Abstract
N-acylethanolamine-hydrolyzing acid amidase (NAAA) is a lysosomal enzyme that primarily degrades palmitoylethanolamine (PEA), a lipid amide that inhibits inflammatory responses. We developed a HEK293 cell line stably expressing the NAAA pro-enzyme (zymogen) and a single step chromatographic purification of the protein from the media. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry MALDI-TOF MS analysis of the zymogen (47.7 kDa) treated with Peptide-N-Glycosidase F (PNGase F) identified 4 glycosylation sites, and acid cleavage of the zymogen into α- and β-subunits (14.6 and 33.3 kDa) activated the enzyme. Size exclusion chromatography estimated the mass of the active enzyme as 45 ± 3 kDa, suggesting formation of an α/β heterodimer. MALDI-TOF MS fingerprinting covered more than 80% of the amino acid sequence, including the N-terminal peptides, and evidence for the lack of a disulfide bond between subunits. The significance of the cysteine residues was established by their selective alkylation resulting in almost complete loss of activity. The purified enzyme was kinetically characterized with PEA and a novel fluorogenic substrate, N-(4-methyl coumarin) palmitamide (PAMCA). The production of sufficient quantities of NAAA and a high throughput assay could be useful in discovering novel inhibitors and determining the structure and function of this enzyme.
Keywords: Endocannabinoid, Glycosylation, Lysosomal enzyme, Mannose-6-phosphate, N-acylethanolamine, N-acylethanolamine-hydrolyzing acid amidase (NAAA), N-terminal nucleophile hydrolase, Palmitoylethanolamine (PEA)
Introduction
Interest in the recently cloned lysosomal enzyme NAAA has increased of late,1-3 driven in part by studies showing that PEA has significant anti-inflammatory, analgesic, and neuroprotective properties.4-6 These effects have been traced at least partially to its potent endogenous agonism of the peroxisome proliferator-activated receptor-α (PPAR-α).7, 8
The potential of blocking N-acylethanolamines (NAEs) like PEA or N-arachidonlyethanolamine (anandamide) from enzymatic degradation via inhibition as a strategy for pain treatment has received considerable interest of late.3 For example, another enzyme known to hydrolyze PEA, fatty acid amide hydrolase (FAAH), primarily hydrolyzes the endocannabinoid anandamide, and inhibitors of this enzyme have been the subjects of clinical trials.9
Although functionally very similar to FAAH in its ability to hydrolyze the biologically significant NAEs anandamide and PEA, NAAA has no homology with it and is most similar in primary amino acid sequence with acid ceramidase (30% homology and 70% similarity), also a lysosomal enzyme, an enzyme that hydrolyzes ceramide to fatty acid and sphingosine.3 Possibly due to the lethal consequences of a rare congenital condition where acid ceramidase is non-functioning (Farber disease), and the identification of acid ceramidase several decades ago, acid ceramidase is better characterized than NAAA at present and appears to be biochemically very similar.10, 11 Both acid ceramidase and NAAA are glycoproteins that undergo removal of an N-terminal signal peptide after biosynthesis, are believed to be transported by the mannose-6-phosphate pathway to the acidic late endosomes and/or lysosomes, and are proteolytically activated by an autocatalytic step under acidic conditions where the polypeptide is cleaved into two chains;10-12 one chain the shorter so-called α-subunit (for simplicity sake hereafter we refer to this chain for NAAA in the analogous manner), and the longer chain the so-called β-subunit (again we prefer to adopt this nomenclature for NAAA) containing the catalytic nucleophile and N-terminal residue cysteine. The other two residues for both enzymes that make up the catalytic triad, along with cysteine, are aspartate and arginine.
NAAA, also like acid ceramidase, suffers from a complete lack of three-dimensional structural information, as no crystal or NMR derived structures with significant homology to either enzyme are available. Although homology models have been published in an attempt to map the active site residues for these enzymes,2, 11 in our opinion they cannot be relied upon as good structural models because of such low homology (< 20% amino acid identity) with the structures available for other members of the choloylglycine hydrolase family, which is part of the N-terminal nucleophile hydrolase superfamily. One major obstacle in the pursuit of a three-dimensional structure for either of these enzymes is the difficulty in producing sufficient quantities of highly pure protein.
In order to produce NAAA in sufficient quantity for an initial proteomic based study, we used mammalian cells and modified expression and purification protocols previously developed for acid ceramidase10 and for NAAA transiently expressed in HEK293T cells.12, 13 We have established a HEK293 mammalian system stably expressing NAAA as a gateway for initial proteomic based enzyme characterization and for assay development. This robust NAAA expression system can be further used to obtain more detail on the post-translational protein modifications, identification of enzyme inhibitors, determination of the mechanisms of their action, and for structural studies.
Experimental section
Materials
Standard laboratory chemicals, buffers, culture media and media components were purchased from Sigma-Aldrich (St. Louis, MO) and Fisher Chemical (Pittsburg, PA). Restriction enzymes, DNA ligase, and other molecular biology chemicals were obtained from New England Biolabs (Beverly, MA).
Vector construction
A full-length cDNA of human NAAA inserted into pcDNA 3.1(+) was kindly provided by Dr. Natsuo Ueda of the Department of Biochemistry, Kagawa University School of Medicine, Kagawa, Japan. The forward primer 5′-TTAAGCTTGAGCCCGAGCC-3′ and the reverse primer 5′-TCCTCGAGGATCCTTTCTACTCGGGTTTCT-3′ containing incorporated HindIII and XhoI restriction sites (underlined respectively) necessary for cloning, were used for PCR amplification of hNAAA cDNA using Pfu DNA polymerase (Stratagene, La Jolla, CA). The fragment was cleaved with the restriction enzymes HindIII and XhoI and inserted by ligation into the pcDNA 3.1/myc-His C vector treated with the same enzymes. The ligated construct was transformed into One Shot Top10 competent E. coli cells, colonies were screened by PCR for the correct insert size and DNA sequence was confirmed.
Stable transfection
HEK293 wild-type cells (American Type Culture Collection, Manassas, VA) were cultured at 37 °C in a humidified incubator (5% CO2) using Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin (P/S). The day before transfection approximately 1 × 106 HEK293 cells were split into two 75 cm2 culture plates. The purified plasmid pcDNA 3.1/myc-His C construct containing NAAA (48 μg) was linearized with PvuI and added to 120 μL Lipofectamine 2000, then transferred into the two culture plates of HEK293 cells according to the manufacturers protocol (Invitrogen). Transfected cells were selected with 600 μg/ml G418 according to a previously determined sensitivity of non transfected HEK293 cells to this concentration of antibiotic,14 and after approximately 14 days individual colonies were harvested, passed to new culture flasks, and tested for NAAA activity. These colonies with relatively high enzymatic activity were eventually cryopreserved under liquid nitrogen and used for stable expression of NAAA as detailed below.
