Abstract
Olfactory ensheathing cells (OECs) migrate from the olfactory epithelium towards the olfactory bulb during development. However, the guidance mechanism for OEC migration remains a mystery. Here we show that migrating OECs expressed the receptor of the repulsive guidance factor Slit-2. A gradient of Slit-2 in front of cultured OECs first caused the collapse of the leading front, then the reversal of cell migration. These Slit-2 effects depended on the Ca2+ release from internal stores through inositol (1,4,5)-triphosphate receptor channels. Interestingly, in response to Slit-2 stimulation, collapse of the leading front required the activation of the F-actin severing protein cofilin in a Ca2+-dependent manner, whereas the subsequent reversal of the soma migration depended on the reversal of RhoA activity across the cell. Finally, the Slit-2-induced repulsion of cell migration was fully mimicked by co-application of inhibitors of F-actin polymerization and RhoA kinase. Our findings revealed Slit-2 as a repulsive guidance factor for OEC migration and an unexpected link between Ca2+ and cofilin signaling during Slit-2-triggered repulsion.
Key words: Ca2+, Cofilin, Migration, Olfactory ensheathing cells, RhoA, Slit-2
Introduction
Olfactory ensheathing cells (OECs) are a unique type of glial cells in the olfactory system, and have been discovered to promote the growth of olfactory sensory axons during development and the regeneration of injured axons after being transplanted into nerve injury sites (Cao et al., 2004; Li et al., 1998; Raisman and Li, 2007; Ramon-Cueto et al., 1998; Vincent et al., 2005). Derived from the olfactory placode, OECs migrate out of the olfactory epithelium (OE) together with growing olfactory sensory axons from the lamina propria (LP) and accumulate as a superficial mass upon reaching the telencephalic vesicle at embryonic day (E) E13–E18 in rat, contributing to the formation of the presumptive olfactory nerve layer (Chuah and West, 2002; Valverde et al., 1992). However, how the migration of OECs is guided during development remains unclear.
Slit and Robo are a pair of conserved repulsive ligands and receptors for axon pathfinding (Dickson and Gilestro, 2006). Slit was first identified in Drosophila as a molecule secreted by midline cells, and was later shown to repel the extension of axons expressing Robo receptors (Brose et al., 1999; Kidd et al., 1999; Seeger et al., 1993) and to regulate the migration of cells such as neuronal precursors (Wu et al., 1999). In the olfactory system, members of the Slit and Robo families are expressed in a specific spatio-temporal pattern and play important roles in the guidance of olfactory sensory axons (Cho et al., 2007; Marillat et al., 2002; Nguyen-Ba-Charvet et al., 2008; Yuan et al., 1999). Whether OECs, which have the same developmental origin as olfactory sensory neurons, are also responsive to Slits is unknown.
The signal transduction underlying the guidance of axon pathfinding and neuronal migration has been studied in cell culture. In dissociated culture of cerebellar granule cells, a frontal gradient of Slit-2 triggers the elevation of the intracellular concentration of Ca2+ ([Ca2+]i) in the leading growth cone, and the subsequent propagation of a Ca2+ wave from the growth cone to the soma mediates the reversal of soma translocation by inhibiting the activity of the small GTPase RhoA (Guan et al., 2007; Xu et al., 2004). However, the molecular mechanism underlying the Slit-2-induced collapse of neuronal growth cones is unclear.
In the present study, we tested the guidance effect of Slit on the migration of cultured OECs. We found that a Slit-2 gradient in front of migrating OECs triggered a Ca2+-dependent collapse and reversal of OEC migration. Furthermore, Slit-2 triggered the activation of the F-actin severing protein cofilin and the inhibition of RhoA, leading to the collapse of the leading front and the subsequent reversal of the polarity of OEC migration.
Results
Robo1 is expressed in OECs both in vitro and in vivo
To explore the potential effects of Slits on OEC migration, we examined the expression of Robos in cultured OECs that exhibit active cell migration (Huang et al., 2008). In purified OEC culture, RT-PCR experiments revealed that mRNAs of both Robo1 and Robo2 were highly expressed. However, mRNA of Robo3 was undetectable (Fig. 1A). Robo1 protein could also be detected in cultured OECs by western blotting, and its expression could be knocked down by transfection with specific short interfering RNA (siRNA) against Robo1 (Fig. 1B; supplementary material Fig. S1C,D). To examine the subcellular distribution of Robo receptors in OECs, we transfected OECs with a construct encoding Robo2 fused with GFP. As shown in Fig. 1C, GFP fluorescence (normalized by membrane marker Dil) was present mostly in the cell surface and enriched at the leading front. The distribution of Robo2–GFP at the leading front suggests that migrating OECs might be responsive to Slit. To examine the expression of Slit in developing OE, we stained the brain sections from E16 mice using the anti-Slit-2 antibody. Western blotting and immunocytochemistry confirmed the specificity of this antibody by detecting the specific Slit-2 signal in a HEK293 cell line stably expressing Slit-2, but not in the control cell line (supplementary material Fig. S1A,B). Consistent with previous reports (Nguyen-Ba-Charvet et al., 2008), immunostaining in E16 brain sections from mice showed that Slit-2 was highly expressed in apical cells of the OE (Fig. 1D). However, at the LP, Robo1 was highly expressed in cells that could be labeled by the antibody against the specific OEC marker p75; presumably these cells were migrating OECs (Fig. 1E). These results suggest that the Slit receptor Robo1 is expressed in migrating OECs both in vitro and in vivo.
Fig. 1.
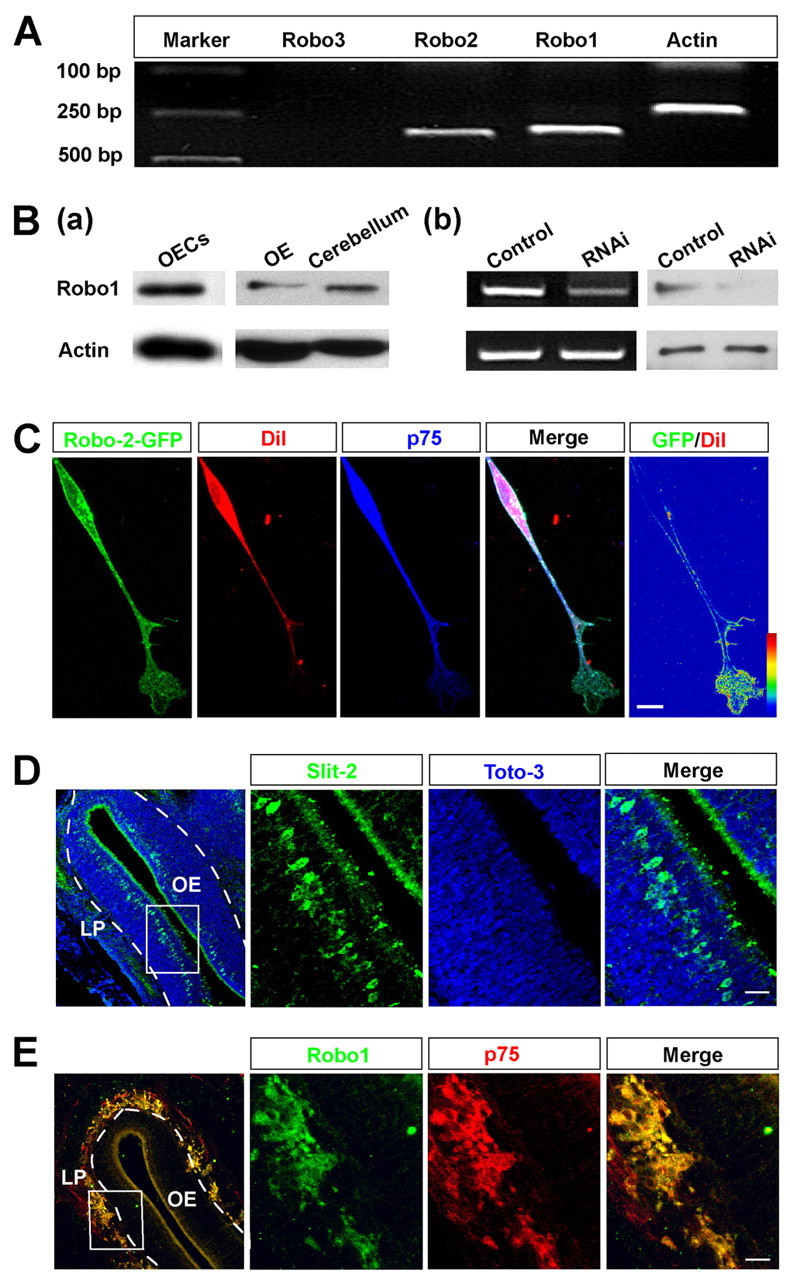
Robo1 is expressed in OECs in vitro and in vivo. (A) RT-PCR analysis of the expression of mRNAs encoding Robo1, Robo2 and Robo3 in cultured OECs. (B) Western blotting detected the expression of Robo1 protein in cultured OECs, OE and cerebellum tissue (a).The expression of Robo1 at both mRNA level (left) and protein level (right) was downregulated by RNA interference (RNAi) in cultured OECs (b). (C) Images of example cell showing the distribution of Robo2–GFP in OECs. The ratio of fluorescence intensity of GFP to Dil represents the Robo2–GFP density, coded by pseudocolors in a linear scale. (D,E) Sagittal brain sections of OE from E16 mice were immunostained for Slit-2 (green) and Toto-3 to visualize nuclei (blue) (D), and for Robo1 (green) and p75 (red) (E). Images of selected regions are shown at high magnification. Scale bars: 20 μm.
