SUMMARY
The presence of foreign DNA in the cytosol of mammalian cells elicits a potent antiviral interferon response. Recently, cytosolic DNA was proposed to induce the synthesis of cyclic-GMP-AMP (cGAMP) upon binding to an enzyme called cGAMP synthase (cGAS). cGAMP activates an interferon response by binding to a downstream receptor called STING. Here we identify natural variants of human STING that are poorly responsive to cGAMP, yet unexpectedly, are normally responsive to DNA and cGAS signaling. We explain this paradox by demonstrating that the cGAS product is actually a non-canonical cyclic-di-nucleotide, cyclic[G(2′ -5′)pA(3′ -5′)p], which contains a single 2′ -5′ phosphodiester bond. Cyclic[G(2′ -5′)pA(3′ -5′)p] potently activates diverse human STING receptors and may therefore be a useful adjuvant or immunotherapeutic. Our results indicate that human STING variants have evolved that can distinguish conventional (3′ -5′) cyclic-di-nucleotides, known only to be produced by bacteria, from the non-canonical cyclic-di-nucleotide produced by mammalian cGAS.
INTRODUCTION
Recognition of pathogen-derived nucleic acid is a major mechanism by which innate immune responses are initiated in mammals (Barbalat et al., 2011). Several families of germ-line encoded nucleic acid sensors have been described, including the Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) (Palm and Medzhitov, 2009; Takeuchi and Akira, 2010). Upon binding nucleic acids, these sensors initiate signaling cascades that lead to the production of cytokines and other immune effector proteins that provide host defense.
The cytosolic presence of foreign double-stranded (ds) DNA triggers a potent antiviral response dominated by the production of type I interferons (IFNs) (Ishii et al., 2006; Stetson and Medzhitov, 2006). An ER-resident host protein called Stimulator of IFN Genes (STING; also called TMEM173, MITA, ERIS and MPYS) was shown to be required for the IFN response to cytosolic dsDNA (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Sun et al., 2009; Zhong et al., 2008). Bacterially-derived second messenger molecules called cyclic-di-nucleotides (CDNs) can also induce an IFN response (McWhirter et al., 2009) that depends on STING (Jin et al., 2011a; Sauer et al., 2011). CDNs are secreted or released into the cytosol by certain bacterial pathogens (Kranzusch et al., submitted; Woodward et al., 2010) and bind directly to STING (Burdette et al., 2011). We previously identified a mutant allele of mouse STING (mSTING), encoding an alanine in place of arginine 231 (R231A), that abolished responsiveness to CDNs, but did not appreciably affect the IFN response to cytosolic dsDNA (Burdette et al., 2011). Thus, although the IFN responses to cytosolic CDNs and dsDNA both require STING, the responses to these chemically distinct ligands can be genetically uncoupled.
Two recent papers (Sun et al., 2013; Wu et al., 2013) unified our understanding of the cytosolic response to CDNs and DNA by proposing that the cytosolic presence of dsDNA leads to the production of a CDN, cyclic-GMP-AMP (cGAMP), by a DNA-dependent sensor enzyme called cGAMP synthase (cGAS). cGAMP was shown to bind and activate STING, but it remained unclear how the mSTING R231A mutant could still initiate responses to dsDNA while lacking responsiveness to CDNs. We therefore sought to investigate the mechanism by which cGAS activates STING.
RESULTS AND DISCUSSION
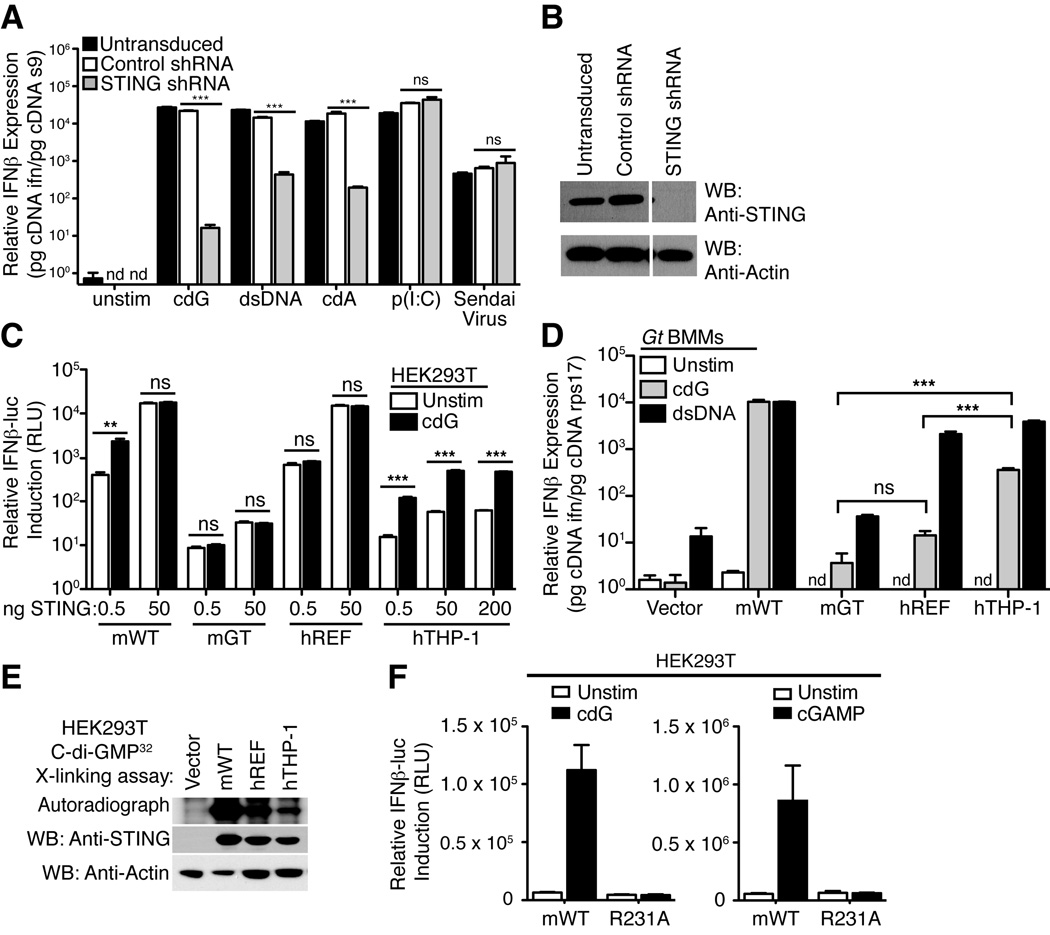
