Abstract
Bovine papillomavirus (BPV) is recognized as a causal agent of benign and malignant tumors in cattle. Thirteen types of BPV are currently characterized and classified into three distinct genera, associated with different pathological outcomes. The described BPV types as well as other putative ones have been demonstrated by molecular biology methods, mainly by the employment of degenerated PCR primers. Specifically, divergences in the nucleotide sequence of the L1 gene are useful for the identification and classification of new papillomavirus types. On the present work, a method based on the PCR-RFLP technique and DNA sequencing was evaluated as a screening tool, allowing for the detection of two relatively rare types of BPV in lesions samples from a six-year-old Holstein dairy cow, chronically affected with cutaneous papillomatosis. These findings point to the dissemination of BPVs with unclear pathogenic potential, since two relatively rare, new described BPV types, which were first characterized in Japan, were also detected in Brazil.
1. Introduction
The Bovine papillomavirus (BPV) is recognized as the causal agent of benign and malignant tumors in cattle, such as cutaneous papillomas, benign fibroplasias, urinary bladder, and esophagus cancer, causing significant economic losses. This oncogenic virus has a double-stranded circular DNA genome of approximately eight kilobases [1].
Currently, the Papillomaviridae family is divided into 16 genera according to their genomic organization [2, 3]. The papillomavirus (PV) genome codified functional, early (E) proteins and structural, late (L) proteins, expressed at different stages of the viral cycle. The L1 is the most conserved gene within a PV genome and has therefore been used for the identification of new PVs: one PV isolate is recognized as a new type if the complete genome has been cloned and the DNA sequence of L1 differs by more than 10% from the closest known PV type. Differences between 2% and 10% define a subtype and less than 2%, a variant [3]. Thirteen types of BPVs are currently well characterized and classified into three distinct genera—Delta, Epsilon, and Xi—each one associated with epithelia lesions of specific histological nature [4].
The BPVs-1 and -2 are classified as Delta papillomaviruses [5]. Characteristically, these types induce the appearance of fibropapillomas, associated with the recruitment of the subepithelial fibroblasts [6]. As far as concerned, both types are also unique in their ability to infect different host species, not only bovines, causing the equine sarcoid [7]. Lately, the genome of a new Delta-BPV type (BPV-13) was fully sequenced [8].
A larger number of BPV types (-3, -4, -6, -9, -10, -11 and -12) belong to the Xipapillomavirus genus. These viruses are considered exclusively epitheliotropic, inducing the formation of “true papillomas,” without the involvement of fibroblasts [9–11]. On the other hand, the BPVs-5 and -8 have the potential to induce both fibropapillomas and true papillomas in the course of their infectious cycle, being classified into a third genus, Epsilonpapillomavirus [12, 13]. The BPV-7 represents an exception and is classified separately (unsigned genus). This virus was first isolated from a cutaneous papilloma lesion and also from healthy teat skin samples [14].
The thirteen described BPV types as well as other putative ones have been demonstrated by molecular biology methods, since papillomaviruses are not prone to be replicated or recovered in cell cultures [15–19]. According to the guidelines outlined by the Papil omavirus Nomenclature Committee (14th International Papillomavirus Conference, Quebec City, QC, Canada), it has been specified that the amplified sequences isolated from novel papillomaviruses isolates could indicate only putative new PV types—instead of PV types—since the PCR amplicons represent only part of the L1 gene [15].
The use of PCR assays with degenerated primers, followed by sequencing has allowed the identification of several PV types in human and other animal hosts [15, 20]. The PCR primer FAP set was designed from two relatively conserved regions found in the L1 gene and has been shown to amplify PVs DNA from both papillomas and healthy tissue of many animal species, including BPVs in bovines [15, 16, 20].
Brazil has a cattle herd of approximately 210 million, being a major exporter of meat, milk and leather. BPVs have been previously detected in Brazil [21], but the extent of the impact of BPVs associated diseases, both in dairy and cattle herds, needs further studies. Available reports in different regions of the country indicate a significant diversity of viral types among the Brazilian herd, implying an evident disease burden [18, 22–25]. Thus, the improvement of knowledge concerning the diagnosis and related clinical aspects of different BPV types among the Brazilian herd should be considered in the development of new sanitary measures, aiming to the prevention of BPV infection and its consequences.
