Abstract
Background
Pseudomonas aeruginosa is an important nosocomial pathogen that exhibits multiple resistances to antibiotics with increasing frequency, making patient treatment more difficult. The aim of the study is to ascertain the population structure of this clinical pathogen in the Hospital Son Llàtzer, Spain.
Results
A significant set (56) of randomly selected clinical P. aeruginosa isolates, including multidrug and non-multidrug resistant isolates, were assigned to sequence types (STs) and compared them with their antibiotic susceptibility profile classified as follows: extensively drug resistant (XDR), multidrug resistant (MDR) and non-multidrug resistant (non-MDR). The genetic diversity was assessed by applying the multilocus sequence typing (MLST) scheme developed by Curran and collaborators, and by the phylogenetic analysis of a concatenated tree. The analysis of seven loci, acsA, aroE, guaA, mutL, nuoD, ppsA and trpE, demonstrated that the prevalent STs were ST-175, ST-235 and ST-253. The majority of the XDR and MDR isolates were included in ST-175 and ST-235. ST-253 is the third in frequency and included non-MDR isolates. The 26 singleton sequence types corresponded mainly to non-MDR isolates. Twenty-two isolates corresponded to new sequence types (not previously defined) of which 12 isolates were non-MDR and 10 isolates were MDR or XDR.
Conclusions
The population structure of clinical P. aeruginosa present in our hospital indicates the coexistence of nonresistant and resistant isolates with the same sequence type. The multiresistant isolates studied are grouped in the prevalent sequence types found in other Spanish hospitals and at the international level, and the susceptible isolates correspond mainly to singleton sequence types.
Keywords: Pseudomonas aeruginosa, Multilocus sequence typing, Multiresistant, Clinical isolates, Population structure
Background
Pseudomonas aeruginosa is a non-fermenting Gram-negative bacterium that is widely distributed in nature. The minimum nutritional requirements, tolerance to a wide variety of physical conditions and intrinsic resistance against many antibiotics explain its role as an important nosocomial pathogen. Certain bacterial clones have been distributed worldwide and, in most cases, associated with multiresistance patterns [1-3]. Because the number of active antibiotics against P. aeruginosa is limited, it is a priority to perform a strict and regular follow up of the resistance patterns in individual hospitals.
In the microbiology laboratory of the Hospital Son Llàtzer (Mallorca, Spain) the number of isolates of P. aeruginosa is increasing annually. In 2010, the number of isolates of P. aeruginosa was 1174, being the second pathogen isolated after Escherichia coli. When the P. aeruginosa resistance pattern of the P. aeruginosa isolates from this hospital were compared with the latest Spanish surveillance study of antimicrobial resistance [4], it was revealed that the resistance levels of the isolates in our hospital were higher against all of the antibiotics commonly used in the treatment of infections caused by P. aeruginosa, contributing to therapeutic difficulties.
The introduction of molecular techniques has led to significant progress in both bacterial identification and typing. In P. aeruginosa, several schemes for molecular typing have been used, such as ribotyping [5], PCR-based fingerprinting [6], or pulsed-field gel electrophoresis (PFGE) [7], which is considered the ‘gold standard’ technique. Curran et al. (2004) developed a multilocus sequence typing (MLST) scheme that discriminates P. aeruginosa isolates by differences in the sequences of seven genes: acsA, aroE, guaA, mutL, nuoD, ppsA and trpE, providing a good comprehensive database that allows the comparison of results obtained in different locations for different sample types [8]. Since this work, MLST has been applied in several studies of P. aeruginosa to better understand the epidemiology of infections in patients with cystic fibrosis and to study multiresistant clones.
The main objective of our study is to characterise the isolates of P. aeruginosa analysed routinely in the Hospital Son Llàtzer at the molecular level. A significant set of randomly selected clinical isolates (fifty-six), including multidrug and non-multidrug resistant isolates, was further studied to determine the population structure of this clinical pathogen in our hospital and to compare it with other Spanish and international multicentre surveillance studies.
Methods
P. aeruginosa culture collection
A total of 56 isolates of P. aeruginosa from 53 specimens recovered from 42 patients of the Hospital Son Llàtzer were randomly selected between January and February 2010. Three samples showed two distinct colony morphologies, and both types of each isolate were studied by MLST to establish possible differences between them (these morphologies are labelled by the number of the isolate, followed by the letters a or b). Isolates from different origins were taken as part of standard care (Table 1). The hospital is a tertiary teaching hospital with 377 beds and serves a catchment population of approximately 250,000 inhabitants. All of the P. aeruginosa isolates were isolated and cultured on Columbia agar with 5% sheep blood (bioMérieux, Marcy d’Etoile, France). The cultivation and incubation times of the plates were performed under routine laboratory conditions (24 h at 37°C). The study was approved by the research board of our hospital. Individual patient’s consent was not sought as isolates were derived from routine diagnostics and as data were processed anonymously.
Table 1.
