Abstract
Mutations in SOD1 cause FALS. The Cu binding capacity of SOD1 has spawned hypotheses that implicate metal-mediated production of reactive species as a potential mechanism of toxicity. In past experiments, we have tested such hypotheses by mutating residues in SOD1 that normally coordinate the binding of Cu, finding that such mutants retain the capacity to induce motor neuron disease. We now describe the lack of disease in mice that express a variant of human SOD1 in which residues that coordinate the binding of Cu and Zn have been mutated (SODMD). SODMD encodes 3 disease-causing and 4 experimental mutations that ultimately eliminate all histidines involved in the binding of metals; and includes one disease-causing and one experimental mutation that eliminate secondary metal binding at C6 and C111. We show that the combined effect of these mutations produces a protein that is unstable but does not aggregate on its own, is not toxic, and does not induce disease when co-expressed with high levels of wild-type SOD1. In cell culture models, we determine that the combined mutation of C6 and C111 to G and S, respectively, dramatically reduces the aggregation propensity of SODMD and may account for the lack of toxicity for this mutant.
Keywords: superoxide dismutase 1, motor neuron disease, transgenic mouse models, protein aggregation
Introduction
Mutations in superoxide dismutase 1 cause a subset of cases of familial amyotrophic lateral sclerosis (FALS) (Rosen et al. 1993). The enzymatic function of superoxide dismutase 1 (SOD1, EC 1.15.1.1) is to catalyze the antioxidant reaction that converts superoxide radicals (O2·-) into hydrogen peroxide (H2O2) and oxygen (O2) (McCord and Fridovich 1969). This enzymatic activity is dependent upon the presence of bound Cu ions (Forman and Fridovich 1973). SOD1 also binds Zn, which functions primarily to increase structural stability (Elam et al. 2003;Potter and Valentine 2003). In the functionally mature enzyme, two SOD1 proteins dimerize with each protein binding one atom of Cu and one atom of Zn. Histidine residues at four positions coordinate the binding of Cu (H46, H48, H63, and H120) with one of these histidines (H63) and three other residues (H71, H80, D83) coordinating the binding of zinc. The two monomers of SOD1 protein in the homodimer bind through non-covalent forces, while an intramolecular disulfide bond between cysteines 57 and 146 confers structural stability within each monomer (Tainer et al. 1982;Hart et al. 1998). Of the 145+ mutations in SOD1 that have been associated with ALS, only a small portion reduces the binding of Cu or Zn in manner that would reduce activity or severely destabilize the protein (Valentine and Hart 2003). Indeed disease-causing mutations do not necessarily have obvious effects on enzymatic activity, leading to the conclusion that disease-associated mutations in the protein cause the protein to acquire a toxic property (Borchelt et al. 1994;Borchelt et al. 1995) .
In vitro studies suggest that normally folded SOD1 can misfold and aggregate upon the loss of metals and/or reduction of its intramolecular disulfide bond (Chattopadhyay et al. 2008;Furukawa et al. 2008). Further studies demonstrate that metal binding prevents the dissociation of the intramolecular disulfide bond, suggesting that metal binding is required to prevent aggregation (Tiwari et al. 2005). Transgenic mouse models expressing SOD1 mutant proteins that abolish either two (H46R/H48Q) or four (H46R/H48Q/H63G/H120G or QUAD) of the normal copper binding sites develop motor neuron disease, which is accompanied by the characteristic formation of detergent-insoluble SOD1 aggregates (Wang et al. 2002;Wang et al. 2003). These mutants are unable to bind copper in their catalytic site; thus, copper binding does not appear to be required for aggregation (Wang et al. 2007). However, additional studies suggest that copper can interact with free cysteines, in particular cysteine 111 (Watanabe et al. 2007). More recently, Kishigami and colleagues suggested that FALS mutations in SOD1 promote monomerization of the enzyme, exposing an adventitial binding site in the dimer interface that does not appear to include cysteine 111 (Kishigami et al. 2010). Thus, the potential for Cu mediated free radical chemistry in the toxicity of mutant SOD1 cannot be completely excluded as the metal may bind adventitially at other sites.
In order to determine the role of Cu and Zn binding by mutant SOD1 in inducing motor neuron disease, we created mice expressing a SOD1 protein (termed SODMD) in which we mutated all of the histidine residues that are normally involved in the binding of Cu and Zn (H46R, H48Q, H63G, H71R, H80R, H120G); additionally we mutated histidine H43 (H43R, an FALS mutation) and the two free cysteine residues located at positions 6 and 111 (C6G, C111S). The mutations at positions 6, 43, 46, 48, and 80 are substitutions associated with ALS. The mutations at 63, 71, 111, and 120 are experimental mutations. This construct, termed SODMD, effectively abolished the binding of copper and zinc in their normal binding sites, and should abolish secondary binding that could occur at a histidines residue near the normal Cu binding site and the two free cysteines. We here describe mice that express SODMD at levels that are equivalent to existing lines of mice that develop ALS-like paralysis; however, finding no evidence of motor neuron disease or associated pathology. Additionally, we demonstrate that SODMD protein possesses properties more similar to WT SOD1, exhibiting a low tendency to aggregate. In cell culture models, we determine that the combined mutation of cysteines 6 and 111 to G and S, respectively, dramatically reduces the aggregation propensity of the SODMD protein and may account for the lack of toxicity for this mutant.
Materials and Methods
Creation of genomic SODMD construct
Mutations in the genomic sequence of human SOD1 were introduced using PCR strategies similar to those previously described (Wang et al. 2003). The starting construct for SODMD was a previously described version of genomic human SOD1 DNA that encodes the following mutations (H46R/H48Q/H63G/H120G) (Wang et al. 2003). The final collection of mutations is indicated in Supplementary Material, Fig. S1. Following the introduction of mutations, all coding exons were sequenced along with adjacent intronic regions. The resulting 12 kb genomic mutant DNA fragment was then injected into mouse embryos, as previously described (Wang et al. 2002). Mice were genotyped as described in Supplementary Materials. All procedures involving animals were approved by the University of Florida Institutional Animal Care and Use Committee.