Unfortunately, BPV epidemiological surveys are still limited by the availability of high-throughput diagnostic techniques that could discriminate different BPV sequences at the same time in co infected samples [16, 26]. In this context, the present work represents an effort to identify BPV types employing an alternative screening method based on the PCR-RFLP technique and correlating the histological data of the analyzed lesions with the diagnosed viral type.
2. Material and Methods
In silico generation of RFLPs: the L1 FAP segment digestion profiles of the BPVs-1 to -13 could be generated with NEB cutter 2.0 [27] from all L1 complete nucleotide sequences available in Genbank (http://www.ncbi.nlm.nih.gov/genbank/). The restriction enzymes sites were chosen both by its presence (or absence) as well as the generated digestion fragments sizes, in order to differentiate those thirteen different BPV types. Histopathological analysis: wart biopsies were obtained from the trunk of a six-year-old Holstein dairy cow chronically affected with cutaneous papillomatosis. Samples from three different lesions were submitted to macroscopic, histological (hematoxylin and Eosin staining) and molecular analyses. DNA extraction and PCR: DNA was extracted from warts for viral typing (Illustra tissue and cells genomic Prep Mini Spin GE Healthcare) and an approximately 470, base-pairs L1 gene segment was amplified using the following primer sequences: forward: FAP59 (5′-TAA CWG TIG GIC AYC CWT ATT-3′) reverse: FAP64 (5′-CCWATATCWVHCATITCICCATC-3′). The PCR, were performed with slight modifications of a previously described protocol [16]. In detail the amplification reactions were performed in a Corbett CG1-96 thermocycler (Corbett Life Science, Sydney, Australia), with GoTaq Master Mix (Promega, Madison, USA), under the following conditions: 5 min at 95°C, followed by 35 cycles of 1 min and 30s at 95°C, 2 min at 52°C and 1 min and 30s at 72°C and a final extension step of 5 min at 72°C. Restriction analysis: an aliquot of PCR fragments were submitted to digestion reactions for RFLP analysis with four different restriction enzymes (DdeI, HinfI, HindIII, MsLI), following manufacturer's instructions (New England Biolabs, Ipswich, USA). Cloned BPV-1 and BPV-2 genomes as well as a known typed clinical sample (Mg-19, BPV-2 typed) were used as positive controls. PCR-RFLP products were analyzed in 2.0% agarose gel electrophoresis stained with ethidium bromide (0.5 μg/mL) in TAE buffer and visualized under UV light. Sequencing: An aliquot of all generated PCR fragments were purified with extraction columns (Illustra GFX PCR DNA and Gel Band Purification Kit GE Healthcare). DNA concentration and purity were determined in a spectrophotometer (Eppendorf BioPhotometer, Hamburg, Germany) and submitted to sequencing reactions: three independent sequencing reactions were done for each PCR fragment in an ABI377 PRISM Genetic Analyzer (Life Applied Biosystems, USA). The quality of DNA sequences was checked and overlapping fragments were assembled using the BioEdit package 7.0.9.0 [28]. Assembled sequences with high quality were aligned using ClustalW 1.83 [29] with default gap penalties. Homology analyses were performed with the NCBI database and BLAST [30]. BioEdit software was used to identify the equivalent amino acid sequences. The sequence alignments were performed using the MEGA 5.0 software [31], using full alignment and 2000 total replications on the bootstrap, in order to ensure a higher level of confidence to our analysis [32]. Phylogenetic Analysis: phylogenetic relationship comparing nucleotide sequences was performed with MEGA. Neighbor-joining trees were drawn using TreeView version 1.6.6 [33]. Nucleotide and amino acid sequences from other BPV types and of a HPV-16 were retrieved from the GenBank (http://www.ncbi.nlm.nih.gov/) for comparison with the obtained sequences here.
Ethics Statement. The protocols used in this study were approved by the Ethical Committee for Animal Experimentation of the Instituto de Zootecnia (Protocol no. 109, on July 06th, 2009) assigned by the President of this Committee. All efforts were made to minimize animal suffering.
