Abstract
We report here a 4-year-old boy with global developmental delay who was referred for karyotyping and fragile X studies. A small interstitial deletion on chromosome 7 at band 7q21 was detected in all cells examined. Subsequent molecular karyotype analysis gave the more detailed result of a 6.3 Mb heterozygous deletion involving the interstitial chromosome region 7q21.11. In this relatively gene-poor region, the presynaptic cytomatrix protein, Piccolo (PCLO) gene appears to be the most likely candidate for copy number loss leading to a clinical phenotype. G-banded chromosome analysis of the parents showed this deletion was inherited from the father. Molecular karyotype analysis of the father’s genome confirmed that it was the same deletion as that seen in the son; however, the father did not share the severity of his son’s phenotype. This cytogenetically-visible deletion may represent another example of a chromosomal rearrangement conferring a variable phenotype on different family members.
Keywords: PCLO protein, human; Haploinsufficiency; Chromosome7, trisomy 7q; Case report; New Zealand
Molecular karyotype analysis is becoming the standard tool for interrogating the genome of individuals with referrals of global developmental delay and related conditions.1,2 In this rapidly expanding field, the interpretation of detectable copy number losses and gains is becoming more challenging. We report here the first case of the paternal inheritance of a 6.3 Mb interstitial deletion in 7q21 that appears to be associated with global developmental delay. The data suggest that haploinsufficiency of one or more genes that lie in this region may be playing a role in the proband’s phenotype.
Case Report
The proband, weighing 3 Kg at birth, was the second child of a non-consanguineous couple. His motor milestones were slightly delayed; he walked at 18 months, at which time he was also able to feed himself with a spoon. He said his first words soon after his first birthday and was combining words before two years of age. Challenging behaviours and symptoms of hyperactivity began to appear in the third and fourth years of life. The Conners’ Teacher Rating Scale, administered at 4 years of age, showed T-scores of 60 for conduct, 85 for hyperactivity, 66 for inattention/passivity, and 62 for hyperactivity. He was a non-dysmorphic child with a completely normal physical examination, growing along the 75th centile for height.
The proband’s 7-year-old sister had no behavioural or developmental problems and was performing at an above-average level at school. The proband’s father had had learning difficulties throughout his childhood which were referred to as dyslexia. He had particular difficulties in mathematics and English and required extra help and tuition. He was able to complete his high school studies and went on to obtain a training certificate and a diploma from a technical college.
Peripheral blood was taken for both routine G-banding and molecular karyotype analysis. In respect to the latter, genomic deoxyribonucleic acid (DNA) was isolated from the peripheral blood using the Gentra Puregene® blood kit (QIAGEN Genomics, Bothell, Washington, USA) according to the manufacturer’s instructions. Using the Affymetrix® Cytogenetics Reagent Kit (Affymetrix, Santa Clara, California, USA) 0.1 micrograms of genomic DNA were labelled. The labelled DNA was applied to an Affymetrix® Cytogenetics Array (2.7 million probes) according to the manufacturer’s instructions, and the array was scanned. The data were analysed using the Affymetrix® Chromosome Analysis Suite (ChAS), Version 1.0.1 and interpreted with the aid of the University of California Santa Cruz genome browser (hg18 assembly).3
Routine G-banded chromosome analysis was undertaken in duplicate for the proband and his parents, which identified a subtle interstitial deletion in the long arm of one chromosome 7, both in the proband and his father [Figure 1] 46,XY,del(7) (q21.1q21.1). Molecular karyotype analysis of the proband confirmed a 6.3 Mb interstitial heterozygous deletion in the chromosome region 7q21.11 (hg18 coordinates chr7: 79,573,975-85,833,865) [Figure 2], as well as a 458 kb terminal duplication in the chromosome region Xp22.33 (hg18 coordinates chrX:858,113-1,316,852). This terminal X chromosome region carries the CRFL2 gene (cytokine receptor-like factor 2), which is a receptor for thymic stromal lymphopoietin and so would not appear to play any role in the phenotype of the proband. Molecular karyotyping of the father showed the same 6.3 Mb heterozygous deletion in 7q21.11 (hg18 coordinates chr7:79,573,967-85,835,041); the small X chromosome duplication event detected in the proband was not found in the father. The 6.3 Mb deletion was not detected in the paternal grandparents of the proband; hence, the deletion arose de novo in the father. Table 1 summarises the genes that are localised to the chromosome 7 region which were deleted in the proband.