Slit-2 gradient repels the migration of cultured OECs
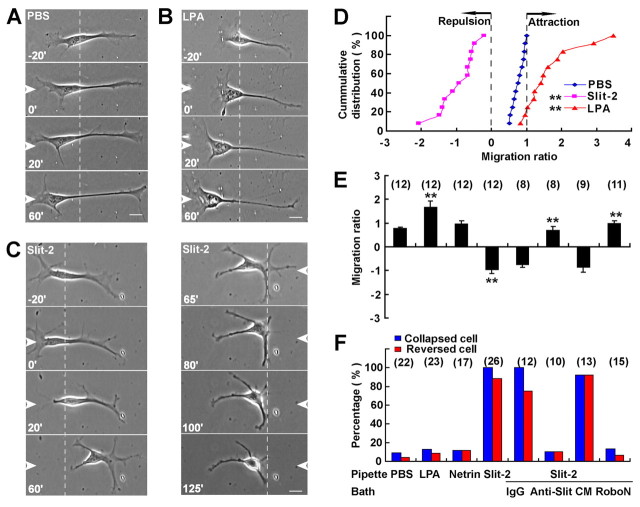
To test whether OECs are responsive to Slit-2, single-cell migration assays were first performed in a low-density culture of OECs obtained from olfactory bulb (OB) tissue. A gradient of Slit-2 was produced in front of isolated migrating OECs by using a micropipette loaded with Slit-2 with repetitive injection by air pressure (Huang et al., 2008). As shown in Fig. 2C,F and supplementary material Movie 1, after the application of a Slit-2 gradient, the leading front of migrating OECs with elaborated lamellipodia was inhibited in their motility and showed collapse and retraction within 20 minutes. For 23 out of 26 of tested cells, the soma later reversed their direction of translocation, with the original trailing tail becoming a new leading front. Interestingly, after OEC migration had been reversed by the Slit-2 gradient, when the Slit-2 gradient was applied to the same OEC from the reverse direction, OEC migration was reversed again (Fig. 2C). By contrast, OECs appeared to have enhanced motility toward the gradient of lysophophatidic acid (LPA), a known attractant for OECs (Huang et al., 2008; Yan et al., 2003) (Fig. 2B,F); whereas OEC migration was not affected by the gradient of another axon guidance molecule Netrin-1, or by phosphate-buffered saline (PBS) (Fig. 2A,F).
Fig. 2.
Slit-2 gradient induces the collapse and reversal of migration of OECs in single-cell migration assay. (A–C) Images of OECs before and after frontal application of a gradient of PBS (A), 500 μM LPA (B) or 4 μg/ml Slit-2 (C). White arrowheads indicate the direction of the micropipette. Scale bars: 20 μm. (D) Cumulative distribution of migration ratios of OECs exposed to a gradient of PBS, Slit-2 or LPA. Migration ratio is the migration rate after loading of the factor gradient divided by that of control period (after/before). Each point represents the result from one OEC (n=12). A ratio >1 and <0 means that the treatment attracts and repels OEC migration, respectively. **P<0.01, Kolmogorov–Smirnov test. (E) Average migration ratios under various conditions, as shown in F. Data are mean + s.e.m.; **P<0.01, Student's t-test. (F) Histograms show the percentages of collapsed or reversed cells in total observed cells under various conditions. Netrin-1, 50 μg/ml; IgG, normal IgG; Anti-Slit, antibody against Slit-2 (1:200); CM, control medium; RoboN, medium containing ectodomain of Robo1.
To quantify the effect of tested factors on OEC migration, we measured migration rates of each cell during the period before and after the application of factors and calculated their ratio (after/before). A ratio >1 implies accelerated cell migration in response to the treatment. Conversely, a ratio <1 implies inhibition of migration, and a ratio <0 implies a reversal of migration (Huang et al., 2008). As shown by the cumulative distribution graph (Fig. 2D), ratios of migration rate for most OECs under Slit-2 gradient were <0, as indicated by a significant left shift in the cumulative distribution curve of migration ratios compared with that of the PBS-treated group (P<0.001, Kolmogorov–Smirnov test), with the average migration ratio under Slit-2 gradient markedly decreased (Fig. 2E). By contrast, ratios of migration rate for most OECs under LPA gradient were >1, and there was a significant right shift in the distribution curve of migration ratios under LPA treatment compared with that of the PBS-treated group (P<0.001, Kolmogorov–Smirnov test) (Fig. 2D). The average migration ratio was markedly increased under LPA gradient, but not changed under Netrin-1 gradient (Fig. 2E). Furthermore, in the presence of anti-Slit-2 antibody or the soluble protein of the Robo1 ectodomain (RoboN), a competitive inhibitor for Slit–Robo interaction (Wu et al., 1999), spontaneous migration of OECs was not affected. However, the Slit-2-induced collapse and reversal of migration of OECs were both blocked, compared with treatment with a normal IgG or control media, respectively (Fig. 2E,F). These results strongly suggest that Slit-2 repels the migration of these cultured OECs.
Similar collapse and reversal of migration of OECs in response to a Slit-2 gradient were observed in cultured OECs obtained from postnatal day 0 (P0) OE tissue (supplementary material Fig. S2A–D). Moreover, in the high-density culture, when Slit-2 gradient was applied toward a group of OECs, cells exhibited the obvious collapse and retraction and escaped from the tip of the micropipette within 35 minutes in a distance-dependent manner (supplementary material Fig. S2E). Taken together, these results show that Slit-2 is a repulsive guidance factor for the migration of cultured OECs.
Ca2+ release from Ins(1,4,5)P3 receptor channels is essential for the effect of Slit-2
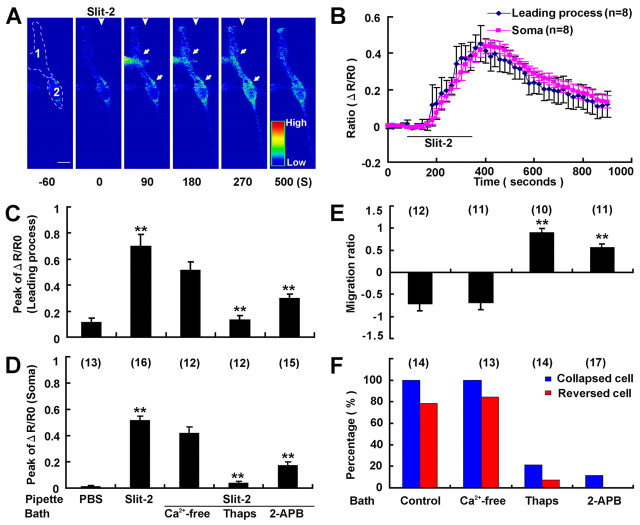
We next examined whether Ca2+ signaling was involved in Slit-2-induced repulsion of OECs. In OECs loaded with the Ca2+-sensitive fluorescent dyes Fluo-4 and Fura-Red, we observed a transient elevation in the ratio of the fluorescent intensity (Fluo-4/Fura-Red) both in the leading process and in the soma in response to the Slit-2 gradient, indicating that Slit-2 triggers the elevation of [Ca2+]i in these cultured OECs (Fig. 3A,B). By contrast, when a PBS gradient was applied at a similar distance from the OEC, no [Ca2+]i elevation was observed in either the leading process or the soma (Fig. 3C,D). The elevation of [Ca2+]i was abolished by pretreatment with thapsigargin, a drug that depletes the intracellular Ca2+ stores, or with 2-APB, an inhibitor of inositol (1,4,5)-trisphosphate [Ins(1,4,5)P3] receptors at the internal stores, but not by removing the extracellular Ca2+ (Fig. 3C,D). Furthermore, the repulsive effect of Slit-2 on OEC migration was totally blocked by bath application of thapsigargin or 2-APB, but not by removing the extracellular Ca2+ (Fig. 3E,F). Thus, Ca2+ release from internal stores through Ins(1,4,5)P3 channels is essential for both Slit-2-induced collapse and reversal of migration of OECs.
Fig. 3.
Intracellular Ca2+ release through the Ins(1,4,5)P3 channel is essential for Slit-2-induced collapse and reversal of migration of OECs. (A) Representative images showing [Ca2+] at various time points (in seconds) before and after frontal application of a Slit-2 gradient (arrowheads). The [Ca2+i]i was determined by the ratio of Fluo-4 to Flura-Red fluorescence and coded by pseudocolors in a linear scale. White arrowheads and arrows indicate the gradient direction and leading process and soma, respectively. Note the [Ca2+]i rise in response to Slit-2 at both the soma and the leading process. The selected sample regions indicate the leading process (1) and the soma (2) for quantification of [Ca2+]i signal. Scale bar: 20 μm. (B) Changes in [Ca2+]i at the leading process and the soma of OECs before and after exposure to a frontal Slit-2 gradient. The traces are based on averaging of [Ca2+]i signal of 8 cells at each time point. (C,D) Summary of average peak levels of Ca2+ signal based on the maximum [Ca2+]i signal from individual cells in the leading process (C) and the soma (D) under various conditions. (E,F) Average migration ratios of OECs (E) and the percentages of collapsed or reversed cells in total observed cells (F) in response to Slit-2 gradient under various conditions. Thaps, thapsigargin. Data are mean + s.e.m.; **P<0.01, Student's t-test.