Overexpression and purification
HEK293 cells stably expressing NAAA (with C-terminal hexa-histidine tag) were cultured at 37 °C in a humidified incubator (5% CO2) on 500 cm2 plates in DMEM with 10% FBS, P/S, and 0.6 mg/mL Geneticin to approximately 90% confluency. Then the culture medium was exchanged for serum-free DMEM with P/S, 0.6 mg/mL Geneticin, and 10 mM NH4Cl and allowed to incubate for 48 hours before harvest of the medium. The harvested medium was centrifuged at 1000 xg for 10 min to remove contaminating cells and ammonium sulfate was added to 60% saturation in four aliquots at 4 °C over a period of one hour. The culture medium was then centrifuged at 15000 xg for 15 min at 4 °C and the pellet resuspended in 2% original volume 40 mM phosphate buffer (pH 6.5), 500 mM NaCl (buffer A), and dialyzed into buffer A with two changes at 4 °C. The dialyzed solution was centrifuged at 15000 xg for 15 min at 4 °C, and incubated with approximately 1 mL of Talon affinity resin per mg total protein for one hour at 4 °C. This mixture was centrifuged at 300 xg for 5 min at 4 °C, and the resin washed twice for 15 min with ten times the resin volume of buffer A containing 25 mM imidazole. The resin pellet was then transferred to a column and NAAA eluted with buffer A containing 150 mM imidazole. The eluted fraction was dialyzed using 40 mM phosphate buffer (pH 6.5), 150 mM NaCl and 1 mM EDTA (buffer B) with two changes at 4 °C. The purified protein concentration was determined by the Bradford dye-binding microassay (Bio-Rad), and was concentrated with Amicon Ultra-0.5 Centrifugal Filters, 10 kDa membrane (Millipore), to approximately 2.5 mg/mL (~50 μM) and stored at 4 °C.
Buffer exchange procedure
The buffer containing NAAA protein was exchanged to another buffer by re-concentrating 3 times to original volume after 25 fold dilution with exchange buffer using 10kDa membrane Amicon Ultra-0.5 Centrifugal Filters (Millipore).
Converting zymogen to active mature NAAA enzyme by acid treatment
100 mM citrate-phosphate buffer, pH 4.5, was added to purified NAAA at a 4:1 v/v ratio of buffer to protein solution, and incubated for 2 hours at 37 °C. For enzymatic assays the acidified NAAA was used directly.
Size exclusion chromatography
In order to evaluate the molecular weight of the active enzyme, we performed size exclusion chromatography using a Sephacryl-100 column (25 × 1 cm). 100 μg purified human NAAA was acidified, dialyzed into buffer B containing 2 mM DTT and concentrated to a volume of 50 μL as described in the previous sections. The concentrated enzyme was manually loaded onto the column, and buffer B with 2 mM DTT was run through the column at a flow rate of 0.1 ml/min. The molecular weight was determined under similar conditions according to Andrews,15 using the Bio-Rad Gel Filtration Standards: thyroglobulin (670 kDa), γ-globulin (158 kDa), bovine serum albumin (66 kDa, added to standards), ovalbumin (44 kDa), myoglobin (17 kDa), and Vitamin B12 (1750 Da).
SDS-PAGE
Protein samples were denatured at 95 °C for 5 min in Laemmli buffer, and were resolved in SDS-PAGE using Any kD (Bio-Rad) gels. After staining, a FluorChem Imaging System (Alpha Innotech Corp., San Leandro, CA) was used to photograph the gels.
N-(4-methyl coumarin)palmitamide (PAMCA) synthesis
Palmitic acid (50 mg, 0.195 mmol) was dissolved in a 1:1 mixture of anhydrous DMF/THF (5mL) at 0 °C. This solution was treated with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (45 mg, 0.26 mmol), 4-dimethylaminopyridine (48 mg, 0.39 mmol), and 7-amino-4-methyl-coumarin (45 mg, 0.26 mmol). The solution was allowed to stir under argon for 24h. The reaction mixture was then diluted with ether (10 mL), washed with water (5 mL), brine (5 mL); and dried over MgSO4. The organic layer was concentrated under reduced pressure and the resulting residue was chromatographed on silica gel (13:7 acetone:hexanes) to yield N-(4-methyl coumarin)palmitamide (49.5 mg, 61%) as a white solid. Mp = 184-186 °C. 1H NMR (500 MHz, CHLOROFORM-d) δ ppm 7.59 (dd, J = 2.0, 8.8 Hz, 1 H) 7.54 (d, J=8.79 Hz, 1 H) 7.51 (d, J=1.46 Hz, 1 H) 7.29 (br. s., 1 H) 6.21 (s, 1 H) 2.42 (s, 3 H) 2.36 (t, J=7.57 Hz, 1 H) 1.70 - 1.78 (m, 2 H) 1.24 - 1.33 (m, 24 H) 0.88 (t, J = 6.8 Hz, 3 H).
NAAA Assay
In order to have an assay method more conducive to high-throughput screening than those published for measuring the NAE hydrolyzing activity of NAAA,2, 16, 17 we developed the fluorogenic PEA analog PAMCA, which is hydrolyzed to fluorescent 7-amino-4-methyl coumarin (AMC) and palmitic acid. The assay procedure is similar to that used in the fluorescence-based assays for FAAH and MGL with each point done in triplicate; to each well of a 96 well plate 180 μL of 100 mM citrate-phosphate buffer (pH 4.5) containing 3 mM DTT, 0.1% Triton X-100, 0.05% BSA, 150 mM NaCl (assay buffer used by Tsuboi et. al.) with 100 ng of purified, acid treated enzyme was added, followed by 20 μL of a dimethyl sulfoxide (DMSO) solution containing PAMCA (final concentration ranging from 0.5 to 80 μM for the saturation curve experiment), and then 10 minutes incubation on a shaking platform. To analyze the activity of the enzyme fractions after the gel filtration chromatography 10 μL aliquots were added in the same manner as above with PAMCA at a final concentration of 5 μM. After shaking, the reaction was allowed to proceed at 37 °C for 30 minutes, with fluorescence readings taken every 10 minutes at a wavelength of 460 nm (using an excitation wavelength of 360 nm) on a Synergy HT Plate Reader using Gen5 software from Bio-Tek. The enzyme activity was calculated by converting the relative fluorescence units to AMC formed, using a standard curve of AMC dissolved in 10% DMSO-assay buffer. In order to determine enzymatic activity with the native substrate PEA, the purified NAAA was first activated by acid treatment as previously described. The acid treated NAAA (20 ng) was incubated with various concentrations of PEA (ranging from 2.5 to 120 μM) in 100 μL 10% DMSO-assay buffer for 30 minutes at 37 °C; each concentration was run in duplicate. Blanks, containing assay buffer without enzyme, for each PEA concentration were run simultaneously in duplicate. The reaction was terminated and extracted with previously published methods.18-20 In short, a mixture of acetone and ethanol (2:1, v/v) containing deuterated PEA as the internal standard was added to each sample, mixed and centrifuged at 4 °C for 5 min at 16,000 xg. The supernatants were collected and evaporated under nitrogen stream. Phosphate buffered saline (pH 7.4), methanol and chloroform (100:100:200 μL) were added to each sample followed by vigorous vortexing and centrifugation. The bottom chloroform fraction was collected and dried under nitrogen stream. Samples were reconstituted in 500 μL ethanol and analyzed using LC-MS/MS according to our previously published methods,18-20 using a TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Electron, San Jose, CA) with an Agilent 1100 HPLC on the front end (Agilent Technologies, Wilmington, DE). Separation was achieved using an Agilent Zorbax SB-CN column (2.1 × 50 mm, 5 μm) with gradient elution using 10 mM ammonium acetate (solvent A, pH 7.3) and 100% methanol (solvent B). Eluted peaks were ionized via atmospheric pressure chemical ionization (APCI) and detected by their respective selected reaction monitoring (SRM) transitions. Enzyme activity was calculated from the decrease in PEA concentration, comparing the blank samples to the enzyme samples. Michaelis-Menten constants were calculated using pro Fit software (Quantum Soft, Uetikon am See, Switzerland) and a Levenberg-Marquardt algorithm.