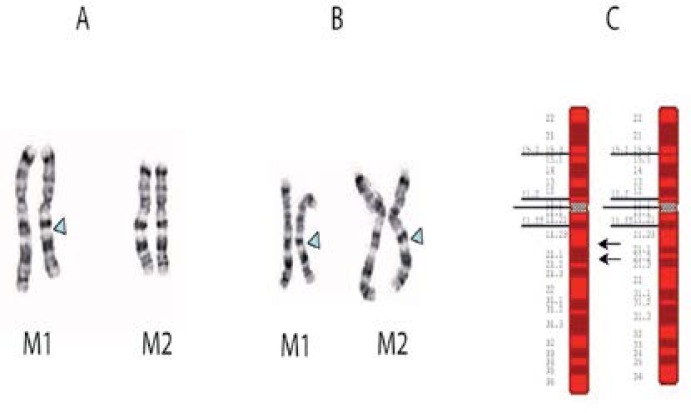
Figure 1 Panels A–C:
Partial karyotype and ideograms of chromosomes 7 of proband and father. Panel A: The homologous chromosomes 7 from two metaphase spreads (M1 and M2) of the proband. Panel B: The homologous chromosomes 7 from two metaphase spreads (M1 and M2) of the proband’s father. Panel C: An ideogram of the location of the deletion on chromosome 7 (left), and a normal chromosome 7 (right).
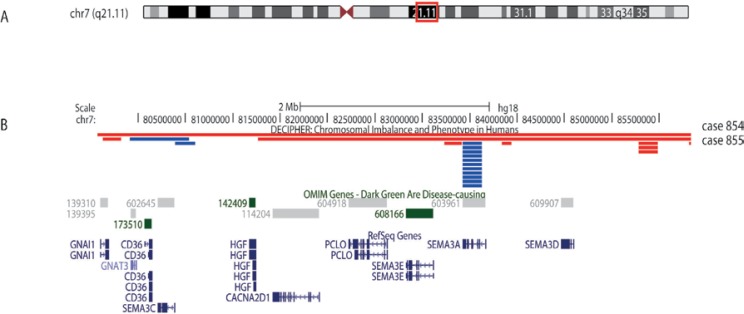
Figure 2 Panels A&B:
Schematic of the chromosome 7 region that is deleted in the proband. Panel A: Ideogram of chromosome 7, together with the location and extent of the deletion detected in the proband (7q21.11; hg18 coordinates chr7: 79,573,975–85,833,865; boxed in red). Panel B: Shows the Refseq genes that are localised to the deleted region, those genes that are curated in the Online Mendelian Inheritance in Man (OMIM) database4 and cases reported in the DECIPHER database.13 The images were taken from the UCSC genome browser.3
Table 1:
Genes that lie in the chromosome 7 region: 79,573,975–85,833,865 (hg18 coordinates)
| Gene | Protein | OMIM | Description |
|---|---|---|---|
| GNAI1 | Guanine nucleotide binding protein G(i) subunit alpha-1 | 139310 | The encoded protein is part of a complex that responds to beta-adrenergic signals by inhibiting adenylate cyclase. |
| CD36 | CD36 molecule (thrombospondin receptor) | 173510 | This protein may have important functions as a cell adhesion molecule. Mutations in this gene cause platelet glycoprotein deficiency. |
| SEMA3C | Semaphorin 3C | 602645 | This protein is a non-multidrug resistance gene of human cancers |
| HGF | Hepatocyte growth factor (hepapoietin A; scatter factor) | 142409 | Hepatocyte growth factor regulates cell growth, cell motility, and morphogenesis by activating a tyrosine kinase signaling cascade after binding to the proto-oncogenic c-Met receptor. |
| CACNA2D1 | Voltage-dependent calcium channel subunit alpha-2/delta-1 | 114204 | This gene encodes a member of the alpha-2/delta subunit family, which is a protein in the voltage-dependent calcium channel complex. |
| PCLO | Piccolo (presynaptic cytomatrix protein) | 604918 | The protein encoded by this gene is part of the presynaptic cytoskeletal matrix, which is involved in establishing active synaptic zones and in synaptic vesicle trafficking. Variations in this gene have been associated with bipolar disorder and major depressive disorder. |
| SEMA3E | Semaphorin 3E | 608166 | Semaphorins serve as axon guidance ligands via multimeric receptor complexes. Screening of patients with CHARGE syndrome for mutations in the SEMA3E gene revealed a de novo missense mutation in an unrelated patient. |
| SEMA3A | Semaphorin 3A | 603961 | This secreted protein can function as either a chemorepulsive agent, inhibiting axonal outgrowth, or as a chemoattractive agent, stimulating the growth of apical dendrites. In both cases, the protein is vital for normal neuronal pattern development. Increased expression of this protein is associated with schizophrenia. |
| SEMA3D | Semaphorin 3D | 609907 | Knockdown of the zebrafish orthologue of semaphorin 3D (SEMA3D) reduces the number of peripheral axons, and expression in the chick suggests that this protein may play an important role in heart development. |
OMIM = Online Mendelian Inheritance in Man; CHARGE = coloboma, heart defect, atresia choanae, retarded growth and development, genital abnormality, and ear abnormality.