List of the alleles and sequence type, origin of the sample, antibiotic pattern and number of patient for each isolate
| Isolates | Patient | Sample origina | STs | acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | Antibiotic patternb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PaC1 |
1 |
W |
ST-1138* |
5 |
3 |
79 |
5 |
1 |
7 |
47 |
XDR |
| PaC49 |
1 |
RS |
ST-1191* |
45 |
148* |
15 |
35 |
53 |
106* |
3 |
MDR |
| PaC51 |
1 |
RS |
ST-1147* |
17 |
3 |
5 |
3 |
1 |
6 |
26 |
non-MDR |
| PaC52 |
1 |
W |
ST-1138* |
5 |
3 |
79 |
5 |
1 |
7 |
47 |
XDR |
| PaC2 |
2 |
W |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC3 |
3 |
R |
ST-235 |
38 |
11 |
3 |
13 |
1 |
2 |
4 |
non-MDR |
| PaC4 |
4 |
R |
ST-800 |
17 |
22 |
11 |
3 |
3 |
15 |
3 |
non-MDR |
| PaC5 |
5 |
W |
ST-108 |
39 |
5 |
20 |
5 |
1 |
6 |
31 |
XDR |
| PaC7 |
6 |
R |
New-1 |
28 |
5 |
36 |
- |
3 |
13 |
7 |
MDR |
| PaC16 |
6 |
R |
New-2 |
28 |
5 |
20 |
- |
3 |
13 |
7 |
MDR |
| PaC9 |
7 |
U |
ST-274 |
23 |
5 |
11 |
7 |
1 |
12 |
7 |
XDR |
| PaC10 |
8 |
U |
ST-1139* |
28 |
22 |
20 |
3 |
3 |
14 |
19 |
XDR |
| PaC19 |
8 |
U |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC32 |
8 |
RS |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC40 |
8 |
R |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC11 |
9 |
U |
ST-235 |
38 |
11 |
3 |
13 |
1 |
2 |
4 |
XDR |
| PaC24 |
9 |
U |
ST-235 |
38 |
11 |
3 |
13 |
1 |
2 |
4 |
XDR |
| PaC12 |
10 |
R |
ST-1140* |
38 |
11 |
20 |
13 |
1 |
2 |
4 |
XDR |
| PaC14 |
11 |
R |
ST-1141* |
113 |
4 |
5 |
67 |
1 |
17 |
26 |
non-MDR |
| PaC15 |
12 |
R |
ST-1142* |
35 |
11 |
112 |
16 |
1 |
6 |
42 |
MDR |
| PaC17 |
13 |
R |
ST-1186* |
39 |
5 |
20 |
59* |
1 |
20 |
15 |
non-MDR |
| PaC18a |
14 |
U |
ST-1143* |
8 |
5 |
9 |
3 |
1 |
4 |
9 |
non-MDR |
| PaC18b |
14 |
U |
ST-1143* |
8 |
5 |
9 |
3 |
1 |
4 |
9 |
non-MDR |
| PaC20 |
15 |
U |
ST-1144* |
39 |
10 |
1 |
3 |
4 |
6 |
7 |
non-MDR |
| PaC21 |
16 |
R |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC23 |
17 |
U |
ST-1187* |
35 |
22 |
5 |
3 |
1 |
104* |
4 |
non-MDR |
| PaC25a |
18 |
U |
ST-207 |
47 |
4 |
5 |
33 |
1 |
6 |
40 |
non-MDR |
| PaC25b |
18 |
U |
ST-207 |
47 |
4 |
5 |
33 |
1 |
6 |
40 |
non-MDR |
| PaC26 |
19 |
R |
ST-235 |
38 |
11 |
3 |
13 |
1 |
2 |
4 |
XDR |
| PaC27 |
20 |
R |
ST-274 |
23 |
5 |
11 |
7 |
1 |
12 |
7 |
non-MDR |
| PaC28 |
21 |
U |
ST-1188* |
69* |
147* |
5 |
11 |
2 |
15 |
2 |
MDR |
| PaC29 |
22 |
R |
ST-1145* |
15 |
5 |
1 |
1 |
1 |
12 |
1 |
non-MDR |
| PaC30 |
23 |
PS |
ST-244 |
17 |
5 |
12 |
3 |
14 |
4 |
7 |
non-MDR |
| PaC31 |
24 |
U |
ST-499 |
11 |
5 |
7 |
27 |
2 |
7 |
33 |
MDR |
| PaC33 |
25 |
R |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC34 |
26 |
R |
ST-179 |
36 |
27 |
28 |
3 |
4 |
13 |
7 |
MDR |
| PaC35 |
27 |
U |
ST-235 |
38 |
11 |
3 |
13 |
1 |
2 |
4 |
MDR |
| PaC36a |
28 |
R |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC36b |
28 |
R |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
MDR |
| PaC37 |
29 |
B |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC38 |
29 |
B |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC39 |
30 |
U |
ST-508 |
15 |
5 |
11 |
3 |
2 |
4 |
3 |
non-MDR |
| PaC41 |
31 |
U |
ST-1189* |
72* |
5 |
11 |
72 |
3 |
105* |
3 |
non-MDR |
| PaC42 |
32 |
W |
ST-253 |
4 |
4 |
16 |
12 |
1 |
6 |
3 |
MDR |
| PaC56 |
32 |
B |
ST-253 |
4 |
4 |
16 |
12 |
1 |
6 |
3 |
MDR |
| PaC43 |
33 |
R |
ST-235 |
38 |
11 |
3 |
13 |
1 |
2 |
4 |
XDR |
| PaC44 |
34 |
U |
ST-316 |
13 |
8 |
9 |
3 |
1 |
6 |
9 |
non-MDR |
| PaC45 |
35 |
U |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC46 |
36 |
R |
ST-1190* |
45 |
148* |
15 |
35 |
53 |
106* |
19 |
MDR |
| PaC47 |
37 |
R |
ST-175 |
28 |
22 |
5 |
3 |
3 |
14 |
19 |
XDR |
| PaC48 |
38 |
R |
ST-253 |
4 |
4 |
16 |
12 |
1 |
6 |
3 |