MALDI-TOF MS Analysis
Before MS analysis or tryptic digestion of active mature NAAA the acid buffer was exchanged to 50 mM ammonium bicarbonate buffer as described earlier. For analysis of the intact protein masses, 0.5 μL (1.25 μg) of the protein was mixed with 0.5 μL sinapinic acid matrix solution (5 mg/mL dissolved in 50% acetonitrile, 50% water, and 0.1% trifluoroacetic acid) and spotted onto an Opti-TOF 384-well plate insert. For the trypsin digested protein samples, 0.5 μL (1.25 μg) of the digest was mixed with 0.5 μL α-cyano-4-hydroxycinnaminic acid matrix solution (5 mg/mL dissolved in 50% acetonitrile, 50% water, and 0.1% trifluoroacetic acid) and spotted onto an Opti-TOF 384-well plate insert. MALDI-TOF MS spectra were acquired on a 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA) fitted with a 200-Hz solid state UV laser (wavelength 355 nm). Spectra of the intact proteins were acquired in linear mode, and spectra of the peptides were acquired in reflectron mode. The conditions used for the MS experiments and instrument calibration were performed as described by Zvonok et. al.21
Deglycosylation of zymogen and active mature NAAA protein for MALDI TOF MS analysis
To obtain samples of deglycosylated NAAA compatible with MALDI TOF MS analysis, the purified zymogen (10 μg) was mixed with 1500 units of PNGase F (New England Biolabs, Ipswich, MA) in 50 mM phosphate buffer, pH 7.5, and incubated for 48 hours at 37 °C. Deglycosylated NAAA samples were exchanged to 50 mM ammonium bicarbonate buffer as described earlier, and spectra were obtained in linear mode using MALDI TOF MS average mass measurement. For trypsin digestion the mature NAAA protein was deglycosylated using PNGase F according to the manufacturer’s protocol as follows: 10 μg of purified, acid treated enzyme was exchanged into 50 mM phosphate buffer, pH 7.5, concentrated to 10 μL, denatured by heating for 10 min at 90 °C, followed by addition of non-ionic detergent NP-40 (final concentration 1%) and 500 units of PNGase F and incubation for two hours at 37 °C. After deglycosylation the proteins were resolved by SDS-PAGE, coomassie stained, and the bands were excised for trypsin in-gel digestion as described below.
Tryptic digestion
10 μg of purified, acid treated NAAA was exchanged to 50 mM ammonium bicarbonate buffer, concentrated to 10 μL, and incubated overnight at 37 °C with MS-grade trypsin (“Trypsin Gold”, Promega) at a NAAA:trypsin mass to mass ratio of 100:1. For the PNGase treated samples, in-gel trypsin digestion was performed. Protein bands were excised from comassie stained SDS-PAGE gels (10 μg protein loaded into the lane), washed five times with 1 ml ammonium bicarbonate buffer with shaking for 10 min at 37 °C, washed twice with 100% acetonitrile for 5 min at room temperature, followed by removal of the acetonitrile, and then drying the gel pieces for one hour at room temperature. The gel pieces were rehydrated with 20 μL 50 mM ammonium bicarbonate buffer containing an appropriate amount of trypsin and incubated overnight at 37 °C. To test the oxidation state of the cysteines, 20 μg of purified, acid treated enzyme was exchanged to 50 mM ammonium bicarbonate buffer and concentrated to 20 μL. NAAA was split into two samples and in one an aliquot of N-ethylmaleimide (NEM) was added to a final concentration of 10 mM, or nothing, and incubated for one hour at room temperature. To the NEM treated sample dithiothreitol (DTT) was added to a final concentration of 25 mM and incubated at 55 °C for one hour. Then an aliquot of iodoacetamide (IAM) was added to the NEM/DTT treated sample to a final concentration of 50 mM and incubated for one hour at room temperature. The NEM/DTT/IAM treated sample along with the untreated control sample were digested overnight at 37 °C with trypsin at a 100:1 NAAA:trypsin ratio.
Results and Discussion
Overexpression, purification, and kinetic characterization of recombinant human NAAA
Our initial attempts to overexpress and purify a sufficient amount of NAAA protein for a mass spectrometric characterization and assay development by transient transfection of HEK293T cells were not very efficient. However, stable transfection of HEK293 cells with NAAA cDNA cloned into the pcDNA 3.1 myc-His vector yielded significantly greater quantities of enzyme. The purification of the zymogen from lysed cells and from the culture media after ammonium chloride treatment stimulated secretion of the lysosomal proteins from the HEK293 cells, similar to the method previously reported,12, 13 were simultaneously pursued. The latter method13 was optimized and found to provide a greater quantity and quality of the lysosomal NAAA, compared to cell lysate that contained along with zymogen all cellular components. Virtually all zymogen (95%) was precipitated with 60% ammonium sulfate following secretion into media, providing partially enriched samples of NAAA. Because the isoelectric point of the pro-enzyme with a hexa-histidine tag was estimated to be almost neutral (pI 7.3), mildly acidic conditions (pH 6.5) were chosen to avoid protein precipitation and decrease non-specific binding of other proteins to the IMAC resin. This was deemed important as we found albumin (abundant in the media) non-specifically binding to the Talon resin (identified by excising the 65 kDa band from SDS-PAGE and performing in-gel tryptic digestion and analysis by MALDI-TOF-MS, data not shown). Extensive washing of the resin with phosphate buffer containing a relatively high concentration of imidazole (25 mM) removed the rest of albumin, and after elution with phosphate buffer containing 150 mM imidazole we calculated an overall 75% yield of the enzyme from the NAAA protein secreted into the media.