Cytoskeletal reorganization induced by Slit-2
Because cytoskeletal reorganization is essential for cell motility (Suetsugu and Takenawa, 2003), we next examined the cytoskeletal reorganization in OECs after Slit-2 exposure by using immunofluorescent staining. As shown in Fig. 4, in untreated OECs, a thin layer of F-actin was evenly distributed in the peripheral region of the leading front, and F-actin bundles spanned the central region of the fan-like lamellipodia of the leading front and at the soma. By contrast, microtubules localized to the central domain of lamellipodia, parallel to the F-actin bundles, with a few microtubules penetrating the peripheral region of the leading front. After Slit-2 stimulation, F-actin in the peripheral region of the leading front disappeared rapidly (within 10 minutes), and F-actin density in the central domain of the lamellipodia reduced gradually. However, microtubules remained extending in the central and peripheral region of the lamellipodia before the full collapse of the leading front, even though F-actin had disappeared in the peripheral region (Fig. 4B,C). Thus, the F-actin in the peripheral region is more sensitive to the Slit-2 stimulation than microtubules, and the disruption of F-actin in the peripheral region of the leading front is implicated in initiating the collapse of leading front in OECs.
Fig. 4.
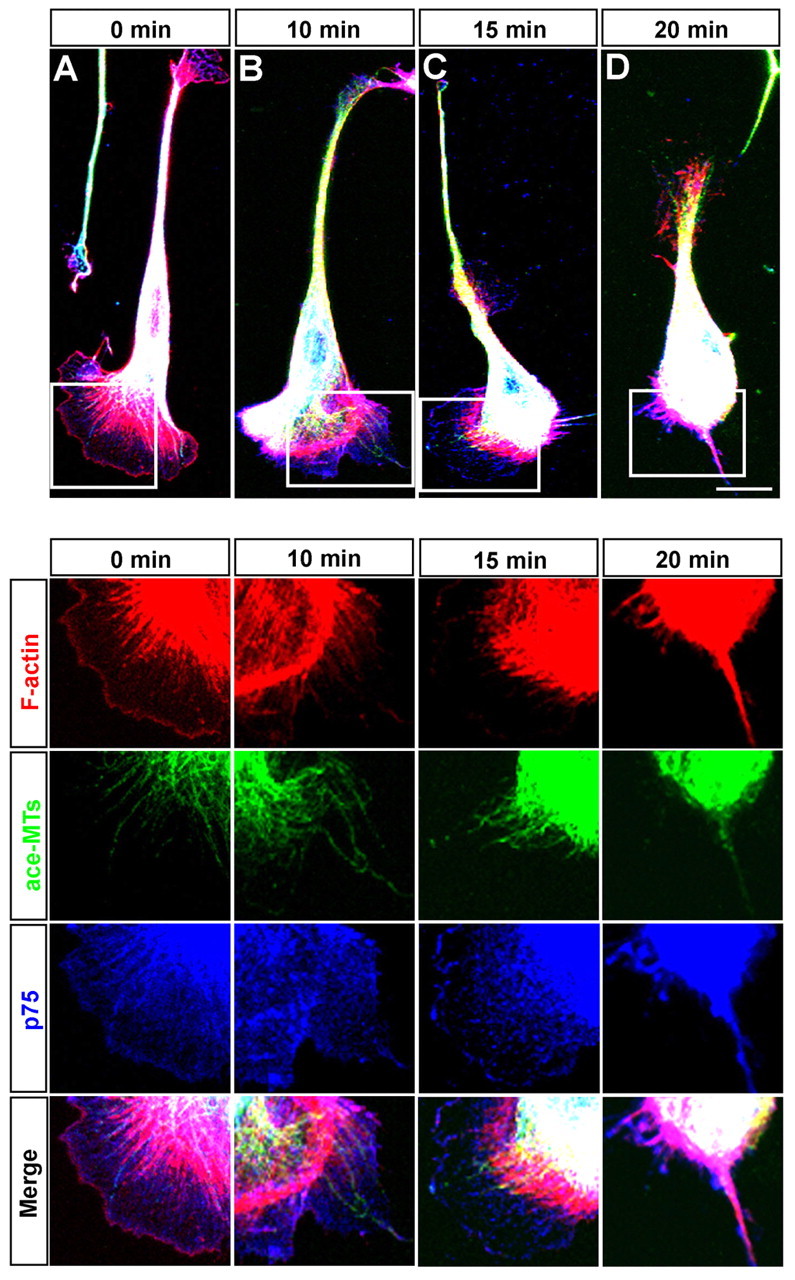
Slit-2 induces cytoskeletal reorganization in the leading front of OECs. OECs stimulated by Slit-2 for indicated times were labeled for F-actin (red), acetylated microtubules (ace-MTs) (green) and p75 (blue). Images of selected regions are shown below at higher magnification. Scale bar: 20 μm.
Cofilin activation through Ca2+–calcineurin signaling is required for Slit-2-induced collapse of the leading front and reversal of migration
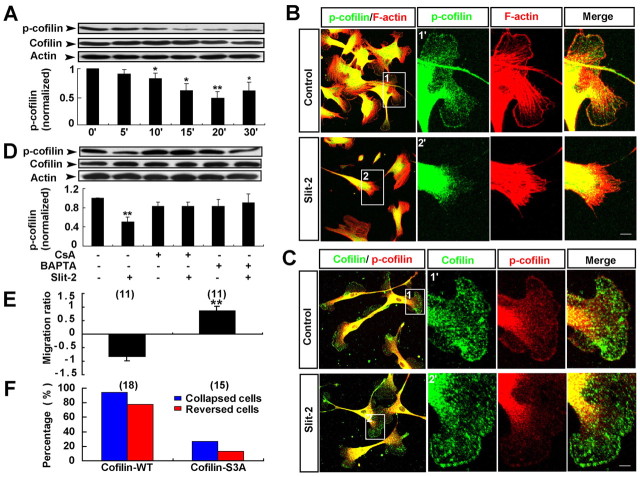
What causes the loss of F-actin in the peripheral region of the cell in response to Slit-2? Because cofilin is a key F-actin severing protein (Theriot, 1997), we therefore examined whether cofilin is involved in the Slit-2-induced loss of F-actin in the leading front of OECs. As shown in Fig. 5A, western blot analysis showed that stimulation of OECs with Slit-2 markedly decreased the level of phosphorylated cofilin (p-cofilin, the inactive form of cofilin) in a time-dependent manner, without affecting the total cofilin level. Immunofluorescent staining showed that in untreated OECs, the inactive p-cofilin was mainly localized at the soma and colocalized with F-actin at the peripheral region of the leading front. After Slit-2 incubation, the p-cofilin level in the whole cell dramatically decreased, and its signal in the peripheral region of the leading front disappeared, a process correlated with the loss of F-actin in the peripheral region (Fig. 5B). Furthermore, anti-p-cofilin and anti-cofilin double-staining showed that the p-cofilin signal rapidly decreased at the peripheral ruffles in Slit-2-stimulated OECs (10 minutes) before the full collapse of the leading front; however, cofilin signal still localized at the peripheral ruffles (Fig. 5C). These results suggest that Slit-2 activates cofilin by dephosphorylation at the peripheral region, which could be responsible for the disruption of F-actin at the peripheral region in response to Slit-2.
Fig. 5.
Activation of cofilin through Ca2+–calcineurin signaling is required for Slit-2-induced collapse and reversal of migration of OECs. (A) Western blot analysis for Slit-2-induced cofilin dephosphorylation and quantitative analysis of the ratio of p-cofilin to cofilin, normalized to the baseline level (0 minutes). (B) Immunocytochemical analysis of the distribution of p-cofilin (green) and F-actin (red) after Slit-2 treatment. Images of selected regions (1) and (2) are shown as (1′) and (2′) at higher magnification. (C) Immunocytochemical analysis of the distribution of cofilin (green) and p-cofilin (red) after Slit-2 treatment. (D) BAPTA-AM or CsA blocked the cofilin dephosphorylation triggered by Slit-2. (E,F) Average migration ratios (E) and the percentages of collapsed or reversed cells (F) in response to Slit-2 in total observed OECs transfected with GFP–cofilin-WT or GFP–cofilin-S3A. Data are mean + s.e.m.; *P<0.05, **P<0.01, Student's t-test. Scale bars: 20 μm.
To further examine whether the activation of cofilin depends on the Ca2+ signal, we added the intracellular Ca2+ chelator BAPTA-AM, or cyclosporin A (CsA), a specific inhibitor of the Ca2+- and calmodulin-dependent phosphatase (calcineurin), to the bath before Slit-2 treatment. Interestingly, the Slit-2-induced dephosphorylation of cofilin in OECs was blocked by either BAPTA-AM or CsA (Fig. 5D). These results suggest that Slit-2 activates cofilin in a Ca2+- and calcineurin-dependent manner.
To further examine the direct role of cofilin in Slit-2-induced collapse of the leading front, we transfected GFP-tagged wild-type cofilin (GFP–cofilin-WT) and a non-phosphorylatable and constitutively active mutant of cofilin (GFP–cofilin-S3A) (Arber et al., 1998) into OECs and analyzed the morphology and motility of OECs transfected with these constructs. After transfection of OECs with either GFP–cofilin-WT or GFP–cofilin-S3A, we did not observe obvious changes in cell morphology and F-actin organization, but observed marked elevation in migration speed compared with cells transfected with GFP alone (supplementary material Fig. S3A–D). When Slit-2 gradient was applied in front of migrating OECs transfected with GFP–cofilin-S3A, it failed to induce the collapse and reversal of migration of these cells. However, Slit-2 still induced the collapse and reversal of migration in OECs transfected with GFP–cofilin-WT (Fig. 5E,F; supplementary material Fig. S3E,F). Taken together, these results indicate that cofilin dephosphorylation in response to Slit-2 through the Ca2+–calcineurin signaling pathway is essential for Slit-2-induced collapse of the leading front.
