MMP14, MMP15, and MMP16 are the most likely to degrade BM in vivo (Hotary et al., 2006). To determine the expression pattern of these three MMPs in breast tumor progression, we analyzed our SAGE data on multiple cell types isolated from normal and neoplastic breast tissue (Allinen et al., 2004). This analysis showed that MMP14 was highly expressed in myoepithelial cells and myofibroblasts, and its expression increased in DCIS-associated myoepithelial cells compared to normal ones (data not shown). To confirm the SAGE data, we analyzed the expression of MMP14 by real-time PCR in bulk tumor samples as well as in purified epithelial and myoepithelial cells, and myofibroblasts. Highest MMP14 expression was detected in myofibroblasts and in DCIS-associated myoepithelium (Figure 2E). High MMP14 levels in bulk invasive tumor tissue were likely due to its expression in myofibroblasts since tumor epithelial cells had low levels of MMP14. Next, we tested the expression of MMP14 in MUC1+ and ITGB6+ cells isolated from MCFDCIS xenografts. Correlating with human primary tumor data, MMP14 was highly expressed in ITGB6+ myoepithelial cells (Figure 2F) strengthening the similarity between our model and human DCIS. The expression pattern of MMP14 in human breast tissue and in xenografts was consistent with the hypothesis that it may be involved in BM degradation and in invasive progression. However, due to the effect of MMP14 on multiple growth factors (including TGFβ) and other MMPs (e.g., MMP9 and MMP2), the consequences of increased MMP14 expression are not easy to predict and its mRNA levels may not always correlate with protease activity (Labbe et al., 2007).
The Effect of Coinjected Normal Myoepithelial Cells and Various Fibroblasts on Tumor Growth and Progression
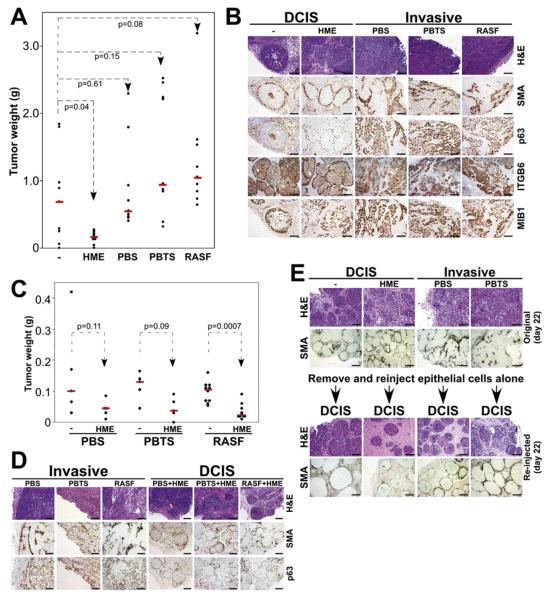
Following the verification that the MCFDCIS xenograft model reproduces main aspects of human basal-like DCIS tumors, we used this model to test the role of nonepithelial cells in tumor progression. We injected MCFDCIS cells into nude mice alone or together with normal primary cultured or immortalized myoepithelial cells (HME), or primary cultured fibroblasts derived from normal breast tissue (PBS), invasive breast carcinomas (PBTS), and rheumatoid arthritis (RASF). All xenografts were analyzed at early (3 to 4 weeks) time points after injection in order to avoid spontaneous progression to invasive tumors. HME statistically significantly suppressed tumor weight, PBS had no measurable effect, and PBTS and RASF increased tumor weight (Figure 3A). Microscopic examination revealed dramatic differences in histology among the different coinjection groups. MCFDCIS cells alone or coinjected with normal myoepithelial cells formed DCIS, whereas coinjection of any fibroblasts resulted in invasive carcinomas (Figure 3B). The DCIS and invasive histology was confirmed by the immunohistochemical analysis of myoepithelial markers. Similar to primary human tumors, MMP14 was highly expressed in bulk invasive xenografts (data not shown). Analysis of the expression of MIB1 (Ki67) revealed increased proliferation in invasive tumors (Figure 3B), correlating with their faster growth rate (data not shown). iFISH analyses of xenografts revealed lack of human fibroblasts in the stroma of the tumors (Figure S6). Thus, despite their inability to persist long-term in immunodeficient mice, coinjected fibroblasts exert a long-lasting effect on tumor weight and histology. All of these experiments were performed at least three times with essentially the same results using myoepithelial cells and fibroblasts derived from multiple independent patients.