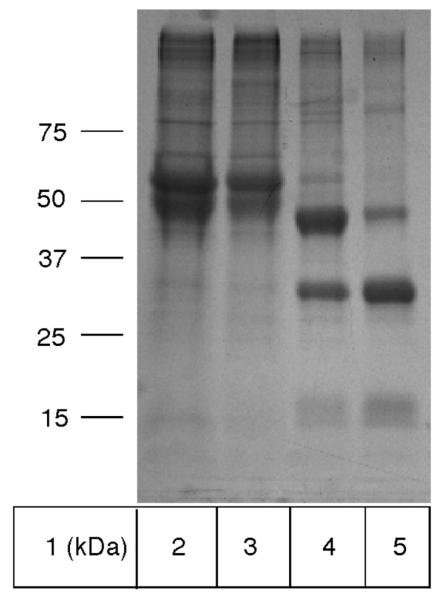
As demonstrated by SDS-PAGE and the MALDI-MS spectra shown in Figure 1 and 2A, the protein obtained was nearly pure and mainly in the proenzyme form, although a significant amount of the α- and β-subunits were present. This is similar to previous results reported for NAAA and acid ceramidase purified from the culture media,10, 13 and the ratio of proenzyme to α- and β-subunits in the purified protein varied somewhat from batch to batch. Taking into account that conversion of the zymogen into heterodimer, consisting of α- and β-subunits, was ultimately necessary to obtain active enzyme for biochemical studies, we did not pursue to control or prevent a cleavage of some of the zymogen during purification.
Figure 1.
SDS-PAGE analysis of human NAAA purification overexpressed by HEK293 cells. 10 μg total protein was loaded into each lane. Lane 1, molecular weight markers; lane 2, proteins from the media precipitated with 60% ammonium sulfate; lane 3, 25 mM imidazole IMAC wash fraction; lane 4, 150 mM imidazole IMAC elution fraction; lane 5, 2 hours acid treatment of purified NAAA at 37 °C.
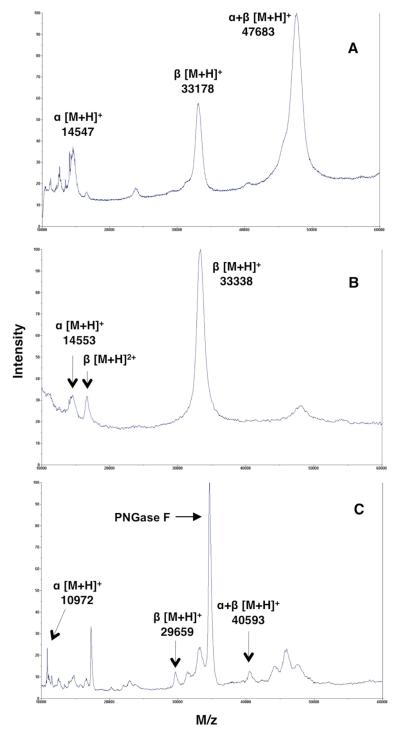
Figure 2.
Linear mode MALDI-TOF mass spectra of purified human NAAA containing the α-subunit, β-subunit, and pro-enzyme (α+β) (A). The majority of the pro-enzyme was converted into the α- and β-subunits after 2 hours acid treatment at 37 °C (B). Purified enzyme after 48 hours treatment with PNGase F at 37 °C under native conditions; intense peak at 34810 m/z is that of PNGase F (C).
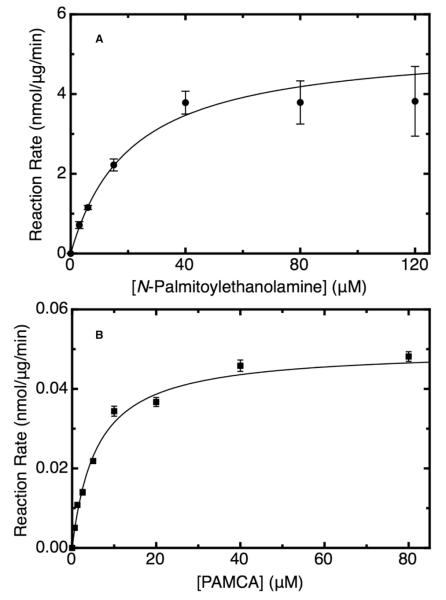
Kinetic characterization of the purified active mature hNAAA enzyme was performed by using an LC-MS methodology previously reported by our laboratory,18-20 where we measured enzymatic activity by determining the decrease in PEA concentration. The purified enzyme displayed Michaelis-Menten kinetics, as shown in Figure 3A, with a Km for PEA of 21 ± 3 μM and a Vmax of 5.4 ± 0.6 nmol/μg/min, similar to the previously reported values of a Km of 35 μM and Vmax of 1.8 nmol/μg/min for the NAAA enzyme purified from rat lung.22 The maximal velocity of the enzyme appears to be somewhat limited by the relatively low solubility of PEA in 10% DMSO aqueous assay buffer, as the specific activity did not increase above a concentration of approximately 40 μM. We observed this sharp leveling off of the activity at higher PEA concentrations every time we repeated the saturation curve experiment. To simplify and accelerate NAAA activity determination in enzyme assays, particularly the screening of inhibitors, the novel fluorogenic compound PAMCA was synthesized and tested as a substrate for enzyme. The enzyme displayed Michaelis-Menten kinetics for PAMCA hydrolysis, with an observed of Km of 6.2 ± 0.7 μM, as shown in Figure 3B, indicative that the binding affinity of NAAA for this compound is similar to or slightly greater than that for PEA. The maximal velocity was two orders of magnitude lower, similar to the slower rate of hydrolysis observed for FAAH and MGL with fluorogenic substrates as compared to the natural substrates.21, 23 However, the sensitivity, precision, minimal sample handling, and high throughput capacity of the fluorescent assay more than make up for the relatively low rate of catalysis with the fluorogenic substrate.
Figure 3.
Kinetic studies of purified mature human NAAA enzyme with the native substrate palmitoylethanolamine, performed by using LC-MS/MS assay (A) and with the fluorogenic substrate N-(4-methyl coumarin)palmitamide in fluorescent assay (B). Assays were performed at 37 °C in pH 4.5 buffer, and data were fit to the Michaelis-Menten equation using a Levenberg-Marquardt algorithm. Mean values ± S.D. are shown.