Immunohistochemistry of spinal cord sections
A detailed description of the methods used in immunostaining is provided in Supplemental Materials.
Creation of SODMD and SODMD cDNA variants
The SODMD cDNA was obtained from the genomic version of this mutant protein by RT-PCR of mRNA isolated from cells transfected with the genomic construct used to make transgenic mice. The RT-PCR product was then reamplified with primers that introduced XhoI sites at the 3’ and 5’ ends of the cDNA, which allowed for subcloning into a previously described version of pEF-BOS vector (Wang et al. 2003).To create variants of SODMD that restored cysteine residues at positions 6 and 111 or altered position 111 to encode tyrosine, the SODMD cDNA was used as a template and new mutations were introduced by PCR techniques, in which the primers contain the desire mutation. Additionally, we added to this new versions new restriction sites for cloning into our most updated version of the pEF-BOS vector: NcoI at 5’ end and SalI at 3’ end (Prudencio et al. 2009b). All cDNA variants of SODMD and derivatives were sequenced to confirm mutations and sequence integrity.
Cell Transfections and analysis of SOD1 solubility
Human embryonic kidney HEK293FT cells, which express the SV40 large T-antigen and permit episomal replication of the pEF-BOS plasmid as well as strong enhancement of transcription, were purchased from Invitrogen (Carlsbad, CA, USA). Cells, cultured in 60 mm lysine-coated dishes (BD Biosciences Bedford, MA, USA), were transiently transfected, at 90-95% confluency, with either one SOD1 construct (4 μg) or equimolar amounts of two SOD1 constructs (~2 μg each). Transfections were performed using Lipofectamine 2000, following the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). Cells were harvested 24 or 48 hours after transfection and extracted in nonionic detergent by a procedure described in detail by Karch and Borchelt (Karch and Borchelt 2008). The procedure generated two fractions termed S1 (detergent-soluble cellular protein) and P2 (detergent-insoluble cellular protein) which were then analyzed by immunoblotting.
Protein concentrations for the detergent-soluble proteins (S1 fraction) and detergent-insoluble proteins (P2 fraction) were obtained using the bicinchoninic acid assay (Pierce Biotechnology, Rockford, IL, USA). Five micrograms of S1 fractions and twenty micrograms of P2 fractions were analyzed in 18% Tris-Glycine polyacrylamide gels (Invitrogen, Carlsbad, CA, USA) and transferred onto nitrocellulose membranes (Optitran BA-S 85, Whatman Inc., New Jersey, USA). Membranes were blocked in 5% nonfat milk in PBS before incubating them with m/hSOD1 antibody or the hSOD1 antibody at a 1:2500 dilutions for one hour at room temperature or overnight at 4°C. Followed by 3 washes in PBS-T (1x PBS, 0.1% Tween 20) for 10 minutes each, primary antibodies were detected by incubations with goat anti-rabbit IgG, 1:5000 (KPL, Gaithersburgh, MD, USA) for an hour at room temperature. After another 3 washes in PBS-T, secondary chemiluminescence was visualized with a Fujifilm imaging system (FUJIFILM Life Science, Stamford, CT, USA).
Estimation of aggregation propensity
The relative aggregation propensity of the SOD1 mutants was assess by comparing the ratio of immunolabeled SOD1 protein in the P2 vs. S1 fractions as previously described (Prudencio et al. 2009a). The intensities of the SOD1 immunoreactive bands in the S1 and P2 fractions establish a ratio value for a particular mutant in a particular immunoblot. To normalize the data from different experiments, each immunoblot that was quantified included a positive control (A4V SOD1) which was used to normalize the data by setting the ratio of S1 to P2 for A4V to 1. Differences in aggregation propensity were assessed by paired Student t-test (GraphPad Prism 4.0, San Diego, CA, USA), and each experiment was repeated at least three times.
Analyses of SOD1 solubility in transgenic mouse tissues
Spinal cords from transgenic animals were extracted in buffers containing non-ionic detergents as previously described above [for a detailed description see (Karch and Borchelt 2008)]. S1 and P2 fractions were analyzed by immunoblotting with hSOD1 antibodies.
SOD1 antibodies
Two antibodies specific to SOD1 were used. The hSOD1 antibody was raised against the human SOD1 sequence between amino acids 24-36 (CYASGEPVVLSGQIT) and is specific for human SOD1 (Bruijn et al. 1997). The m/hSOD1 antibody was raised against peptide sequences 124-136 (CYDDLGKGGNEESTK) of human SOD1, which is conversed between mouse and human SOD1 and thus recognizes both species (Pardo et al. 1995).
Statistical analyses
All statistical analyses were performed in GraphPad Prism Software 5.0 as explained in figure legends.
Results
To study the role of metal binding in aggregation and disease, we created transgenic mice expressing SODMD from the 12 kb human genomic SOD1 construct. In order to produce this construct, mutations were introduced in all 5 coding exons (Supplementary Material, Fig. S1). Mutations that target each of the histidine residues known to be involved in metal binding (H46, H48, H63, H71, H80, H120) were introduced by building from a previously described genomic construct in which histidines at positions 46, 48, 63, and 120 were mutated as follows H46R/H48Q/H63G/H120G (termed the SOD-Quad) (Wang et al. 2003). The additional mutations added were H71R and H80R (charge conserving experimental mutation at H71 and FALS mutation at H80). Additionally H43, which is located adjacent to the normal Cu binding site, was mutated to H43R, a FALS mutation. Finally, we mutated the two free cysteine residues at positions 6 and 111. Cysteine 6 was mutated to Gly (G) to encode an FALS mutation. Cysteine 111 was mutated to Ser (S), which is the amino acid at this position in mouse SOD1 and most other mammals (Wang et al. 2006). Collectively these mutations eliminate all known primary and secondary Cu and Zn binding sites.