Discussion
The deleted region of chromosome 7 encompasses 10 genes [Table 1]. Of these, the Online Mendelian Inheritance in Man (OMIM)4 database shows that 3 are disease-causing: OMIM 173510 (platelet glycoprotein IV deficiency), OMIM 142409 (hepatic growth factor), and OMIM 608166 (semaphorin 3E/coloboma, heart defect, atresia choanae, retarded growth and development, genital abnormality, and ear abnormality [CHARGE] syndrome). In terms of the latter, disruption of, or mutations in, the SEMA3E gene are rare in patients with CHARGE syndrome, which is associated with mental retardation but usually occurs in combination with eye defects (coloboma) and heart anomalies that were not detected in the case reported here.5 Only one CHARGE patient has been described with a mutation in the SEMA3E gene, and this was a missense mutation resulting in a serine to leucine substitution at amino acid position 243.5 Of the remaining genes located in the deleted region, the presynaptic cytomatrix protein, Piccolo (PCLO), gene appears to be a likely candidate to be associated with developmental delay.
The PCLO gene expresses Piccolo (presynaptic cytomatrix protein) which is a component of the presynaptic cytoskeletal matrix that is involved in maintaining the neurotransmitter release site in register with the postsynaptic reception apparatus.6–9 Piccolo appears be involved in the cycling of synaptic vesicles at presynaptic nerve terminals of glutamatergic and gamma aminobutyric acid-ergic (GABAergic) central nervous system synapses.
Overexpression of the PCLO gene has been implicated in bipolar/mood disorders in humans.10 Interestingly, knockdown studies in mice show that Piccolo controls the extracellular levels of glutamate in the hippocampus when stimulated, and appears to play a pivotal role in synaptic plasticity in area CA1 and in hippocampus-dependent learning.11 It is tempting to speculate that the semaphorin genes that are localised to the deleted chromosome 7 region may also play a role, together with the PCLO gene, in the proband’s phenotype. The semaphorins have roles in axonal guidance and haploinsufficiency for these proteins may affect normal brain development; however, animal modelling data are limited in terms of heterozygous knockdown studies of these genes. At least for semaphorin 3C, targeted disruption of the mouse orthologue leads to cardiovascular defects although only in the homozygous state; heterozygous mice are indistinguishable from their wild-type littermates.12
Significantly, the Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER) database13 contains two patients’ entries, cases 854 and 855 carrying deletions of 35.4 Mb and 17 Mb, respectively, that largely overlap the region identified in the case reported here; these patients exhibited a developmental delay/mental retardation phenotype. In addition, Courtens et al. reported a patient with moderate developmental delay and mild dysmorphic features with maternal monosomy in chromosome 7q21 (between 75.82 Mb and 92.59 Mb).14 Also of relevance is the almost complete absence of large genomic variation among unaffected individuals within this region of chromosome 7 suggesting that copy-number variations (CNVs) in this region are not benign.
If Piccolo contributed to the phenotype observed in our case study, then a number of mechanisms might explain why the same deletion that was present in the proband’s father resulted in a less severe clinical phenotype. Clinical variability may be due to a two hit model, or allele-specific expression.15,16 In respect of the former, an additional CNV, or a mutation below our detection threshold, may underlie differing clinical severity. In respect of the latter, it is unclear if there are parent-of-origin expression differences of the PCLO gene. Interestingly, imprinting of chromosome 7 is well-documented, but appears to be limited to those genes that lie distally in 7q23.1.17–19 This case represents another example of a chromosomal rearrangement conferring a variable phenotype on different family members.20
Conclusion
This rare case is the first to report a small but detectable interstitial deletion in 7q11.23 that is paternally-inherited and appears to be implicated in developmental delay. Haploinsufficiency of the PCLO gene seems to be the most likely explanation for the clinical phenotype. Further cases of deletions that lie in this region of chromosome 7 should help confirm the role of Piccolo in developmental delay, and the mechanism underpinning clinical variability.