Evaluation of the molecular weight of the active mature enzyme by size exclusion chromatography
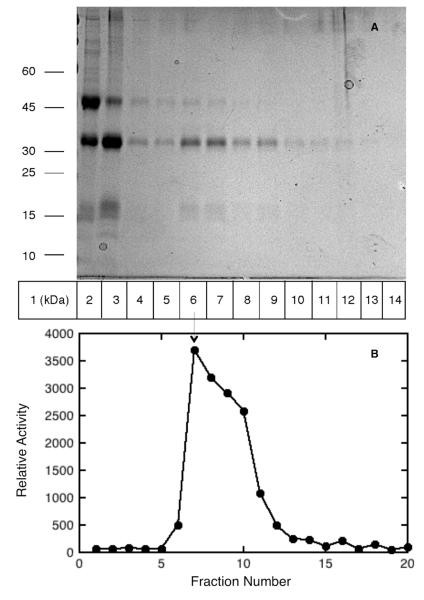
Human acid ceramidase, the closest known homolog to NAAA with 70% similarity and 33% identity in amino acid sequence, is a heterodimer composed of α- and β-subunits;10, 11 however, NAAA purified from rat lung was found to have only the β-subunit.22 Western blot and immunoprecipitation analysis of FLAG tag purified active and inactive mature hNAAA suggested that α- and β-subunits involved in formation of complex.12 To obtain more evidence confirming the subunit structure of the active mature hNAAA enzyme we performed size exclusive chromatography studies using a Sephacryl S-100 column calibrated by running a mixture of protein standards. The purified zymogen after cleavage by acid treatment, dialysis, and concentration was loaded on calibrated Sephacryl S-100 column and elution fractions were collected. The fractions activities were measured with the PAMCA substrate hydrolysis assay, while the protein composition of the fractions were determined using SDS-PAGE analysis. As shown in lane 3 of Figure 4A, the purified zymogen was mostly cleaved, forming α- and β-subunits after the incubation at acidic pH. The highest amount of α- and β-subunits and peak of hydrolyzing activity was observed in elution fractions 7-8 (lane 6-7 of Figure 4A and B). The molecular weight of protein in fractions 7-8 was estimated based on calibrated standard curve as 45 ± 3 kDa, suggesting that the active mature NAAA enzyme is a heterodimer.
Figure 4.
The molecular weight of active mature NAAA estimated by size exclusive chromatography using a Sephacryl-100 column. SDS-PAGE of Sephacryl-100 column load and elution fractions; lane 1, molecular weight markers; lane 2, purified NAAA; lane 3, purified NAAA after 2 hours acid treatment that was loaded onto column; lanes 4-14, eluted column fractions 5-15 (A). Relative activity of column fractions measured with fluorogenic assay using 10 μL aliquots of each fraction (B). Peak activity observed in column fraction 7 (lane 6), corresponding to a calculated molecular weight of 45 ± 3 kDa.
MALDI-TOF-MS analysis of zymogen, it’s processing and deglycosylation
As shown in Figure 2A, the linear mode MALDI-TOF mass spectrum of the purified enzyme sample confirmed the previous SDS-PAGE results that NAAA had been purified to near homogeneity. The majority of purified enzyme was in the zymogen form, with an average mass of 47.7 kDa, however the α- and β-subunits also were observed (14.5 and 33.2 kDa, respectively). In addition, the relative broadness of the peaks suggests that the purified enzyme pool is heterogeneous due to the variable modifications of the oligosaccharide side chains in N-glycosylated protein. After incubation of the zymogen under acidic conditions, the linear mode MALDI-TOF mass spectrum shown in Figure 2B confirmed that the enzyme had been nearly completely converted into the active form comprising the two main peaks belonging to the α- and β-subunits with an average mass of 14.6 and 33.3 kDa, respectively.
We encountered significant difficulty using both enzymatic and chemical approaches in obtaining a fully deglycosylated sample of NAAA in order to determine the molecular weight of the enzyme without the oligosaccharide chains. The glycoamidase PNGase F, that cleaves between the innermost N-acetylglucosamine (GlcNAc) and asparagine resides in N-glycosylated proteins, is the most effective with denatured samples, and heat denaturation of NAAA without detergents caused the protein to precipitate. Alternatively, the added SDS kept the protein in solution, however it completely suppressed MS signal detection. Chemical deglycosylation using trifluoromethanesulfonic acid24 treatment of lyophilized NAAA resulted in the loss of the protein. Therefore we used a relatively high concentration of PNGase F and incubated with non denatured NAAA over extended periods (1-3 days) at 37 °C. After 48 hours incubation we observed partial deglycosylation (Figure 2C), while beyond this time the NAAA proteins could not be detected by MALDI or by SDS-PAGE. It is likely that NAAA is much more stable in its glycosylated form, similar to previous observations with other N-glycosylated proteins.25-27 The spectrum in Figure 2C, obtained at 48 hours incubation with PNGase F, shows peaks representing both the fully de- and glycosylated forms, as well as partially deglycosylated NAAA intermediates. The fully deglycosylated forms were observed in linear mode MALDI-TOF MS as: α-subunit with an average mass of 10972 Da, which is relatively close to the theoretical mass of 10966 Da, the β-subunit with an average mass of 29659 Da, which is close to the theoretical mass of 29662 Da, and the pro-enzyme with an average mass of 40593 Da, also close to the theoretical mass of 40610 Da. The partially deglycosylated β-subunit and zymogen intermediates, as shown in Figure 2C, are comprised of groups of two and four individual peaks, respectively. The distance between peaks in each group is approximately 1500 Da, corresponding to the incremental removal of the N-linked oligosaccharide chains from glycosylation sites, which is in agreement with the typical mass of a high mannose type oligosaccharide that is phosphorylated (~1500 Da).28 Therefore, there are a total of 4 and 2 glycosylation sites in the zymogen and in each of the subunits, respectively. Our result agrees with a previous site-directed mutagenesis study of NAAA that identified 4 actual N-glycosylation sites (Asn37, Asn107, Asn309, and Asn333) out of 6 predicted.12
MALDI-TOF MS fingerprinting of tryptic digest of glycosylated and deglycosilated mature NAAA
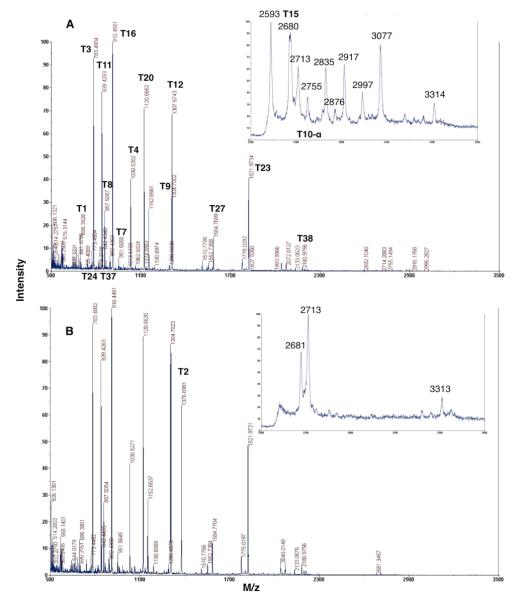
We performed the trypsin digestion of glycosylated and deglycosylated mature NAAA and characterized the obtained peptides by MALDI-TOF MS fingerprinting (Figure 5). The majority of the tryptic peptides with a mass exceeding 500 Da were identified (Table 1) with good mass accuracy (the error typically less than 10 ppm and at most 45 ppm between the calculated and measured masses). We were unable to directly observe the T5 peptide (1031.5302 m/z), most likely because T4 (1030.5316 m/z) was the more prominent peptide and its +1 Da isotope peak coincided with monoizotopic mass of T5. Analysis of the expected and observed isotope distribution curves for the T4 and T5 peptides revealed their overlapping and confirmed the presence of both peptides in the tryptic digest.
