Abstract
Unlike random heteropolymers, natural proteins fold into unique ordered structures. Understanding how these are encoded in amino-acid sequences is complicated by energetically unfavourable non-ideal features—for example kinked α-helices, bulged β-strands, strained loops and buried polar groups—that arise in proteins from evolutionary selection for biological function or from neutral drift. Here we describe an approach to designing ideal protein structures stabilized by completely consistent local and non-local interactions. The approach is based on a set of rules relating secondary structure patterns to protein tertiary motifs, which make possible the design of funnel-shaped protein folding energy landscapes leading into the target folded state. Guided by these rules, we designed sequences predicted to fold into ideal protein structures consisting of α-helices, β-strands and minimal loops. Designs for five different topologies were found to be monomeric and very stable and to adopt structures in solution nearly identical to the computational models. These results illuminate how the folding funnels of natural proteins arise and provide the foundation for engineering a new generation of functional proteins free from natural evolution.
For proteins to fold, the interactions favouring the native state must collectively outweigh the non-native interactions, resulting in funnel-shaped energy landscapes1–3. However, it is not obvious how the ubiquitous non-covalent interactions that stabilize proteins—van der Waals interactions, hydrogen bonding and hydrophobic packing—can selectively favour the biologically relevant unique native structure over the vastly larger number of non-native conformations. Protein design provides an opportunity to investigate this problem: hypotheses about how unique folded structures are encoded in amino-acid sequences can be evaluated by designing proteins de novo and experimentally assessing how well they fold4–6.
Previous work on protein fold design has focused on stabilizing the desired folded state7–13. However, robustly designing protein structures with funnel-shaped energy landscapes may require not only the stabilization of a unique folded state7–13 (positive design), but also the destabilization of non-native states14–16 (negative design). Protein design methodology has been developed to find sequences that stabilize a desired folded state and destabilize specific non-native states14–20. However, the challenge of disfavouring the vast number of non-native states quite generally remains an unsolved problem.
We hypothesized that funnel-shaped energy landscapes can be robustly generated by requiring that the local interactions between residues close along the linear sequence, which determine protein secondary structure, and the non-local interactions between residues distant along the chain, which stabilize protein tertiary structure, consistently favour the same folded conformation21. We sought principles for designing ‘ideal’ proteins that have this property. To disfavour non-native states systematically by negative design, we focused on the local interactions because non-local interactions vary strongly with even small changes in tertiary structure. We began by investigating the mapping between local interactions favouring specific secondary structure patterns and protein tertiary structure motifs, seeking local structure patterns that strongly favour single tertiary motifs over all others.
We focused on a basis set of tertiary structure motifs consisting of two or three secondary structure elements adjacent in the linear sequence, which make extensive intramotif interactions. We investigated the mapping from secondary structure patterns to these tertiary structure features using a combination of de novo folding calculations with the Rosetta program22 and analyses of naturally occurring protein structures in the Protein Data Bank. Multiple protein folding simulations were carried out for each motif for a range of different lengths of the strands, helices and loops, using a sequence-independent backbone model. For each choice of lengths, we computed the fraction of trajectories that arrived at the desired motif topology. These calculations revealed that the extent of folding to a particular motif is very strongly dependent on the lengths of the secondary structures. Detailed study of these dependencies identified three fundamental rules, which are described in the following section.
Rules relating local structures to tertiary motifs
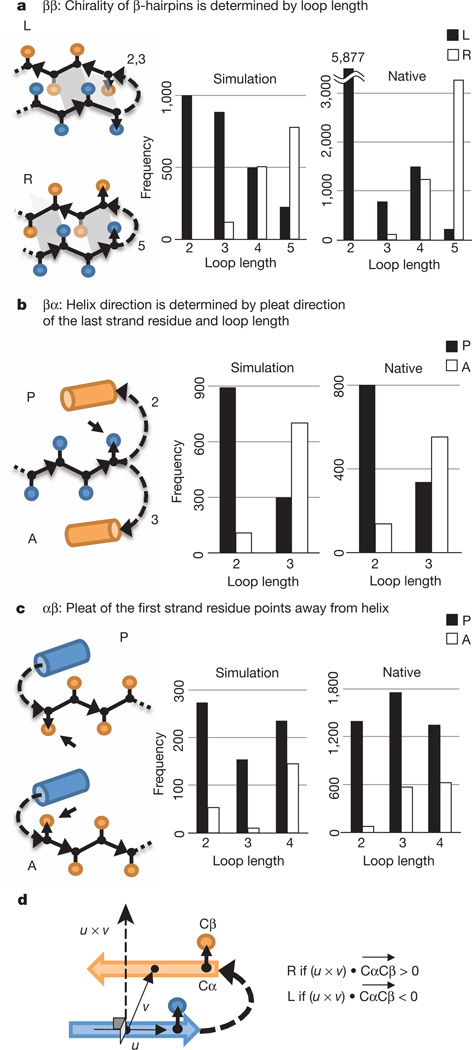
The fundamental rules describe the junctions between adjacent secondary structure elements (Fig. 1). There are three distinct junction classes in the αβ-folds we sought to design—ββ, βα and αβ—and three corresponding rules.
Figure 1. Fundamental rules.
a, ββ-rule. L (left-handed) and R (right-handed) ββ-units are illustrated (see Fig. 1d for chirality definition). The dependence of chirality on loop length is shown in the histograms. b, βα-rule. P (parallel) and A (antiparallel) βα-units are illustrated. The dependence of orientation (P versus A) on loop length is shown in the histograms. c, αβ-rule. d, Chirality (L versus R) of a ββ-unit. The chirality is defined on the basis of the orientation of the Cα-to-Cβ vector, , of the strand residue preceding or following the connecting loop. u is a vector along the first strand and v is a vector from the centre of the first strand to the centre of the second strand.