Figure 3. The Effect of Nonepithelial Cells on MCFDCIS Xenografts.
(A) The effect of coinjection of different cells on tumor weight. Normal myoepithelial cells (HME) significantly suppressed tumor weight, fibroblasts from normal breast (PBS) had no effect, and fibroblasts from breast tumors (PBTS) and rheumatoid arthritis synovium (RASF) increased tumor weight. (B) Histological and immunohistochemical analyses of MCFDCIS xenografts from coinjection experiments.
(C and D) The dominant effect of normal myoepithelial cells on tumor weight (C) and histology (D). Tumors resulting from the injection of MCFDCIS cells together with PBS+HME, PBTS+HME, and RASF+HME were significantly smaller and had DCIS histology compared to xenografts initiated from MCFDCIS cells coinjected with fibroblasts (PBS, PBTS, or RASF).
(E) Tumors of various coinjections were removed and analyzed 22 days after injection (original day 22). Reinjection of MCFDCIS cells isolated from these tumors (without any coinjection) resulted in DCIS-like xenografts (reinjected day 22).
Scale bars correspond to 100 μm in all panels.
To define whether the normal myoepithelial cells can overcome the tumor promoting effects of fibroblasts, we also analyzed xenografts obtained from the coinjection of all three cell types: MCFDCIS tumor cells, normal myoepithelial cells, and fibroblasts. Interestingly, inclusion of normal myoepithelial cells was able to reverse the tumor growth and progression-promoting effects of all fibroblasts since tumors derived from triple injections were smaller and had DCIS histology (Figures 3C and 3D).
To exclude the possibility that the tumor growth and progression-promoting effects of coinjected human fibroblasts were due to the preferential outgrowth of a subpopulation of MCFDCIS cells with preexisting or acquired invasive properties, we isolated tumor epithelial cells from xenografts formed from MCFDCIS cells alone and from different coinjections and reinjected them (without adding any nonepithelial cells) into new (naive) nude mice. All reinjected tumors had DCIS histology, suggesting that the MCFDCIS epithelial cells were similar in all tumors and that fibroblasts have to be present at the time of injection to exert their progression-promoting effects (Figure 3E). SNP array analysis of xenografts formed from MCFDCIS cells at different time points (3–8 weeks) and from different coinjection groups also demonstrated that acquisition of additional genetic alterations in tumor epithelial cells is not necessary for progression to invasion (Figure S3C).
Signaling Network in MCFDCIS Cells
Our gene expression data of epithelial and myoepithelial cells purified from MCFDCIS xenografts and human primary tissue suggested the preferential activation of the TGFβ1, Hh, cell adhesion, and p63 pathways in ITGB6+ myoepithelial cells (Table 1; Figure 2A; Figure S5). Recent studies have demonstrated significant crosstalk among these signaling pathways (Di Marcotullio et al., 2007; Lauth and Toftgard, 2007). Thus, we investigated the activity of these pathways and their role in regulating myoepithelial cell phenotypes in MCFDCIS cells.
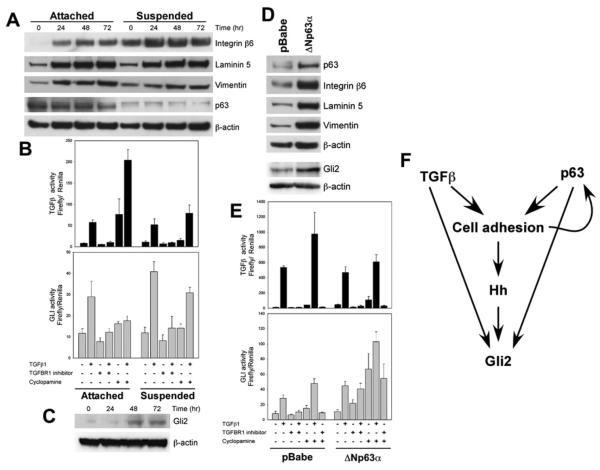
To determine if TGFβ1 influences cell adhesion or p63, MCFDCIS cells grown in 2D or in suspension cultures were analyzed at different time points following TGFβ1 treatment for the expression of p63 and for targets of the two pathways. TGFβ1 did not affect p63 in any conditions analyzed, but upregulated the expression of integrin β6, laminin 5, and vimentin, whereas p63 protein levels decreased in suspension (Figure 4A), consistent with prior studies in MCF10A cells (Carroll et al., 2006).