3. Results
In silico analyses of the restrictionsites in FAP L1 fragments of 20 deposited BPVs sequences (from BPV-1 to BPV-13) revealed no intratype variation associated with the relative cut positions for the four enzymes employed (Table 1). Macroscopic and histological evaluations identified the collected lesions as cutaneous fibrous papillomas (Figures 1 and 2). From all the three lesions studied, two- (IZ1 and IZ3) RFLP profiling suggests the presence of BPV-11. On the other hand, IZ2 sample gene rated a profile which could not match with any of the thirteen characterized viral types (Figure 3, Table 1). In accordance with these results, DNA sequencing and BLAST analysis of IZ2 indicated a rare putative type (BAPV-3), originally described in Japan [16]. Furthermore, the DNA sequencing of other two samples confirms them as the recently described BPV-11 [10]. These sequences were deposited in GenBank (access numbers: HQ435675 and HQ612180). The phylogenetic reconstruction using this partial genomic sequence allowed its comparison with other sequences of Papillomaviridae family members (Figure 4).
Table 1.
L1 FAP segment digestion profiles of the BPVs-1 to -13 for Dde I, Hinf I, MsL I and HindIII.
| L1 FAP Fragment | DdeI | HinfI | MsLI | HindIII | |
|---|---|---|---|---|---|
| BPV-1 | 475 bp | 264 | 329 | 301 | 475 |
| 159 | 146 | 174 | |||
| 52 | |||||
|
| |||||
| BPV-2 | 475 bp | 316 | 329 | 475 | 475 |
| 159 | 146 | ||||
|
| |||||
| BPV-3 | 473 bp | 319 | 473 | 373 | 473 |
| 154 | 87 | ||||
| 13 | |||||
|
| |||||
| BPV-4 | 469 bp | 324 | 469 | 469 | 230 |
| 145 | 182 | ||||
| 87 | |||||
|
| |||||
| BPV-5 | 469 bp | 469 | 254 | 469 | 469 |
| 145 | |||||
| 70 | |||||
|
| |||||
| BPV-6 | 472 bp | 321 | 384 | 373 | 472 |
| 151 | 88 | 99 | |||
|
| |||||
| BPV-7 | 484 bp | 403 | 271 | 484 | 484 |
| 81 | 119 | ||||
| 94 | |||||
|
| |||||
| BPV-8 | 469 bp | 469 | 317 | 469 | 322 |
| 152 | 147 | ||||
|
| |||||
| BPV-9 | 469 bp | 316 | 344 | 469 | 469 |
| 153 | 125 | ||||
|
| |||||
| BPV-10 | 472 bp | 319 | 381 | 373 | 472 |
| 90 | 91 | 99 | |||
| 63 | |||||
|
| |||||
| BPV-11 | 475 bp | 420 | 407 | 370 | 475 |
| 55 | 68 | 105 | |||
|
| |||||
| BPV-12 | 469 bp | 351 | 317 | 469 | 469 |
| 118 | 152 | ||||
|
| |||||
| BPV-13 | 475 bp | 316 | 329 | 475 | 475 |
| 109 | 146 | ||||
| 50 | |||||
Reference Sequences:
BPV-1_NC_001522.1, BPV-1_X02346.1, BPV-2_M20219.1, BPV-2_X01768.1,
BPV-3_AF486184.1, BPV-3_AJ620207.1, BPV-3_NC_004197.1, BPV-4_X05817.1,
BPV-5_AF457465.1, BPV-5_NC_004195.1, BPV-6_AJ620208.1, BPV-7_DQ217793.1,
BPV-7_NC_007612.1, BPV-8_EB_DQ098917.1, BPV-8_NC_009752.1, BPV-9_AB331650.1,
BPV-10_AB331651.1, BPV-11_AB543507, BPV-12_JF834523, BPV-13_JQ798171.
Figure 1.