Statement of the rules requires the definition of the chirality (L versus R) of a ββ-unit and the orientation (P versus A) of βα- and αβ-units (Fig. 1). The chirality of a ββ-unit is defined on the basis of the orientation of the Cα-to-Cβ vector, , of the strand residue preceding or following the connecting loop: letting u be a vector along the first secondary structure element and v be a vector from the centre of the first secondary structure element to the centre of the second secondary structure element, if (u × v) • (where a cross denotes vector product and a dot denotes scalar product) is positive the unit is right handed (R), and if it is negative the unit is left handed (L) (Fig. 1d). For βα- and αβ-units in which the β-strand is in a β-sheet that the helix packs against, the vectors in the strand are roughly collinear with the vector between the centres of the strand and the helix. We define the orientation of a βα-unit to be parallel (P) if the vector from strand to helix is parallel to the vector of the last residue in the strand, and to be antiparallel (A) if the two are antiparallel (Fig. 1b). The orientation of an αβ-unit is P if the vector of the first residue in the strand is parallel to the vector from helix to strand, and is A if the two are antiparallel (Fig. 1c) (see Supplementary Methods 4 and 5 for details).
ββ-rule
The chirality of β-hairpins is determined by the length of the loop between the two strands. Rosetta folding simulations of a peptide with two equal-length β-strands connected by a variable-length loop were carried out on a sequence-independent backbone model (Methods Summary, Methods and Supplementary Methods 1). The chirality (Fig. 1d) of the end points of multiple independent Monte Carlo trajectories was computed. The results (Fig. 1a, left) are quite striking: two- and three-residue loops almost always give rise to L-hairpins, whereas five-residue loops give rise primarily to R-hairpins. These results suggest that the chirality of β-hairpins is determined by the chirality (l-amino acids versus d-amino acids) and local structural preferences of the polypeptide chain; indeed, only a restricted set of loop types have been found to be compatible with ββ-junctions23. Analysis of ββ-units in known protein structures (Supplementary Methods 3) shows that the chirality of ββ-units in native structures is correlated with loop length in a manner very similar to the simulations (Fig. 1a, right). Consistent with the idea that torsional strain is responsible for the trends, the calculated torsion energies of loops in native structures for two- and three-residue loops are lower for L-hairpins, and those for five-residue loops are lower for R-hairpins (Supplementary Fig. 2). This rule allows control over the pleating of β-hairpins.
βα-rule
The preferred orientation of βα-units is P for two-residue loops and A for three-residue loops. Secondary-structure-constrained folding simulations similar to those described in the previous paragraph strongly show this trend, and it is also observed in native protein structures (Fig. 1b). The rule arises in part from the bendability of the protein backbone (Supplementary Fig. 3). This rule is very useful for both positive and negative design, as it allows control of the side of a β-sheet that a helix will pack onto.
αβ-rule
The preferred orientation of αβ-units is P. In secondary-structure-constrained folding simulations, this trend is observed strongly for loops two residues in length and for longer lengths when the loop provides a hydrogen-bonded capping interaction to stabilize the helix and does not extend the strand (Fig. 1c, left, and Supplementary Fig. 4). A very similar trend is again observed in native protein structures (Fig. 1c, right).
It must be emphasized that the three rules are largely independent of the amino-acid sequence of the secondary structures or connecting loops. As such, they must arise from the intrinsic chirality and local structural preferences of the polypeptide chain rather than from sequence-specific contributions. Whereas local sequence–structure relationships have been extensively studied24–27, there has been much less work on sequence-independent properties (the cataloguing of the discrete sets of loops compatible with junctions between secondary structure elements is a notable exception23). These rules provide a powerful way to perform negative design at the backbone level.
Emergent rules
The next level of complexity in αβ-proteins beyond two secondary structure elements is segments of three consecutive secondary structure elements. Secondary-structure-constrained Rosetta folding simulations revealed strong dependencies of the chirality (Supplementary Fig. 1d) of ββα- and αββ-units and the foldability of βαβ-units on the lengths of the connecting loops and the secondary structure elements. These dependencies are formulated in emergent rules (Supplementary Fig. 1 and Supplementary Discussion 1), which follow from the fundamental rules described in the previous section. The rules specify how to choose the lengths of secondary structure elements and the connecting loops to favour a desired conformation of a ββα-, αββ- or βαβ-unit.
Rule-based design of funnelled folding landscapes
The fundamental and emergent rules make possible the encoding of funnel-shaped energy landscapes. We can sculpt energy landscapes to be strongly funnelled by designing secondary structure patterns that favour the tertiary motifs present in the desired topology and disfavor non-native motifs. The desired structure is then further stabilized by using RosettaDesign8 to obtain sequences with favourable non-local interactions such as complementary hydrophobic core packing. The latter step involves purely positive design because the energy of the desired structure is optimized without regard to competing states, whereas the design of sequences that favour specific secondary structure patterns also has elements of negative design because non-native conformations are disfavoured by the local structural preferences of the protein backbone captured by the rules.
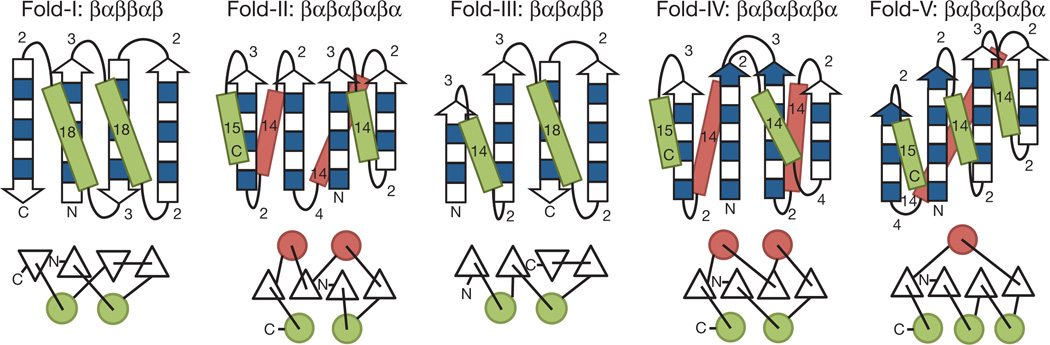
We tested this approach by attempting to design strongly funneled landscapes for five different folds (Fig. 2 and Supplementary Discussion 2). The first step is to choose secondary structure lengths that favour the desired fold and disfavour alternatives. We illustrate how to choose the secondary structure lengths that favour a desired topology with Fold-I, the classic ferredoxin-like fold (Fig. 2, leftmost fold). The secondary structure elements are, in order, β1α1β2β3α2β4. To assign the lengths of the loops and strands, we apply the emergent rules to the αββ- and ββα-triples and the βα- and αβ-rules to the two βαβ-units: (β1α1)A(α1β2)P(α1β2β3)L(β2β3α2)R(β3α2)A(α2β4)P. Reading directly from Fig. 1 and Supplementary Fig. 1, we find that for strand length 7 the ideal loop lengths between successive secondary structure elements are 3, 2, 2, 3 and 2 (from the amino to the carboxy terminus). To assign the lengths of the helices, we find from Supplementary Fig. 10 that for strand length 7 the optimal helix length is 18. We can apply the same procedure to each of the other folds to obtain the corresponding ideal secondary structure lengths (Fig. 2): for Folds-II, -IV and -V, we treat (αβα) as (αβ)P(βα)P/A and apply the corresponding two fundamental rules.