RhoA is required for the reversal of migration triggered by Slit-2
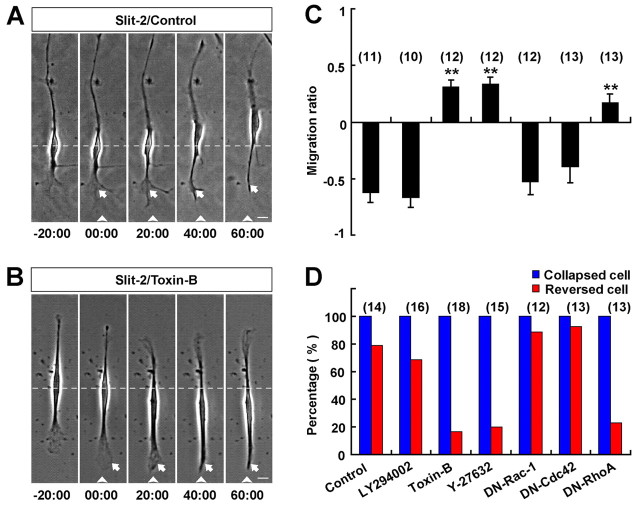
What causes the reversal of migration of OECs in response to Slit-2? Because RhoGTPases (RhoA, Cdc42, Rac1) play a key role in directional cell migration (Fukata et al., 2003) and the signal transduction of Slit in axon guidance (Wong et al., 2001), we next tested whether RhoGTPases are involved in the Slit-2-induced reversal of migration of OECs. In the presence of a general RhoGTPase inhibitor, toxin-B (10 ng/ml), the spontaneous migration of OECs was not affected (Fig. 6B). Frontal application of the Slit-2 gradient on these toxin-B-treated OECs still triggered the collapse of the leading front, but failed to reverse the migration of the soma, which stopped their forward migration after the full collapse of the leading front (Fig. 6B–D). By contrast, pretreatment with the specific phosphoinositide 3-kinase (PI3K) inhibitor LY-294002 (20 μM) did not affect Slit-2-induced repulsion of OECs (Fig. 6C–D). These results suggest that RhoGTPase activity is essential for the reversal of soma migration of OECs, but not for the collapse of leading process triggered by Slit-2.
Fig. 6.
RhoA activity is required for the reversal of migration triggered by Slit-2. (A,B) Images of migrating OECs before and after exposure to a Slit-2 gradient without (A) or with (B) toxin-B (10 ng/ml) incubation. White arrowheads and arrows indicate the direction of micropipette and the leading front of OECs, respectively. Scale bars: 20 μm. (C,D) Summary of average migration ratios (C) and the percentages of collapsed or reversed cells in total observed cells (D) in response to Slit-2 gradient under various conditions (see text for details). Data are mean + s.e.m.; **P<0.01, Student's t-test.
Further experiments were carried out in OECs transfected with constructs expressing fusion proteins of GFP and various dominant-negative (DN) forms of RhoGTPases. We selected single OECs of similar level of GFP fluorescence intensity for cell migration assays (supplementary material Fig. S4A,B). We found that in cultured OECs expressing DN-RhoA–GFP, frontal application of Slit-2 gradient failed to induce the reversal of migration, although the leading process still exhibited collapse and retraction (Fig. 6C,D; supplementary material Fig. S4E). By contrast, Slit-2-induced collapse and reversal were not affected in cells expressing DN-Rac1–GFP or DN-Cdc42–GFP (Fig. 6C,D; supplementary material Fig. S4C,D). Thus, RhoA is specifically required for the soma reversal. In support of this notion, we found that Slit-2-induced reversal of migration was abolished by bath incubation with Y-27632 (20 μM), a specific inhibitor of RhoA-dependent kinase (Rho kinase) (Fig. 6C,D), with the spontaneous migration of these OECs being unaffected.
To further elucidate the role of RhoA in the reversal of migration, we used pull-down assays to examine the activity of RhoA in cultured OECs. We found that treatment with either serum (10%) or the RhoA activating agent LPA (10 μM) (Yan et al., 2003) resulted in marked elevation of RhoA activity in these cultured OECs (Fig. 7A,B). Interestingly, bath application of Slit-2 caused a reduction of RhoA activity in a time-dependent manner (Fig. 7C,D), suggesting that the reversal in the direction of soma translocation might be related to the inhibition, rather than the activation, of RhoA. This notion was further supported by photometric analysis of the active RhoA in migrating OECs by using a FRET (fluorescence resonance energy transfer)-based biosensor pRichu-RhoA, which was constructed by linking CFP-conjugated RhoA and the YFP-conjugated RhoA-binding domain (RBD) of Rhotekin, a configuration sensitive to RhoA activation by guanine nucleotide exchange factor (Yoshizaki et al., 2003). As shown in Fig. 7E, the FRET signal for the active RhoA displayed a polarized distribution in a migrating OEC, with the leading front exhibiting higher activity than the soma and the trailing end. Application of a Slit-2 gradient in front of migrating OECs transfected with pRichu-RhoA led to a reduction in the FRET signal of RhoA activity in the leading front and a reversal of the polarity of active RhoA in this OEC (Fig. 7E,F). Taken together, these results suggest that downregulation of RhoA signaling is specifically required for the soma reversal of OECs triggered by Slit-2.
Fig. 7.
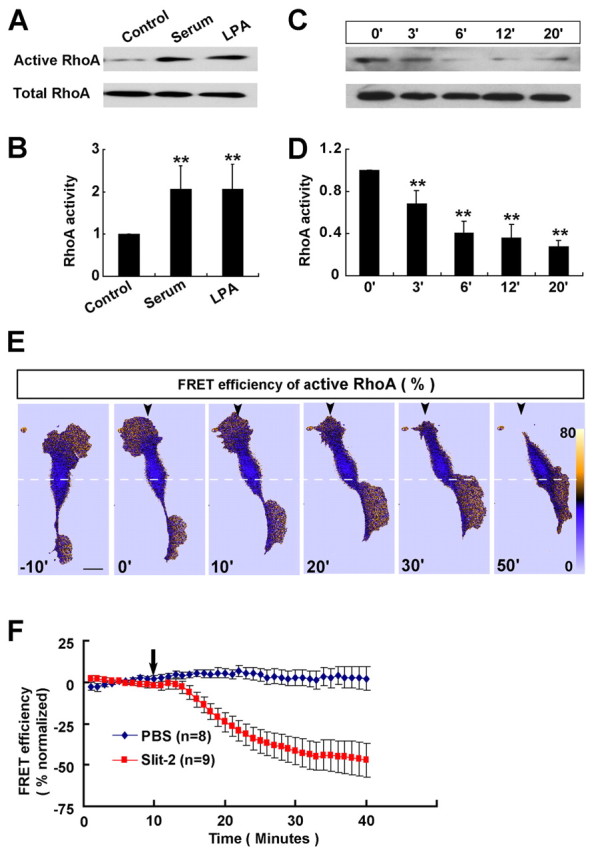
RhoA inhibition by Slit-2 is required for reversal of migration. (A–D) Representative western blots (A,C) and quantitative results (B,D) showing RhoA activity in response to treatment with serum (10%, 30 minutes), LPA (10 μM, 30 minutes) and Slit-2 (4 μg/ml, 0–20 minutes) in a pull-down assay. For quantitative analysis, the ratio of active RhoA to total RhoA was normalized to the baseline level (untreated control or 0 minutes). Data are mean + s.e.m.; **P<0.01, Student's t-test. (E) Representative FRET images (in pseudocolors) showing the inhibition and redistribution of active RhoA in response to a Slit-2 gradient. Scale bar: 20 μm. (F) Changes in the FRET signal at leading process of cultured OECs before and after the application of PBS or Slit-2 gradients.
Collapse of the leading front with simultaneous inhibition of RhoA triggers the reversal of soma translocation
To further examine how the coordination between the collapse of the leading front and the reversal of soma translocation in response to Slit-2 gradient is achieved, we applied a gradient of latrunculin A (LA) or cytochalasin D (CD), to mimic the Slit-2-triggered collapse of the leading front. After the application of a gradient of LA or CD (50 μM and 5 mM, respectively, in the pipette), the leading front of most OECs (15 out of 16 in CD; 14 out of 16 in LA) was inhibited in their motility and showed obvious collapse within 20 minutes. Only a few of these cells (4 out of 16 in 5 mM CD; 4 out of 16 in 50 μM LA) later reversed their soma translocation (Fig. 8B,E,F). Thus the collapse of the leading front does not seem to be sufficient to trigger the reversal of soma translocation, and other signals triggered by Slit-2 should be required.
Fig. 8.