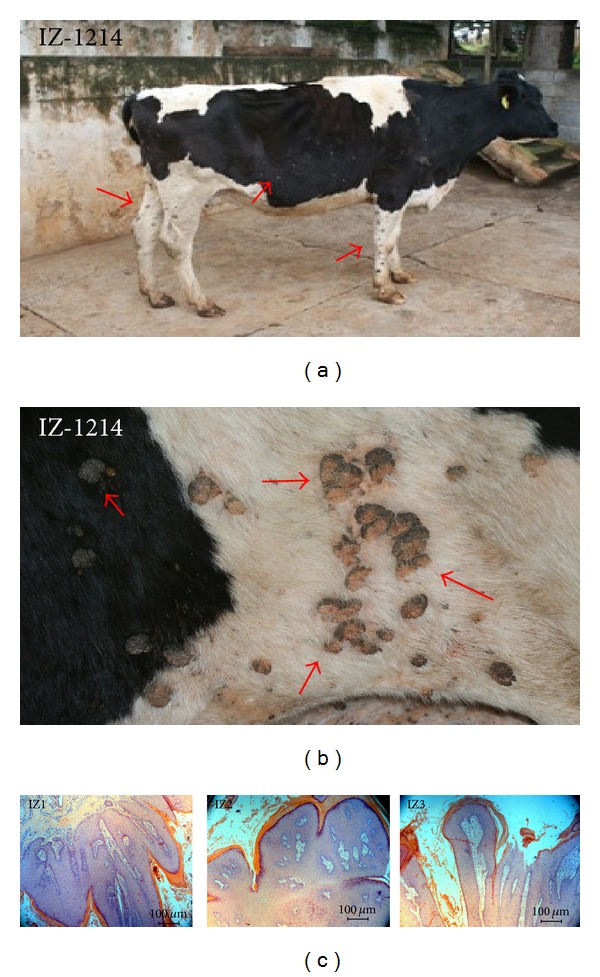
(a) Analyzed animal IZ-1214 with disseminated cutaneous papillomatosis. (b) Gross aspect of the papilloma lesions with a cauliflower-like appearance. (c) Histological preparation (hematoxylin and eosin staining, or HE) of the collected lesions (IZ1, IZ2, and IZ3) indicating characteristic hyperkeratosis, acanthosis, and papillomatosis with dermis proliferation in HE preparation (100x).
Figure 2.
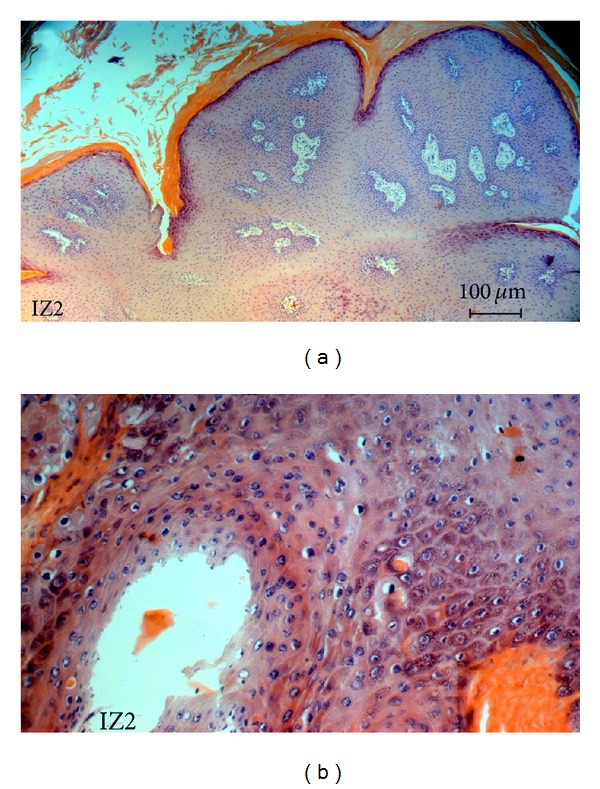
(a) Histopathology of a wart biopsy: detailed aspect of the IZ2 lesion exhibiting characteristic hyperkeratosis, acanthosis and dermal proliferation, indicated by arrows (100x). (b) Presence of koilocytosis.
Figure 3.