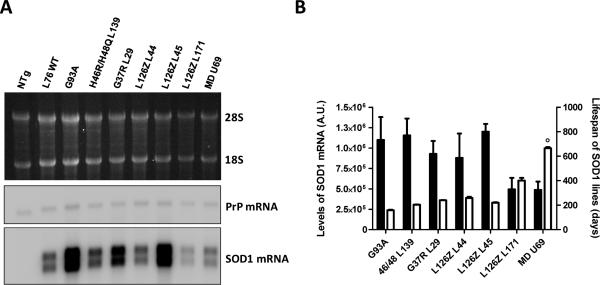
A total of seven founder lines were initially produced, from which we selected the highest expressing line (U-69) to expand (Supplementary Material, Fig. S2). To compare the expression of mutant SOD1 in the U-69 SODMD mice to previously established lines of mutant SOD1 mice, we determined the levels of transgene mRNA by Northern blotting (Fig. 1). For each line of mice we harvested 3 animals of similar ages. Mice expressing the mutations G93A [Line Gur 1 (Gurney et al. 1994)] or double histidine (H46R/H48Q) mutants [Line 139 (Wang et al. 2002)] showed the highest mRNA levels, followed by WT L76, mice expressing the G37R mutant [Line 29 (Wong et al. 1995)] and two different lines of L126Z SOD1 mice [Lines 44 and L45 (Wang et al. 2005a)]. A line of L126Z mice [L171 (Wang et al. 2005a)] expresses mutant SOD1 at levels that were equivalent to SODMD Line U69 mice (Figs. 1A and B).
Figure 1. Comparison of transgene mRNA levels in spinal cords of mutant SOD1 transgenic mouse lines.
(A) Northern blot showing the mRNA levels of different lines of mice expressing WT or mutant human SOD1 proteins. Top, ethidium bromide stained gel to visualize the 28S and 18S RNAs. Middle, Northern blot of the same RNA samples probed with cDNA for mouse PrP mRNA, which served as a loading control. Bottom, Northern blot probed with human SOD1 cDNA. (B) Quantifications of mRNA levels of three different Northern blots are represented by the black bars. White bars indicate the mean survival times of SOD1 lines calculated from data collected in our lab from animals harvested over more than 4 years (see Supplemental Material, Fig. S3). The ‘o’ symbol over the survival time of SODMD line indicates that the data bar represents mean of lifespan to sacrifice, however no disease symptoms were noted in these mice.
Previous studies have observed that the levels of transgene expression relate to disease development in mutant SOD1 mice (Wong et al. 1995;Wang et al. 2002;Wang et al. 2005b). To relate expression levels to onset we compiled data on the age at which different lines of mice reach endstage –defined by paralysis of the hindlimbs. When we compared such data with transgene expression levels, we observed that mice reaching end-stage at earlier ages, present higher levels of transgene expression (Fig. 1B). For example, G93A mice express the highest levels and develop paralysis at the earliest times, around 5 months old [159.1 ± 4.03 days; data from our colony (Supplementary Material, Fig. S3)], while the lowest expressing mice (L126Z L171) did not develop paralysis until they were 13 months old (400.3 ± 22.56 days; Fig. 1B; Supplementary Material, Fig. S3). Additionally, correlation analysis demonstrated that the age to paralysis in transgenic mice is dependent on the level of mutant SOD1 mRNA (Supplementary Material, Fig. S4). Thus, although the levels of transgene mRNA in the U-69 SODMD mice was on the lower end of the spectrum, we concluded that SODMD mice from line U-69 expressed the mutant within the range to induce an ALS phenotype if the mutation is pathogenic.
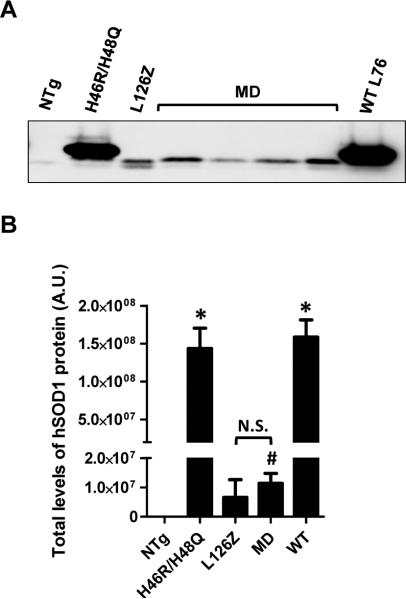
In our experience the levels of transgene mRNA in mouse spinal cords do not necessarily correlate with the steady-state levels of mutant SOD1 protein present in spinal cords because some mutants are relatively unstable. For example, the L126Z mutant accumulates to lower levels than predicted by mRNA levels (Wang et al. 2005a). Thus, to determine protein levels in the U-69 mice relative to other mutants, we evaluated total SOD1 protein levels by immunoblotting of spinal cord extracts from young animals. Compared to the L126Z Line 171 mice, the levels of SODMD protein were similar (Fig. 2). Note, however, that the levels are much lower than that of WT SOD1 mice (Line 76) or mice that express the H46R/H48Q mutant (Line 139). These results, together with our analysis in mRNA SOD1 levels, suggest that disease development in SODMD would be predicted to occur around the age of onset of L126Z L171 mice. However, unexpectedly, the SODMD mice remained free of ALS-like symptoms, such as obvious hind limb weakness, gait disturbances, or paralysis out to 2 years of age. As the mice approached 2 years of age, we began to observe death due to other morbidities, but no symptoms of weakness or paralysis were noted. To alleviate un-necessary stress to the aged animals, no animals were aged longer than 24 months and no SODMD mice developed ALS-like symptoms by this age.
Figure 2. Protein levels in the spinal cord of SODMD mice resemble those of L126Z SOD1 mice.