non-MDR |
| PaC54 |
38 |
R |
ST-253 |
4 |
4 |
16 |
12 |
1 |
6 |
3 |
non-MDR |
| PaC50 |
39 |
US |
ST-1146* |
5 |
11 |
57 |
33 |
1 |
6 |
3 |
non-MDR |
| PaC53 |
40 |
U |
ST-1148* |
16 |
5 |
12 |
3 |
2 |
1 |
18 |
non-MDR |
| PaC55 |
41 |
R |
ST-179 |
36 |
27 |
28 |
3 |
4 |
13 |
7 |
non-MDR |
| PaC57 | 42 | R | ST-1149* | 18 | 11 | 57 | 5 | 1 | 20 | 26 | non-MDR |
Asterisk mark (*) indicates the new alleles, new alleles combinations, and new sequence types described in this study. -, gene not amplified/sequenced.
aSample origin: blood (B), wound (W), urine (U), respiratory (R), rectal smear (RS), urethral smear (US), prepuce smear (PS).
bThe isolates were classified according to the resistance pattern, as multidrug-resistant (MDR, non-susceptible to ≥1 agent in ≥3 antibiotic categories), extensively drug-resistant (XDR, non-susceptible to ≥ 1 agent in all but ≤2 antibiotic categories) and non-multidrug resistant (non-MDR).
Phenotypic and antibiotic susceptibility characterisations
The 56 isolates were biochemically and phenotypically characterised using the authomatized VITEK®2 GN method (bioMériux, Marcy d’Etoile, France) and the oxidase reaction test. Their antibiogram profiles were established by the disk diffusion method on Mueller-Hinton agar plates (bioMérieux, Marcy d’Etoile, France) following CLSI recommendations for all antibiotics, except for fosfomycin which followed the French Microbiology Society recommendations [9,10]. Borderline values were assessed by the E-test method (bioMérieux, Marcy d’Etoile, France). The antibiotics tested were amikacin, aztreonam, cefepime, ceftazidime, ciprofloxacin, colistin, gentamicin, fosfomycin, imipenem, levofloxacin, meropenem, piperacillin-tazobactam and tobramycin. For the isolates resistant to imipenem and/or meropenem, the determination of metallo-β-lactamases (MBLs) using E-test strips with Imipenem-EDTA was performed (bioMérieux, Marcy d’Etoile, France). The classification of multiresistance was performed according to Magiorakos et al. [11]. The isolates were classified according to the resistance pattern as multidrug resistant (MDR, non-susceptible to at least one agent in three or more antimicrobial categories), extensively drug resistant (XDR, non-susceptible to at least one agent in all but two or fewer antimicrobial categories; i.e. bacterial isolates remain susceptible to only one or two categories), pandrug-resistant (PDR, non-susceptible to all agents in all antimicrobial categories), and non-multidrug resistant (non-MDR).
DNA extraction: PCR amplification and DNA sequencing
Bacterial genomic DNA for PCR amplification was obtained as previously described [12]. The housekeeping genes acsA, aroE, guaA, mutL, nuoD, ppsA and trpE were amplified and sequenced for the 56 isolates using the primers described previously [8]. The PCR conditions have been slightly modified. The reactions were performed using an Eppendorf thermocycler, with an initial denaturation step at 96°C 2 min, followed by 35 cycles of denaturation at 96°C for 1 min for all of the genes, a primer annealing temperature, depending on the gene (55–58°C for aroE and nuoD; 58°C for acsA and guaA; and 58–60°C for mutL, ppsA and trpE), for 1 min and a primer extension at 72°C for 1 min for all of the genes, with the exception of aroE (1.5 min). A final elongation step was performed at 72°C for 10 min. The PCR amplification reactions were performed as previously described [12]. The amplified products were purified with Multiscreen HTS PCR 96-well filter plates (Millipore). Sequence reactions were carried out using the ABI Prism BigDye Terminator version 3.1 and the sequences were read with an automatic sequence analyser (3130 genetic analyzer; Applied Biosystems).