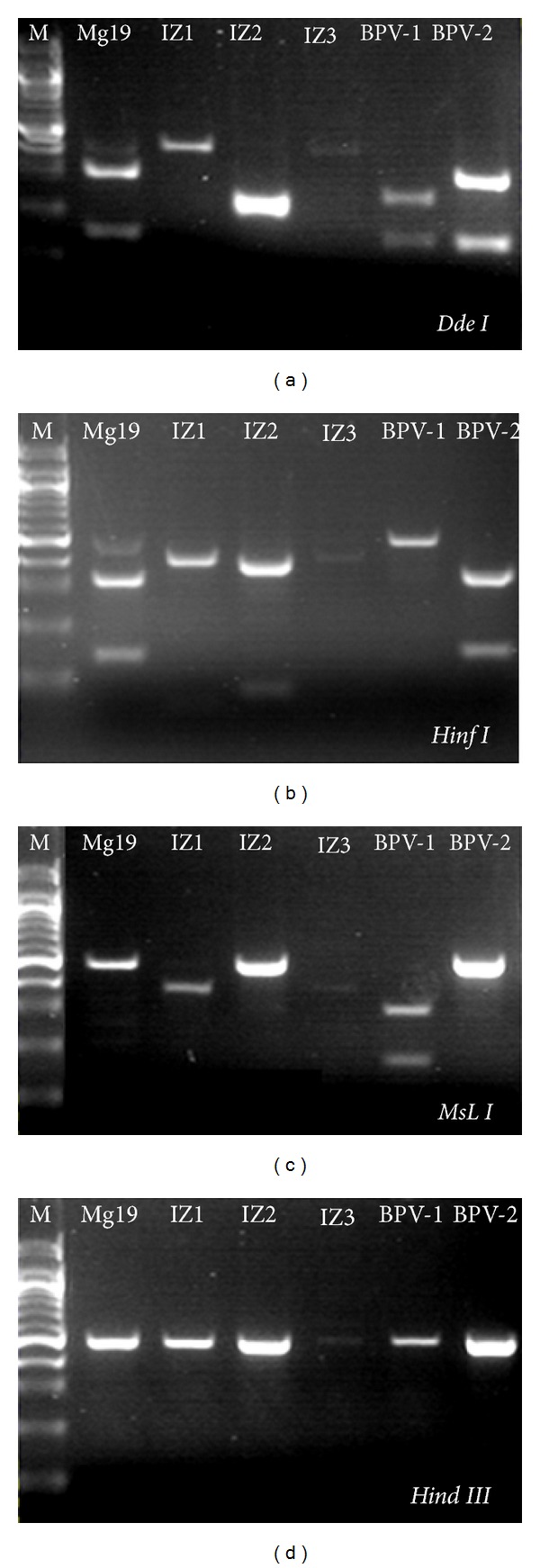
RFLP performed for restriction enzymes DdeI, HinfI, MsLI, and HindIII. As BPV-1 and BPV-2 are the commonly associated with fibropapillomas, their L1 FAP amplicons were generated and digested as positive controls. IZ1, IZ2, IZ3 amplicons are from three different lesions of the same animal (IZ-1214). Mg-19 sample is from another bovine, clinically affected with cutaneous papillomatosis. Molecular marker: 100 bp ladder (New England Biolabs, Ipswich, UK).
Figure 4.
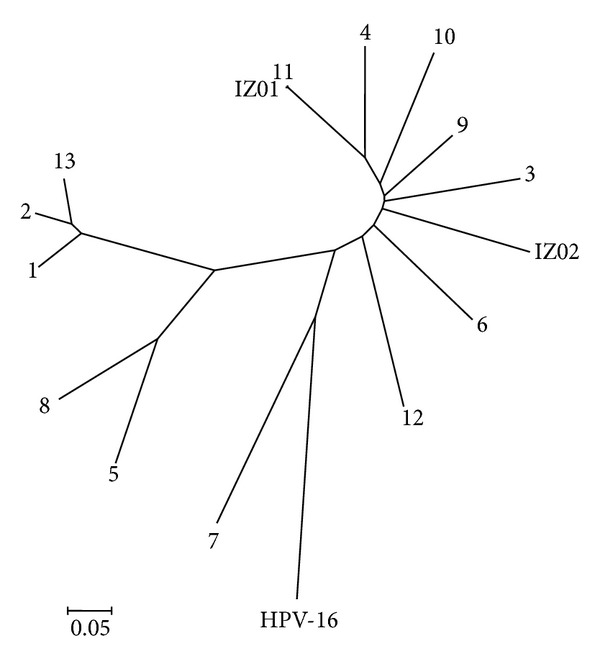
Neighbor-joining phylogenic tree constructed with 2000 bootstrap replications, using partial L1 nucleotide sequences, indicating the putative type BAPV-3 in IZ2 sample, originally described in Japan (accession: AY300819). The analysis of the other two samples, IZ1 and IZ3, demonstrated the presence of an identical sequence in both lesions, sharing 99% percent of similarity with the recently described BPV-11 type (accession: AB543507.1). As IZ3 sequence is identical to IZ1, it will not be shown here.