Figure 4. Pathways Regulating Myoepithelial Cell Phenotypes.
(A) Immunoblot analysis of MCFDCIS cells treated with TGFβ1 for the indicated time in attached or suspension culture. Integrin β6, laminin 5, and vimentin levels are increased following TGFβ1 treatment. p63 is not affected by TGFβ1, but it is downregulated in suspension.
(B) TGFβ and Hh pathway activity in MCFDCIS cells following the indicated treatments determined using luciferase reporters. Error bars represent mean ± SD.
(C) Immunoblot analysis of Gli2 protein levels in MCFDCIS cells treated with TGFβ1 for the indicated time in attached culture.
(D) Immunoblot analysis of MCFDCIS cells infected with control (pBabe) and ΔNp63α overexpressing retroviruses. ΔNp63α overexpression increases integrin β6, laminin 5, vimentin, and Gli2 levels.
(E) TGFβ and Hh pathway activity in control and ΔNp63α overexpressing MCFDCIS cells following the indicated treatments determined using luciferase reporters. Error bars represent mean ± SD.
(F) Summary of interactions among TGFβ, Hh, cell adhesion, and p63 pathways.
Next, we analyzed the transcriptional activity of the TGFβ and Hh pathways in MCFDCIS cells using luciferase reporter constructs. As expected, strong TGFβ transcriptional activation was observed following TGFβ1 treatment, which was eliminated by treatment with a TGFBR1 inhibitor (Figure 4B). Treatment with TGFβ1 also increased Gli transcriptional activity and Gli2 protein levels (Figures 4B and 4C). Unexpectedly, cyclopamine (an inhibitor of SMO activation) treatment alone increased the activity of the TGFβ responsive promoter, which was further augmented by TGFβ1 treatment (Figure 4B). The mechanism of this induction is unknown, but it appears to involve SMO and TGFβ receptors, since another SMO inhibitor had similar effects and a TGFBR1 inhibitor abolished it (data not shown).
p63 has been shown to play an essential role in the regulation of epithelial stem cell function and differentiation (McKeon, 2004). To determine the effect of constitutive overexpression of p63 on TGFβ and Hh signaling and luminal and myoepithelial cell differentiation, we generated derivatives of MCFDCIS cells overexpressing ΔNp63α (the predominant isoform in MCFDCIS cells). Overexpression of ΔNp63α was confirmed by immunoblot analysis of its expression as well as that of its transcriptional targets (Figure 4D). Control and ΔNp63α overexpressing cells responded similarly to TGFβ1 treatment, whereas the induction of the Gli reporter by cyclopamine and cyclopamine+TGFβ1 were more significant in ΔNp63α overexpressing than in control cells presumably due to the induction of Gli2 expression by ΔNp63α (Figures 4D and 4E). In summary, there appears to be extensive crosstalk among the TGFβ, Hh, cell adhesion, and p63 signaling pathways in MCFDCIS cells (Figure 4F), and the combined action of these pathways may be required for the maintenance of the bipotential progenitor and differentiated myoepithelial cell phenotypes.
Pathways Regulating Myoepithelial Differentiation and Progression to Invasion
To determine if any of the pathways we identified as candidate regulator of myoepithelial cell differentiation and progression to invasion play an essential role in these processes, we generated derivatives of MCFDCIS cells with altered expression of the selected genes. Specifically, using lentiviral shRNA, we downregulated SMAD4, TGFBR2, GLI2, and MMP14 levels (decreasing p63 resulted in cell death), whereas using retroviral constructs, we overexpressed ΔNp63α and MMP14 in MCFDCIS cells. For each of the genes tested, 5–13 shRNAs were evaluated for efficient downregulation of mRNA and the best two clones were further validated for their effects on protein and signaling activity (Figures S7 and S8). No significant phenotypic change was observed in any of the MCFDCIS derivatives in culture (data not shown).