Sequence analysis and allele and nucleotide diversity
Sequence analysis was performed as described previously [12]. Individual phylogenetic trees and concatenated analyses of the sequenced gene fragments were constructed [12]. The allelic and nucleotide diversities were calculated from the gene sequences using the DnaSP package, version 3.51 [13]. For each isolate, the combination of alleles obtained at each locus defined its allelic profile or sequence type (ST). The ST and allele assignment were performed at the P. aeruginosa MLST website (http://pubmlst.org/paeruginosa/). If a sequence did not match with an existing locus in the database, it was designated as a “new” allele. Moreover, the new STs that did not match any allele combination in the database were also numbered as “new”. The clustering of the STs and the split decomposition were performed as previously described [12]. The new nucleotide sequences of each different allele of each locus determined in this study and the new sequence types were sent to curator Eleanor Pinnock for introduction into the P. aeruginosa MLST website (http://pubmlst.org/paeruginosa/).
The diversity and rarefaction indexes for the statistical analysis were calculated using the PAST v.2.0 program [14]. The coverage index (C) was calculated as C = 1 - (n/N), with n being the number of sequence types, and N the number of strains analysed.
Results
Description of the bacterial isolates
In total, 227 P. aeruginosa isolates were obtained from 145 patients between January and February 2010. The antibiotic resistance patterns for the isolates were 21.4% XRD, 17.2% MRD and 61.4% non-MRD. In total, 56 P. aeruginosa isolates from 53 specimens were randomly chosen from the different groups of antibiotic resistance and further studied. Three of them showed two different colony morphologies, and both types were studied by MLST. The isolates were classified according to the resistance pattern as 21.4% MDR, 37.5% XDR and 41.1% non-MDR. The antibiotic pattern and the individual profiles are shown in Table 2.
Table 2.
Susceptibility antibiotic pattern for each Pseudomonas aeruginosa isolate analysed
| |
|
ANTIBIOTICSb |
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Antibiotic patterna | AMK | GEN | TOB | IMP | MEM | CAZ | FEP | CIP | LVX | ATM | TZP | COL | FOS | IMP-EDTA |
| PaC1 |
XDR |
S |
S |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC49 |
MDR |
S |
S |
S |
R |
R |
I |
R |
S |
S |
R |
S |
S |
S |
Neg |
| PaC51 |
non-MDR |
S |
S |
S |
R |
R |
S |
S |
S |
S |
S |
S |
S |
R |
Neg |
| PaC52 |
XDR |
S |
I |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC2 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
I |
R |
S |
R |
Neg |
| PaC3 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
R |
R |
S |
S |
S |
R |
- |
| PaC4 |
non-MDR |
S |
I |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
- |
| PaC5 |
XDR |
S |
S |
S |
R |
R |
R |
R |
S |
I |
R |
R |
S |
R |
Neg |
| PaC7 |
MDR |
S |
S |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
Neg |
| PaC16 |
MDR |
S |
S |
S |
R |
I |
R |
R |
R |
R |
R |
R |
S |
S |
Neg |
| PaC9 |
XDR |
S |
I |
S |
R |
R |
S |
R |
R |
R |
R |
S |
S |
R |
Neg |
| PaC10 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
I |
R |
S |
R |
Neg |
| PaC19 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
R |
Neg |
| PaC32 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
I |
R |
S |
R |
Neg |
| PaC40 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
R |
Neg |
| PaC11 |
XDR |
R |
R |
R |
S |
S |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC24 |
XDR |
R |
R |
R |
S |
S |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC12 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Pos |
| PaC14 |
non-MDR |
S |
I |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC15 |
MDR |
S |
I |
S |
S |
S |
S |
S |
S |
S |
I |
S |
S |
R |
- |
| PaC17 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC18a |
non-MDR |
S |
I |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC18b |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC20 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC21 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC23 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
- |
| PaC25a |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC25b |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC26 |
XDR |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC27 |
non-MDR |
S |
S |
S |
R |
S |
S |
S |
S |
S |
S |
S |
S |
R |
Neg |
| PaC28 |
MDR |
S |
S |
S |
S |
S |
S |
S |
R |
S |
R |
S |
S |
R |
Neg |
| PaC29 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC30 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
S |
- |
| PaC31 |
MDR |
S |
S |
S |
R |
R |
S |
S |
R |
R |
R |
S |
S |
R |
Neg |
| PaC33 |
XDR |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC34 |
MDR |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
S |
Pos |
| PaC35 |
MDR |
I |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
S |
Pos |
| PaC36a |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC36b |
MDR |
S |
R |
R |
R |
R |
S |
S |
R |
R |
S |
R |
S |
R |
Neg |
| PaC37 |
XDR |
S |
R |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC38 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Pos |
| PaC39 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC41 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
- |
| PaC42 |
MDR |
S |
S |
S |
R |
R |
R |
R |
S |
S |
R |
R |
S |
R |
Neg |
| PaC56 |
MDR |
S |
S |
S |
R |
R |
R |
R |
S |
S |
R |
S |
S |
R |
Neg |
| PaC43 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
R |
Pos |
| PaC44 |
non-MDR |
S |
I |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC45 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
Neg |
| PaC46 |
MDR |
S |
S |
S |
R |
R |
I |
S |
S |
R |
S |
S |
S |
S |
Neg |
| PaC47 |
XDR |
S |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
S |
R |
Neg |
| PaC48 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC54 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC50 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
Neg |
| PaC53 |
non-MDR |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
- |
| PaC55 |
non-MDR |
S |
S |
S |
R |
R |
I |
S |
S |
S |
S |
S |
S |
S |
Neg |
| PaC57 | non-MDR | S | S | S | S | S | S | S | S | S | S | S | S | R | - |
aThe isolates were classified according to the resistance pattern, as multidrug-resistant (MDR, non-susceptible to ≥1 agent in ≥3 antibiotic categories), extensively drug-resistant (XDR, non-susceptible to ≥1 agent in all but ≤2 antibiotic categories) and non-multidrug resistant (non-MDR).