Our previous work (Burdette et al., 2011; Sauer et al., 2011) focused primarily on mouse STING and it is not yet clear whether all human STING variants can respond to CDNs (Conlon et al., 2013; Jin et al., 2011b). In agreement with previous reports (Sun et al., 2013; Wu et al., 2013), we found that the human THP-1 cell line responds robustly to CDNs in a manner dependent on STING (Figure 1A, B). We cloned a hSTING allele from THP-1 cells and compared its amino acid sequence to the previously widely studied reference allele (NP_938023.1; denoted here as hSTINGREF) (Ishikawa and Barber, 2008) (Supplementary Figure 1). We found that hSTINGREF and hSTINGTHP-1 differ at four amino acid positions. Notably, hSTINGTHP-1 encodes an arginine (R) at position 232, corresponding to R231 in mSTING; in contrast, hSTINGREF encodes a histidine (H) at position 232. We decided to test the functionality of individual STING alleles by expressing these alleles in 293T cells that lack endogenous STING. As previously observed (Burdette et al., 2011), overexpression of mSTING in 293T cells induces ligand-independent activation of an IFN-luciferase reporter construct, but expression of lower amounts of mSTING renders 293T cells responsive to CDNs (Figure 1C). Similarly, 293T cells over-expressing hSTINGREF spontaneously activated an IFN-luciferase reporter (Figure 1C). However, unlike mSTING, cells expressing low amounts of hSTINGREF were poorly responsive to stimulation with cyclic-di-GMP (Figure 1C). In contrast, cells expressing hSTINGTHP-1 were responsive to cyclic-di-GMP, and exhibited ~10-fold induction of the IFN-luciferase reporter, similar to what is seen with mSTING (Figure 1C).
Figure 1. Variable responsiveness of human STING variants to cyclic-di-nucleotides maps to arginine 232.
(A) THP-1 cells were transduced with vectors encoding an shRNA targeting STING or a control shRNA. Cells were then stimulated with cyclic-di-GMP (cdG), dsDNA, cyclic-di-AMP (cdA), poly-inosine:cytosine (pI:C), or Sendai Virus, and induction of human interferon-β mRNA was assessed by quantitative reverse transcriptase PCR. (B) Western blotting confirmed that knockdown of STING was effective. (C) HEK293T cells were transfected with the indicated amounts of various mouse (m) or human (h) STING expression plasmid and then stimulated 6h later by transfection with synthetic cdG (5µM). GT denotes the null I199N allele of STING from Goldenticket (Gt) mice. STING activation was assessed by use of a cotransfected IFN-luciferase reporter construct. (D) Gt (STING-null) macrophages were transduced with retroviral vectors encoding the indicated STING alleles and were then stimulated 48h later by transfection with cdG (5µM) or dsDNA 70-mer oligonucleotide (0.5µg/mL). IFN induction was measured by qRT-PCR. ND, not detected. (E) Binding assay of STING to 32P-c-di-GMP. STING proteins were expressed in HEK293T cells and cell lysates were subjected to UVcrosslinking with 32P-cdG, and resolved by SDS-PAGE. Binding was quantified by autoradiography. Western blots of cell lysates with an anti-STING polyclonal antibody confirmed similar expression of the various STING proteins. (F) Responsiveness of mSTING to cGAMP is affected by mutations of R231. The indicated mutants were tested as in C. Data are representative of at least three independent experiments and are presented as mean. Error is represented as standard error of the mean. ***, P < 0.0001; **. P < 0.005. See also Figure S1.