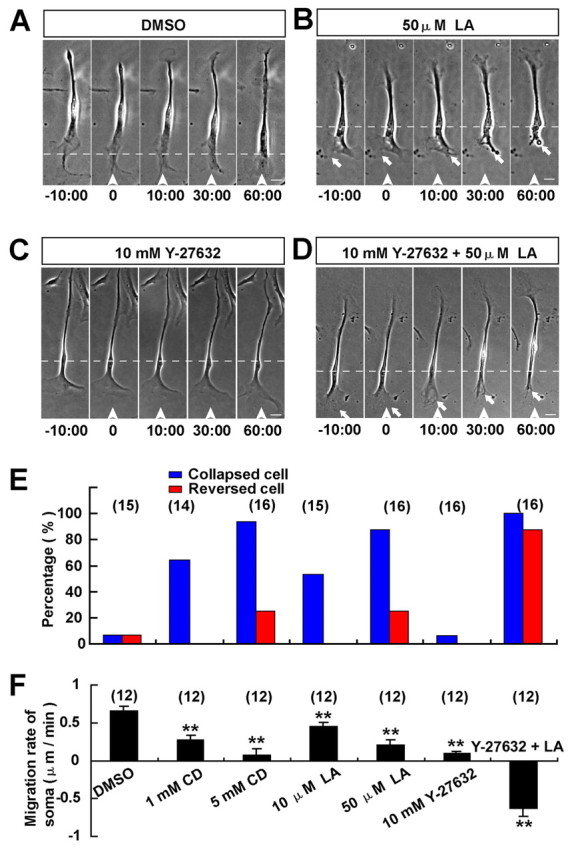
Collapse of leading front with simultaneous inhibition of RhoA triggers reversal of migration. (A–D) Images of migrating OECs before and after the frontal application of a gradient of DMSO (A), LA (B), Y-27632 (C), or Y-27632 and LA mixture (D). Scale bars: 20 μm. (E,F) Percentages of collapsed or reversed cells in total observed cells (E) and migration rates of the soma (F) after the frontal application of various drug gradients. Data are mean + s.e.m.; **P<0.01, Student's t-test.
We have shown that the Slit-2 gradient inhibits RhoA activity in OECs. To further examine whether the downregulation of RhoA activity in the leading front is sufficient to trigger the reversal of soma translocation, we applied a gradient of the Rho kinase inhibitor Y-27632 (10 mM in the pipette) in front of migrating OECs to mimic a gradient of RhoA inhibition across the cell. As shown in Fig. 8C,E,F, the Y-27632 gradient did not cause the collapse of the leading front nor the reversal of soma migration, but dramatically reduced the soma motility. Surprisingly, when we applied a gradient of the mixture of Y-27632 and LA in front of migrating OECs, most cells (14 out of 16) quickly reversed their migration after the collapse of the leading front (Fig. 8D–F), a process reminiscent of the collapse and reversal of migration triggered by Slit-2 (Fig. 2C). These results suggest that Slit-2 triggers a coordinated collapse of the leading front along with a gradient of RhoA inhibition across the cell to reverse OEC migration.
Discussion
It has been shown that the OB secretes some soluble factors that might attract OEC migration from the OE towards the OB (Liu et al., 1995). Another potential guidance mechanism for OEC migration is that the OE might secrete some repellants to promote OEC migration toward the OB during early development. Recent studies have shown that glial cell migration can also be directed by some axon guidance molecules (Tsai and Miller, 2002). Because OECs share a common origin (olfactory placode) with olfactory sensory neurons and migrate out from the OE to the OB together with olfactory sensory axons during the same developing period (Valverde et al., 1992), it is likely that guidance cues for olfactory sensory axons, like Slit (Cho et al., 2007; Nguyen-Ba-Charvet et al., 2008), might also guide the migration and distribution of OECs.
In the present study, we have shown that Robo proteins are expressed in cultured OECs and exhibit enriched distribution at the leading edge. A Slit-2 gradient indeed strongly repelled the migration of these cultured OECs. To our knowledge, this is the first guidance factor discovered to repel OEC migration. Because Slit-2 is highly expressed in the apical cells of OE, it is likely that it might help Robo-expressing OECs and olfactory axons migrate out of the OE through chemorepulsion during early development. Slits expressing in the OB might also regulate the stop and scattering of OECs that have arrived at the surface of the OB. OECs have been reported to pioneer the olfactory sensory nerves and provide a conductive substrate for the growth of olfactory sensory axons during development (Tennent and Chuah, 1996; Tisay and Key, 1999). An intriguing possibility is that the guidance of OECs by Slits might contribute to the guidance of axons because of the close interaction between neurons and glia. Extensive future studies using Slit and Robo mutant mice are needed to address these possibilities. Because multiple Slit and Robo family members are expressed in the developing olfactory system, double or even triple mutants of Slits and Robos might be required to clarify whether and how Slits guide the migration and distribution of OECs in developing olfactory system. Conditional knockout mutants with OEC-specific deletion of Robos are required to address whether Slit proteins could directly guide the migration of OECs in vivo and whether this guidance of OEC migration by Slit contributes to the pathfinding of olfactory sensory axons.
The intracellular signal transduction for Slit-2-induced collapse of the leading front of migrating cells has been largely unclear. Cultured OECs provide a good experimental system for addressing this issue because of their large cell size, active migration and high responsiveness to Slit-2. In the present work, we found that cofilin is a major downstream target of Slit-2 in triggering the collapse of the leading front of migrating OECs. ADF/cofilin family members are key regulators of F-actin dynamics and mediate the rapid turnover of F-actin by severing F-actin near the pointed ends (Bamburg, 1999; Moon and Drubin, 1995; Theriot, 1997). Recent studies have shown that cofilin has complicated regulation of F-actin dynamics and diverse effects on F-actin polymerization (Van Troys et al., 2008). Whether cofilin promotes F-actin assembly or disassembly depends upon the concentration of cofilin relative to actin and the relative concentrations of other actin-binding proteins. A low cofilin to actin ratio promotes actin disassembly, whereas a high ratio promotes actin assembly in vitro (Andrianantoandro and Pollard, 2006; Chan et al., 2009; Van Troys et al., 2008). During chemotaxis, spatial and temporal regulation of cofilin activity is required for cell directional migration (Mouneimne et al., 2006; Nishita et al., 2005). A previous study has shown that Slit-2 could induce the local synthesis of cofilin in Xenopus retinal growth cones (Piper et al., 2006). In our studies, we observed that stimulation of OECs with Slit-2 significantly decreased the p-cofilin level, without affecting the total cofilin level. These results suggest that Slit-2-induced collapse of OECs is mediated by changes of the ratio of p-coflin to non-p-cofilin, but not by an increase of total cofilin proteins in these migratory glial cells. We also found that cofilin proteins were mainly the phosphorylated inactive form at the peripheral ruffle of normal migratory OECs. Upon Slit-2 stimulation, cofilin was quickly activated at cell peripheral ruffles, a process that might be responsible for the disruption of F-actin in this region and the subsequent collapse of leading front. In support of this notion, F-actin at cell peripheral ruffles was lost upon Slit-2 stimulation (Fig. 4) and a gradient of CD or LA, which promotes F-actin de-polymerization, fully mimicked the Slit-2-induced collapse of the leading front (Fig. 8). Furthermore, overexpression of the constitutive active cofilin-S3A mutant could block the Slit-2-triggered collapse of OECs. The global presence of cofilin-S3A might set a new balance of F-actin turnover at a very high speed, override any local regulation of cofilin activity triggered by Slit-2 gradient, and thus abolish the Slit-2-induced collapse of the leading front of OECs. It is likely that similar signaling mechanisms might happen during Slit-triggered collapse and repulsion of neuronal growth cones. Interestingly, other repulsive guidance molecules, such as Semaphorin-3A, Semaphorin-3F, bone morphogenic protein (BMP) and myelin-associated inhibitors including Nogo-66, have been shown to trigger the collapse and repulsion through cofilin activation (Aizawa et al., 2001; Hsieh et al., 2006; Shimizu et al., 2008; Wen et al., 2007). Thus, activation of cofilin and the consequent fast severing of F-actin at the leading front might be a general molecular mechanism for the collapse of the leading front in response to different repulsive factors.
The F-actin-severing activity of cofilin can be regulated by reversible phosphorylation at the Ser3 residue, with the loss of activity upon phosphorylation. It has been shown that LIM kinases (LIMK) specifically phosphorylate cofilin at Ser3, whereas phosphatases Slingshot and calcineurin are capable of dephosphorylating it (Arber et al., 1998; Huang et al., 2006). Previous studies have shown that regulation of cofilin activity by LIMK and Slingshot is involved in regulating growth cone motility and morphology in response to extracellular cues, including Semaphorin-3A, BMP, brain-derived neurotrophic factor, and myelin-associated inhibitors (Aizawa et al., 2001; Gehler et al., 2004; Hsieh et al., 2006; Meberg and Bamburg, 2000; Wen et al., 2007). It is also shown that the Ca2+–calcineurin signaling pathway can dephosphorylate cofilin and mediate BMP-triggered repulsion of growth cones. Interestingly, calcineurin also promotes the activity of another cofilin-dephosphorylation enzyme Slingshot, which also directly de-phosphorylates and suppresses LIMK activity (Soosairajah et al., 2005; Wang et al., 2005; Wen et al., 2007). Thus, inhibition of LIMK activity is one potential mechanism for the reduction in cofilin phosphorylation upon activation of the Ca2+–calcineurin signaling pathway. Consistent with these previous reports, the present study showed that the Ca2+–calcineurin signaling pathway activated cofilin and mediated the collapsing effect of Slit-2 to the leading front of OECs. How Ca2+–calcineurin signaling pathway is activated and whether Slingshot and LIMK are involved in the cofilin activation by Slit remains to be further clarified.
RhoGTPases including Cdc42, Rac1 and RhoA can also regulate cofilin through regulation of LIMK activity by phosphorylation of its kinase activation loop, with Rac/Cdc42 acting through p21-activated kinases 1 and 4, Cdc42 through mytotonic-dystrophy-related cdc42-binding kinase, and RhoA though ROCK (Dan et al., 2001; Edwards et al., 1999; Ohashi et al., 2000; Sumi et al., 2001). However, our study showed that none of the dominant negative mutants of Cdc42, Rac1 and RhoA could block Slit-2-triggered collapse of the leading front in OECs. Thus, RhoGTPases do not play a significant role in cofilin regulation and the leading front collapse in response to Slit-2 in these OECs. Instead, the Ca2+–calcineurin signaling pathway should be the major downstream event that leads to the cofilin activation and the consequent collapse of the leading front in response to Slit-2 stimulation.