Figure 2. Derivation of secondary structure lengths from the rules for five protein topologies.
Fold-I: Ferredoxin-like fold. Fold-II: Rossmann2×2 fold. Fold-III: IF3-like fold. Fold-IV: P-loop2×2 fold. Fold-V: Rossmann3×1 fold. In the upper illustrations, numbers represent the secondary structure lengths following from the rules described in Fig. 1 and Supplementary Fig. 1. Strand lengths are represented by filled and open boxes. The filled boxes represent pleats coming out of the page, and the open boxes represent pleats going into the page. In the lower illustrations, the design topologies are represented with circles (helices) and triangles (strands) connected by solid lines (loops).
To build tertiary backbone structures from the two-dimensional representations of protein folds depicted in Fig. 2, we carry out multiple independent Rosetta folding simulations using the secondary structure strings obtained from the rules. For Folds-I, -III and -V, a significant fraction of trajectories produced the desired topology because the secondary structure lengths were chosen specifically to encode it. Folds-II and -IV are not distinguished by the rules, and to resolve this ambiguity we varied the secondary structure lengths and used folding simulations to select lengths strongly favouring one or the other fold. For larger proteins, such degeneracies are likely to increase and additional rules may need to be identified to resolve them. Within the population of structures with the desired topology, there is still considerable variety in the distances and angles between the secondary structure elements, the loop conformations and the twist of the β-sheet. This variation is important because it provides a range of starting points for designing sequence-structure pairs with very low energy as described in the next paragraph.
Up to this point, specific sequence information has not been introduced; the representations are of the protein backbone alone. For each backbone in the ensemble, we then use Monte Carlo simulated annealing to identify amino acids and side-chain conformations that give rise to very low-energy structures. This is carried out using fixed-backbone RosettaDesign8 calculations followed by relaxation of the structure of the backbone and the side chains in the Rosetta all-atom energy function28. These sequence design and structure refinement calculations are then iterated8 to generate a tightly packed hydrophobic core with a packing density approaching that of close-packed crystals. Larger hydrophobic amino acids (Ile, Leu and Phe) are favoured in the core to create a strong driving force for folding29. Negative design is applied to the edge β-strands and the protein surface to destabilize non-native conformations and disfavour oligomerization: inward-pointing polar residues are introduced in the strands and hydrophobic patches are removed from the surface. The designed structures are then filtered according to energy, packing (as assessed by RosettaHoles30) and the local sequence–structure compatibility (Methods) to disfavour other structures (this last criterion is effectively a negative design step). Finally, for each sequence passing these filters, 200,000–400,000 independent Rosetta ab initio structure prediction simulations starting from an extended chain22 are performed to map out the folding energy landscapes. Roughly 10% of the designs have funnel-shaped energy landscapes leading into the designed structures (Fig. 3a; compare with Supplementary Fig. 11) and these are selected for experimental characterization. Proteins designed with this protocol (summarized in Supplementary Fig. 12) by construction have consistent local and non-local interactions. Notably, the only globular protein designed de novo before this work, Top7 (ref. 8), also satisfies our rules and has consistent local and non-local interactions.
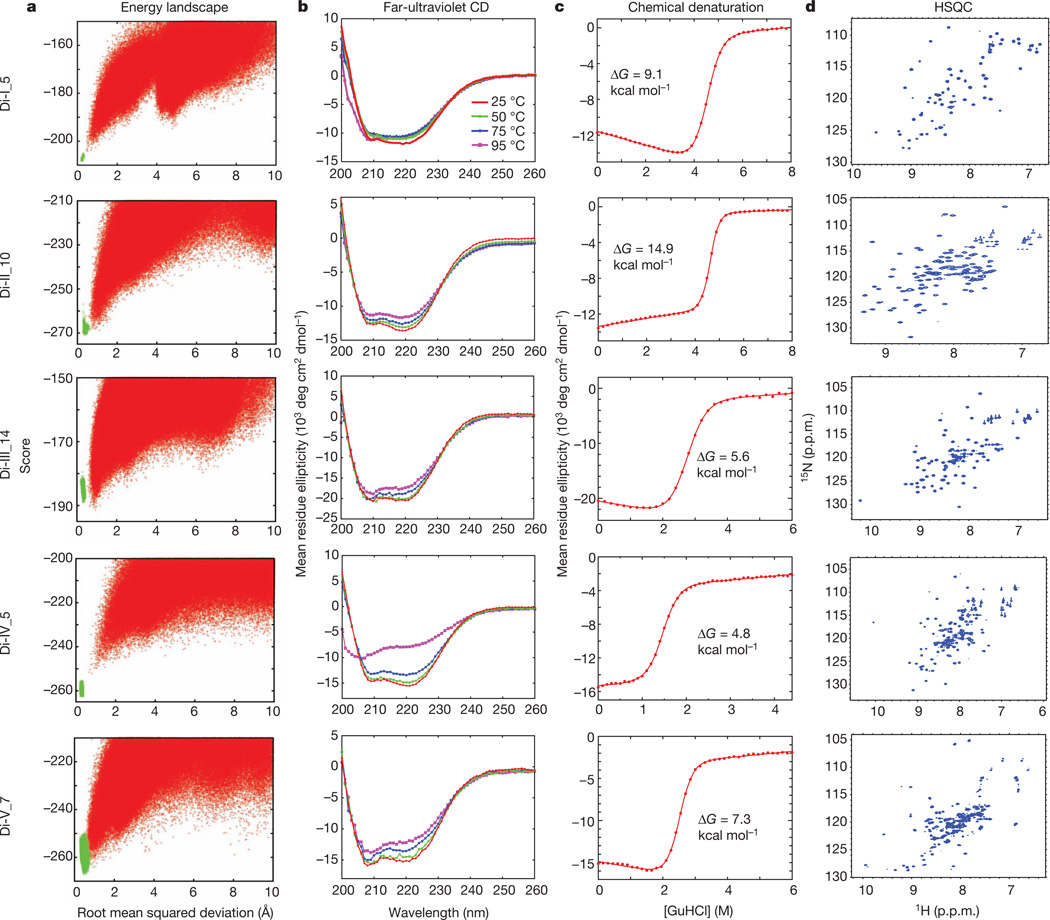
Figure 3. Characterization of design for each of the five folds.