293T cells expressing STING do not respond to stimulation by dsDNA, presumably due to lack of expression of cGAS (Sun et al., 2013) or perhaps other dsDNA sensors. Therefore, to test whether the hSTING variants could respond to dsDNA stimulation, cGAS+ STING-null (‘goldenticket’) (Sauer et al., 2011) macrophages were transduced with hSTING expression vectors. Even the hSTINGREF variant that responds poorly to CDNs conferred responsiveness to dsDNA (Figure 1D). hSTINGREF therefore phenocopies mSTINGR231A and uncouples responsiveness to CDNs and dsDNA (Burdette et al., 2011).
Consistent with the above results with hSTING variants, an R231H mutant of mSTING responded poorly to CDNs, as did R232A or R232H variants of hSTINGTHP-1 (Supplementary Figure S2A). Arginine 231/232 thus appears critical for responsiveness to CDNs in mouse/human STING. Introduction of an H232R mutation in hSTINGREF was not sufficient to restore responsiveness to CDNs; indeed, we found that a second substitution (G230A) was also required (Supplementary Figure S3). All the variant STING alleles we tested bound cyclic-di-GMP (Figure 1E; Supplementary Figure S2B) (Huang et al., 2012; Ouyang et al., 2012; Yin et al., 2012), consistent with the fact that residues 230 and 232 are located in loops that cover but do not form the CDN binding pocket.
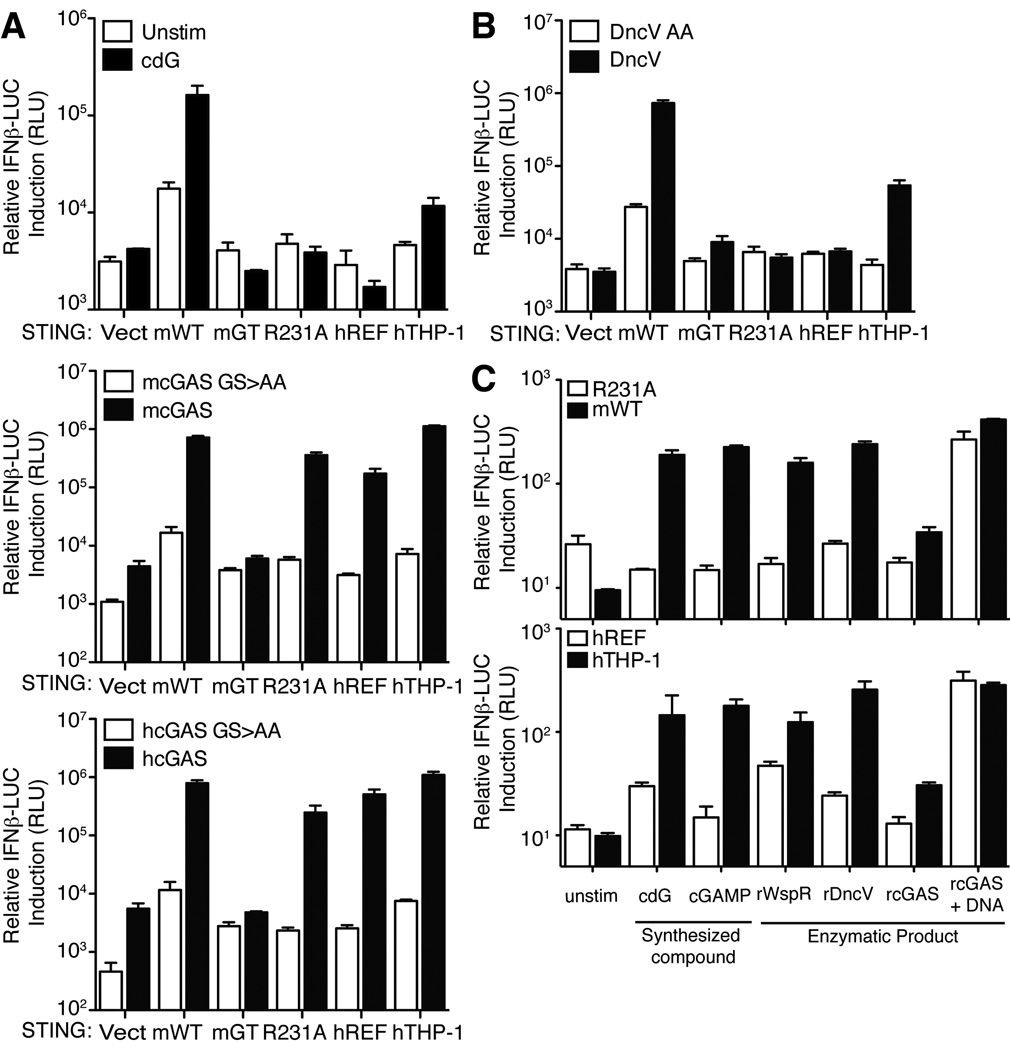
Importantly, mSTINGR231A also failed to respond to chemically synthesized cGAMP (Figure 1F) (Kellenberger et al., 2013). This observation raised the question of whether R231A/R232H variants of STING would respond to the cGAS enzyme that is believed to activate STING via production of cGAMP. Surprisingly, we found that human or mouse cGAS expression robustly activates hSTINGREF and mSTINGR231A variants (Figure 2A). We considered several explanations for this puzzling result. One possibility is that the response is due simply to overexpression of the cGAMP synthase in mammalian cells; however, overexpression of DncV, a bacterial cGAMP synthase from V. cholerae (Davies et al., 2012), did not activate hSTINGREF or mSTINGR231A but did activate wild-type mSTING and hSTINGTHP-1 (Figure 2B). An alternative hypothesis is that cGAS might activate STING by a direct physical interaction and in a manner independent of cGAMP production. However, this explanation also appears to be incorrect. As previously demonstrated (Sun et al., 2013), overexpression of catalytically dead mutants of human or mouse cGAS (GS>AA; Figure 2A) failed to activate STING variants, arguing that cGAS signaling depends on the production of a second messenger rather than on a direct physical interaction with STING. To confirm this interpretation, we produced the enzymatic product of cGAS by providing ATP, GTP and dsDNA to purified recombinant cGAS in vitro. As a negative control, dsDNA (required to stimulate cGAS activity) was omitted from a parallel reaction. The resulting cGAS products were then purified and transfected into 293T cells expressing STING variants. In contrast to synthetic cGAMP, the cGAS product was able to activate hSTINGREF and mSTINGR231A (Figure 2C). This experiment argues against a model in which cGAS activates hSTINGREF via a direct physical interaction.