(A) Western blot of total protein levels of indicated mouse lines using an antibody that specifically recognizes human SOD1 protein (hSOD1 antibody). Note that the L126Z truncation mutant and SODMD proteins run slightly faster than other SOD1 proteins in SDS-PAGE gels. (B) Quantification of the total human SOD1 protein levels. No significant differences in protein levels in spinal cord were found between SODMD and L126Z mice. NTg denotes non-transgenic animals, N.S. non-significant differences, statistical differences between NTg and the different lines of mice are also indicated: *p ≤ 0.05, #p ≤ 0.005. Protein levels were quantified from 3 independent immunoblots.
As noted above, the levels of transgene expression are critical in transgenic animal models. For example, mice expressing the G37R mutation under the mouse prion promoter do not develop motor neuron disease unless levels of expression are raised through breeding to homozygosity (Wang et al. 2005b). Similar examples occur for a line of mice that express the D90A mutant of SOD1 (Jonsson et al. 2006) and the G93A mutant via a vector that utilizes a Thy1 promoter (Jaarsma et al. 2008). Thus, we self-crossed SODMD mice with the intention to increase SOD1 protein levels through homozygosity, which should translate into a more rapid disease development. A total of 20 transgene positive mice were produced from homozygous matings with the expectation that approximately 33% of the transgene positive animals (25% overall) should be homozygous. The number of transgene positive animals from these matings was not suggestive of any loss of viability of homozygous animals. However, we still failed to observe disease symptoms in any of these mice out to 24 months of age.
Several previous studies have established that co-expressing high levels of WT SOD1 with mutant SOD1 can accelerate disease onset and, moreover, induce disease in animals that express mutant protein at levels that are below threshold to develop ALS-like symptoms (Deng et al. 2006;Jaarsma et al. 2008;Deng et al. 2008;Prudencio et al. 2010). In an attempt to induce disease development in SODMD mice, we mated these mice to the Gur WT strain of mice that express WT SOD1 at very high levels. Out of 59 animals that resulted from such mating, 11 harbored both WT and SODMD transgenes. Animals were aged out to 24 months with none developing paralysis. Mice from the Gur WT strain develop a gait disturbance late in life and we observed this phenotype, but none progressed to overt motor neuron disease and paralysis. For comparison, we also mated our low expressing L126Z (line 171) mice to Gur WT mice, generating 4 mice that harbored both transgenes. Compared to mice expressing only the L126Z Line 171 transgenes, mice harboring with the WT and L126Z transgenes developed motor neuron disease 20-25 weeks earlier (Supplementary Material, Fig. S5). Thus, the low level of expression of the SODMD transgene cannot explain the lack of effect by the co-expression of WT SOD1. Therefore, we conclude that, although the MD and L126Z variants exhibit similar stability profiles (ratio of steady-state protein to transgene mRNA is similar), the SODMD mutant is much less pathogenic than the L126Z mutant.
Histologic analysis of the SODMD mice
To determine whether the SODMD mice may have early pathologic signs of ALS that are known to occur at time points previous to obvious symptoms, we conducted a histologic analyses of tissues from old SODMD animals. Axonal degeneration is an early disease feature observed in G93A SOD1 transgenic mice (Fischer et al. 2004). A consequence of this phenomenon is axon demyelination. Thus, we performed myelin staining in sciatic nerve sections from non-transgenic, symptomatic H46R/H48Q, and old SODMD mice (Supplementary Material, Fig. S6). In transverse sciatic nerve sections, only symptomatic H46R/H48Q mice appeared to present a lower number of myelinated axons as well as smaller diameter of myelin sheaths, while nerve sections from the SODMD mice did not differ from those of non-transgenic mice (Supplementary Material, Fig. S6, A to I). Similarly, longitudinal sciatic nerve sections showed reduced numbers of myelinated fibers in paralyzed H46R/H48Q SOD1 mice, while old SODMD mice appeared largely normal (Supplementary Material, Fig. S6, J to R). Thus, even at relatively old ages, SODMD mice did not present any obvious abnormality in myelinated fibers of sciatic nerve.
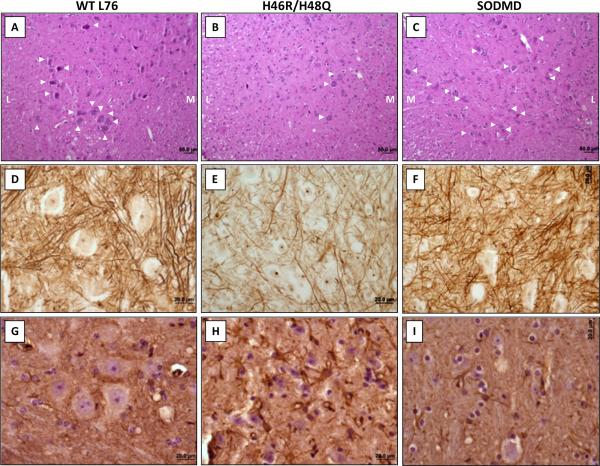
Within the spinal cord, hematoxylin and eosin staining indicated reduced presence of motor neurons in the ventral horn of symptomatic H46R/H48Q SOD1 mice (Figs. 3A and 3B). However, the number of motor neurons in WT and SODMD mice did not obviously differ (Figs. 3A and 3C). Silver staining demonstrated that, symptomatic mice expressing mutant SOD1 proteins present reduced number of fibers compared to WT or SODMD mice (Figs. 3D to 3F). Gliosis is another of the earliest characterized abnormalities in SOD1 transgenic mice (Wong et al. 1995). However, GFAP immunoreactivity was only markedly increased in symptomatic H46R/H48Q SOD1 mice with no obvious difference observed between WT and SODMD mice (Figs. 3G to 3I). Thus, our histological analysis demonstrated that mice expressing the SODMD variant are free of any of the preclinical pathologic changes characteristic mutant SOD1 mice that develop paralysis.
Figure 3. SODMD mice do not develop gliosis or show evidence of motor neuron abnormalities.