bAntibiotics tested: Amikacin (AMK), Gentamicin (GEN), Tobramycin (TOB), Imipenem (IMP), Meropenem (MEM), Ceftazidime (CAZ), Cefepime (FEP), Ciprofloxacin (CIP), Levofloxacin (LVX), Aztreonam (ATM), Piperacilin/Tazobactam (TZP), Colistin (COL), Fosfomycin (FOS), Imipenem-EDTA (IMP-EDTA). Results are indicated as S sensitive, I intermediate, R non-susceptibility, Neg negative, Pos positive, - not applicable.
MLST analysis
A total of 2,882 nucleotides were analysed for the 56 isolates: 390 bp for acsA, 498 for aroE, 373 bp for guaA, 442 bp for mutL, 366 bp for nuoD, 370 bp for ppsA and 443 bp for trpE. The number of polymorphic sites in the seven loci studied varied from 69 (aroE) to 11 (nuoD) in the 56 isolates studied. The number of alleles oscillated from 20 for acsA to 6 for nuoD. The guaA and trpE genes exhibited 15 different alleles, mutL and ppsA exhibited 14 different alleles and aroE exhibited 10 different alleles. The allelic and nucleotide diversity are shown in Table 3. The acsA, aroE, mutL and ppsA genes displayed new alleles not previously described. The three isolates with two different colony morphologies presented the same allelic profile, although one of them (PaC36a and PaC36b) had different antibiotic susceptibility profiles. The allelic profile for the different isolates and for each gene analysed are given in Table 1. The new alleles and the new sequence types not previously described are indicated with an asterisk mark. The MLST analysis of the 56 isolates showed 32 different sequence types.
Table 3.
Genetic diversity of the selected loci among the Pseudomonas aeruginosa isolates analysed in this study
| No of isolates | Locus | Fragment length (bp) | No. of alleles | Gene diversity ± SD | No. of polymorphic sites | Avg. number of nucleotide differences | Nucleotide diversity ± SD |
|---|---|---|---|---|---|---|---|
| 56 |
acsA |
390 |
20 |
0.904 ± 0.027 |
23 |
5.15325 |
0.01321 ± 0.00095 |
| 56 |
aroE |
498 |
10 |
0.827 ± 0.025 |
69 |
9.10909 |
0.01829 ± 0.00507 |
| 56 |
guaA |
373 |
15 |
0.868 ± 0.034 |
14 |
2.62013 |
0.00702 ± 0.00062 |
| 54 |
mutL |
442 |
14 |
0.764 ± 0.055 |
28 |
3.16702 |
0.00717 ± 0.00169 |
| 56 |
nuoD |
366 |
6 |
0.642 ± 0.048 |
11 |
1.52922 |
0.00418 ± 0.00081 |
| 56 |
ppsA |
370 |
14 |
0.879 ± 0.024 |
39 |
4.61364 |
0.01247 ± 0.00347 |
| 56 | trpE | 443 | 15 | 0.876 ± 0.023 | 19 | 4.50260 | 0.01016 ± 0.00076 |
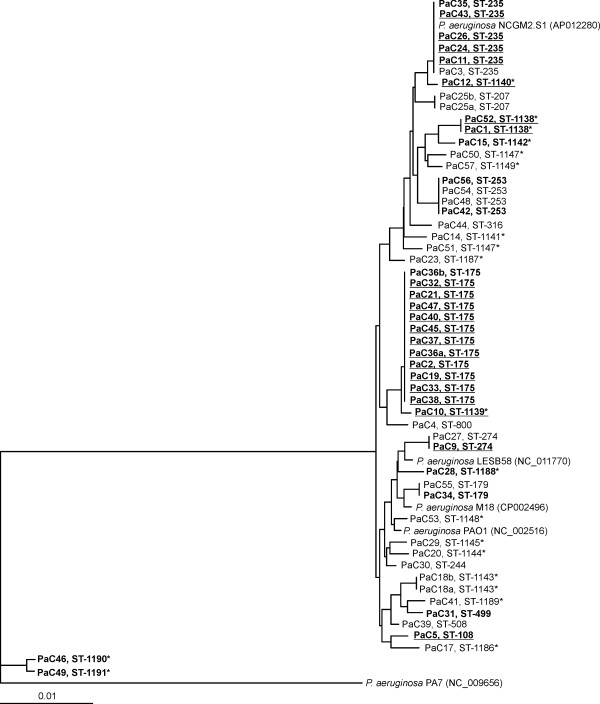
Individual phylogenetic trees for each gene were constructed and, to build a more robust phylogeny, a concatenated analysis considering the seven genes was also performed (Figure 1). Two isolates with mucoid phenotype, PaC7 and PaC16, both isolated from the same patient (number 6), were not included in the analysis because we were unable to amplify and sequence the mutL gene. All of the clinical isolates studied, except PaC46 and PaC49, were related with a similarity between 98.5 - 100%. PaC46 and PaC49, belonged to the same clonal complex and shared a 99.8% similarity between them, less than 95.8% with the other clinical isolates and 95.7% with P. aeruginosa PA7, considered to be an outlier of the species [15]. The corresponding genes of P. aeruginosa PA7 and PAO1 have a similarity of 91.6%, and this percentage is lower when other species of the genus were considered. A SplitsTree was constructed with all of the isolates analysed (Figure 2), and recombination was observed. The most abundant sequence types observed were ST-175, ST-235 and ST-253.