Figure 2. STING variants are responsive to cGAS.
(A) HEK293T cells were transfected with the indicated STING alleles and with human and mouse cGAS (wt and GS>AA mutants) (Sun et al., 2013) as indicated. STING activation was assessed by a co-transfected IFN-luciferase reporter construct. (B) HEK293T cells were transfected with the indicated STING alleles and with a mammalian expression vector encoding a cGAMP synthase (DncV) from V. cholerae. STING activation was assessed as in A. (C) In vitro enzymatically generated products of rWspR, rDncV and rcGAS were transfected into digitonin permeabilized HEK293T cells expressing the indicated mouse and human STING proteins. Chemically synthesized cyclic-di-GMP (cdG) and cGAMP were included as controls. STING activation was assessed as in A and B. Data are representative of at least three independent experiments and are presented as mean. Error is represented as standard error of the mean. See also Figure S2.
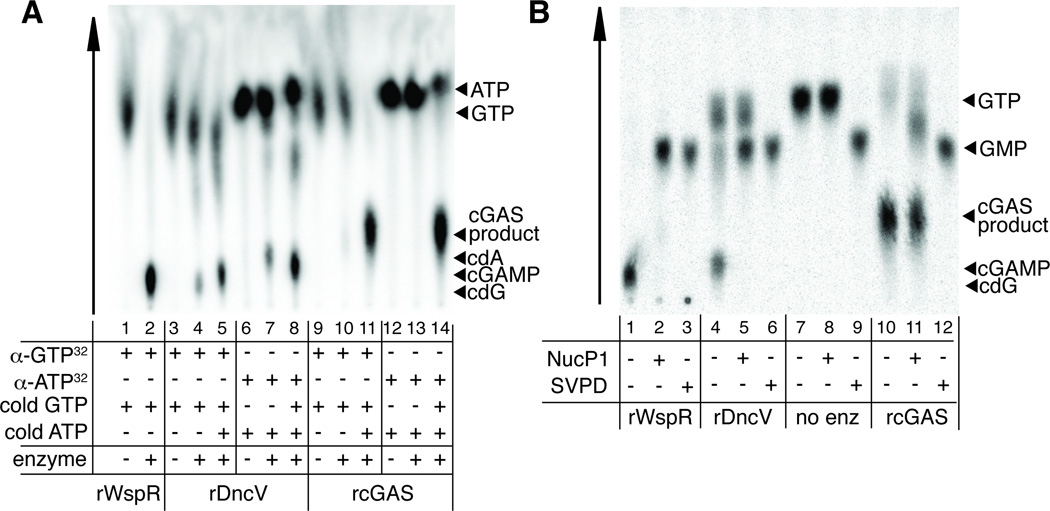
cGAS is structurally homologous to another innate immune sensor called oligoadenylate synthase (OAS1), which produces a non-canonical oligoadenylate polymer containing 2′ -5′ phosphodiester bonds (Kranzusch et al., 2013; Sun et al., 2013). Therefore we hypothesized that cGAS might not produce a canonical CDN as previously proposed (Sun et al., 2013; Wu et al., 2013), but might instead produce a novel CDN, containing 2′ -5′ phosphodiester bond(s), that could stimulate variant STING alleles. Such a non-canonical CDN would have a mass identical to the canonical 3′ -5′ phosphodiester-linked CDN, and thus the two products would not have been easy to distinguish by previously published mass spectrometric analyses of the cGAS product (Sun et al., 2013; Wu et al., 2013). We tested our hypothesis by providing radiolabelled α32P-GTP or α32P-ATP to recombinant purified cGAS or V. cholerae DncV and analyzing the products by thin-layer chromatography. As reported previously, DncV can produce some c-di-AMP if provided only ATP, and some c-di-GMP if provided only GTP, but prefers to make cGAMP when provided both ATP and GTP (Davies et al., 2012) (Figure 3A). cGAS requires both ATP and GTP substrates, and the resulting product migrates significantly differently than any of the canonical CDNs produced by DncV, suggesting that cGAS produces a novel, noncanonical CDN (Figure 3A).