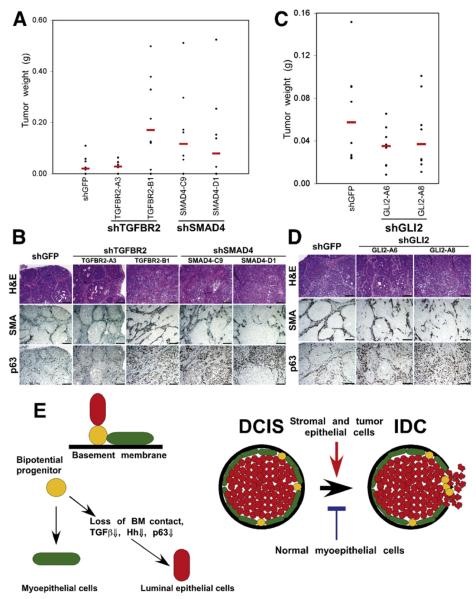
Next, xenograft assays were performed to analyze gain-of-function and loss-of-function effects on tumor weight and histology. Downregulation of TGFBR2 and SMAD4 increased tumor weight and promoted invasive histology due to a decrease in myoepithelial cells (Figures 5A and 5B). Downregulation of Gli2 similarly promoted progression to invasion, but decreased tumor weight (Figures 5C and 5D). Thus, the invasive tumor phenotype was independent of tumor size but was determined by the presence of myoepithelial cells. Overexpression of MMP14 did not have significant effect on MCFDCIS xenografts (Figures S8A and S8B; data not shown). In contrast, xenografts derived from MCFDCIS cells expressing MMP14 shRNA showed loss of myoepithelial cells and invasive phenotype, but had no difference in tumor weight compared to shGFP controls (Figures S8C–S8D; data not shown). Overexpression of ΔNp63α had no effect on MCFDCIS xenografts (Figure S8E; data not shown).
Figure 5. Pathways Regulating In Situ to Invasive Carcinoma Progression.
(A and B) The effect of loss of TGFβ signaling in MCFDCIS cells on tumor weight (A) and histology (B). Downregulation of TGFBR2 (in one of the clones) or SMAD4 increased tumor weight (TGFBR2-A3, p = 0.7899; TGFBR2-B1, p = 0.02996; SMAD4-C9, p = 0.09847; SMAD4-D1, p = 0.2379) and resulted in invasive tumors.
(C and D) The effect of decreased Gli2 expression in MCFDCIS cells on tumor weight (C) and histology (D). Downregulation of Gli2 decreased tumor weight (GLI2-A6, p = 0.1605; GLI2-A8, p = 0.2786), but increased invasive histology. Scale bars correspond to 100 μm in all panels.
(E) Hypothetical model summarizing our results and explaining in situ to invasive breast carcinoma progression. Bipotential mammary epithelial progenitor cells can give rise to myoepithelial and luminal cells. Detachment from the basement membrane (BM) and subsequent downregulation of TGFβ, Hh, integrin-ECM, and p63 pathways lead to luminal epithelial differentiation. Myoepithelial cells are necessary for the formation of DCIS. Stromal fibroblasts promote while normal myoepithelial cells inhibit tumor progression in part through their effects on the BM.
DISCUSSION
The DCIS to invasive carcinoma transition is a clinically important yet poorly understood step of breast tumorigenesis (Allred et al., 2001; Burstein et al., 2004). Others and we have analyzed the gene expression and genetic profiles of tumor epithelial cells isolated from DCIS and invasive tumors but have not been able to define a tumor stage-specific molecular event (Chin et al., 2004; Ma et al., 2003; Porter et al., 2003; Yao et al., 2006). At the same time, the importance of changes in the microenvironment during tumor progression has been increasingly recognized (Bissell et al., 2005; Tlsty and Hein, 2001; Weinberg and Mihich, 2006). Focal disruption of basement membrane appears to coincide with the disappearance of myoepithelial cells and stromal changes in human DCIS with high risk of progression to invasive carcinoma (Man et al., 2003). Correlating with the disappearance of the myoepithelial cells during the in situ to invasive carcinoma transition, the gene expression and epigenetic profiles of myoepithelial cells associated with DCIS become distinct from those in normal breast (Allinen et al., 2004; Hu et al., 2005). The signals that initiate these changes are unknown, although paracrine interactions with neoplastic epithelial and a variety of stromal cells are potential candidates. In contrast to normal myoepithelial cells, tumor-associated fibroblasts and myofibroblasts have been shown to promote tumorigenesis (Bissell et al., 2005; Tlsty and Hein, 2001; Weinberg and Mihich, 2006). We therefore tested the hypothesis that myoepithelial cells and fibroblasts regulate the in situ to invasive carcinoma transition using a xenograft model of human DCIS and primary human breast tumors.