4. Discussion
Originally, the recently characterized BPV-11 was described with the employment of a Xipapillomavirus consensus primer [10]. Here, the same type using the FAP generic primer could be detected, indicating this set as an effective alternative for the identification of BPVs. Other than that, we described the simultaneous presence of two BPV types in three different wart samples, obtained from a chronically affected animal with disseminated papillomatosis.
The typed BPV-11 and BAPV-3 samples have a fibrous aspect, with a similar fibropapilloma histology that is not commonly associated with the exclusive infection of the keratinocytes, commonly attributed to the Xi BPVs. In a previous report [10], Hatama and others discussed the “uncertain nature of BPV-11 tumorigenicity” since BPV-11 was first diagnosed in a fibropapilloma lesion in which the BPV-1 was also detected. Despite the eventual limitations of the diagnostic methods employed, we could detect the same viral type in at least two different cauliflower-like lesions, with a suggestive fibrous core. In addition to Hatama's report, our data also link the BPV-11 with cutaneous papillomas, indicating it as a pathogenic type. Furthermore, the putative type BAPV-3, described by Ogawa and others [16], was detected just once from a skin papilloma sample, without histological description, being genetically associated with the BPV-3, -4, and -6, or Xi BPVs.
The L1 gene has taxonomical relevance due to its high degree of conservation, which can be accessed with the use of generic primer sets. In accordance with this, the restriction sites located in FAP fragments appears to be maintained, without intratype variations for the restriction enzymes employed, indicating that phylogenetic studies comparing BPVs and other members of the Papillomaviridae family are possible using a relatively short DNA sequence.
Since the early nineties, methods based on PCR-RFLP analysis within the L1 gene of the human papillomavirus (HPV) have been used for viral typing and infection diagnosis from a variety of sources, including cervical samples, fresh and paraffin-embedded tissues [34, 35]. Particularly, the PCR-RFLP method is useful to identify coinfections due to its sensitivity and specificity [36].
5. Conclusion
As an easy, rapid, and cost-effective assay, the PCR-RFLP represents a less-laborious approach than DNA cloning and sequencing, being an alternative as a first-line screening test, both for the diagnosis of an already classified virus type as to indicating the needing of DNA sequencing due to mixed and/or unknown digestion profiles. In a previous survey in Paraná State, Brazil, Claus and others [26] suggest that the occurrence of multiple or mixed BPVs infection may be widespread throughout Brazilian cattle herds and may occur in other Brazilian geographical regions. In agreement, our findings support these results and reinforce the notion that multiple papillomaviruses infections, with a significant pathogenic potential, can be as frequent in cattle as in human hosts [26].
To the best of our knowledge, the restriction map employed here is the first to be produced specifically for the screening and typing of BPVs. Our findings also point to the ubiquity of BPVs dissemination since two relatively rare, new described BPV types, which were first characterized in Japan, were also detected in Brazil.
Acknowledgments
The authors thank the Ministério de Ciência, Tecnologia e Inovação/Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq Proc. 402539/2011-7), Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES), and Fundação do Desenvolvimento Administrativo (PAP-FUNDAP) for the financial support and Nadia Jorge Berriel, Carolina da Paz Sabino, and Jim Hesson (http://www.AcademicEnglishSolutions.com/) for the editorial support.