Figure 5.
MALDI-TOF MS spectra of trypsin digest of mature NAAA (A) and deglycosylated mature NAAA (B) obtained in reflectron mode. Insets - zoom scan area 2500-3500 Da obtained in linear mode.
Table 1.
MALDI-TOF MS fingerprinting of the purified mature human NAAA digested with trypsin.
| Position | Peptide sequences with mass exceeding 500 Da |
m/z calculated |
m/z measured |
Error (ppm) |
|---|---|---|---|---|
| T1/29-35 | SPPAAPR | 695.3835 | 695.3828 | −0.99 |
| T2/36-47 | FNVSLDSVPELR | 1376.7056** | 1376.698 | −5.52 |
| T3/48-53 | WLPVLR | 783.4876 | 783.4904 | 3.6 |
| T4/54-61 | HYDLDLVR | 1030.5316 | 1030.5302 | −1.41 |
| T5/62-71 | AAMAQVIGDR | 1031.5302 | 1031.5302*** | 0 |
| T7/75-82 | WVHVLIGK | 951.5774 | 951.5658 | −12.19 |
| T8/83-89 | VVLELER | 857.5091 | 857.5087 | −0.43 |
| T9/90-100 | FLPQPFTGEIR | 1304.6997 | 1304.7002 | 0.38 |
| T10-α*/101-125 | GMCDFMNLSLADCLLVNLAYESSVF | 2755.2486 | ||
| T10-β*/126-135 | CTSIVAQDSR | 1079.515 | 1079.5109 | −3.8 |
| T10-β+IAM | C(IAM)TSIVAQDSR | 1136.5364 | 1136.533 | 2.992 |
| T10-β+NEM | C(NEM)TSIVAQDSR | 1204.5627 | 1204.5626 | 0.083 |
| T11/136-142 | GHIYHGR | 839.4271 | 839.4293 | 2.65 |
| T12/143-153 | NLDYPFGNVLR | 1307.6743 | 1307.6743 | 0 |
| T14/155-163 | LTVDVQFLK | 1062.6194 | 1062.6024 | −15.98 |
| T15/164-188 | NGQIAFTGTTFIGYVGLWTGQSPHK | 2680.3518 | 2680.3652 | 5.01 |
| T16/189-196 | FTVSGDER | 910.4265 | 910.4551 | 31.39 |
| T18/199-211 | GWWWENAIAALFR | 1619.8118 | 1619.7942 | −10.87 |
| T20/213-221 | HIPVSWLIR | 1120.6626 | 1120.6652 | 2.29 |
| T21/212-236 | ATLSESENFEAAVGK | 1552.7489 | 1552.7358 | −8.41 |
| T23/240-256 | TPLIADVYYIVGGTSPR | 1821.9745 | 1821.9734 | −0.61 |
| T24/257-263 | EGVVITR | 773.4516 | 773.4504 | −1.5 |
| T26/266-283 | DGPADIWPLDPLNGAWFR | 2039.9974 | 2040.0183 | 10.25 |
| T27/284-296 | VETNYDHWKPAPK | 1584.7805 | 1584.7699 | −6.69 |
| T28/297-300 | EDDR | 534.2154 | 534.2266 | 21.01 |
| T30/302-306 | TSAIK | 519.3137 | 519.2904 | −44.8 |
| T31/307-348 | ALNATGQANLSLEALFQILSVVPVYNNLTIYTTVMSAGSPDK | 4458.2901 | ||
| T32/349-352 | YMTR | 570.2704 | 570.2725 | 3.61 |
| T37/363-369 | GHPFEQK | 842.4155 | 842.4346 | 22.67 |
| T38/370-387 | LISEEDLNMHTGHHHHHH | 2180.9791 | 2180.9756 | −1.6 |
The T10 peptide is split into two peptides belonging to different subunits after acid treatment, which was performed prior to trypsin digest.
The calculated molecular weight of the T2 peptide was corrected for conversion of asparagine to aspartic acid after deglycosylation by PNGaseF.
The T5 monoizotopic peak coincides with more abundant the T4+1 Da isotope peak.
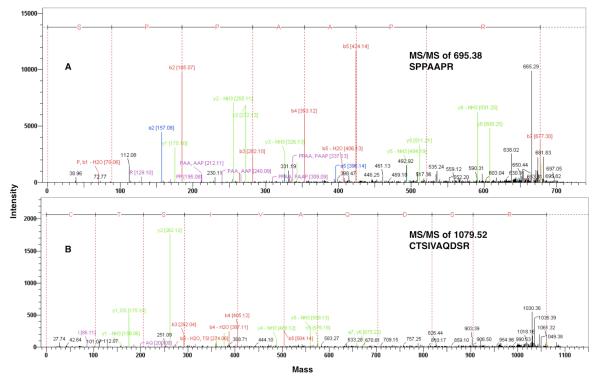
It was previously suggested12 that either the first 27 or 28 amino acids represent a signal sequence of nascent NAAA protein, necessary for the translocation of a pro-enzyme into the endoplasmic reticulum. We identified in the tryptic digest of mature NAAA an ion at 695.4 m/z that after MS/MS fragmentation was confidently assigned to the sequence SPPAAPR (amino acid 29-35 in pre-pro-enzyme) corresponding to the T1 peptide (Figure 6A). Therefore, the signal peptidase cleaves pre-pro-enzyme after signal peptide at position between 28th and 29th amino acid residues resulting in an N-terminal serine in α-subunit.
Figure 6.
MALDI-TOF MS/MS analysis of the NAAA tryptic peptides containing the N-terminal sequences of α- (A) and β-subunits (B).
The Cys126 of human NAAA was presumed to be located at the N-terminus of the β-subunit, similar to Cys131 in purified rat lung NAAA,14and shown to be essential for the proteolytic cleavage of the pro-enzyme.12 The cysteine 126 to alanine mutant was unable to undergo self-proteolysis and convert from the pro-enzyme to the active mature form,12 leaving some ambiguity of it’s importance for activity. Here we showed the significance of the cysteine residue(s) for enzyme activity by their selective alkylation in mature NAAA, as alkylation by either IAM or NEM resulted in an almost complete loss of activity. We were able to detect the 1079.5 m/z ion corresponding to the T10-β peptide and perform its MS/MS analysis, confirming that the N-terminal residue of the β-subunit of the human enzyme is a cysteine (Figure 6B).