The MCFDCIS cells form DCIS-like xenografts that spontaneously progress to invasive carcinomas. Based on our molecular analyses, this xenograft model resembles human basal-like DCIS. We demonstrated that in the absence of normal myoepithelial cells, coinjection of fibroblasts (regardless of their tissue of origin) promoted progression of DCIS to invasive tumors. Additionally, breast tumor-associated and, even more dramatically, inflammatory fibroblasts from rheumatoid arthritis, also increased tumor growth. Coinjection of normal myoepithelial cells overcame the tumor progression-promoting effects of fibroblasts and effectively suppressed tumor weight. Most importantly, these differences in tumor growth and histology were not caused by permanent genetic changes in the epithelial cells of the tumors. Furthermore, MCFDCIS cells retrieved from invasive tumors were still able to form DCIS when reinjected into naive mice in the absence of additional fibroblasts.
We found that the unique ability of the MCFDCIS cells to form DCIS is due to their bipotential progenitor property that enables their differentiation into luminal and myoepithelial cells in vivo. Differentiation to the myoepithelial cell phenotype is required for DCIS histology. Based on our data, we hypothesized that this process is influenced by coinjected normal myoepithelial cells and fibroblasts, presumably via paracrine factors.
Based on our immunohistochemical analyses and gene expression profiling of luminal and myoepithelial cells isolated from DCIS xenografts and human breast tissue, we identified TGFβ, Hh, cell adhesion, and p63 signaling pathways as potential regulators of the luminal and myoepithelial cell phenotypes. We found extensive crosstalk among these pathways in MCFDCIS cells and demonstrated that decreasing TGFβ and Hh pathway activity via downregulating TGFBR2/SMAD4 and Gli2 expression, respectively, resulted in the loss of myoepithelial cells and accelerated progression to invasion. These findings suggest a critical role for TGFβ and Hh signaling in breast tumor progression.
Many of the targets of these signaling pathways encode ECM proteins and receptors, and several of these are regulated by more than one signaling pathway. For example, both p63 and TGFβ1 upregulate ITGB6 expression, which in turn can activate latent TGFβ1, generating a positive feedback loop. At the same time, p63 protein levels are regulated by cell adhesion, and luminal epithelial differentiation may be initiated by detachment from basement membrane (BM) and subsequent downregulation of p63.
Gene ontology analysis of genes differentially expressed between epithelial and myoepithelial cells isolated from primary human tissue as well as from MCFDCIS xenografts demonstrated that myoepithelial cells play important roles in BM synthesis and maintenance. The phenotypic changes that occur in these cells in DCIS may lead to progressive degradation of BM, which eliminates the barrier between the epithelial and stromal cell compartments and also results in the loss of myoepithelial cells. Thus, the integrity of BM may be key to the maintenance of the basal/myoepithelial cell layer. Proteases secreted by tumor epithelial and other cells in the microenvironment may promote tumor progression via destroying the BM, resulting in the loss of bipotential progenitors and myoepithelial cells. Consistent with these, we found dramatic upregulation of MMP14 in DCIS-associated myoepithelial cells of human tissue and in the ITGB6+ myoepithelial cells of MCFDCIS xenografts. However, when we tested the effect of MMP14 downregulation in the MCFDCIS model, we found that decreased MMP14 levels led to decreased number of myoepithelial cells and invasive histology. This seemingly paradoxical effect is likely due to the intricate interactions between TGFβ and MMP14. Specifically, MMP14 activates TGFβ by releasing it from its latent form, and TGFβ upregulates MMP14 (Labbe et al., 2007; Mu et al., 2002). Therefore, modulating MMP14 expression influences TGFβ signaling, which we showed was required for the myoepithelial cell phenotype. Thus, the effects of MMP14 overexpression or loss are context dependent.