Figure 1.
Concatenated phylogenetic tree showing the molecular evolutionary relationships of the seven genes analysed (acsA, aroE, guaA, mutL, nuoD, ppsA and trpE) between the studied clinical Pseudomonas aeruginosa isolates. The antibiotic profile is indicated in the figure: the MDR isolates are labelled in bold and the XDR isolates are indicated in bold and underlined. Clinical strains PaC7 and PaC16 are not included in the phylogenetic tree. Asterisk mark (*) indicates the new sequence types described in this study.
Figure 2.
SplitsTree showing the distribution of all of the sequence types obtained for the clinical Pseudomonas aeruginosa isolates studied. The SplitsTree was based on the analysis of the allelic profiles of the acsA, aroE, guaA, mutL, nuoD, ppsA and trpE genes. The MDR isolates are labelled in bold and the XDR isolates are indicated in bold and underlined. The sequence types represented by more than one isolate are indicated in italic font. Asterisk mark (*) indicates the new sequence types described in this study.
Patients and antibiotic resistance pattern
Thirty-five isolates were single isolates (one per patient), and, in seven patients, more than one isolate of P. aeruginosa was obtained during the two-month period studied (patients 1 and 8, four isolates each; patients 6, 9, 29, 32 and 38, two isolates each) (see Table 1). In two patients (9 and 38), all of the isolates studied belonged to the same ST and had the same antibiotic resistance profile. Isolates with different STs were isolated from three patients (patients 1, 6 and 8). Four isolates were isolated from patient 1 during the time of the study: PaC1 and PaC52 from a wound sample, and PaC49 and PaC51 from rectal smear. PaC1 and PaC52, were isolated with one month of difference, and belonged to the same ST and showed the same antibiotic resistance profile with the exception of gentamicin (intermediate susceptibility). PaC49 and PaC51 were assigned to different STs and showed differences in the antibiotic resistance profile. Patient 6 showed the same antibiotic profile (with the exception of meropenem). Four isolates with slight differences in the antibiotic profile were recovered from patient 8 (PaC10 and PaC19 from urine samples were isolated with three days of difference, PaC32 from a rectal smear and PaC40 was of respiratory origin). Isolate PaC10 was assigned to a different ST based on differences in guaA allele, although it belonged to the same clonal complex. Two isolates were isolated the same day from patient 29 from two different samples (catheter and blood); both of the isolates showed the same ST but presented differences in their antibiotic profile and in the production of MBLs, as detected by phenotypic methods. Two isolates of patient 32 obtained from different origins with two weeks of difference showed differences in piperacilin/tazobactam-susceptibility, but belonged to the same ST (see Table 1 and 2).
Population structure and susceptibility to antibiotics
From the 56 isolates analysed, 23 were non-MDR and 33 were multiresistant (MDR or XDR). The non-MDR isolates were singleton STs, with the exception of ST-235 and ST-253. From the 56 isolates, 32 isolates were carbapenem-non-susceptible (57.1%) and 15.6% of them were MBL-positive. From those isolates, one was non-susceptible to only imipenem, and thirty-one were non-susceptible to both (isolate PaC16 showed intermediate resistance to meropenem). The 32 carbapenem-non-susceptible isolates were distributed into 15 sequence types: ST-175 (12 isolates), ST-235 (3), ST-179 (2), ST-253 (2), ST-274 (2), ST-108 (1), and ST-499 (1), and eight new sequence types (seven singletons and one with two isolates). Only four of these types (ST-175, ST-235, ST-253 and ST-274) were also described previously in the study of 16 Spanish hospitals [16].