Figure 3. cGAS produces a non-canonical cyclic dinucleotide.
(A) Purified recombinant WspR, DncV and cGAS were mixed with α32P-GTP or α32P-ATP and the indicated unlabeled nucleotides. Reactions were mixed with TLC running buffer and nucleic acid species were resolved on a PEI-Cellulose TLC plate. (B) WspR, DncV and cGAS products labeled with α32PGTP were digested with nuclease P1 or Snake Venom Phosphodiesterase (SVPD) and resolved on a PEI-Cellulose TLC plate. See also Figure S3.
We analyzed the cGAS and DncV products by specific nuclease digestion. The cGAS product is partially cleaved by nuclease P1, which selectively digests 3′ -5′ phosphodiester linkages (Pino et al., 2008), suggesting that the cGAS product contains at least one 3′ -5′ phosphodiester linkage (Figure 3B). Nuclease P1 digestion is incomplete, as it does not lead to generation of GMP, in contrast to what is observed upon treatment of the DncV product with nuclease P1 (Figure 3B). As a control, digestion of the cGAS or DncV products with snake venom phosphodiesterase, which cleaves both 2′ -5′ and 3′ -5′ phosphodiester linkages (Pino et al., 2008), led to complete digestion (Figure 3B). Taken together these results suggested that the cGAS product might also contain a 2′ -5′ phosphodiester linkage.
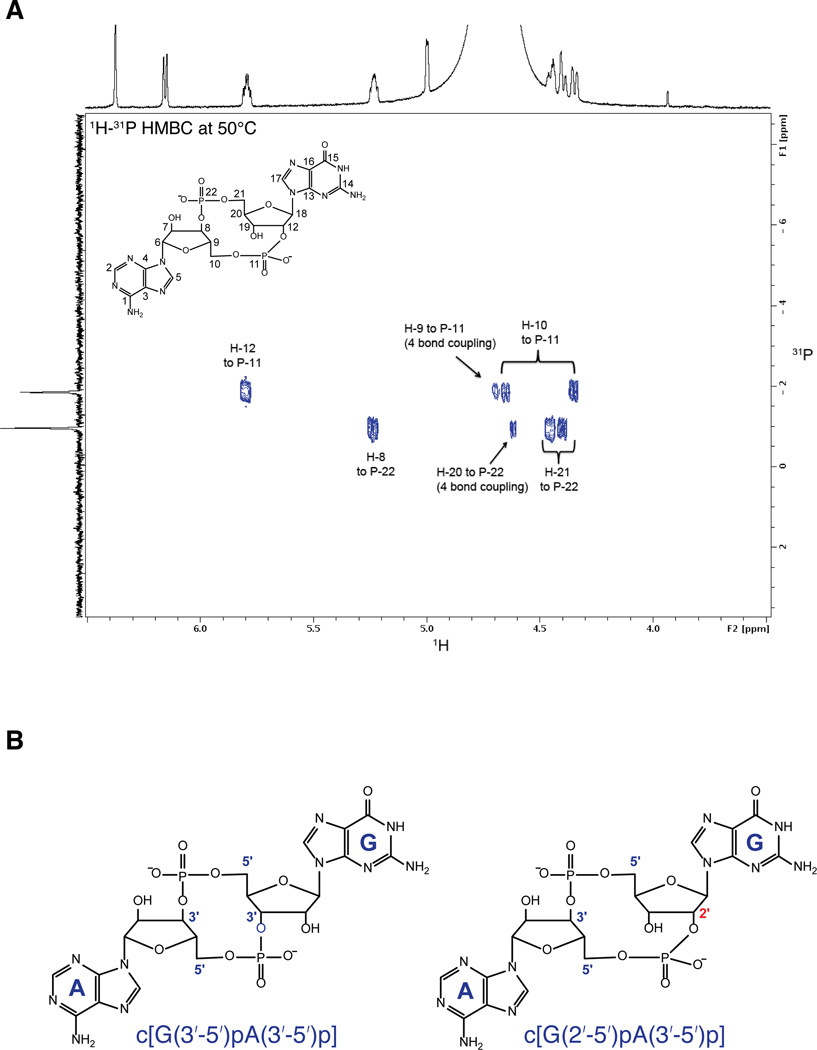
To identify the nature of the cGAS product, and in particular to ascertain the regiochemistry of the phosphodiester linkages, we analyzed the cGAS product by NMR (Figure 4A, Supplementary Figure S4). These results indicate that the product of cGAS is a noncanonical CDN containing a single 2′ -5′ phosphodiester linkage, and its chemical structure is assigned as cyclic[G(2′ -5′)pA(3′ -5′)p] (Figure 4B; see Supplementary Extended Discussion). This second messenger appears to be a robust activator of STING. A recently published crystallographic and enzymatic study of cGAS independently identified this noncanonical CDN as the product of cGAS (Gao et al., 2013) but did not address its unique signaling properties. Our results suggest that the unique ability of cyclic[G(2′ -5′)pA(3′ -5′)p] to stimulate diverse hSTING alleles may explain why cGAS synthesizes this unusual molecule. Although the bacterial cGAMP synthase DncV is also a distant homolog of OAS and cGAS (Davies et al., 2012), our IFN-reporter and thin-layer chromatography data strongly suggest that DncV produces a canonical CDN with two 3′ -5′ linkages (Figure 4B). It remains to be determined whether any bacterial enzymes produce non-canonical CDNs, or whether perhaps non-canonical CDNs are unique to mammals.