In an MS based investigation of acid ceramidase all 6 cysteines were found to be involved in the formation of 3 disulfide bridges, one linking the two subunits, while the two other are located in the β-subunit.10 Unexpectedly, the N-terminal cysteine of β-subunit that was supposed to be the catalytic nucleophile is involved in formation of an intrasubunit disulfide bridge.10 There are a total of three cysteines in the primary amino acid sequence of human NAAA (two in the α–subunit, Cys103 and Cys113, and one in the β-subunit, Cys126) and disulfide bridge formation between two subunits possible only if Cys126 of β-subunit were involved. To determine the oxidation state of Cys126 we performed a series of cysteine alkylation and reduction experiments of mature NAAA followed by trypsin digestion and MALDI TOF MS analysis. The treatment of the enzyme with alkylation reagent (IAM or NEM), followed by reduction with DTT and exposure to the second alkylation reagent (NEM or IAM) should determine whether the cysteine is in a reduced (labeled with the first reagent) or an oxidized state (labeled after reduction with the second reagent). We performed the experiment by reversing the order in which IAM and NEM were used to ensure the results were not dependent on the reactivity of these reagents. When IAM was the first reagent in mature NAAA treatment, followed by DTT reduction and NEM alkylation, in trypsin digest we observed a peptide with a molecular weight of 1136.64 m/z that was not present in the non-alkylated control. Analysis by MS/MS fragmentation revealed this peptide to have the sequence CTSIVAQDSR, with the carbamidomethylated cysteine (data not shown). We observed neither non-alkyated (1079.52 m/z), nor NEM modified peptide (1204.56 m/z) with that sequence. When NEM was added first, followed by DTT reduction and IAM treatment, only the NEM alkylated peptide (1204.56 m/z) was identified in tryptic digested mature NAAA. In addition, as previously noted nearly all enzymatic activity was extinguished after alkylation by either IAM or NEM, strongly suggesting that cysteine 126 at the N-terminus of the β-subunit is the catalytic nucleophile and not involved in disulfide bridge formation with α-subunit.
There are four actual N-glycosylation sites in mature NAAA that should be found after trypsin digest in three predicted N-glycosylated peptides – T2NGlc, T10-αNGlc, and T312NGlc. The group of peaks starting at 2593 m/z and ending at 3077 m/z was detected using MALDI TOF instrument (Figure 5A, insert) in mature NAAA digested with trypsin. In the tryptic digest of the deglycosylated mature NAAA those peaks are not present, while we clearly observe the 1376 m/z T2 peptide (Figure 5B, insert). The difference in mass between peptides in this group and the T2 peptide is in the range 1200-1700 Da, close to the average mass of the oligosaccharide chains we determined from the linear mode MALDI-TOF MS analysis of the intact mature and deglycosylated enzyme. The most intense 2593 and 3077 m/z peaks were subjected MS/MS analysis, however we were unable to obtain good quality fragmentation data to identify the structure and composition of the oligosaccharide chains attached to glycosylation site in these peptides. The detailed posttranslational mature NAAA characterization is beyond the scope of this initial proteomic analysis, and will be undertaken later.
We were unable to observe the two other glycosylated or deglycosylated T10-α and T31 peptides in the trypsin digest of mature or PNGase F treated NAAA using MALDI TOF MS. The detection of glycosylated peptides was compromised likely due to heterogeneous glycosylation producing several glycoforms as is typical in N-glycosylated proteins,29 resulting in signal intensity spreading into a group of peaks and their relatively high masses. In addition, extraction of deglycosylated peptides after in gel trypsin digestion was possibly not efficient enough to observe them in the MALDI TOF MS spectra.
Overall, 26 of the 28 total tryptic peptides with masses higher than 500 Da, covering more than 80% of the mature NAAA amino acid sequence, were identified using MALDI TOF MS fingerprinting.
Conclusion
We established a HEK293 cell line stably expressing human NAAA and developed a relatively simple single chromatographic step purification that together are capable of yielding milligram quantities of pure enzyme. We kinetically characterized the purified human enzyme, and developed a fluorescent assay based on a novel fluorogenic substrate PAMCA, that has comparable affinity to the native substrate PEA and now makes high throughput screening of enzyme inhibitors more affordable and rapid. We confirmed the number of glycosylation sites and determined the molecular weights of the fully glycosylated and deglycosylated forms of the enzyme, along with the approximate molecular weights of the oligosaccharide chains. The structure of the active mature form of the enzyme appears to be a heterodimer, consisting of a non-covalent complex of α- and β-subunits. MALDI TOF MS fingerprinting covered more than 80% of the mature NAAA amino acid sequence including the N-terminal peptides of the α- and β-subunits. The putative catalytic nucleophile, which is the N-terminal cysteine 126 residue of the β-subunit, was demonstrated to be in a reduced state. The loss of virtually all enzymatic activity of mature NAAA treated with IAM or NEM was correlated with cysteine 126 alkylation, determined by MS/MS analysis of the tryptic digest. This established HEK293 NAAA expression and purification system provides sufficient enzyme for more detailed future proteomic analysis, pharmacological characterization and structural studies.
Acknowledgments
We thank Prof. Natsuo Ueda, Prof. Kazuhito Tsuboi, and Dr. Toru Uyama for the kind gift of the full length human NAAA clone, a NAAA antibody, and for the valuable advice provided. We thank Dr. Yazen Jmiean and Mahmoud Nasr for experimental assistance. This work was supported by grants DA003801, DA007312, and DA009158 from the National Institutes of Health/National Institute on Drug Abuse. The views expressed in this publication do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention by trade names, commercial practices or organizations imply endorsement by the U.S. Government.