a, Energy landscapes obtained from Rosetta ab initio structure prediction simulations on Rosetta@home. Red points represent the lowest-energy structures obtained in independent Monte Carlo structure prediction trajectories starting from an extended chain for each sequence; the y axis shows the Rosetta all-atom energy and the x axis shows the Cα root mean squared deviation from the design model. Green points represent the lowest-energy structures obtained in trajectories starting from the design model. Less sampling around the designed minima is observed for the higher-contact-order topology, Fold-IV44. b, The far-ultraviolet circular dichroism (CD) spectra at various temperatures. c, Chemical denaturations with GuHCl (square brackets denote concentration) at 220nmand 25 °C. The data were fitted to a two-state model (red solid line) to obtain the free energy of unfolding ΔG. d, Two-dimensional 1H–15N HSQC spectra at 25 °C and 600 MHz. p.p.m., parts per million.
Experimental characterization of designed proteins
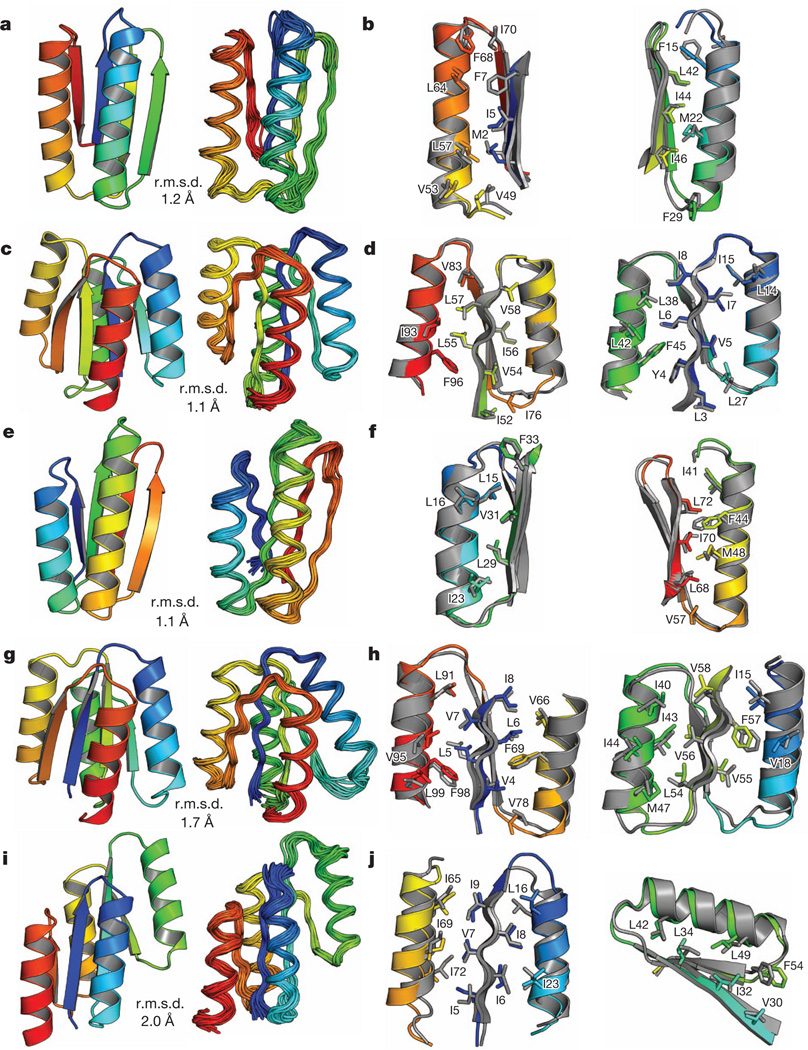
We obtained synthetic genes encoding 11 designs for Fold-I, 12 for Fold-II, 14 for Fold-III, 5 for Fold-IV and 12 for Fold-V (Supplementary Table 8). None of these proteins is homologous to any known protein (BLAST E-value <0.02 against the NCBI nr database of non-redundant protein sequences). The proteins were expressed, purified and characterized by circular dichroism spectroscopy, size-exclusion chromatography combined with multi-angle light scattering (SECMALS), and NMR spectroscopy. For all five folds, most of the designs are expressed and soluble and many are extremely stable (Table 1 and Supplementary Tables 1–5). Data for the most stable monomeric design for each fold that had a well-resolved NMR spectrum (Di-I_5, Di-II_10, Di-III_14, Di-IV_5 and Di-V_7; ‘Di’ indicates designed ideal protein, the Roman numeral is the fold type and the number is the identifier of the design) are shown in Fig. 3, Supplementary Fig. 13 and Supplementary Table 6. These five proteins are soluble at concentrations of 0.9–1.6 mM, have far-ultraviolet circular dichroism spectra characteristic of αβ-proteins and have cooperative unfolding transitions with a free energy of unfolding of >5 kcal mol−1 (Fig. 3b, c and Supplementary Table 6). The designed proteins were found to be monomeric by SEC-MALS (Supplementary Fig. 13). The two-dimensional 1H–15N heteronuclear single quantum coherence (HSQC) spectra show the expected number of well-dispersed sharp peaks (Fig. 3d), indicating that the designed proteins are well packed. The solution structures of all five designs were determined by solution-state NMR spectroscopy (Fig. 4). Extensive validation analyses, including excellent agreement between back-calculated and measured NMR data (Supplementary Table 7), suggest that the NMR structures are quite high quality. The structures are remarkably consistent with the computational design models for both the protein backbone and the core side chains (Fig. 4, Supplementary Fig. 14 and Supplementary Table 6).