References
- 1.Campo MS. Papillomavirus and disease in humans and animals. Veterinary and Comparative Oncology. 2003;1:3–14. doi: 10.1046/j.1476-5829.2003.00001.x. [DOI] [PubMed] [Google Scholar]
- 2.Chan S-Y, Delius H, Halpern AL, Bernard H-U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. Journal of Virology. 1995;69(5):3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Villiers E-M, Fauquet C, Broker TR, Bernard H-U, Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 4.Borzacchiello G, Roperto F. Bovine papillomaviruses, papillomas and cancer in cattle. Veterinary Research. 2008;39(5, article 45) doi: 10.1051/vetres:2008022. [DOI] [PubMed] [Google Scholar]
- 5.Campo MS, Jarrett WFH, Barron R, O’Neil BW, Smith KT. Association of bovine papillomavirus type 2 and bracken fern with bladder cancer in cattle. Cancer Research. 1992;52(24):6898–6904. [PubMed] [Google Scholar]
- 6.Jelínek F, Tachezy R. Cutaneous papillomatosis in cattle. Journal of Comparative Pathology. 2005;132(1):70–81. doi: 10.1016/j.jcpa.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Nasir L, Campo MS. Bovine papillomaviruses: their role in the aetiology of cutaneous tumours of bovids and equids. Veterinary Dermatology. 2008;19(5):243–254. doi: 10.1111/j.1365-3164.2008.00683.x. [DOI] [PubMed] [Google Scholar]
- 8.Lunardi M, Alfieri AA, Otonel RA, et al. Genetic characterization of a novel bovine papillomavirus member of the Deltapapillomavirus genus . Veterinary Microbiology. 2012;162(1):207–213. doi: 10.1016/j.vetmic.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Jarrett WFH, Campo MS, O’Neil BW, Laird HM, Coggins LW. A novel bovine papillomavirus (BPV-6) causing true epithelial papillomas of the mammary gland skin: a member of a proposed new BPV subgroup. Virology. 1984;136(2):255–264. doi: 10.1016/0042-6822(84)90162-4. [DOI] [PubMed] [Google Scholar]
- 10.Hatama S, Ishihara R, Ueda Y, Kanno T, Uchida I. Detection of a novel bovine papillomavirus type 11 (BPV-11) using Xipapillomavirus consensus polymerase chain reaction primers. Archives of Virology. 2011;156(7):1281–1285. doi: 10.1007/s00705-011-0970-7. [DOI] [PubMed] [Google Scholar]
- 11.Zhu W, Dong J, Shimizu E, et al. Characterization of novel bovine papillomavirus type 12 (BPV-12) causing epithelial papilloma. Archives of Virology. 2012;157(1):85–91. doi: 10.1007/s00705-011-1140-7. [DOI] [PubMed] [Google Scholar]
- 12.Bloch N, Sutton RH, Spradbrow PB. Bovine cutaneous papillomas associated with bovine papillomavirus type 5. Archives of Virology. 1994;138(3-4):373–377. doi: 10.1007/BF01379140. [DOI] [PubMed] [Google Scholar]
- 13.Tomita Y, Literak I, Ogawa T, Jin Z, Shirasawa H. Complete genomes and phylogenetic positions of bovine papillomavirus type 8 and a variant type from a European bison. Virus Genes. 2007;35(2):243–249. doi: 10.1007/s11262-006-0055-y. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa T, Tomita Y, Okada M, Shirasawa H. Complete genome and phylogenetic position of bovine papillomavirus type 7. Journal of General Virology. 2007;88(7):1934–1938. doi: 10.1099/vir.0.82794-0. [DOI] [PubMed] [Google Scholar]
- 15.Antonsson A, Hansson BG. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. Journal of Virology. 2002;76(24):12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa T, Tomita Y, Okada M, et al. Broad-spectrum detection of papillomaviruses in bovine teat papillomas and healthy teat skin. Journal of General Virology. 2004;85(8):2191–2197. doi: 10.1099/vir.0.80086-0. [DOI] [PubMed] [Google Scholar]