Mice expressing WT (A, D, G), H46R/H48Q (B, E, H) and MD (C, F, I) SOD1 proteins were characterized histologically. (A-C) Hematoxylin and eosin stain on paraffin embedded sections a low power (10x) demonstrates significantly lower numbers of motor neurons (indicated with white arrowheads) in the ventral horn of symptomatic H46R/H48Q (B) mice than in WT (A) and MD (C) mice. M: medial portion of the spinal cord, L: lateral portion of the spinal cord). (D-F) Silver staining of paraffin embedded sections indicates reductions in fiber density in the spinal cord of symptomatic H46R/H48Q (E) mice, while the cords of WT (D) and MD (F) mice appear largely normal. Note that all staining was done simultaneously for all sections. (G-I) GFAP immunostaining demonstrates low GFAP immunoreactivity in WT (G) and MD (I) spinal cords, while strong gliosis is apparent in symptomatic H46R/H48Q (H) SOD1 mice. Bars represent 50 μm in A-C and 20 μm in D-I, and pictures were taken in the ventral portion of the spinal cord. The images shown are representative of an analysis of at least 3 animals per genotype. SODMD and NTg SOD1 mice were harvested at 2 years of age, while H46R/H48Q mice were harvested at symptomatic stages (~10 months).
Lastly, we looked for biochemical signatures of disease onset in the SODMD mice by using detergent extraction, sedimentation, and immunoblotting to look for detergent-insoluble aggregates of SODMD protein in spinal cord of aged mice (Supplementary Material, Fig. S7). Similar to the data from immunoblot analysis of total spinal cord protein of young SODMD mice, the levels of SODMD protein in the detergent-soluble (S1) fraction of older SODMD were low. Moreover, we did not observe accumulation of SODMD protein in the detergent-insoluble (P2) fraction. For comparison, detergent-insoluble L126Z mutant protein was readily detected in spinal cords of paralyzed L126Z mice (Supplementary Material, Fig. S7) and mice expressing H46R/H48Q mutant (Supplementary Material, Fig. S7). Although the level of detergent insoluble L126Z protein in the spinal cords of paralyzed L126Z mice was much lower than that of H46R/H48Q mice, the level of detergent-insoluble mutant protein in spinal cords of old SODMD mice was below the level of detection.
Cell culture models of SOD1 aggregation
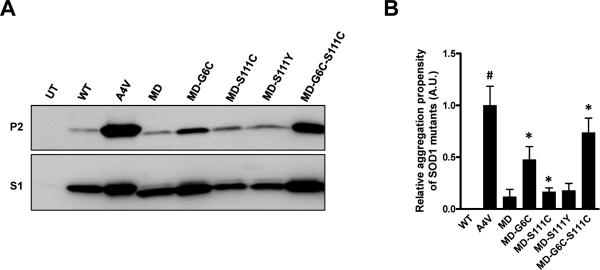
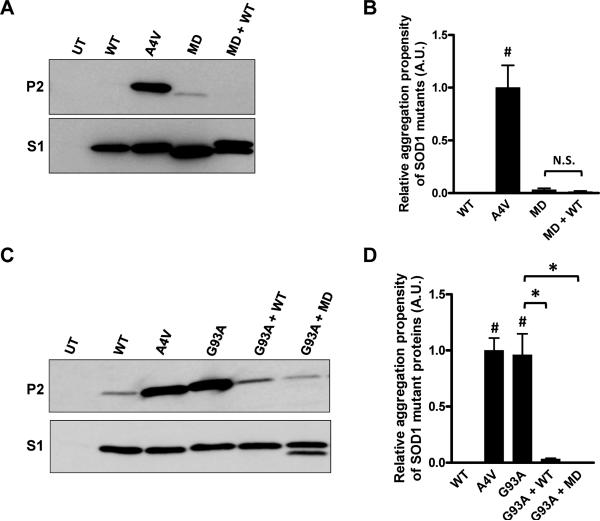
One question that might rise from these studies is whether the 9 single-point mutations in the SODMD protein render the protein a completely disordered molecule. To address this issue we created expression plasmids for SODMD cDNA genes for transfection of cultured cells, using vectors that produce high levels of expression. In cell culture, SODMD is more highly expressed such that it is clearly detected in the detergent-soluble S1 fractions (Fig. 4A, lower panel); however, as in our mouse model, we do not observe significant accumulation of detergent-insoluble forms of SODMD protein (Figs. 4A and B); no difference from WT at 48 hours following transfection (Fig. 4B). Based on previous study (Karch and Borchelt 2008), we suspected that the mutations in the cysteine residues of SODMD (C6G, C111S) might be responsible for lowering aggregation propensity. To define the role of mutations in cysteine 6 and 111 in SODMD in reducing aggregation propensity, we systematically changed each residue in a series of constructs. Changing position 6 from glycine back to cysteine increased aggregation propensity, but the level remained lower than that of the A4V mutant, which served as a positive control (Figs. 4A and B). Changing position111 from serine back to cysteine had minimal impact on aggregation propensity (Figs. 4A and B). Similarly, changing position 111 from serine to an FALS mutation (C111Y [http://alsod.iop.kcl.ac.uk/Index.aspx], had minimal impact (Figs. 4A and B). Changing positions 6 and 111, together, back to cysteine produced a protein with an aggregation propensity similar to A4V SOD1 (Figs. 4A and B). We also tested, in cultured cells, whether co-expression of WT SOD1 with SODMD would induce aggregation of either protein. As predicted by the mouse mating studies above, SODMD was not induced to aggregate when co-expressed with WT SOD1 (Figs. 5A and B). Collectively, these data demonstrate that the mutations introduced at positions 6 and 111 of the SODMD construct reduced its propensity to aggregate.
Figure 4. Restoration of cysteine at amino acids 6 and 11 in the context of SODMD mutations increases the aggregation propensity.