Table 1.
Summary of experimental results for designed proteins
| Designs tested | Expressed* | Soluble* | αβ-protein circular dichroism spectrum |
Stable†(Tm ≥ 95 °C) | Monomeric‡ | Well-resolved NMR§ | Success | |
|---|---|---|---|---|---|---|---|---|
| Fold-I | 11 | 9 | 8 | 6 | 3 | 2 | 3 | 1 (9%) |
| Fold-II | 12 | 12 | 12 | 10 | 10 | 4 | 4 | 4 (33%) |
| Fold-III | 14 | 13 | 11 | 9 | 7 | 6 | 5 | 3 (21%) |
| Fold-IV | 5 | 4 | 4 | 4 | 2 | 4 | 3 | 2 (40%) |
| Fold-V | 12 | 11 | 10 | 3 | 3 | 1 | 1 | 1 (8%) |
The second column shows the number of designs experimentally tested for the fold in the leftmost column. The subsequent columns give the number of designs that satisfy each criterion. The successful designs are defined as those that satisfy all criteria. The details of the results are shown in Supplementary Tables 1–5.
Expression and solubility were assessed by SDS–polyacrylamide gel electrophoresis and mass spectrometry.
Stability was measured by thermal denaturation; Tm is the melting temperature.
SEC-MALS was used to determine oligomerization state. We counted the number of designs in which the main peak of the absorbance at 280nm corresponds to the monomeric state.
For Folds-I and -II, one-dimensional NMR spectra were collected, and for Folds-III, -IV and -V, two-dimensional 1H–15N HSQC spectra were collected.
Figure 4. Comparison of computational models with experimentally determined structures.
a, c, e, g, i, Comparison of overall topology. Design models (left) and NMR structures (right); the Cα root mean squared deviation (r.m.s.d.) between them is indicated. b, d, f, h, j, Comparison of core side-chain packing in superpositions of design models (rainbow) and NMR structures (grey). The left and right panels show close-up views of the core packing and correspond to the left and right portions of the structures shown in a, c, e, g and i. a, b, Di-I_5 (Protein Data Bank code, 2KL8); c, d, Di-II_10 (2LV8); e, f, Di-III_14 (2LN3); g, h,Di-IV_5 (2LVB); i, j, Di-V_7 (2LTA). The design models and NMR structures are available from http://psvs-1_4-dev.nesg.org/ideal_proteins/.
Concluding remarks
We have demonstrated that strongly funnelled landscapes can be designed by encoding consistency between the local and non-local interactions using rules that relate secondary structure lengths to tertiary structure patterns. The rules, which arise from the chirality and local structural preferences of the polypeptide chain, make possible the simultaneous positive design of interactions favouring the desired structure and negative design against competing alternatives. It is plausible that the same principles shape the folding landscapes of naturally occurring proteins, which are more frustrated but still exhibit the remarkable property of having unique native states considerably lower in energy than the vast number of alternative topologies. This idea is supported by the fact that the relationships between secondary structure patterns and tertiary structure motifs we identified in simulations are also observed in native structures (Fig. 1 and Supplementary Figs 1, 5, 7, 9 and 10); as in our design strategy, the disfavouring of the myriad alternative states may be achieved by naturally occurring sequences through the stabilization of local structures that disfavour non-native topologies31,32.
The design principles and methodology we have described should allow the ready design of a wide range of robust and stable protein building blocks for the next generation of engineered functional proteins33–41. Almost all protein design and engineering efforts so far have repurposed naturally occurring proteins that evolved for some other, often unrelated, function35–41. It should now become possible to custom-design protein scaffolds ideal for the desired function, and to build larger assemblies42,43 and materials from robust ideal building blocks.
METHODS
Rosetta folding simulations
Rosetta folding simulations using a sequence-independent backbone model were carried out in the studies of the fundamental rules (Fig. 1), the emergent rules (Supplementary Fig. 1) and the building of tertiary backbone structures in the rule-based designs. These simulations are referred to as secondary-structure-constrained folding simulations in the main text because the phi and psi angles of each residue are limited to the region of the Ramachandran plot compatible with the assigned secondary structure. We first introduce the backbone model and then describe the fragment assembly method45 used for simulating the backbone model.
The backbone model consists of main-chain atoms (N, NH, Cα,C and CO) and Cβ atoms with a pseudo-atom representing a generic side chain (the centroid model of Rosetta22). The Rosetta potential function terms and weights are as follows: steric repulsion (vdw = 1.0), overall compaction (rg = 1.0), secondary structure pairings (ss_pair = 1.0, rsigma = 1.0 and hs_pair = 1.0) and hydrogen bonds (hbond_sr_bb = 1.0, hbond_lr_bb = 1.0). For the steric radius of the side-chain pseudo-atom, the radius of Val was used.
Fragment assembly45 was used for sampling conformations of the backbone model. Backbone fragment sets consisting of 1, 3 or 9 consecutive residue fragments were prepared in advance from a non-redundant set of X-ray structures46; the fragments have information only on the phi, psi and omega torsion angles. We performed Monte Carlo simulations in which in each attempted Monte Carlo trial, a new conformation is generated by replacing the torsion angles (phi, psi and omega) of a randomly selected frame consisting of 1, 3 or 9 consecutive residues with the torsion angles of a randomly selected fragment compatible with the assigned secondary structure. Importantly, in the calculations for the fundamental rules, we used only one-residue fragments to avoid the possibility that the evolutional history of natural protein structures would bias the simulation results. Because we found that the fundamental rules are observed both in the simulations and in the natural proteins (Fig. 1), we used all fragment lengths in the simulations relating to the emergent rules and the rule-based designs. In the calculations for the fundamental rules, the total number of Monte Carlo steps in one trajectory was 500×(length of a simulated chain), and the temperature was 1.0. In the emergent rules and rule-based designs, the total number of Monte Carlo steps in one trajectory was 300×(length of a simulated chain), and the temperature was 1.5.
Sequence design protocol
Sequence design was performed using the RosettaDesign approach8 with several extensions.