Figure 4. cGAS produces a cyclic dinucleotide containing a 2′ -5′ phosphodiester linkage.
(A) 1H-31P HMBC of HPLC-purified cGAS product acquired at 600 MHz and 50° C. Critical through-bond correlations for the phosphodiester bonds are indicated. NMR elucidated structure of cGAS product is also shown. (B) Chemical structures of canonical c[G(3′ -5′)pA(3′ -5′)p] cGAMP (left) and non-canonical c[G(2′ -5′)pA(3′ -5′)p] cGAMP (right). See also Figure S4.
We identified the R231A mutant of mSTING after a thorough site-directed mutagenesis of STING (Burdette et al., 2011). We were surprised to discover that the corresponding arginine (R232) is a site of natural polymorphism in human STING. It is tempting to speculate that R232H variant of hSTING may confer a selective advantage by reducing responses to bacterial CDNs, while still retaining responsiveness to endogenous non-canonical CDNs produced by cGAS in response to viral dsDNA. Indeed, although production of type I IFNs is essential for control of most viruses, type I IFNs have often been found to be detrimental in the response to bacterial infections (Monroe et al., 2010). It is not yet clear why the R231A/R232H STING variants can be stimulated by the non-canonical CDN but not by canonical CDNs. An explanation awaits high resolution structures of these variants bound to cyclic[G(2′ -5′)pA(3′ -5′)p].
CDNs have been proposed to be useful as vaccine adjuvants or immunotherapeutics (Chen et al., 2010). In addition, a synthetic STING activator, DMXAA, has been tested in human clinical trials as a novel chemotherapeutic agent. DMXAA was not found to be effective in humans, likely because it is unable to stimulate hSTING (Conlon et al., 2013). In this context, our results may be significant as they suggest that non-canonical 2′ -5′ linked CDNs might have clinical value as potent pan-agonists of diverse STING variants, including those variants that respond poorly to canonical CDNs or DMXAA.
EXPERIMENTAL PROCEDURES
Mice and cell lines
THP-1 cells were grown in RPMI 1640 supplemented with 10% FBS, penicillin-streptomycin and L-glutamine. HEK293T cells were grown in DMEM supplemented with 10% FBS, penicillin-streptomycin and L-glutamine. GP2 retroviral packaging cell lines were maintained in DMEM supplemented with 10% FBS, penicillin-streptomycin and L-glutamine. Animal protocols were approved by the University of California, Berkeley Animal Care and Use Committee.
STING Knockdown
Knockdown of human STING (clone ID NM_198282.1-901s1c1) was achieved using pLKO.1 (The RNAi Consortium). The sequence for knockdown of human STING is 5′ -GCA GAG CTA TTT CCT TCC ACA which correspond to 5′ -CCG GGC AGA GCT ATT TCC TTC CAC ACT CGA GTG TGG AAG GAA ATA GCT CTG CTT TTT G forward oligo and 5′ -AAT TCA AAA AGC AGA GCT ATT TCC TTC CAC ACT CGA GTG TGG AAG GAA ATA GCT CTG C reverse oligo. Oligos were annealed and cloned into AgeI and EcoRI digested pLKO.1 (Addgene) and retrovirally transduced into THP-1 cells in parallel with scramble shRNA control constructs. Stable cell lines were selected with puromycin. THP-1 cells were differentiated with 1µg/mL PMA for 24 hours. Cells were allowed to rest for 24 hours and then restimulated for 6 hours with the indicated ligands. IFN induction was measured by qRT-PCR as described below.
Cell Stimulation and reagents
Bone marrow macrophages and HEK293T cells were stimulated using Lipofectamine 2000 (Invitrogen). Unless otherwise specified, cyclic-di-GMP, cyclic-di-AMP, polyI:C and Vaccinia Virus 70mer DNA was prepared as described previously (Burdette et al., 2011) and used at similar concentrations. Sendai virus was purchased from Charles River Laboratories. cGAMP was synthesized as previously described (Kellenberger et al., 2013).