(A) Immunoblots of detergent-extracted HEK293FT cells expressing the indicated SOD1 proteins for 48 hours. (B) Quantification of the aggregation propensity. All values are normalized to the aggregation propensity of A4V (set to 1). Student t-tests were performed to compare aggregation propensities of SOD1 proteins to WT SOD1: *p ≤ 0.05, #p ≤ 0.005. The graphed data represent quantification of at least 3 measurements of distinct immunoblots similar to the image in panel A.
Figure 5. Co-expression of the SODMD variant with G93A mutant SOD1 suppresses aggregation.
Co-expression of SODMD with WT SOD1 does not induce a rapid increase of aggregation of either protein (A, B). Additionally, SODMD can reduce aggregation of G93A when co-expressed; a property shared with WTSOD1 (C, D). (A, C) Immunoblots of detergent-insoluble (P2) and soluble (S1) fractions of indicated SOD1 proteins expressed in HEK293T cells for 48 (A) or 24 (C) hours. UT denotes untransfected cells. (B, D) Quantification of the aggregation propensity indicates that, in this set of transfections, only A4V and G93A SOD1 proteins are able to significantly aggregate. Student t-tests were performed to evaluate significant differences: *p ≤ 0.05, #p ≤ 0.005; N.S.: non-significant differences. The graphed data represent quantification of at least 3 measurements of distinct immunoblots similar to the images shown in panels A and C.
In previous studies of mutant SOD1 aggregation in cell culture models, we have shown that co-expression of WT SOD1 with mutant protein slows the aggregation rates of the mutant protein (Prudencio et al. 2009a). When two mutant SOD1 proteins are co-expressed, both readily aggregate (Prudencio et al. 2009a). Therefore, we asked whether SODMD behaved like WT or mutant protein in co-transfection with the previously characterized G93A mutant (Fig. 5). Cells were transfected with vectors for WT and A4V mutants to provide negative and positive controls, along with vectors for G93A, G93A+WT, G93A+MD, (Figs. 5C and D). Similar to WT SOD1, co-expression of SODMD with G93A resulted in less aggregation of the G93A mutant. Thus, SODMD mutant proteins conserve features of the WT SOD1 protein, suggesting that the amino acid substitutions in SODMD have not resulted in a completely disordered protein.
Discussion
We present here an experimental SOD1 mutation that eliminates both primary and secondary Cu and Zn sites in SOD1. Despite the presence of 5 amino acid substitutions associated with FALS, mice expressing this mutant did not develop motor neuron disease. Moreover, we found no evidence of insidious disease in aged SODMD mice such as reductions in axon numbers in sciatic nerve, spinal cord gliosis, or accumulation of detergent-insoluble mutant protein in spinal cord. Attempts to induce disease by mating SODMD mice to homozygosity or by mating the SODMD mice to mice that over-express WT SOD1 also failed to induce disease. In cell culture models, we show that SODMD protein possesses a low propensity to aggregate and through additional mutagenesis we attribute the low aggregation of SODMD to changes of cysteine residues at position 6 and 111 to glycine and serine respectively. Thus, the SODMD mice represent a model in which we have expressed an experimental SOD1 mutant protein that encodes intragenic mutations that suppress aggregation and simultaneously render the protein non-toxic.
SODMD mice, expression of mutant SOD1, and motor neuron disease
It is well established that the appearance of disease in mice that express mutant SOD1 is highly dependent upon levels of transgene expression. There are examples in which two-fold changes in transgene mRNA expression have a major impact on age to paralysis; strains of mice have been described in which disease is not present when mice are heterozygous for a transgene, but develop disease when homozygous (Wang et al. 2005b;Jaarsma et al. 2008). As compared to lines of mice that develop disease when heterozygous for mutant SOD1 transgenes, the expression levels of the SODMD variant were in the lower end of expression. However, the levels of transgene mRNA in the SODMD line U-69 mice were not statistically different from those of L126Z Line 171 mice, which develop disease at ~1 year of age. Additionally the levels of mutant protein in SODMD and L126Z Line 171 were quite similar. Thus, we can confidently conclude that the SODMD variant is significantly less toxic than the L126Z mutant, which shares the characteristic of being relatively unstable.
To increase levels of expression, we intercrossed transgenic SODMD mice to obtain homozygous SODMD mice. Although we did not specifically identify the homozygous animals from this cross, we estimate from the frequency of transgene positive animals in these litters that we should have had ~5 homozygous SODMD mice out of the transgene positive offspring that were produced. However, none of the mice in this cohort developed disease symptoms. Additionally, we examined all 20 of these mice pathologically to look for early pathologic signs of disease (Kennel et al. 1996;Borchelt et al. 1998;Watanabe et al. 2001;Fischer et al. 2004;Hegedus et al. 2007), such as gliosis (Wong et al. 1995). However, we found no evidence of gliosis, no obvious change in motor neuron numbers or appearance, and no obvious change in axonal myelination or axonal numbers in sciatic nerve. Thus, we can find no evidence that SODMD protein is neurotoxic.
An additional means to raise total SOD1 levels and induce disease in mutant SOD1 mice is to produce bigenic mice that co-express high levels of WT SOD1 with mutant protein (Deng et al. 2006). Thus we attempted to use this tactic to elaborate toxicity of SODMD protein, using the Gur WT strain that has a more robust impact on mutant SOD1 toxicity (Prudencio et al. 2010). However, again, we failed to detect motor neuron disease in SODMD mice that co-expressed WT SOD1.
Although a negative study can be subject to mis-interpretation because an absence of phenotype is decidedly less desirable than induction of disease, we have done all that we can to elaborate toxicity by the SODMD variant and yet fail to observe even the earliest pathologic features of motor neuron disease. With the disclaimer that it remains possible that a much higher level of SODMD expression might induce disease, we can say with confidence that the SODMD mutant is significantly less toxic that the L126Z mutant. We can also say that the SODMD mutant does not interact with WT SOD1 in a manner to increase toxicity and induce disease.