The environment for each residue position was classified into one of three layers, core, boundary or surface, on the basis of the solvent-accessible surface area (SASA) of main-chain and Cβ atoms and the secondary structure type, and only designated amino-acids for each layer were allowed for design. The core was defined with SASA ≤15Å for helices and strands and SASA ≤25Å for loops; the boundary was defined with 15Å <SASA < 60 Å for helices and strands and 25Å < SASA < 40 Å for loops; and the surface was defined with SASA ≥60Å for helices and strands and SASA ≥40 Å for loops. The amino acids V, I, L, M, F, Y and W were used in the core; V, I, L, Y, W, D, E, N, Q, K, R, S and T were used at the boundary; and D, E, N, Q, K, R, S, T and H were used on the surface. To favour larger hydrophobic amino acids in the interior of protein structures, Ala was allowed only for helices and loops in the core and at the boundary, and Gly was allowed only for loops in all layers. Pro was allowed in loops and at the beginning of helices and strands. The loop residue immediately before a helix is one of D, N, S and T to provide the helix capping. This method was applied to the design of Folds-II to -V.
To favour larger hydrophobic amino acids (I, L and F) in the core, we modified the reference energy8 of each amino acid.
To introduce inward-pointing polar residues in the edge strands (in most cases charged residues in the middle of the edge strands), we used a resfile, by which designated amino acids can be assigned at a specified residue position.
For aromatic residues of F, Y and H, we limited the χ2 angle to range from 70° to 110°, the range frequently observed in nature. This restriction was applied to the design of Folds-III to -V.
After designing sequences, we relaxed the backbone and side chains of the designed structures28. These sequence design and structure refinement calculations were iterated. The designed structures were then filtered on the basis of their Rosetta all-atom energy22, packing as assessed by RosettaHoles30, and the local sequence and structure compatibility. Finally, we visually inspected the designed structures and mutated buried polar and exposed hydrophobic residues using Foldit47.
Rosetta command lines are provided in Supplementary Data to perform the design protocol.
Local sequence–structure compatibility
To evaluate the compatibility between the local sequence and the local structure, we collected 200 fragments for each nine-residue frame in the designed sequence from a non-redundant set of X-ray structures based on the sequence similarity and secondary structure prediction22 (the standard Rosetta fragment generation protocol for ab initio structure prediction). Then, for each frame, we calculated the root mean squared deviation between the designed local structure and each of the 200 fragments. Designs were ranked on the basis of the total number of fragments for which the root mean squared deviation was less than 1.0Å, and those with high values were selected.
Protein expression and purification
For all designed sequences, a Gly–Ser was added at the C terminus to give a spacer between the designed region and the C-terminal 6×His tag. The genes encoding the designed sequences, which were cloned into plasmid pET29b, were obtained from GenScript. The designed proteins were expressed in Escherichia coli BL21 Star (DE3) cells as non-labelled proteins for all designs for Folds-I and -II, and as uniformly U-15N-labelled proteins for all designs for Folds-III to -V. The non-labelled proteins were expressed using auto-induction media48, and the U-15N-labelled proteins were expressed using MJ9 minimal media49, which contain 15Nammonium sulphate as the sole nitrogen source and 12C glucose as the sole carbon source. The expressed proteins with a 6×His tag at the C terminus were purified through a nickel affinity column. The purified proteins were then dialysed against typical PBS buffer, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4, at pH 7.4; this buffer was used for all the experiments except NMR structure determination. The expression, solubility and purity of the designed proteins were assessed by SDS–polyacrylamide gel electrophoresis and mass spectrometry (TSQ LC/MS, Thermo Scientific).
Circular dichroism
All circular dichroism data were collected on an Aviv 62A DS spectrometer. Far-ultraviolet circular dichroism spectra of designed proteins were measured from 260 to 200nm for 14–25 µM protein samples in PBS buffer (pH 7.4) at various temperatures of 25, 50, 75 and 95 °C in a 1-mm-path-length cuvette. The protein concentrations were determined from the absorbance at 280nm (ref. 50) using an ultraviolet spectrophotometer (NanoDrop, Thermo Scientific). Tm is the melting temperature where the number of folded proteins is equal to the number of unfolded proteins during temperature denaturation. Chemical denaturations with GuHCl were monitored at 220nm for 3–4 µM protein samples in PBS buffer (pH 7.4) at 25 °C in a 1-cm-path-length cuvette. The GuHCl concentration was automatically controlled by a Microlab titrator (Hamilton). The chemical denaturation curves were fitted by nonlinear least-squares analysis using a two-state unfolding and linear extrapolation model51. The free-energy change for the unfolding transition, ΔG, and the value representing its dependency on the denaturant, the m-value (of which a higher value indicates higher cooperativity), were obtained from the fitting.
Size-exclusion chromatography combined with multi-angle light scattering
SEC-MALS experiments were performed using a miniDAWN TREOS static light-scattering detector (Wyatt Technology) combined with a HPLC system (LC 1200 Series, Agilent Technologies). One hundred microlitres of 400–700 µM protein samples in PBS buffer (pH 7.4) was injected into a Superdex 75 10/300 GL column (GE Healthcare) equilibrated with PBS buffer at a flow rate of 0.5 ml min−1. The protein concentrations were calculated from the absorbance at 280 nm detected by the HPLC system. Static light-scattering data were collected at three different angles, 41.4°, 90.0° and 138.6°, at 658 nm. These data were analysed using ASTRA software (version 5.3.4, Wyatt Technology) with a change in the refractive index with concentration (dn/dc value) of 0.185 ml g−1.
Nuclear magnetic resonance
To assess the core packing of designed proteins, one-dimensional 1H NMR spectra were measured for the designs for Folds-I and -II, and two-dimensional 1H-15N HSQC spectra were measured for the designs for Folds-III to -V. The spectra were collected for 0.5–1.5 mM protein samples in 90% 1H2O/10% 2H2O PBS buffer (pH 7.4) at 25 °C on a Varian INOVA 600 MHz spectrometer. The most stable monomeric design with a well-resolved NMR spectrum for each fold (Di-I_5, Di-II_10, Di-III_14, Di-IV_5 and Di-V_7) was selected for NMR structure determination.