No relations statistically significant could be established in our study between antibiotic resistance and other variables as sex, age of patients, sample origin or STs, probably because the low sampling potential. However, a statistically significant association was observed between the prevalent ST (ST-175) and multiresistant isolates (p = 0.003).
Diversity analysis
To assess the extent of the diversity analysed in the study, a rarefaction curve was constructed. Despite the high diversity of the sequence types, the number of different sequence types referred to the number of isolates analysed did not reach a saturation curve, indicating that the diversity was higher than detected, a finding that was confirmed when the coverage index (C) was calculated (51%). Additional isolates should be analysed to ascertain the population structure of clinical P. aeruginosa present in our hospital completely. Diversity was evaluated using the Dominance (D), Shannon (H), Simpson and Evenness indices, and the values obtained for each index (0.075, 3.087, 0.925 and 0.684, respectively) indicate a highly diverse sample. However, when only the diversity of the multiresistant isolates (MDR and XDR) were considered, a softer saturation curve was detected and the coverage index was higher (62.5%), indicating that the diversity was better screened. This result was also supported by the diversity indices (D of 0.1621, H of 2.303, Simpson of 0.8379 and Evenness of 0.6255).
Discussion
The role of P. aeruginosa as a pathogen and its implication in nosocomial outbreaks has been widely studied. The present study was focused on the analysis of the population structure and diversity of P. aeruginosa clinical isolates randomly chosen from their different patterns of antibiotic resistance in a single hospital. The isolates include different antibiotic non-susceptibility profiles (21.4% MDR, 37.5% XDR and 41.1% non-MDR).
The MLST analysis showed a high diversity, as reported in other previous studies. The 56 isolates were grouped into 32 different sequence types, 12 sequence types that were previously described (including 34 isolates) and 20 new ones (including 22 isolates). The singleton sequence types (26 isolates) corresponded mainly to the non-MDR isolates (16 isolates). Twenty-two of the isolates corresponded to new sequence types (not previously defined) of which 12 isolates were non-MDR, 6 isolates were MDR and 4 isolates were XDR. The clinical isolates studied showed a variable number of polymorphic sites and alleles, indicating the variability of the isolates selected. It is remarkable that we found the presence of new alleles (not previously described) of four genes, acsA, aroE, mutL and ppsA.
The analysis of the seven loci demonstrated that the prevalent STs were ST-175, ST-235 and ST-253. ST-175 is widely distributed worldwide [17] and is the isolate most frequently isolated in this study, with twelve isolates obtained from eight patients. This ST is also the most prevalent in the studies of García-Castillo et al. and Cholley et al. [16,17]. ST-175 has been reported as a contaminant of the hospital environment, a coloniser of respiratory secretions in cystic fibrosis patients, and has been associated with the multiresistant isolates of P. aeruginosa. All of the isolates included in this group were multiresistant (eleven XDR isolates and one MDR) and were sensitive to colistin, 90% to amikacin, 37% to aztreonam and nearly 10% to ceftazidime and cefepime. All of the isolates were resistant to the other antibiotics tested, and only one of them was MBL positive.
ST-235 is the second most frequently isolated sequence type, with six isolates (from five patients); four isolates were XDR, one isolate was MDR, and another was non-MDR. This ST has been involved in the dissemination of the genes encoding MBLs and has been associated to multiple resistance mechanisms [18], although Cholley et al. described strains with the same ST and none of them was MBL-positive [17]. Three of our five isolates were non-MBLs producers. In a previous study performed in another Majorcan hospital, ST-235 has been described as a VIM-13 producing β-lactamase [19]. ST-179, previously described in Mallorca as VIM-2 producer, was also MBL-positive [19].
The third most abundant sequence type was ST-253, with four isolates. These isolates were isolated from two patients; two isolates were MDR, and two were non-MDR.
Only one isolate was colistin-resistant, corresponding to ST-244 reported previously in Korea as the isolate most frequently colistin non-susceptible and sensitive to other antibiotics [20]. Our isolate was isolated in a mixed culture with Morganella morganii and Serratia marcescens, both inherently resistant to colistin.
The high discriminatory power of the MLST profiling allowed the differentiation among isolates obtained from the same patient at different dates and sampling sites. When the specimen was associated with the site of infection, the sequence type or clonal complex obtained and the antibiotic resistance profiles were the same.
Conclusions
The present results indicate that P. aeruginosa isolates revealed a significant frequency of recombination and a panmictic net-like population structure, as was suggested by Kiewitz and Tümler [21]. The population structure of clinical P. aeruginosa present in our hospital indicates the coexistence of nonresistant and resistant isolates with the same sequence type. The multiresistant isolates studied are grouped in the prevalent sequence types found in other Spanish hospitals and at the international level, and the susceptible isolates correspond mainly to singleton sequence types.
Competing interests
The authors declare no competing interests; financial or otherwise.
Authors’ contributions
MG carried out the molecular genetic studies, participated in the sequence analysis and drafted the manuscript. MP carried out the molecular genetic analysis. MCG, VFB, and PDA carried out the isolation and phenotypic and the antibiogram analysis. AP performed the statistical analysis. MG, MCG, EGV and JL conceived the study. All co-authors participated in the design of the study and coordination and helped to the draft manuscript. All authors read and approved the final manuscript.