- 17.Maeda Y, Shibahara T, Wada Y, et al. An outbreak of teat papillomatosis in cattle caused by bovine papilloma virus (BPV) type 6 and unclassified BPVs. Veterinary Microbiology. 2007;121(3-4):242–248. doi: 10.1016/j.vetmic.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 18.Claus MP, Lunardi M, Alfieri AF, Ferracin LM, Fungaro MHP, Alfieri AA. Identification of unreported putative new bovine papillomavirus types in Brazilian cattle herds. Veterinary Microbiology. 2008;132(3-4):396–401. doi: 10.1016/j.vetmic.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 19.Hatama S, Nobumoto K, Kanno T. Genomic and phylogenetic analysis of two novel bovine papillomaviruses, BPV-9 and BPV-10. Journal of General Virology. 2008;89(1):158–163. doi: 10.1099/vir.0.83334-0. [DOI] [PubMed] [Google Scholar]
- 20.Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. Journal of General Virology. 1999;80(9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 21.Stocco Dos Santos RC, Lindsey CJ, Ferraz OP, et al. Bovine papillomavirus transmission and chromosomal aberrations: an experimental model. Journal of General Virology. 1998;79(9):2127–2135. doi: 10.1099/0022-1317-79-9-2127. [DOI] [PubMed] [Google Scholar]
- 22.Wosiacki SR, Claus MP, Alfieri AF, Alfieri AA. Bovine papillomavirus type 2 detection in the urinary bladder of cattle with chronic enzootic haematuria. Memorias do Instituto Oswaldo Cruz. 2006;101(6):635–638. doi: 10.1590/s0074-02762006000600009. [DOI] [PubMed] [Google Scholar]
- 23.Yaguiu A, Carvalho C, Freitas AC, et al. Papillomatosis in cattle: in situ detection of Bovine Papillomavirus DNA sequences in reproductive tissues. Brazilian Journal of Morphological Sciences. 2006;23(3):129–136. [Google Scholar]
- 24.Diniz N, Melo TC, Santos JF, et al. Simultaneous presence of bovine papillomavirus in blood and in short-term lymphocyte cultures from dairy cattle in Pernambuco, Brazil. Genetics and Molecular Research. 2009;8(4):1474–1480. doi: 10.4238/vol8-4gmr668. [DOI] [PubMed] [Google Scholar]
- 25.Lunardi M, Claus MP, Alfieri AA, Fungaro MHP. Phylogenetic position of an uncharacterized Brazilian strain of bovine papillomavirus in the genus Xipapillomavirus based on sequencing of the L1 open reading frame. Genetics and Molecular Biology. 2010;33(4):745–749. doi: 10.1590/S1415-47572010005000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claus MP, Lunardi M, Alfieri AA, et al. Multiple bovine papillomavirus infections associated with cutaneous papillomatosis in Brazilian cattle herds. Brazilian Archives of Biology and Technology. 2009;52:93–98. [Google Scholar]
- 27.Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Research. 2003;31(13):3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symposium Series. 1999;41:95–98. [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(23):13429–13434. doi: 10.1073/pnas.93.23.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D.m.page R. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 34.Lungu O, Wright TC, Jr., Silverstein S. Typing of human papillomaviruses by polymerase chain reaction amplification with L1 consensus primers and RFLP analysis. Molecular and Cellular Probes. 1992;6(2):145–152. doi: 10.1016/0890-8508(92)90059-7. [DOI] [PubMed] [Google Scholar]
- 35.Astori G, Arzese A, Pipan C, De Villiers E-M, Botta GA. Characterization of a putative new HPV genomic sequence from a cervical lesion using L1 consensus primers and restriction fragment length polymorphism. Virus Research. 1997;50(1):57–63. doi: 10.1016/s0168-1702(97)00054-3. [DOI] [PubMed] [Google Scholar]
- 36.Nobre RJ, de Almeida LP, Martins TC. Complete genotyping of mucosal human papillomavirus using a restriction fragment length polymorphism analysis and an original typing algorithm. Journal of Clinical Virology. 2008;42(1):13–21. doi: 10.1016/j.jcv.2007.11.021. [DOI] [PubMed] [Google Scholar]