Intragenic suppression of SODMD toxicity
To eliminate all histidines involved in the binding of Cu or Zn required mutations at 7 positions. In making the SODMD mutant, the starting construct was a genomic DNA construct that we generated to eliminate the Cu binding site of SOD1 (H46R/H48Q/H63G/H120G) that we have previously used to produce mice that develop motor neuron disease (Wang et al. 2003). In this mutant H46 and H48 carry FALS mutations, whereas the mutations at 63 and 120 are experimental. In the SODMD mutant we add additional FALS mutations at H43 (H43R) and H80 (H80R) with an experimental mutation of H71 (H71R). Collectively, these mutations obliterate the Cu and Zn binding sites; the H43R mutation removes a histidine that is adjacent to the Cu-binding histidines at positions 46 and 48. In cell culture, we show that this mutant (which is designated MD-G6C-S111C in Fig. 4) shows a relatively high aggregation propensity. Thus, mutant SOD1 lacking all the normal Cu and Zn binding sites retains the ability to aggregate.
To remove two potentially ancillary sites of metal binding, we included in the SODMD mutant changes of cysteines 6 and 111 to glycine and serine. Because these studies were initiated several years ago, the SODMD mutant was constructed and injected into mice before studies from our laboratory and others demonstrated that mutation of cysteine 111 to serine has a marked effect on mutant SOD1 aggregation (Banci et al. 2007;Niwa et al. 2007;Karch and Borchelt 2008;Cozzolino et al. 2008). In cell culture, we demonstrate that mutating positions 6 and 111 of SODMD cDNA to revert back to cysteine at these positions converts the SODMD variant from low to high aggregation propensity. Thus, we suspect that the lack of toxicity by SODMD could be due, at least partially, to intragenic suppression of aggregation by mutations of the two cysteine residues. We also note that the lack of toxicity could also be due to some other structural changes in the protein induced by the loss of cysteine 6 and 111. Studies by Kishigami and colleagues suggest that modification of cysteine 111 produces structural changes in mutant SOD1 that promote monomerization and the binding of a reactive Cu to the newly exposed dimer interface (Kishigami et al. 2010). Such binding may not be possible in the SODMD variant.
These findings clearly indicate the need to further probe the role of t cysteines 6 and 111 in human SOD1 in toxicity. Overall, these studies provide a new starting point from which to decipher mechanisms of mutant SOD1 toxicity by defining a mutant that possesses low toxicity with low aggregation propensity.
Metal binding, aggregation and disease
The original intent of our study was to test the toxicity of a variant of mutant SOD1 that lacked both primary and secondary Cu and Zn binding sites. We verified that SODMD does not bind Cu or Zn by analyzing the metal content of protein extracted from transfected cell cultures (Supplementary Material, Fig. S8). When over expressed in cell culture, WT SOD1 shows low incorporation of Cu. However, SODMD was found to lack both Cu and Zn as one would expect. Thus, we are confident that the SODMD variant binds Cu and Zn poorly.
In previous work, we have established that eliminating the copper ligands at positions 46, 48, 63, and 120 creates a mutant that retains a high propensity to aggregate when expressed at high levels in culture cells and produces disease with mutant SOD1 aggregates in transgenic mice. The binding of zinc is known to confer structural stability, thus abolishing zinc binding could in theory increase the potential of the protein to misfold and aggregate. In cultured cells, the SODMD-C6C111, in which positions 6 and 111 were reverted to cysteine, showed high aggregation propensity. Thus, metal-deficient forms of mutant SOD1 can produce aggregates. However, since the SODMD mutant neither binds metal nor aggregates, we cannot be certain whether the lack of metal binding, changes in structural features of the protein, or the lack of aggregation reduced toxicity. It seems that it will be of interest to determine whether experimental derivatives of the SODMD construct that retain high aggregation propensity while also retaining poor binding of Cu and Zn can induce motor neuron disease.
Supplementary Material
Acknowledgements
M.P. and D.R.B. designed research, M.P., H.H.B. and H.L. performed research, M.P. and D.R.B analyzed data and wrote the manuscript. All authors read and approved the final manuscript. Authors do not have conflicts of interest.
Funding
This work was funded by a grant from the National Institutes of Neurologic Disease and Stroke (P01 NS049134 – Program Project award to Drs. Joan S. Valentine, P. John Hart, D.R. Borchelt, and J.P. Whitelegge).
Abbreviations
- ALS
amyotrophic lateral sclerosis
- FALS
familial amyotrophic lateral sclerosis
- SOD1
superoxide dismutase 1
- WT
wild-type
References
- Banci L, Bertini I, Durazo A, Girotto S, Gralla EB, Martinelli M, Valentine JS, Vieru M, Whitelegge JP. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Guarnieri M, Wong PC, Lee MK, Slunt HS, Xu Z-S, Sisodia SS, Price DL, Cleveland DW. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J. Biol. Chem. 1995;270:3234–3238. doi: 10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HH, Guarnieri M, Xu Z-S, Wong PC, Brown RH, Jr., Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Wong PC, Becher MW, Pardo CA, Lee MK, Xu ZS, Thinakaran G, Jenkins NA, Copeland NG, Sisodia SS, Cleveland DW, Price DL, Hoffman PN. Axonal transport of mutant superoxide dismutase 1 and focal axonal abnormalities in the proximal axons of transgenic mice. Neurobiol. Dis. 1998;5:27–35. doi: 10.1006/nbdi.1998.0178. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, Borchelt DR, Price DL, Cleveland DW. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Durazo A, Sohn SH, Strong CD, Gralla EB, Whitelegge JP, Valentine JS. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18663–18668. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M, Amori I, Pesaresi MG, Ferri A, Nencini M, Carri MT. Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:866–874. doi: 10.1074/jbc.M705657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Jiang H, Fu R, Zhai H, Shi Y, Liu E, Hirano M, Dal Canto MC, Siddique T. Molecular dissection of ALS-associated toxicity of SOD1 in transgenic mice using an exon-fusion approach. Hum. Mol. Genet. 