In the present study, we found that collapse of the leading front is necessary, but not sufficient, to trigger the reversal of soma translocation, suggesting that other signals independent of the collapse of the leading front should be activated by Slit-2 to reverse the soma translocation. As discussed below, this signal might be a gradient of RhoA inhibition. RhoGTPases act spatially and temporally to control cell migration by regulating cytoskeletal reorganization. Rac1 is known to promote the extension of lamellipodia, and the dominant negative mutant of Rac1 is often used to abolish the formation of lamellipodia. Cdc42 is known to promote the extension of filopodia. By contrast, RhoA is commonly considered to regulate cell contractility at the cell body and contributes to the retraction of the trailing end of migrating cells (Etienne-Manneville and Hall, 2002). However, increasing evidence has shown that active RhoA distributes in the leading front. In free migrating neurons, fibroblasts and MDCK cells, the active RhoA displays a polarized distribution with highest activity in the front part of the cell rather than in the rear (Guan et al., 2007; Kurokawa and Matsuda, 2005; Pertz et al., 2006) and might be essential for the re-orientation and stabilization of the direction of cell migration (Palazzo et al., 2001; Wen et al., 2004). Some studies report that Slit activates the activity of RhoA and Rac1, but inactivates Cdc42 (Fan et al., 2003; Wong et al., 2001; Yang and Bashaw, 2006). However, other studies show that Slit inhibits the activity of RhoA, Rac1 and Cdc42 (Guan et al., 2007; Liu et al., 2006; Prasad et al., 2007; Tole et al., 2009; Wong et al., 2001).
RhoGTPases are known to be regulated by a large group of GTPase activating proteins (GAPs), guanine nucleotide exchange factors (GEFs) and guanine nucleotide dissociation inhibitors (GDIs) that selectively target different GTPases (Etienne-Manneville and Hall, 2002). It is likely that different type of cells express different combination of GAPs, GEFs and GDIs, resulting in the differential effects on RhoGTPase activity and cell motility in response to Slit. In the present study, we found that RhoA was inhibited by Slit-2 in cultured OECs. Because inhibition of RhoA activity was specifically required for Slit-2-induced reversal of migration, but not for the collapse of leading front, the collapse of the leading front and the reversal of soma translocation are probably two related events that involve distinct intracellular signals. We found that free migrating OECs exhibited a front-high and back-low polarity of active RhoA. In response to a frontal gradient of Slit-2, this polarity of active RhoA was gradually reversed. Moreover, a gradient of inhibition of RhoA signaling across the cell by a gradient of Y-27632 suppressed the soma motility, but was not sufficient to trigger the reversal of soma translocation. Interestingly, a gradient of Y-27632 together with LA in front of migrating OECs fully mimicked the collapse and reversal of migration of OECs trigged by Slit-2. These results suggest that a gradient of RhoA inhibition across the cell in response to a Slit-2 gradient might reverse the intrinsic polarity of the cell, but the reversal of soma migration depends on the coordination of a reversal of the cell polarity and a simultaneous collapse of the leading process. These two coordinated processes are likely to allow intracellular proteins that are anchored at the original leading front to be released and redistributed to the original tail (Fig. 9). This could be a general mechanism for the repulsive guidance of the migration of different cell types, because frontal application of Y-27632 together with LA also reliably triggered the collapse and reversal of migration of another cell type, cultured Schwann cells (supplementary material Fig. S5).
Fig. 9.
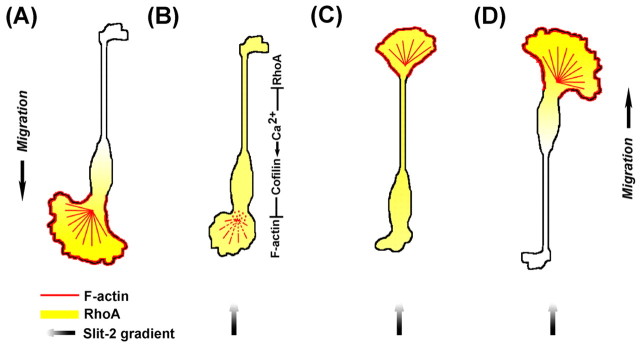
Model of Slit-2-induced collapse and reversal of migration of OECs. (A) Spontaneously migrating OEC exhibits a front–rear gradient in the distribution of RhoA. (B–D) Frontal exposure of a Slit-2 gradient triggers an elevation of [Ca2+]i at the leading front and soma, leading to the activation of cofilin, whose activity is responsible for the disruption of F-actin and the collapse of leading front. Reversal of the polarity of OECs requires both the gradient of RhoA inhibition across the cell and the collapse of the leading front. Colors indicate F-actin (red), RhoA (yellow) and Slit-2 gradient (black).
Materials and Methods
Primary culture and OEC purification
Primary OEC cultures were prepared from the OB of adult male Sprague-Dawley rats and purified by differential cell adhesiveness as described previously (Huang et al., 2008). Briefly, the olfactory nerve layer was peeled away from the rest of OB, dissociated with 0.25% trypsin (Sigma, St Louis, MO) at 37°C for 15 minutes. Tissues were triturated using a Pasteur pipette and plated on an uncoated 25 cm2 culture flask twice; each sample was then incubated for 36 hours at 37°C in 5% CO2. Non-adhesive cell suspension was collected, seeded onto 35 mm dishes (Corning, NY) coated with poly-L-lysine (0.1mg/ml, Sigma), and incubated with DMEM/F-12 (1:1, vol/vol; DF12, Gibco, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT), 2 μM forskolin (Sigma) and 10 ng/ml bFGF (Sigma). The overall purity of OECs was around 95%. Primary cultures of OECs from OE of P0 Sprague-Dawley rats were prepared as described previously (Au and Roskams, 2003).
RT–PCR and western blotting
For RT-PCR, total RNA was extracted from cultures of purified OECs with Trizol reagent (Invitrogen, Gaithersburg, MD), converted to cDNA using the Revert Aid First Strand cDNA Synthesis Kit (MBI Fermentas, Hanover, MD), and one twentieth of the product was used in 20 μl PCR reactions. Primers for Robos were designed and the specificity of primers was measured by Primer Premier 5.0 software. Robo1, forward, 5′-TGCAGCAGCCGCCGAGTATG-3′, reverse, 5′-ATTATAGGCCGTGCCTCCAG-3′; Robo2, forward, 5′-CAACACTCTGGTAACGTGGA-3′, reverse, 5′-ATCTCAGCCTTCCGAGGA-3′; and Robo3, forward, 5′-CACCATACGTGGAGGAAAGC-3′, reverse, 5′-GGGCTGGGGATCTCCCTGCA-3′.
To detect changes in phosphorylated cofilin, cultures of purified OECs were serum-starved for 8–10 hours before Slit-2 (4 ng/ml) treatments, then were lysed in 0.2 ml lysis buffer containing protease inhibitor mixture set I (Calbiochem, La Jolla, CA). Proteins were separated by 12% SDS-PAGE gel electrophoresis and transferred onto the polyvinylidene difluoride (PVDF) membrane. Blotted membranes were blocked for 3 hours at room temperature in 5% milk and incubated with anti-cofilin (1:1000; Cell Signaling Technology, Beverly, MA) or anti-p-cofilin (Ser3, 1:1000; Cell Signaling) overnight at 4°C, rinsed and incubated for 1 hour at room temperature with a horseradish-peroxidase-conjugated antibody against rabbit IgG (1:10,000; BioRad, Hercules, CA). Chemiluminescent detection was performed with the ECL kit (Pierce, Rockford, IL). β-actin as a loading control was detected alongside the experimental samples (1:3000; Chemicon, Temecula, CA). For detection of Robo1 and Slit-2 by western blotting, 8% SDS-PAGE gel was used, with 1:200 anti-Robo1 antibody (R&D, Cat. AF1749) and 1:500 anti-Slit-2 antibody (Chemicon, Cat. AB5701), respectively. For detection of the expression of mutant RhoGTPases in cultured OECs after transfection, 12% SDS-PAGE gel and 1:1000 anti-GFP antibody (Molecular Probes) were used.
Pull-down assay of active RhoA
For analyzing RhoA activity in cell lysates, an activated RhoA pull-down kit was used following protocols provided by the manufacturer (Cytoskeleton, Denver, CO). Briefly, OEC cultures were starved overnight, then stimulated by Slit-2 before being lysed in 250 μl of the supplied lysis buffer containing protease inhibitor cocktail. About 20 μl of each lysate was used for protein quantification and western blotting analysis of total RhoA. For the rest of lysates, a volume of equal protein amounts from each sample were incubated with Rhotekin-RBD affinity beads for 1 hour at 4°C, followed by two washes in the wash buffer. Bound proteins were collected and examined by 12% SDS-PAGE for western blotting analysis.