The five designs were expressed and purified following the standard, largely automated NESG protocol52. The designs were expressed in E. coli BL21 (DE3) pMGK cells as U-15N,5%13C-enriched proteins, and U-15N,U-13C-enriched proteins using MJ9 minimal media49. The U-15N,5%13C-labelled proteins were generated for stereospecific assignments of methyl groups of Val and Leu53 and for measurements of residual dipolar couplings54. The expressed proteins were purified using an ÄKTAxpress (GE Healthcare) two-step protocol consisting of IMAC (HisTrap HP column, GE Healthcare) and gel filtration chromatography (HiLoad 26/60 Superdex 75 column, GE Healthcare). The purified proteins were dissolved in 90% 1H2O/10% 2H2O buffer containing 20 mM MES, 200 mM NaCl, 10 mM DTT, 5 mM CaCl2 and 0.02% NaN3 at pH 6.5 for Di-I_5 and Di-II_10; 100 mM NaCl, 5.6 mM Na2HPO4, 1.1 mM KH2PO4 and 3 mM DTT at pH 7.5 for Di-III_14; and 100 mM NaCl, 5 mM DTT, 0.02% NaN3, 10 mM Tris-HCl at pH 7.5 for Di-IV_5 and Di-V_7. The expression, solubility and purity of the five proteins were assessed by SDS–polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization–time of flight mass spectrometry.
Experimental NMR structure determination was carried out without any knowledge of the design model. For NMR structure determination, all NMR spectra were recorded at 25 °C using cryogenic NMR probes. Triple-resonance NMR data were collected on Varian INOVA 600 MHz or Bruker AVANCE 800 MHz spectrometers, and simultaneous three-dimensional 15N/13Caliphatic/13Caromatic-edited nuclear Overhauser enhancement spectroscopy (NOESY55; mixing time, 100 ms) and three-dimensional 13C-edited aromatic NOESY (mixing time, 100 ms) spectra were acquired on the Bruker AVANCE 800 MHz spectrometer. Two-dimensional constant-time 1H-13C HSQC spectra, with 28-ms and 42-ms constant-time delays, were recorded for the U-15N,5%13C-enriched samples on the Varian INOVA 600 MHz spectrometer to obtain stereospecific assignments of methyl groups of Val and Leu53. Backbone 15N-1H residual dipolar couplings in two alignment media, PEG and phage, were determined from J-modulated spectra54 for Di-II_10, Di-III_14 and Di-V_7. All NMR data were processed using the program NMRPIPE56 and analysed using the program XEASY57. Spectra were referenced to external DSS. Sequence-specific resonance assignments were determined as described previously58. Chemical shift data were deposited in the Biological Magnetic Resonance Data Bank with BMRB IDs 16387, 18558, 18145, 18561 and 18465 for Di-I_5, Di-II_10, Di-III_14, Di-IV_5 and Di-V_7, respectively. Initial NOESY peak lists containing expected intraresidue, sequential and α-helical medium-range NOE peaks were generated from the obtained assignments and then manually edited by visual inspection of the NOESY spectra. Subsequent manual peak picking was then used to identify remaining, primarily long-range NOEs58. Backbone dihedral angle constraints were derived from the chemical shifts using the program TALOS+59 for residues located in well-defined secondary structure elements, and were used for structure determination. Residual dipolar couplings were used as orientational constraints for well-defined residues during structure determination for Di-II_10, Di-III_14 and Di-V_7. The program CYANA60,61 was used to assign NOEs automatically and to calculate the structure. The 20 conformers with the lowest target function values were refined in explicit water solvent62 using the program CNS63. RPF analysis of AUTOSTRUCTURE64,65 was used in parallel to guide the iterative cycles of noise/artefact peak removal, peak picking and NOE assignments. The finally obtained structure coordinates were deposited in the Protein Data Bank. The structural statistics and global structure quality factors including VERIFY3D66, PROSAII67, PROCHECK68, and MOLPROBITY69 raw and statistical Z-scores were computed using PDBSTAT and PSVS 1.470. The global goodness-of-fit of the final structure ensembles with the NOESY peak list data was determined using the RPF analysis program71. The NMR data are available from http://psvs-1_4-dev.nesg.org/ideal_proteins/.
Supplementary Material
Acknowledgements
We thank N. Grishin for suggesting target folds for design, P. Rajagopal for one-dimensional NMR measurements of Folds-I and -II, and J. Siegel for measurements by mass spectrometer. We also thank P.-S. Huang and Y.-E. A. Ban for computational tools; J. L. Gallaher for experimental assistance; J. Castellanos for the help with designing Fold-IV; H.-W. Lee, K. Pederson and J. Prestegard for measurements of residual dipolar couplings; and S. Khare, F. DiMaio, I. Andre, S. Fleishman, J. Mills, S. Takada, S. Fuchigami and G. Chikenji for comments on the manuscript. This work was supported by HHMI, DOE, DARPA, DTRA and the National Institutes of General Medical Science Protein Structure Initiative (PSI:Biology) programme, grant U54 GM094597. N.K. was also supported by Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowships for Research Abroad.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions N.K., R.T.-K., G.L., G.T.M. and D.B. designed the research. N.K. performed folding simulations and analysed natural proteins. N.K. wrote program code. N.K. and R.T.-K. performed computational design work: Di-I_5 and Di-IV_5 were designed by N.K., and Di-II_10, Di-III_14 and Di-V_7 were designed by R.T.-K. R.T.-K. expressed, purified and characterized the designed proteins by biochemical assay. R.X. and T.B.A. prepared isotope-enriched protein samples for NMR structure determination. G.L. collected NMR data and determined the solution NMR structures. N.K., R.T.-K., G.L., G.T.M. and D.B. wrote the manuscript.
Author Information The NMR structures of the five designs have been deposited in the RCSB Protein Data Bank under the accession numbers 2KL8 (Di-I_5), 2LV8 (Di-II_10), 2LN3 (Di-III_14), 2LVB (Di-IV_5) and 2LTA (Di-V_7). NMR data have been deposited in the Biological Magnetic Resonance Data Bank under the accession numbers 16387 (Di-I_5), 18558 (Di-II_10), 18145 (Di-III_14), 18561 (Di-IV_5) and 18465 (Di-V_7).