References
- 1.George A, Marquis-Nicholson R, Zhang LT, Love JM, Ashton F, Aftimos S, et al. Chromosome microarray analysis in a clinical environment: new perspective and new challenge. Br J Biomed Sci. 2011;68:100–8. doi: 10.1080/09674845.2011.11730334. [DOI] [PubMed] [Google Scholar]
- 2.Marquis-Nicholson R, Aftimos S, Hayes I, George A, Love DR. Array comparative genomic hybridisation: a new tool in the diagnostic genetic armoury. NZ Med J. 2010;123:50–61. [PubMed] [Google Scholar]
- 3.Genome Browser. University of California Santa Cruz (UCSC). From: http://genome.ucsc.edu Accessed: Apr 2012.
- 4.Online Mendelian Inheritance in Man (OMIM) database. From: http://www.ncbi.nlm.nih.gov/omim Accessed: Apr 2012.
- 5.Lalani SR, Safiullah AM, Molinari LM, Fernbach SD, Martin DM, Belmont JW. SEMA3E mutation in a patient with CHARGE syndrome. J Med Genet. 2004;41:e94. doi: 10.1136/jmg.2003.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenster SD, Chung WJ, Zhai R, Cases-Langhoff C, Voss B, Garner AM, et al. Piccolo, a presynaptic zinc finger protein structurally related to bassoon. Neuron. 2000;25:203–14. doi: 10.1016/s0896-6273(00)80883-1. [DOI] [PubMed] [Google Scholar]
- 7.Fenster SD, Garner CC. Gene structure and genetic localization of the PCLO gene encoding the presynaptic active zone protein Piccolo. Int J Dev Neurosci. 2002;20:161–71. doi: 10.1016/s0736-5748(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 8.Fenster SD, Kessels MM, Qualmann B, Chung WJ, Nash J, Gundelfinger ED, et al. Interactions between Piccolo and the actin/dynamin-binding protein Abp1 link vesicle endocytosis to presynaptic active zones. J Biol Chem. 2003;278:20268–77. doi: 10.1074/jbc.M210792200. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee K, Yang X, Gerber SH, Kwon HB, Ho A, Castillo PE, et al. Piccolo and bassoon maintain synaptic vesicle clustering without directly participating in vesicle exocytosis. Proc Natl Acad Sci USA. 2010;107:6504–9. doi: 10.1073/pnas.1002307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, Webster MJ. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol Psychiatry. 2011;69:353–9. doi: 10.1016/j.biopsych.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibi D, Nitta A, Ishige K, Cen X, Ohtakara T, Nabeshima T, et al. Piccolo knockdown-induced impairments of spatial learning and long-term potentiation in the hippocampal CA1 region. Neurochem Int. 2010;56:77–83. doi: 10.1016/j.neuint.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Feiner L, Webber AL, Brown CB, Lu MM, Jia L, Feinstein P, et al. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development. 2001;128:3061–70. doi: 10.1242/dev.128.16.3061. [DOI] [PubMed] [Google Scholar]
- 13.Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER) database. From: http://decipher.sanger.ac.uk/ Accessed: Apr 2012. [DOI] [PMC free article] [PubMed]
- 14.Courtens W, Vermeulen S, Wuyts W, Messiaen L, Wauters J, Nuytinck L, et al. An interstitial deletion of chromosome 7 at band q21: A case report and review. Am J Med Genet A. 2005;134A:12–23. doi: 10.1002/ajmg.a.30106. [DOI] [PubMed] [Google Scholar]
- 15.Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nord AS, Roeb W, Dickel DE, Walsh T, Kusenda M, O’Connor KL, et al. Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur J Hum Genet. 2011;19:727–31. doi: 10.1038/ejhg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okita C, Meguro M, Hoshiya H, Haruta M, Sakamoto YK, Oshimura M. A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics. 2003;81:556–9. doi: 10.1016/s0888-7543(03)00052-1. [DOI] [PubMed] [Google Scholar]
- 18.Monk D, Wagschal A, Arnaud P, Müller PS, Parker-Katiraee L, Bourc’his D, et al. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 2008;18:1270–81. doi: 10.1101/gr.077115.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dória S, Sousa M, Fernandes S, Ramalho C, Brandão O, Matias A, et al. Gene expression pattern of IGF2, PHLDA2, PEG10 and CDKN1C imprinted genes in spontaneous miscarriages or fetal deaths. Epigenetics. 2010;5:444–50. doi: 10.4161/epi.5.5.12118. [DOI] [PubMed] [Google Scholar]
- 20.Al-Murrani A, Ashton F, Aftimos S, George AM, Love DR. Amino-terminal microdeletion within the CNTNAP2 gene associated with variable expressivity of speech delay. Case Rep Genet. 2012;2012:172408. doi: 10.1155/2012/172408. [DOI] [PMC free article] [PubMed] [Google Scholar]