Single-cell migration assays
Slit-2 was purified as described previously (Guan et al., 2007) from conditioned medium collected from a cultured HEK293 cell line stably expressing human Slit-2–myc. Single OEC migration assay and quantitative analysis were carried out as described previously (Huang et al., 2008). Briefly, purified OECs were replated onto coverslips coated with laminin (10 μg/ml) at a low density (1000 cells per coverslip), and were used for experiments in serum-free Leibovitz's L-15 medium (Gibco) 12–24 hours after plating. Experiments were carried out on a heated stage (37°C) under a phase contrast microscope (CK40, Olympus Optial, Tokyo, Japan). The micropipette tip (1 μm opening) was placed at about 100 μm away from the cell center. A standard pressure pulse of 3 psi was applied (2 Hz, 20 milliseconds duration) (Lohof et al., 1992). Images were recorded in a time-lapse mode (1 picture every 5 minutes) with a CCD camera (JVC TK-1381; Victor Company, Yokohama, Japan) attached to the microscope, using Scion Image Software (Frederock, MD). For pharmacological treatments, cells were pre-incubated with thapsigargin (1 μM; Alamone Labs), 2-APB (100 μM; Calbiochem), Y-27632 (20 μM; Calbiochem), or LY-294002 (20 μM; Calbiochem) for at least 30 minutes, or with toxin-B (10 ng/ml; Calbiochem) overnight. Drugs existed in the medium throughout the migration assay.
Plasmids and cell transfection
GFP-tagged cofilin-WT and cofilin-S3A were constructed by subcloning the full-length cofilin or cofilin-S3A into pEGFP vector (Clontech) between BamHI and HinDIII sites. Robo2–GFP, DN-RhoA–GFP, DN-Rac-1–GFP and DN-Cdc-42–GFP plasmids were described previously (Guan et al., 2007). The Robo1-specific siRNA sequences are: sense, 5′-GCAACAGGATGAATTAGAA-3′ and antisense 5′-CGTTGTCCTACTTAATCTT-3′. For OEC transfection, we used rat Astrocyte Nucleofector Kit (Amaxa, Cologne, Germany) according to the manufacturer's instructions (program T-20).
Immunostaining
For brain tissue staining, embryonic brains were directly removed and fixed with 4% paraformaldehyde (PFA) at E16. Sagittal brain sections of 20 μm were cut on a freezing microtome and immediately processed for immunostaining by 1 hour blocking in 5% BSA plus 0.3% Triton X-100 at room temperature, overnight incubation with primary antibodies at 4°C, and for 1 hour at room temperature incubation with appropriate secondary antibodies (1:1000; Molecular Probes, Eugene, OR). The primary antibodies used were anti-p75 (1:500; Promega, Madison, WI), anti-Slit-2 (1:200; Chemicon) and anti-Robo1 (1:200; R&D) (Farmer et al., 2008; Marlow et al., 2008; Nguyen-Ba-Charvet et al., 2008). Sections were counterstained for Toto-3 (1:1000; Molecular Probes) to visualize the nucleus. For OEC staining, cultured OECs were fixed with fresh 4% PFA for 20 minutes. The primary antibodies were anti-p-cofilin (1:200; Cell Signaling), p75 (1:500; Promega), cofilin (1:200; Santa Cruz Biotechnology), Slit-2 (1:200; Chemicon) or acetylated tubulin (1:1000; Sigma). For visualization of F-actin, cells were incubated with rhodamine-conjugated phalloidin (1:60; Molecular Probes) at room temperature for 1 hour. Images were acquired on an Olympus FV-1000 confocal system using a multitrack configuration and processed using Adobe Photoshop CS 8.0.
Calcium imaging
Calcium imaging was performed as described previously (Guan et al., 2007). Briefly, OECs were rinsed three times before being loaded with Fluo-4 AM and Fura-Red AM (2 μM, Molecular Probes, Eugene, OR), with 0.1% dimethyl sulphoxide (DMSO) in the extracellular medium (ECM), for 30 minutes at 37°C, and then incubated for another 30 minutes after being rinsed three times with ECM. Imaging was performed at room temperature on a Zeiss LSM-510 confocal microscope with a 40× 1.30 NA oil objective (Zeiss Plan-Neofluar). Cells were excited with a laser at 488 nm, and the intensity of the emission between 505 and 550 nm was measured as the Fluo-4 signal; emission >635 nm was simultaneously detected as the Fura-Red signal. Images were acquired at 5-second intervals. The Ca2+ signals were measured as the change in the ratio of Fluo-4 to Fura-Red, relative to the baseline before applying Slit-2 (ΔR/R0), in selected regions using Zeiss LSM-510 software. The leading process and soma in OECs were defined as described in a previous work (Huang et al., 2008). To quantify the Ca2+ signal at the leading process, the fluorescence signal in the whole leading process was measured (Fig. 3A). In some experiments, cells were incubated in ECM containing pharmacological agents for at least 30 minutes before measurement.
FRET-based imaging of active RhoA with three-channel microscopy
The FRET probe pRaichu-RhoA (YFP–RBD-RhoA–CFP) for monitoring the subcellular RhoA activity was kindly provided by Michiyuki Matsuda (Osaka University, Osaka, Japan) (Yoshizaki et al., 2003). Cells transfected with the FRET probe were imaged on a Nikon Ti microscope with a 40× oil lens (1.30 NA) using the Perfect Focus System and were illuminated by a polychrome IV monochromator (TILL Photonics). Filter sets for FRET imaging were CFP (excitation 436 nm; emission, 480/40 nm HQ, DM 455 nm), FRET (excitation 436 nm; emission, 535/30 nm HQ, DM 515 nm) and YFP (excitation 510 nm; emission, 535/30 nm HQ, DM 515 nm). Images of the three channels were recorded simultaneously by using the Cascade 512B CCD (Roper Scientific). Background images were subtracted from the raw images before carrying out FRET calculation. Corrected FRET (FRETC) was calculated on a pixel-by-pixel basis for the image using the following equation: FRETC=FRET–a×YFP–b×CFP, where FRET, CFP and YFP corresponded to background-subtracted images, acquired through the FRET, CFP and YFP channels, respectively. a and b were the fraction of bleed-through of YFP and CFP fluorescence through the FRET channel, respectively, and the two values were determined by using cells transfected with YFP or CFP alone. We used the following equation: E=FRETC /(CFP+FRETC)×100% to quantify the FRET signal by using MetaFluo and NIH ImageJ software (PixFRET Plug-in) (Feige et al., 2005). To quantify the Slit-2 effect, the change in FRET signals at the leading process were measured using ImageJ software and normalized to the baseline level, presented as ΔE/E0 (E0 is the average FRET signal before Slit-2 application).
Statistical analysis
All data presented represent results from at least three independent experiments. Statistical analysis was performed using Student's t-test with two tails, the Kolmogorov–Smirnov test or ANOVA with pair-wise comparisons.
Acknowledgements
We thank C.-b. Guan and Q. Hu for technical support in imaging studies; Yi Rao (Washington University School of Medicine, St Louis, MI) for providing the Slit-2-myc cell line; Pico Caroni (Freidrich Miescher Institute, Basel) for cofilin constructs; and Michiyuki Matsuda (Osaka University, Osaka, Japan) for pRaichu-RhoA probe. This study was supported by National Key Basic Research Program (2006CB500702, 2006CB806600, 2007CB947100), Ministry of Science and Technology of China (2009ZX09311-001) and National Natural Science Foundation (30770657, 30625023).