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc. Natl Acad. Sci. USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onuchic JN, Wolynes PG, Luthey-Schulten Z, Socci ND. Toward an outline of the topography of a realistic protein-folding funnel. Proc. Natl Acad. Sci. USA. 1995;92:3626–3630. doi: 10.1073/pnas.92.8.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nature Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 4.Hill RB, Raleigh DP, Lombardi A, DeGrado WF. De novo design of helical bundles as models for understanding protein folding and function. Acc.Chem. Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterfoss GL, Kuhlman B. Computer-based design of novel protein structures. Annu. Rev. Biophys. Biomol. Struct. 2006;35:49–65. doi: 10.1146/annurev.biophys.35.040405.102046. [DOI] [PubMed] [Google Scholar]
- 6.Samish I, MacDermaid CM, Perez-Aguilar JM, Saven JG. Theoretical and computational protein design. Annu. Rev. Phys. Chem. 2011;62:129–149. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- 7.Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 8.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 9.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J. Mol. Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 10.Calhoun JR, et al. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J. Mol. Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Isogai Y, Ito Y, Ikeya T, Shiro Y, Ota M. Design of lambda Cro fold: solution structure of amonomeric variant of the de novo protein. J. Mol. Biol. 2005;354:801–814. doi: 10.1016/j.jmb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Shah PS, et al. Full-sequence computational design and solution structure of a thermostable protein variant. J. Mol. Biol. 2007;372:1–6. doi: 10.1016/j.jmb.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 13.Hu X, Wang H, Ke H, Kuhlman B. Computer-based redesign of a beta sandwich protein suggests that extensive negative design is not required for de novo beta sheet design. Structure. 2008;16:1799–1805. doi: 10.1016/j.str.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecht MH, Richardson JS, Richardson DC, Ogden RC. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990;249:884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 15.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl Acad. Sci. USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin W, Kambara O, Sasakawa H, Tamura A, Takada S. De novo design of foldable proteins with smooth folding funnel: automated negative design and experimental verification. Structure. 2003;11:581–590. doi: 10.1016/s0969-2126(03)00075-3. [DOI] [PubMed] [Google Scholar]
- 17.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 18.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. Computational de novo design, and characterization of an A(2)B(2) diiron protein. J. Mol. Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 19.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nature Struct. Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 20.Kortemme T, et al. Computational redesign of protein-protein interaction specificity. Nature Struct. Mol. Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 21.Go N. Theoretical studies of protein folding. Annu. Rev. Biophys. Bioeng. 1983;12:183–210. doi: 10.1146/annurev.bb.12.060183.001151. [DOI] [PubMed] [Google Scholar]
- 22.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 23.Street TO, Fitzkee NC, Perskie LL, Rose GD. Physical-chemical determinants of turn conformations in globular proteins. Protein Sci. 2007;16:1720–1727. doi: 10.1110/ps.072898507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bystroff C, Baker D. Prediction of local structure in proteins using a library of sequence-structure motifs. J. Mol. Biol. 1998;281:565–577. doi: 10.1006/jmbi.1998.1943. [DOI] [PubMed] [Google Scholar]
- 25.Hunter CG, Subramaniam S. Protein local structure prediction from sequence. Proteins. 2003;50:572–579. doi: 10.1002/prot.10310. [DOI] [PubMed] [Google Scholar]
- 26.Etchebest C, Benros C, Hazout S, deBrevern AG. A structural alphabet for local protein structures: improved prediction methods. Proteins. 2005;59:810–827. doi: 10.1002/prot.20458. [DOI] [PubMed] [Google Scholar]
- 27.Voelz VA, Shell MS, Dill KA. Predicting peptide structures in native proteins from physical simulations of fragments. PLoS Comput. Biol. 2009;5:e1000281. doi: 10.1371/journal.pcbi.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyka MD, et al. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 2011;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 30.Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming PJ, Gong H, Rose GD. Secondary structure determines protein topology. Protein Sci. 2006;15:1829–1834. doi: 10.1110/ps.062305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chikenji G, Fujitsuka Y, Takada S. Shaping up the protein folding funnel bylocal interaction: lesson from a structure prediction study. Proc.Natl Acad. Sci.USA. 2006;103:3141–3146. doi: 10.1073/pnas.0508195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan J, DeGrado WF. Denovo design of catalytic proteins. Proc.Natl Acad. Sci. USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Correia BE, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc.Natl Acad. Sci. USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 38.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azoitei ML, et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 41.Khare SD, et al. Computational redesign of a mononuclear zinc metalloenzyme for organophosphate hydrolysis. Nature Chem. Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisenbeis S, et al. Potential of fragment recombination for rational design of proteins. J. Am. Chem. Soc. 2012;134:4019–4022. doi: 10.1021/ja211657k. [DOI] [PubMed] [Google Scholar]
- 44.Bonneau R, Ruczinski I, Tsai J, Baker D. Contact order and ab initio protein structure prediction. Protein Sci. 2002;11:1937–1944. doi: 10.1110/ps.3790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 46.Huang PS, et al. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS ONE. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper S, et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Jansson M, et al. High-level production of uniformly N-15- and C-13-enriched fusion proteins in Escherichia coli. J. Biomol. NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 50.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 52.Acton TB, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of themethyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 54.Tjandra N, Grzesiek S, Bax A. Magnetic field dependence of nitrogen-proton J splittings in N-15-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 1996;118:6264–6272. [Google Scholar]
- 55.Shen Y, Atreya HS, Liu GH, Szyperski T. G-matrix Fourier transform NOESY-based protocol for high-quality protein structure determination. J. Am. Chem. Soc. 2005;127:9085–9099. doi: 10.1021/ja0501870. [DOI] [PubMed] [Google Scholar]
- 56.Delaglio F, et al. Nmrpipe - a multidimensional spectral processing systembased on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The program Xeasy for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 58.Liu GH, et al. NMR data collection and analysis protocol for high-throughput protein structure determination. Proc. Natl Acad. Sci. USA. 2005;102:10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Güntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 61.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 62.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 63.Brünger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 64.Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- 65.Huang YJ, et al. An integrated platform for automated analysis of protein NMR structures. Methods Enzymol. 2005;394:111–141. doi: 10.1016/S0076-6879(05)94005-6. [DOI] [PubMed] [Google Scholar]
- 66.Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 67.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 68.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 69.Word JM, Bateman RC, Presley BK, Lovell SC, Richardson DC. Exploring steric constraints on protein mutations using MAGE/PROBE. Protein Sci. 2000;9:2251–2259. doi: 10.1110/ps.9.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 71.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.