2008;17:2310–2319. doi: 10.1093/hmg/ddn131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Shi Y, Furukawa Y, Zhai H, Fu R, Liu E, gorrie GH, Khan MS, Hung WY, Bigio EH, Lukas T, Dal Canto MC, O'Halloran TV, Siddique T. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam JS, Taylor AB, Strange R, Antonyuk S, Doucette PA, Rodriguez JA, Hasnain SS, Hayward LJ, Valentine JS, Yeates TO, Hart PJ. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, Khan J, Polak MA, Glass JD. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Forman HJ, Fridovich I. On the stability of bovine superoxide dismutase. The effects of metals. J. Biol. Chem. 1973;248:2645–2649. [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Yamanaka K, O'Halloran TV, Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng H-X, Chen W, Zhai P, Sufit RL, Siddique T. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hart PJ, Liu H, Pellegrini M, Nersissian AM, Gralla EB, Valentine JS, Eisenberg D. Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis. Protein Sci. 1998;7:545–555. doi: 10.1002/pro.5560070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus J, Putman CT, Gordon T. Time course of preferential motor unit loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2007;28:154–164. doi: 10.1016/j.nbd.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Jaarsma D, Teuling E, Haasdijk ED, De Zeeuw CI, Hoogenraad CC. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J. Neurosci. 2008;28:2075–2088. doi: 10.1523/JNEUROSCI.5258-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson PA, Graffmo KS, Brannstrom T, Nilsson P, Andersen PM, Marklund SL. Motor neuron disease in mice expressing the wild type-like D90A mutant superoxide dismutase-1. J. Neuropathol. Exp. Neurol. 2006;65:1126–1136. doi: 10.1097/01.jnen.0000248545.36046.3c. [DOI] [PubMed] [Google Scholar]
- Karch CM, Borchelt DR. A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennel PF, Finiels F, Revah F, Mallet J. Neuromuscular function impairment is not caused by motor neurone loss in FALS mice: an electromyographic study. Neuroreport. 1996;7:1427–1431. doi: 10.1097/00001756-199605310-00021. [DOI] [PubMed] [Google Scholar]
- Kishigami H, Nagano S, Bush AI, Sakoda S. Monomerized Cu, Zn-superoxide dismutase induces oxidative stress through aberrant Cu binding 1. Free Radic. Biol. Med. 2010;48:945–952. doi: 10.1016/j.freeradbiomed.2010.01.008. [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Niwa J, Yamada S, Ishigaki S, Sone J, Takahashi M, Katsuno M, Tanaka F, Doyu M, Sobue G. Disulfide bond mediates aggregation, toxicity, and ubiquitylation of familial amyotrophic lateral sclerosis-linked mutant SOD1. J. Biol. Chem. 2007;282:28087–28095. doi: 10.1074/jbc.M704465200. [DOI] [PubMed] [Google Scholar]
- Pardo CA, Xu Z, Borchelt DR, Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl. Acad. Sci. USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SZ, Valentine JS. The perplexing role of copper-zinc superoxide dismutase in amyotrophic lateral sclerosis (Lou Gehrig's disease). J. Biol. Inorg. Chem. 2003;8:373–380. doi: 10.1007/s00775-003-0447-6. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Durazo A, Whitelegge JP, Borchelt DR. Modulation of mutant superoxide dismutase 1 aggregation by co-expression of wild-type enzyme. J. Neurochem. 2009a;108:1009–1018. doi: 10.1111/j.1471-4159.2008.05839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M, Durazo A, Whitelegge JP, Borchelt DR. An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Hum. Mol. Genet. 2010;19:4774–4789. doi: 10.1093/hmg/ddq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M, Hart PJ, Borchelt DR, Andersen PM. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum. Mol. Genet. 2009b;18:3217–3226. doi: 10.1093/hmg/ddp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng H-X, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH., Jr. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- Tiwari A, Xu Z, Hayward LJ. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2005;280:29771–29779. doi: 10.1074/jbc.M504039200. [DOI] [PubMed] [Google Scholar]
- Valentine JS, Hart PJ. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc Natl Acad Sci U. S. A. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Caruano-Yzermans A, Rodriguez A, Scheurmann JP, Slunt HH, Cao X, Gitlin J, Hart PJ, Borchelt DR. Disease-associated mutations at copper ligand histidine residues of superoxide dismutase 1 diminish the binding of copper and compromise dimer stability. J. Biol. Chem. 2007;282:345–352. doi: 10.1074/jbc.M604503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Slunt H, Gonzales V, Fromholt D, Coonfield M, Copeland NG, Jenkins NA, Borchelt DR. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum Mol Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Borchelt DR. Mapping superoxide dismutase 1 domains of non-native interaction: roles of intra- and intermolecular disulfide bonding in aggregation. J. Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xu G, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR. Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol Dis. 2002;10:128–138. doi: 10.1006/nbdi.2002.0498. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Li H, Gonzales V, Fromholt D, Karch C, Copeland NG, Jenkins NA, Borchelt DR. Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: {alpha}B-crystallin modulates aggregation. Hum Mol Genet. 2005a;14:2335–2347. doi: 10.1093/hmg/ddi236. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Slunt HH, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR. Coincident thresholds of mutant protein for paralytic disease and protein aggregation caused by restrictively expressed superoxide dismutase cDNA. Neurobiol. Dis. 2005b;20:943–952. doi: 10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. Histological Evidence of Protein Aggregation in Mutant SOD1 Transgenic Mice and in Amyotrophic Lateral Sclerosis Neural Tissues. Neurobiol. Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Nagano S, Duce J, Kiaei M, Li QX, Tucker SM, Tiwari A, Brown RH, Jr., Beal MF, Hayward LJ, Culotta VC, Yoshihara S, Sakoda S, Bush AI. Increased affinity for copper mediated by cysteine 111 in forms of mutant superoxide dismutase 1 linked to amyotrophic lateral sclerosis. Free Radic. Biol. Med. 2007;42:1534–1542. doi: 10.1016/j.freeradbiomed.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, Sisodia SS, Cleveland DW, Price DL. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.