Human DCIS has been shown to have numerous genetic alterations and is almost indistinguishable from invasive tumors (Chin et al., 2004; Yao et al., 2006). As such, additional mutations in the epithelial cells are not necessary for the in situ to invasive transition; loss of the BM, epithelial cell organization and polarity due to disappearance of myoepithelial cells, appears to be sufficient to pave the way for tumor progression and invasion. A simplified view of these dynamic cellular and signaling interactions and their effect on the DCIS to invasive carcinoma progression are summarized in Figure 5E. Breast tumors are very heterogeneous with several distinct molecular subtypes and potentially distinct tumor progression pathways. MCFDCIS cells resemble basal-like breast tumors thought to originate from bipotential stem cells (Yehiely et al., 2006). Specifically, the ITGB6+ myoepithelial cells are derived from MCFDCIS cells; thus, they are genetically abnormal contrary to myoepithelial cells isolated from human nonbasal DCIS (Allinen et al., 2004). However, due to the fact that DCIS extends into normal ducts and lobules, genetically abnormal myoepithelial cells may only be detectable at the site of tumor initiation. Indeed, our careful examination of slides dual stained for p53 and CK5 from basal-like DCIS identified rare double-positive cells potentially reflecting myoepithelial cells with mutant p53. Further molecular analysis of these cells in multiple DCIS tumors is necessary to confirm the presence of genetically abnormal myoepithelial cells and to determine if this influences risk of progression to invasion. However, our model may not be universally true for all breast tumors, and there are possibly other pathways of progression to invasive breast carcinoma.
In summary, our data suggest that the progression of in situ to invasive breast carcinoma may not be due to the intrinsic properties of the tumor epithelial cells acquired during tumor evolution but determined by complex interactions among all the cell types that compose the tumor microenvironment. Our conclusions are based the characterization of a xenograft model of human DCIS and primary human breast tissue samples. Thus, our results not only highlight the importance of the microenvironment in breast tumor progression, but also point to the significance of the myoepithelial cell layer and basement membrane as gatekeepers of DCIS. Furthermore, the results suggest that therapeutic strategies targeting interactions of tumor epithelial cells with their surroundings may be more beneficial to inhibiting tumor progression than focusing on the tumor epithelial cells alone.
EXPERIMENTAL PROCEDURES
Cells and Tissue Specimens
MCF10A-series was obtained from Dr. Fred Miller (Karmanos Cancer Institute, Detroit, MI). HME50 and D920 immortalized myoepithelial cells (Gudjonsson et al., 2002a; Shay et al., 1995) were generously provided by Drs. Jerry Shay and Ole Petersen. RASF were generous gifts by Drs. Steve Goldring and John D. Mountz. Primary normal myoepithelial cells and fibroblasts from normal and neoplastic breast were purified from tissue samples from the Brigham and Women's Hospital (Boston, MA) as previously described (Allinen et al., 2004). All human tissue was collected using protocols approved by the Institutional Review Boards. Cells were grown in the media recommended by the providers. Myoepithelial cells were maintained in MEGM (Cambrex, Walkers-ville, MD), while fibroblasts were kept in DMEM (Invitrogen, Carlsbad, CA) with 10% iron fortified bovine calf serum (Hyclone, Logan, UT). Generation of retroviruses and lentiviruses and infection of MCFDCIS cells were carried out as previously described (Carroll et al., 2006; Moffat et al., 2006).
Xenograft Experiments
For xenograft studies 100,000 MCFDCIS cells were injected subcutaneously into 6- to 9-week-old female nude mice alone or together with 2- to 3-fold excess of HME, RASF, PBS, or PBTS cells in 50% Matrigel (BD Biosciences, Bedford, MA). Tumors were allowed to grow for 3–8 weeks. Xenografts were weighed and then either snap frozen on dry ice and stored at −80°C for DNA/RNA purification, formalin fixed and paraffin embedded, or processed for cell sorting. Animal experiments were conducted following protocols approved by the Harvard Medical School Animal Care and Use Committee.
Immunohistochemistry, Immunocytochemistry, Immunofluorescence, Immunoblot, iFISH, and FACS Analyses
The list of antibodies used is provided in Table S2. Immunoblot, immunocytochemistry, and FACS analyses were performed as recommended by the suppliers essentially as previously described (Allinen et al., 2004). Immunohistochemistry and iFISH procedure are described in the Supplemental Data.
Cell Purification, SAGE, SNP Array, PCR, and Statistical Analyses
Cell purification and SAGE library generation and analyses were performed as described previously (Allinen et al., 2004), except that MUC1 (clone DF3) and ITGB6 (clone 3G9) antibodies were used. Gene ontology enrichment and clustering analyses are described in the Supplemental Data. SNP array analysis was performed by the Dana-Farber Microarray Core using Affymetrix 11K XbaI or 250K StyI SNP arrays as described (Allinen et al., 2004). cDNA synthesis, quantitative RT-PCR, and semiquantitative RT-PCR were carried out as described (Allinen et al., 2004; Hu et al., 2005). A list of primers used is available upon request. Xenograft weights and relative gene expression levels were analyzed using two-sided exact Wilcoxon Rank Sum test stratified by experiment when the data from different experiments were combined. In mouse xenograft experiments when both sides of the mouse got the same injection and the weights of the two tumors correlated, the average of the two tumor weights was used as the end point. There were no corrections for multiple comparisons.