Cloning, mutagenesis and plasmids
The THP-1 STING allele was amplified from cDNA using 5′ hSTING HindIII(5′ – ATC GAA GCT TCC ACC ATG CCC CAC TCC AGC CTG) and 3′ hSTING NotI (5′ – ATC GGC GGC CGC TCA GGC ATA GTC AGG CAC GTC ATA AGG ATA AGA GAA ATC CGT GCG GAG AG). Resulting PCR product was cloned into pCDNA3 using HindIII/NotI digestion. THP-1 STING was amplified and cloned into MSCV2.2 using the 3′ primer listed above and 5′ hSTING XhoI (5′ – ATC GCT CGA GCC ACC ATG CCC CAC TCC AGC CTG) and XhoI/NotI digestion. IFN-luciferase, TK-Renilla and mouse STING plasmids were used as previously described (Burdette et al., 2011). Mutations in human STING were introduced using Quikchange Site Directed Mutagenesis Kit (Stratagene). cDNA clones corresponding to mouse and human cGAS (MGC Fully Sequenced Human MB21D1 cDNA, Accession: BC108714.1, Clone ID: 6015929; EST Fully Sequenced Mouse E330016A19Rik cDNA, Accession: BC145653.1, Clone ID: 40130956) were obtained from Open Biosystems and correspond to those described previously (Sun et al., 2013; Wu et al., 2013). Mouse cGAS was amplified from cDNA clones with an N-terminal flag tag with forward oligo 5′ -mcGAS-KpnI (5′ -ATC GGG TAC CCC ACC ATG GAT TAC AAG GAT GAC GAT GAC AAG GAA GAT CCG CGT AGA AGG) and reverse oligo 3′ -mcGAS-NotI (5′ -ATC GGC GGC CGC TCA AAG CTT GTC AAA AAT TGG). Likewise, hcGAS was amplified with forward oligo 5′ -hcGAS-flag-KpnI (5′ -ATC GGG TAC CCC ACC ATG GAT TAC AAG GAT GAC GAT GAC AAG CAG CCT TGG CAC GGA AAG G) and reverse 3′ -hcGAS-NotI (5′ ATC GGC GGC CGC TCA AAA TTC ATC AAA AAC TGG AAA C). Both PCR products were cloned into pcDNA3 at KpnI and NotI restriction enzyme sites. DncV was amplified using DncV fwd BamHI (5′ – GCA TGG ATC CGC CAC CAT GAC TTG GAA CTT TCA CCA G) and DncV rev NotI (5′ – GCA TGC GGC CGC TCA GCC ACT TAC CAT TGT GCT GC) and cloned into pCDNA3 using BamHI and NotI. For cloning into MSCV2.2, DncV was amplified using DncV fwd XhoI (5′ – GCA TCT CGA GCC ACC ATG ACT TGG AAC TTT CAC CAG) and DncV rev NotI. Resulting DNA was cloned into MSCV 2.2 digested with XhoI/NotI. Constructs for bacterial mcGAS overexpression were constructed as follows. N-terminal His6-SUMO tag amplified by PCR using His6 SUMO Nco (5′ -TAA TAA GGA GAT ATA CCA TGG GCA GCA GCC) and His6 SUMO Sal (5′ -GAA TTC GTC GAC ACC AAT CTG TTC TCT GTG AGC) off of pCDF-Duet2 template (gift from M. Rape lab, UC-Berkeley) and cloned into pET28a using NcoI and SalI to make pET28a-H6SUMO. Full length mcGAS was PCR amplified from the mouse cDNA clone described above using mcGAS fwd Sal (5′ -GAT GTC GAC ATG GAA GAT CCG CGT AGA AGG ACG) and mcGAS rev Xho (5′ -ATC CTC GAG TCA AAG CTT GTC AAA AAT TGG AAA CC) and cloned into pET28a-H6SUMO using SalI and XhoI to make pET28a-H6SUMO-mcGAS that expresses full length mcGAS fused to an N-terminal His6 SUMO tag.
cGAS product purification and structural characterization
The cGAS product was purified using reverse-phase HPLC on an Agilent 1260 Infinity HPLC equipped with an Agilent Polaris C18-A column (5 µm, 250 mm × 10 mm, 180 Å). Purification conditions include a 100% to 0% gradient of solvent A over 20 min at 50°C and a flow rate of 5 mL/min, where solvent A is 100 mM ammonium acetate in water and solvent B is acetonitrile. Purified elution fractions were evaporated multiple times in order to remove excess ammonia. Resonance assignments were made using COSY, 1H-13C HSQC, NOESY, 1H-13C HMBC, and 1H-31P HMBC. Characterization of cGAS product: 1H NMR (900 MHz, D2O, 50°C, δ): 8.44 (1H, s), 8.42 (1H, s), 8.03 (1H, s), 6.31 (1H, s), 6.09 (1H, J = 8 Hz, d), 5.75 (1H, m), 5.18 (1H, m), 4.93 (1H, s), 4.74, 4.62, 4.59 (1H, J = 12 Hz, d), 4.55 (1H, s), 4.38 (1H, m), 4.33 (1H, J = 12 Hz, d), 4.28 (1H, J = 12 Hz, d); 31P {1H decoupled} NMR (600 MHz, D2O, 50°C, δ): (all resonances are singlets) −0.96, −1.86; HRMS (m/z): [M-H]- monoisotopic mass calculated for C20H24N10O13P2, 673.0927; found, 673.0909. [M+Na-2H]− monoisotopic mass calculated for C20H24N10O13P2, 695.0752; found, 695.0728.