Contributor Information
Margarita Gomila, Email: marga.gomila@uib.es.
Maria del Carmen Gallegos, Email: mcgallegos@hsll.es.
Victoria Fernández-Baca, Email: vfernandez-baca@hsll.es.
Antonio Pareja, Email: apareja@hsll.es.
Margalida Pascual, Email: margalidapascual80@hotmail.com.
Paz Díaz-Antolín, Email: mpdiaz@hsll.es.
Elena García-Valdés, Email: elena.garciavaldes@uib.es.
Jorge Lalucat, Email: jlalucat@uib.es.
Acknowledgments
This work was supported by the General Board for Evaluation and Accreditation of the Department of Health and Consume of the Autonomous Community of the Balearic Islands (Dirección General de Evaluación y Acreditación, de la Conselleria de Salut i Consum, de la Comunidad Autónoma de las Islas Baleares). M. Gomila is the recipient of a postdoctoral contract from the Juan de la Cierva Programme of the Spanish Ministerio de Ciencia e Innovación. E. García-Valdés and J. Lalucat want to thank the support of the projects CGL2008-03242 and CSD2009-00006 from the Ministerio de Economia y Competitividad (Spain) and Fondo Europeo de Desarrollo Regional (FEDER) funding. The authors want to thank the critical revision of Dr. A. Oliver. All authors report no conflicts of interest relevant to this article.
References
- Cramer N, Wiehlmann L, Tümmler B. Clonal epidemiology of Pseudomonas aeruginosa in cystic fibrosis. Int J Med Microbiol. 2010;300:526–533. doi: 10.1016/j.ijmm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Renom F, Yáñez A, Garau M. et al. Prognosis of COPD patients requiring frequent hospitalization: role of airway infection. Respiratory Med. 2010;104:840–848. doi: 10.1016/j.rmed.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- Picazo JJ, Betriu C, Rodríguez-Avial I, Culebras E, Gómez M, López F, Grupo VIRA. Vigilancia de resistencias a los antimicrobianos: estudio VIRA 2006. Enferm Infecc Microbiol Clin. 2006;24:617–628. doi: 10.1157/13095373. [DOI] [PubMed] [Google Scholar]
- Denamur E, Picard B, Goullet P, Bingen E, Lambert N, Elion J. Complexity of Pseudomonas aeruginosa infection in cystic fibrosis: combined results from esterase electrophoresis and rDNA restriction fragment length polymorphism analysis. Epidemiol Infect. 1991;106:531–539. doi: 10.1017/S0950268800067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elaichouni A, Verschraegen G, Claeys G, Devleeschouwer M, Godard C, Vaneechoutte M. Pseudomonas aeruginosa serotype O12 outbreak studied by arbitrary primer PCR. J Clin Microbiol. 1994;32:666–671. doi: 10.1128/jcm.32.3.666-671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol. 2007;45:3707–3712. doi: 10.1128/JCM.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran B, Jonas D, Crundmann H, Pitt T, Dowson C. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Wayne, PA: Clinical and Laboratory Standard Institute; 2010. (20th informational supplement, document M100-S20). [Google Scholar]
- Societé Française de Microbiologie. Comité de l’antibiograme de la societe française de microbiologie. Recommandations; 2010. [Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Gomila M, Ramírez A, Lalucat J. Diversity of environmental Mycobacterium isolates from hemodialysis water as shown by a multigene sequencing approach. Appl Environm Microbiol. 2007;73:3787–3797. doi: 10.1128/AEM.02934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Paleontol Electr. 2001;4:9. [Google Scholar]
- Roy PH, Tetu SG, Larouche A. et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One. 2010;5:e8842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Castillo M, Del Campo R, Morosini MI. et al. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J Clin Microbiol. 2011;49:2905–2910. doi: 10.1128/JCM.00753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholley P, Thouverez M, Hocquet D, van der Mee-Marquet N, Talon D, Bertrand X. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol. 2011;49:2578–2583. doi: 10.1128/JCM.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. Dissemination of IMP-6 metallo-β-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother. 2011;66:2791–2796. doi: 10.1093/jac/dkr381. [DOI] [PubMed] [Google Scholar]
- Juan C, Zamorano L, Mena A, Alberti S, Pérez JL, Oliver A. Metallo-β-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J Antimicrob Chemother. 2010;65:474–478. doi: 10.1093/jac/dkp491. [DOI] [PubMed] [Google Scholar]
- Lee JY, Song JH, Ko KS. Identification of nonclonal Pseudomonas aeruginosa isolates with reduced colistin susceptibility in Korea. Microb Drug Resist. 2011;17:299–304. doi: 10.1089/mdr.2010.0145. [DOI] [PubMed] [Google Scholar]
- Kiewitz C, Tümmler B. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol. 2000;182:3125–3135. doi: 10.1128/JB.182.11.3125-3135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]