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/2/186/DC1
References
- Aizawa H., Wakatsuki S., Ishii A., Moriyama K., Sasaki Y., Ohashi K., Sekine-Aizawa Y., Sehara-Fujisawa A., Mizuno K., Goshima Y., et al. (2001). Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat. Neurosci. 4, 367-373 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13-23 [DOI] [PubMed] [Google Scholar]
- Arber S., Barbayannis F. A., Hanser H., Schneider C., Stanyon C. A., Bernard O., Caroni P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805-809 [DOI] [PubMed] [Google Scholar]
- Au E., Roskams A. J. (2003). Olfactory ensheathing cells of the lamina propria in vivo and in vitro. Glia 41, 224-236 [DOI] [PubMed] [Google Scholar]
- Bamburg J. R. (1999). Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185-230 [DOI] [PubMed] [Google Scholar]
- Brose K., Bland K. S., Wang K. H., Arnott D., Henzel W., Goodman C. S., Tessier-Lavigne M., Kidd T. (1999). Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795-806 [DOI] [PubMed] [Google Scholar]
- Cao L., Liu L., Chen Z. Y., Wang L. M., Ye J. L., Qiu H. Y., Lu C. L., He C. (2004). Olfactory ensheathing cells genetically modified to secrete GDNF to promote spinal cord repair. Brain 127, 535-549 [DOI] [PubMed] [Google Scholar]
- Chan C., Beltzner C. C., Pollard T. D. (2009). Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. H., Lepine M., Andrews W., Parnavelas J., Cloutier J. F. (2007). Requirement for Slit-1 and Robo-2 in zonal segregation of olfactory sensory neuron axons in the main olfactory bulb. J. Neurosci. 27, 9094-9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah M. I., West A. K. (2002). Cellular and molecular biology of ensheathing cells. Microsc. Res. Tech. 58, 216-227 [DOI] [PubMed] [Google Scholar]
- Dan C., Kelly A., Bernard O., Minden A. (2001). Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 276, 32115-32121 [DOI] [PubMed] [Google Scholar]
- Dickson B. J., Gilestro G. F. (2006). Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu. Rev. Cell Dev. Biol. 22, 651-675 [DOI] [PubMed] [Google Scholar]
- Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. (1999). Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1, 253-259 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. (2002). Rho GTPases in cell biology. Nature 420, 629-635 [DOI] [PubMed] [Google Scholar]
- Fan X., Labrador J. P., Hing H., Bashaw G. J. (2003). Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron 40, 113-127 [DOI] [PubMed] [Google Scholar]
- Farmer W. T., Altick A. L., Nural H. F., Dugan J. P., Kidd T., Charron F., Mastick G. S. (2008). Pioneer longitudinal axons navigate using floor plate and Slit/Robo signals. Development 135, 3643-3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J. N., Sage D., Wahli W., Desvergne B., Gelman L. (2005). PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 68, 51-58 [DOI] [PubMed] [Google Scholar]
- Fukata M., Nakagawa M., Kaibuchi K. (2003). Roles of Rho-family GTPases in cell polarisation and directional migration. Curr. Opin. Cell Biol. 15, 590-597 [DOI] [PubMed] [Google Scholar]
- Gehler S., Shaw A. E., Sarmiere P. D., Bamburg J. R., Letourneau P. C. (2004). Brain-derived neurotrophic factor regulation of retinal growth cone filopodial dynamics is mediated through actin depolymerizing factor/cofilin. J. Neurosci. 24, 10741-10749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C. B., Xu H. T., Jin M., Yuan X. B., Poo M. M. (2007). Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell 129, 385-395 [DOI] [PubMed] [Google Scholar]
- Hsieh S. H., Ferraro G. B., Fournier A. E. (2006). Myelin-associated inhibitors regulate cofilin phosphorylation and neuronal inhibition through LIM kinase and Slingshot phosphatase. J. Neurosci. 26, 1006-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. Y., DerMardirossian C., Bokoch G. M. (2006). Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18, 26-31 [DOI] [PubMed] [Google Scholar]
- Huang Z. H., Wang Y., Cao L., Su Z. D., Zhu Y. L., Chen Y. Z., Yuan X. B., He C. (2008). Migratory properties of cultured olfactory ensheathing cells by single-cell migration assay. Cell Res. 18, 479-490 [DOI] [PubMed] [Google Scholar]
- Kidd T., Bland K. S., Goodman C. S. (1999). Slit is the midline repellent for the robo receptor in Drosophila. Cell 96, 785-794 [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Matsuda M. (2005). Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol. Biol. Cell 16, 4294-4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Field P. M., Raisman G. (1998). Regeneration of adult rat corticospinal axons induced by transplanted olfactory ensheathing cells. J. Neurosci. 18, 10514-10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Hou J., Hu X., Wang X., Xiao Y., Mou Y., De Leon H. (2006). Neuronal chemorepellent Slit2 inhibits vascular smooth muscle cell migration by suppressing small GTPase Rac1 activation. Circ. Res. 98, 480-489 [DOI] [PubMed] [Google Scholar]
- Liu K. L., Chuah M. I., Lee K. K. (1995). Soluble factors from the olfactory bulb attract olfactory Schwann cells. J. Neurosci. 15, 990-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohof A. M., Quillan M., Dan Y., Poo M. M. (1992). Asymmetric modulation of cytosolic cAMP activity induces growth cone turning. J. Neurosci. 12, 1253-1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillat V., Cases O., Nguyen-Ba-Charvet K. T., Tessier-Lavigne M., Sotelo C., Chedotal A. (2002). Spatiotemporal expression patterns of slit and robo genes in the rat brain. J. Comp. Neurol. 442, 130-155 [DOI] [PubMed] [Google Scholar]
- Marlow R., Strickland P., Lee J. S., Wu X., Pebenito M., Binnewies M., Le E. K., Moran A., Macias H., Cardiff R. D., et al. (2008). SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res. 68, 7819-7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg P. J., Bamburg J. R. (2000). Increase in neurite outgrowth mediated by overexpression of actin depolymerizing factor. J. Neurosci. 20, 2459-2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A., Drubin D. G. (1995). The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol. Biol. Cell 6, 1423-1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. (2006). Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16, 2193-2205 [DOI] [PubMed] [Google Scholar]
- Nguyen-Ba-Charvet K. T., Di Meglio T., Fouquet C., Chedotal A. (2008). Robos and slits control the pathfinding and targeting of mouse olfactory sensory axons. J. Neurosci. 28, 4244-4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., Mizuno K. (2005). Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 171, 349-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K., Nagata K., Maekawa M., Ishizaki T., Narumiya S., Mizuno K. (2000). Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J. Biol. Chem. 275, 3577-3582 [DOI] [PubMed] [Google Scholar]
- Palazzo A. F., Cook T. A., Alberts A. S., Gundersen G. G. (2001). mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3, 723-729 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006). Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440, 1069-1072 [DOI] [PubMed] [Google Scholar]
- Piper M., Anderson R., Dwivedy A., Weinl C., van Horck F., Leung K. M., Cogill E., Holt C. (2006). Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron 49, 215-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Qamri Z., Wu J., Ganju R. K. (2007). Slit-2/Robo-1 modulates the CXCL12/CXCR4-induced chemotaxis of T cells. J. Leukoc. Biol. 82, 465-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisman G., Li Y. (2007). Repair of neural pathways by olfactory ensheathing cells. Nat. Rev. Neurosci. 8, 312-319 [DOI] [PubMed] [Google Scholar]
- Ramon-Cueto A., Plant G. W., Avila J., Bunge M. B. (1998). Long-distance axonal regeneration in the transected adult rat spinal cord is promoted by olfactory ensheathing glia transplants. J. Neurosci. 18, 3803-3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Tear G., Ferres-Marco D., Goodman C. S. (1993). Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron 10, 409-426 [DOI] [PubMed] [Google Scholar]
- Shimizu A., Mammoto A., Italiano J. E., Jr, Pravda E., Dudley A. C., Ingber D. E., Klagsbrun M. (2008). ABL2/ARG tyrosine kinase mediates SEMA3F-induced RhoA inactivation and cytoskeleton collapse in human glioma cells. J. Biol. Chem. 283, 27230-27238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosairajah J., Maiti S., Wiggan O., Sarmiere P., Moussi N., Sarcevic B., Sampath R., Bamburg J. R., Bernard O. (2005). Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J. 24, 473-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S., Takenawa T. (2003). Regulation of cortical actin networks in cell migration. Int. Rev. Cytol. 229, 245-286 [DOI] [PubMed] [Google Scholar]
- Sumi T., Matsumoto K., Shibuya A., Nakamura T. (2001). Activation of LIM kinases by myotonic dystrophy kinase-related Cdc42-binding kinase alpha. J. Biol. Chem. 276, 23092-23096 [DOI] [PubMed] [Google Scholar]
- Tennent R., Chuah M. I. (1996). Ultrastructural study of ensheathing cells in early development of olfactory axons. Brain Res. Dev. Brain Res. 95, 135-139 [DOI] [PubMed] [Google Scholar]
- Theriot J. A. (1997). Accelerating on a treadmill: ADF/cofilin promotes rapid actin filament turnover in the dynamic cytoskeleton. J. Cell Biol. 136, 1165-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisay K. T., Key B. (1999). The extracellular matrix modulates olfactory neurite outgrowth on ensheathing cells. J. Neurosci. 19, 9890-9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S., Mukovozov I. M., Huang Y. W., Magalhaes M. A., Yan M., Crow M. R., Liu G. Y., Sun C. X., Durocher Y., Glogauer M., et al. (2009). The axonal repellent, Slit2, inhibits directional migration of circulating neutrophils. J. Leukoc. Biol. 86, 1403-1415 [DOI] [PubMed] [Google Scholar]
- Tsai H. H., Miller R. H. (2002). Glial cell migration directed by axon guidance cues. Trends Neurosci. 25, 173-175; discussion 175-176 [DOI] [PubMed] [Google Scholar]
- Valverde F., Santacana M., Heredia M. (1992). Formation of an olfactory glomerulus: morphological aspects of development and organization. Neuroscience 49, 255-275 [DOI] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008). Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649-667 [DOI] [PubMed] [Google Scholar]
- Vincent A. J., West A. K., Chuah M. I. (2005). Morphological and functional plasticity of olfactory ensheathing cells. J. Neurocytol. 34, 65-80 [DOI] [PubMed] [Google Scholar]
- Wang Y., Shibasaki F., Mizuno K. (2005). Calcium signal-induced cofilin dephosphorylation is mediated by Slingshot via calcineurin. J. Biol. Chem. 280, 12683-12689 [DOI] [PubMed] [Google Scholar]
- Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004). EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820-830 [DOI] [PubMed] [Google Scholar]
- Wen Z., Han L., Bamburg J. R., Shim S., Ming G. L., Zheng J. Q. (2007). BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. J. Cell Biol. 178, 107-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Ren X. R., Huang Y. Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L., et al. (2001). Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107, 209-221 [DOI] [PubMed] [Google Scholar]
- Wu W., Wong K., Chen J., Jiang Z., Dupuis S., Wu J. Y., Rao Y. (1999). Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature 400, 331-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. T., Yuan X. B., Guan C. B., Duan S., Wu C. P., Feng L. (2004). Calcium signaling in chemorepellant Slit2-dependent regulation of neuronal migration. Proc. Natl. Acad. Sci. USA 101, 4296-4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Lu D., Rivkees S. A. (2003). Lysophosphatidic acid regulates the proliferation and migration of olfactory ensheathing cells in vitro. Glia 44, 26-36 [DOI] [PubMed] [Google Scholar]
- Yang L., Bashaw G. J. (2006). Son of sevenless directly links the Robo receptor to rac activation to control axon repulsion at the midline. Neuron 52, 595-607 [DOI] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R. E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. (2003). Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162, 223-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Zhou L., Chen J. H., Wu J. Y., Rao Y., Ornitz D. M. (1999). The mouse SLIT family: secreted ligands for ROBO expressed in patterns that suggest a role in morphogenesis and axon guidance. Dev. Biol. 212, 290-306 [DOI] [PubMed] [Google Scholar]