Luciferase Assays
Cells were transfected with TGFβ (p3TP-lux) or Gli (pGL3B-8XGliBS-lc-luc) reporter constructs (Cooper et al., 2003; Sasaki et al., 1997; Wrana et al., 1992) together with renilla luciferase. Twenty-four hours posttransfection, cells were treated and incubated for another 24 hr, and then luciferase activity was determined. For cells treated in suspension, transfection was performed in attached culture, and then cells were split into ultra-low-binding 24-well plates and treated.
Supplementary Material
SIGNIFICANCE.
Although there has been a dramatic improvement in our ability to detect DCIS, our understanding of the factors involved in its progression has just begun. Analysis of epithelial cells from DCIS and invasive tumors has failed to identify stage-specific differences. However, the myoepithelial cell layer and basement membrane, present only in DCIS, are key distinguishing and diagnostic features. Here, we show that a key event of tumor progression is the disappearance of the myoepithelial cell layer due to defective myoepithelial cell differentiation regulated by intrinsic and microenvironmental signals. Thus, myoepithelial cells can be considered gatekeepers of the in situ to invasive carcinoma transition; understanding the pathways that regulate their differentiation may open new venues for cancer therapy and prevention.
ACKNOWLEDGMENTS
We greatly appreciate the help of Diana Calogrias with the acquisition of human tissue samples and Natasha Pliss with immunohistochemistry. We thank Drs. John Mountz (University of Alabama, Birmingham, AL), Steven Goldring (Hospital for Special Surgery, New York, NY), Ole Petersen (Panem Institute, Denmark), Jerry Shay (University of Texas Southwestern Medical Center, Dallas, TX) for providing us with cells as described above; Dr. Paul Weinreb (Biogen-Idec, Cambridge, MA) for help with ITGB6 assays; Dr. Jonathan Yingling (Eli Lilly, Indianapolis, IN) for TGFBR inhibitors; Dr. Tae-Hwa Chun (University of Michigan, Ann Arbor, MI) for retroviral constructs encoding MMP14, the Broad Institute TRC for shRNA constructs; Dr. Bert Vogelstein, Dr. Myles Brown, and Lauren Campbell for their critical reading of the manuscript; and the Genome Sciences Centre, British Columbia Cancer Agency, Vancouver Canada for sequencing. This work was supported in part by NIH (CA89393, CA94074, and CA116235), DOD (W8IXWH-04-1-0452), and ACS (RSG-05-154-01-MGO) grants to KP; DOD (W81XWH-07-1-0408) to W.C.H.; Susan G. Komen Foundation fellowship (PDF042234) to M.H.; and by Biogen-Idec. Work from the M.J.B. laboratory was supported by NIH (CA64786) to M.J.B. and Ole Petersen. K.P. and W.C.H. receive research support from and are consultants to Novartis Pharmaceuticals, Inc. K.P. also receives research support from Biogen-Idec and is a consultant to Aveo Pharmaceuticals, Inc.
Footnotes
ACCESSION NUMBERS SAGE and SNP data were deposited to the Cancer Genome Anatomy Project (http://cgap.nci.nih.gov/SAGE) and to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo; accession no. GSE10747), respectively.
SUPPLEMENTAL DATA The Supplemental Data include Supplemental Experimental Procedures, five supplemental tables, and eight supplemental figures and can be found with this article online at http://www.cancercell.org/cgi/content/full/13/5/394/DC1/.