References
- 1.Solorzano C, Antonietti F, Duranti A, Tontini A, Rivara S, Lodola A, Vacondio F, Tarzia G, Piomelli D, Mor M. Synthesis and structure-activity relationships of N-(2-oxo-3-oxetanyl)amides as N-acylethanolamine-hydrolyzing acid amidase inhibitors. J. Med. Chem. 53(15):5770–81. doi: 10.1021/jm100582w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solorzano C, Zhu C, Battista N, Astarita G, Lodola A, Rivara S, Mor M, Russo R, Maccarrone M, Antonietti F, Duranti A, Tontini A, Cuzzocrea S, Tarzia G, Piomelli D. Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation. Proc Natl Acad Sci U S A. 2009;106(49):20966–71. doi: 10.1073/pnas.0907417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda N, Tsuboi K, Uyama T. N-acylethanolamine metabolism with special reference to N-acylethanolamine-hydrolyzing acid amidase (NAAA) Prog. Lipid Res. 49(4):299–315. doi: 10.1016/j.plipres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Calignano A, La Rana G, Giuffrida A, Piomelli D. Control of pain initiation by endogenous cannabinoids. Nature. 1998;394(6690):277–81. doi: 10.1038/28393. [DOI] [PubMed] [Google Scholar]
- 5.Calignano A, La Rana G, Piomelli D. Antinociceptive activity of the endogenous fatty acid amide, palmitylethanolamide. Eur. J. Pharmacol. 2001;419(2-3):191–8. doi: 10.1016/s0014-2999(01)00988-8. [DOI] [PubMed] [Google Scholar]
- 6.Lambert DM, Vandevoorde S, Jonsson KO, Fowler CJ. The palmitoylethanolamide family: a new class of anti-inflammatory agents? Curr. Med. Chem. 2002;9(6):663–74. doi: 10.2174/0929867023370707. [DOI] [PubMed] [Google Scholar]
- 7.Lo Verme J, Fu J, Astarita G, La Rana G, Russo R, Calignano A, Piomelli D. The nuclear receptor peroxisome proliferator-activated receptor-alpha mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 2005;67(1):15–9. doi: 10.1124/mol.104.006353. [DOI] [PubMed] [Google Scholar]
- 8.O’Sullivan SE. Cannabinoids go nuclear: evidence for activation of peroxisome proliferator-activated receptors. Br. J. Pharmacol. 2007;152(5):576–82. doi: 10.1038/sj.bjp.0707423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jhaveri MD, Richardson D, Chapman V. Endocannabinoid metabolism and uptake: novel targets for neuropathic and inflammatory pain. Br. J. Pharmacol. 2007;152:624–632. doi: 10.1038/sj.bjp.0707433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulze H, Schepers U, Sandhoff K. Overexpression and mass spectrometry analysis of mature human acid ceramidase. Biol. Chem. 2007;388(12):1333–43. doi: 10.1515/BC.2007.152. [DOI] [PubMed] [Google Scholar]
- 11.Shtraizent N, Eliyahu E, Park JH, He X, Shalgi R, Schuchman EH. Autoproteolytic cleavage and activation of human acid ceramidase. J. Biol. Chem. 2008;283(17):11253–11259. doi: 10.1074/jbc.M709166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao LY, Tsuboi K, Okamoto Y, Nagahata S, Ueda N. Proteolytic activation and glycosylation of N-acylethanolamine-hydrolyzing acid amidase, a lysosomal enzyme involved in the endocannabinoid metabolism. Biochim. Biophys. Acta. 2007;1771(11):1397–405. doi: 10.1016/j.bbalip.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Zhao LY, Uyama T, Tsuboi K, Tonai T, Ueda N. Amino acid residues crucial in pH regulation and proteolytic activation of N-acylethanolamine-hydrolyzing acid amidase. Biochim. Biophys. Acta. 2008;1781(11-12):710–7. doi: 10.1016/j.bbalip.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Pei Y, Mercier RW, Anday JK, Thakur GA, Zvonok AM, Hurst D, Reggio PH, Janero DR, Makriyannis A. Ligand-Binding Architecture of Human CB2 Cannabinoid Receptor: Evidence for Receptor Subtype-Specific Binding Motif and Modeling GPCR Activation. Chem. Biol. 2008;15(11):1207–1219. doi: 10.1016/j.chembiol.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem. J. 1964;91(2):222–33. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saturnino C, Petrosino S, Ligresti A, Palladino C, De Martino G, Bisogno T, Di Marzo V. Synthesis and biological evaluation of new potential inhibitors of N-acylethanolamine hydrolyzing acid amidase. Bioorganic & Medicinal Chemistry Letters. 20(3):1210–3. doi: 10.1016/j.bmcl.2009.11.134. [DOI] [PubMed] [Google Scholar]
- 17.Tsuboi K, Sun YX, Okamoto Y, Araki N, Tonai T, Ueda N. Molecular characterization of N-acylethanolamine-hydrolyzing acid amidase, a novel member of the choloylglycine hydrolase family with structural and functional similarity to acid ceramidase. J. Biol. Chem. 2005;280(12):11082–11092. doi: 10.1074/jbc.M413473200. [DOI] [PubMed] [Google Scholar]
- 18.Williams J, Wood J, Pandarinathan L, Karanian DA, Bahr BA, Vouros P, Makriyannis A. Quantitative method for the profiling of the endocannabinoid metabolome by LC-atmospheric pressure chemical ionization-MS. Anal. Chem. 2007;79(15):5582–93. doi: 10.1021/ac0624086. [DOI] [PubMed] [Google Scholar]
- 19.Wood JT, Williams JS, Pandarinathan L, Courville A, Keplinger MR, Janero DR, Vouros P, Makriyannis A, Lammi-Keefe CJ. Comprehensive profiling of the human circulating endocannabinoid metabolome: clinical sampling and sample storage parameters. Clin. Chem. Lab. Med. 2008;46(9):1289–95. doi: 10.1515/CCLM.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood JT, Williams JS, Pandarinathan L, Janero DR, Lammi-Keefe CJ, Makriyannis A. Dietary docosahexaenoic acid supplementation alters select physiological endocannabinoid-system metabolites in brain and plasma. J. Lipid Res. 2010;51(6):1416–23. doi: 10.1194/jlr.M002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zvonok N, Williams J, Johnston M, Pandarinathan L, Janero DR, Li J, Krishnan SC, Makriyannis A. Full mass spectrometric characterization of human monoacylglycerol lipase generated by large-scale expression and single-step purification. J. Proteome Res. 2008;7(5):2158–64. doi: 10.1021/pr700839z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda N, Yamanaka K, Yamamoto S. Purification and characterization of an acid amidase selective for N-palmitoylethanolamine, a putative endogenous anti-inflammatory substance. J. Biol. Chem. 2001;276(38):35552–35557. doi: 10.1074/jbc.M106261200. [DOI] [PubMed] [Google Scholar]
- 23.Ramarao MK, Murphy EA, Shen MW, Wang Y, Bushell KN, Huang N, Pan N, Williams C, Clark JD. A fluorescence-based assay for fatty acid amide hydrolase compatible with high-throughput screening. Anal. Biochem. 2005;343(1):143–51. doi: 10.1016/j.ab.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 24.Edge AS. Deglycosylation of glycoproteins with trifluoromethanesulphonic acid: elucidation of molecular structure and function. Biochem. J. 2003;376(Pt 2):339–50. doi: 10.1042/BJ20030673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arakawa T, Prestrelski SJ, Kenney WC, Carpenter JF. Factors affecting short-term and long-term stabilities of proteins. Adv Drug Deliv Rev. 2001;46(1-3):307–26. doi: 10.1016/s0169-409x(00)00144-7. [DOI] [PubMed] [Google Scholar]
- 26.Baker HM, Day CL, Norris GE, Baker EN. Enzymatic deglycosylation as a tool for crystallization of mammalian binding proteins. Acta Crystallogr. D. Biol. Crystallogr. 1994;50(Pt 4):380–4. doi: 10.1107/S0907444993013435. [DOI] [PubMed] [Google Scholar]
- 27.Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009;98(4):1223–45. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varki A, Kornfeld S. Structural studies of phosphorylated high mannose-type oligosaccharides. J. Biol. Chem. 1980;255(22):10847–58. [PubMed] [Google Scholar]
- 29.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 6(10):713–23. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]