Luciferase assay
HEK293T cells were plated in TC-treated 96-well plates at 0.5 × 106 cells ml−1. The next day, the cells were transfected with indicated constructs, together with IFN-β-firefly luciferase and TK-Renilla luciferase reporter constructs. Following stimulation for 6 h with the indicated ligands, the cells were lysed in passive lysis buffer (Promega) for 5 min at 25° C. The cell lysates were incubated with firefly luciferase substrate (Biosynth) and the Renilla luciferase substrate coelenterazine (Biotium), and luminescence was measured on a SpectraMax L microplate reader (Molecular Devices). The relative Ifnb expression was calculated as firefly luminescence relative to Renilla luminescence. Statistical differences were calculated with an unpaired two-tailed Student’s t-test using Prism 5.0b software (GraphPad).
In vitro cyclic-di-nucleotide synthesis
In vitro DncV reactions were carried out in 20mM Tris-Cl, pH=8, 20 Mg(OAc)2, 10% glycerol and 1mM DTT, 0.1mg/mL BSA. Reactions contained 250µM GTP, 250µM ATP or 125µM GTP and 125µM ATP as indicated in Figures. In addition, 33nM α32P-GTP (3000Ci/mmol, Perkin-Elmer) or 33nM α32P-ATP (3000Ci/mmol, Perkin-Elmer) was included in reaction where indicated. Reactions were started by addition of 1µM purified DncV protein. In vitro cGAS reactions were carried out in 40mM Tris-Cl, pH=7.5, 100mM NaCl, 10mM MgCl2. Cold nucleotide and alpha-labeled GTP is at the same concentrations as in DncV reactions. Reactions were started by addition of 200nM purified cGAS. Where indicated, herring testes DNA (Sigma) was added to reactions at a final concentration of 0.1mg/mL. WspR reactions were performed as described previously (Burdette et al., 2011). Reactions were incubated for 1 hour at 37° C, boiled for 5min at 95° C, and spun for 10 minutes at 13,000rpm. Reactions were removed and mixed 1:5 with TLC running buffer (1:1.5 (v/v) saturated NH4SO4 and 1.5M KH2PO4, pH 3.6) and spotted on PEI-cellulose TLC plate (Sigma). Following solvent migration, the TLC plate was exposed to a phosphorimager screen and imaged using Typhoon scanner. For in vitro product transfection into 293T cells, reactions were scaled up, radiolabeled nucleotide was omitted and the concentration of ATP and GTP was increased to 2mM.
Supplementary Material
HIGHLIGHTS.
Human STING variants respond poorly to cGAMP but still respond to cytosolic DNA.
DNA sensing through cGAS leads to production of a non-canonical 2′ -5′ linked cGAMP.
STING variants respond robustly to 2′ -5′ linked cGAMP and not canonical cGAMP.
ACKNOWLEDGEMENTS
This work is supported by NIH grants AI075039 (R.E.V.), AI063302 (R.E.V.), AI080749 (R.E.V.), AI103817 (R.E.V.), and DP2 OD008677 (M.C.H.). R.E.V. is an Investigator in the Pathogenesis of Infectious Disease funded by the Burroughs Wellcome Fund. M.C.H. is a Career Award at the Scientific Interface Investigator funded by the Burroughs Wellcome Fund. E.J.D is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2131-12). D.L.B. is supported by an NIH National Research Service Award fellowship F32 (AI091100). C.A.K. and S.C.W. are supported by an NIH NRSA Training Grant in chemical biology and C.A.K. is supported by a Department of Defense NDSEG fellowship. We are grateful to J. Mekalanos (Harvard Medical School) for the gift of DncV, S. Lory (Harvard Medical School) for the gift of Pseudomonas and Vibrio diguanylate cyclases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kuolee R, Yan H. The potential of 3',5'-cyclic diguanylic acid (c-di-GMP) as an effective vaccine adjuvant. Vaccine. 2010;28:3080–3085. doi: 10.1016/j.vaccine.2010.02.081. [DOI] [PubMed] [Google Scholar]
- Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, et al. Mouse, but not Human STING, Binds and Signals in Response to the Vascular Disrupting Agent 5,6-Dimethylxanthenone-4-Acetic Acid. J Immunol. 2013 doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2',5')pA(3'5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013 doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011a;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011b;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic di-GMP and Cyclic AMP-GMP. J Am Chem Soc. 2013 doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee ASY, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second messenger enzymes in innate immunity. Cell Reports. 2013 doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee ASY, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. doi: 10.1016/j.celrep.2013.05.008. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Induction of type I interferons by bacteria. Cell Microbiol. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Pino S, Ciciriello F, Costanzo G, Di Mauro E. Nonenzymatic RNA ligation in water. J Biol Chem. 2008;283:36494–36503. doi: 10.1074/jbc.M805333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.