REFERENCES
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Allred DC, Mohsin SK, Fuqua SA. Histological and biological evolution of human premalignant breast disease. Endocr. Relat. Cancer. 2001;8:47–61. doi: 10.1677/erc.0.0080047. [DOI] [PubMed] [Google Scholar]
- Barsky SH, Karlin NJ. Myoepithelial cells: Autocrine and paracrine suppressors of breast cancer progression. J. Mammary Gland Biol. Neoplasia. 2005;10:249–260. doi: 10.1007/s10911-005-9585-5. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: The role of extracellular matrix and its degrading enzymes. Cold Spring Harb. Symp. Quant. Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. N. Engl. J. Med. 2004;350:1430–1441. doi: 10.1056/NEJMra031301. [DOI] [PubMed] [Google Scholar]
- Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat. Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, Brugge JS, Ellisen LW. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, et al. In situ analyses of genome instability in breast cancer. Nat. Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- Cooper MK, Wassif CA, Krakowiak PA, Taipale J, Gong R, Kelley RI, Porter FD, Beachy PA. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 2003;33:508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Screpanti I, Gulino A. Multiple ubiquitin-dependent processing pathways regulate hedgehog/gli signaling: Implications for cell development and tumorigenesis. Cell Cycle. 2007;6:390–393. doi: 10.4161/cc.6.4.3809. [DOI] [PubMed] [Google Scholar]
- Fisher ER, Dignam J, Tan-Chiu E, Costantino J, Fisher B, Paik S, Wolmark N. Pathologic findings from the National Surgical Adjuvant Breast Project (NSABP) eight-year update of Protocol B-17: Intraductal carcinoma. Cancer. 1999;86:429–438. doi: 10.1002/(sici)1097-0142(19990801)86:3<429::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J. Cell Sci. 2002a;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002b;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AR, Bissell MJ. The influence of tissue microenvironment (stroma and extracellular matrix) on the development and function of mammary epithelium. Epithelial Cell Biol. 1993;2:79–89. [PubMed] [Google Scholar]
- Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat. Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- Labbe E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- Lauth M, Toftgard R. Non-canonical activation of GLI transcription factors: Implications for targeted anti-cancer therapy. Cell Cycle. 2007;6:2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- Lerwill MF. Current practical applications of diagnostic immunohistochemistry in breast pathology. Am. J. Surg. Pathol. 2004;28:1076–1091. doi: 10.1097/01.pas.0000126780.10029.f0. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, et al. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man YG, Tai L, Barner R, Vang R, Saenger JS, Shekitka KM, Bratthauer GL, Wheeler DT, Liang CY, Vinh TN, Strauss BL. Cell clusters overlying focally disrupted mammary myoepithelial cell layers and adjacent cells within the same duct display different immunohistochemical and genetic features: Implications for tumor progression and invasion. Breast Cancer Res. 2003;5:R231–R241. doi: 10.1186/bcr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon F. p63 and the epithelial stem cell: More than status quo? Genes Dev. 2004;18:465–469. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- Miller F. Xenograft models of premalignant breast disease. J. Mammary Gland Biol. Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J. Natl. Cancer Inst. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. MCF10DCIS.com [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J. Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Hu M. Do myoepithelial cells hold the key for breast tumor progression? J. Mammary Gland Biol. Neoplasia. 2005;10:231–247. doi: 10.1007/s10911-005-9584-6. [DOI] [PubMed] [Google Scholar]
- Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol. Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Shay JW, Tomlinson G, Piatyszek MA, Gollahon LS. Spontaneous in vitro immortalization of breast epithelial cells from a patient with Li-Fraumeni syndrome. Mol. Cell. Biol. 1995;15:425–432. doi: 10.1128/mcb.15.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J. Mammary Gland Biol. Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- Tlsty TD, Hein PW. Know thy neighbor: Stromal cells can contribute oncogenic signals. Curr. Opin. Genet. Dev. 2001;11:54–59. doi: 10.1016/s0959-437x(00)00156-8. [DOI] [PubMed] [Google Scholar]
- Weinberg R, Mihich E. Eighteenth annual pezcoller symposium: Tumor microenvironment and heterotypic interactions. Cancer Res. 2006;66:11550–11553. doi: 10.1158/0008-5472.CAN-06-3149. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol. Med. 2006;12:537–544. doi: 10.1016/j.molmed.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Yokota T, Yoshimoto M, Akiyama F, Sakamoto G, Kasumi F, Nakamura Y, Emi M. Frequent multiplication of chromosomal region 8q24.1 associated with aggressive histologic types of breast cancers. Cancer Lett. 1999;139:7–13. doi: 10.1016/s0304-3835(98)00329-2. [DOI] [PubMed] [Google Scholar]
- Zambruno G, Marchisio PC, Marconi A, Vaschieri C, Melchiori A, Giannetti A, De Luca M. Transforming growth factor-beta 1 modulates beta 1 and beta 5 integrin receptors and induces the de novo expression of the alpha v beta 6 heterodimer in normal human keratinocytes: Implications for wound healing. J. Cell Biol. 1995;129:853–865. doi: 10.1083/jcb.129.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.