Abstract
Virus-specific CD8+ T cells play an important role in controlling HIV/SIV replication. These T cells recognize intracellular pathogen-derived peptides displayed on the cell surface by individual MHC class I molecules. In the SIV-infected rhesus macaque model, five Mamu class I alleles have been thoroughly characterized with regard to peptide binding, and a sixth was shown to be uninvolved. In this study, we describe the peptide binding of Mamu-A1*007:01 (formerly Mamu-A*07), an allele present in roughly 5.08% of Indian-origin rhesus macaques (n=63 of 1240). We determined a preliminary binding motif by eluting and sequencing endogenously bound ligands. Subsequently, we used a positional scanning combinatorial library and panels of single amino acid substitution analogs to further characterize peptide binding of this allele and derive a quantitative motif. Using this motif, we selected and tested 200 peptides derived from SIVmac239 for their capacity to bind Mamu-A1*007:01, 33 were found to bind with an affinity of 500nM or better. We then used PBMC from SIV-infected or vaccinated but uninfected, A1*007:01-positive rhesus macaques in IFN-γ Elispot assays to screen the peptides for T cell reactivity. In all, eleven of the peptides elicited IFN-γ+ T cell responses. Six represent novel A1*007:01-restricted epitopes. Furthermore, both Sanger and ultra-deep pyrosequencing demonstrated the accumulation of amino acid substitutions within four of these six regions, suggestive of selective pressure on the virus by antigen-specific CD8+ T cells. Thus, it appears that Mamu-A1*007:01 presents SIV-derived peptides to antigen-specific CD8+ T cells and is part of the immune response to SIVmac239.
Keywords: SIV, MHC, Macaque, Epitope, Escape
Introduction
The SIV-infected rhesus macaque has long been an important animal model for HIV infections in humans. This model allows researchers to answer questions regarding the early events after infection, viral variation, disease pathogenesis, and to test new vaccine strategies. Having the ability to manipulate variables like viral strain, dose, and route of infection allows scientists to precisely monitor the immune response to SIV. However, despite intensive study, we still do not understand what constitutes an efficacious cytotoxic T-lymphocyte (CTL) response. Since CTL are selected and educated based on the MHC present in an individual, and since different MHC alleles are able to bind different populations of peptides, understanding the MHC alleles and the SIV-derived peptides they present is necessary to understand why some genotypes are better able to control SIV infection. Control of SIV viral replication may be due to differences in the particular peptides bound, or properties of the CTL engendered by MHC alleles in particular genotypes.
Several lines of evidence indicate that CTL are responsible for control of HIV/SIV viral replication. Depletion of CD8+ lymphocytes in SIV-infected macaques results in increased viral loads (Matano et al. 1998; Jin et al. 1999; Schmitz et al. 1999; Metzner et al. 2000; Alexander et al. 2001). The appearance of virus-specific CTL is concurrent with the decline of peak viremia (Yasutomi et al. 1993; Borrow et al. 1994; Koup et al. 1994; Reimann et al. 1994; Ogg et al. 1998). Additionally, it has been established that specific MHC class I alleles are associated with either slow or rapid disease progression (Klein et al. 1995; Kaslow et al. 1996; McNeil et al. 1996; Carrington et al. 1999; Hendel et al. 1999; Migueles et al. 2000; Kaslow et al. 2001; Tang et al. 2002; Yant et al. 2006; Loffredo et al. 2007b). Finally, it is well documented that CTL exert strong enough selective pressure on immunodeficiency viruses sufficient to drive variation both inside and outside the epitopes they target (Phillips et al. 1991; Wolinsky et al. 1996; Borrow et al. 1997; Goulder et al. 1997; Price et al. 1997; Mortara et al. 1998; Van Baalen et al. 1998; Evans et al. 1999; Soudeyns et al. 1999; Wilson et al. 1999; Allen et al. 2000; Chen et al. 2000; Barouch et al. 2002; O’Connor et al. 2002; Vogel et al. 2002; Barouch et al. 2003; Peyerl et al. 2003; Friedrich et al. 2004a; Friedrich et al. 2004b). It is therefore critical to characterize these CTL responses and the MHC class I molecules that restrict them.
To date, five Macaca Mulatta (Mamu) class I alleles (Mamu-A1*001:01, -A1*002:01, -A1*011:01, -B*017:01, and –B*008:01) have been thoroughly characterized with regard to their peptide-binding motifs and the SIV-derived peptides they bind (Allen et al. 2001; Mothe et al. 2002; Loffredo et al. 2004; Sette et al. 2005; Loffredo et al. 2009). Another allele, Mamu-B*001:01, was shown not to be involved in restricting epitopes derived from SIVmac239 (Loffredo et al. 2005). Taking into account the fact that macaques can express between 4 and 12 different MHC Class I alleles (Boyson et al. 1996; Urvater et al. 2000), the identification and study of additional alleles enhances the animal model’s utility.
To fully characterize an MHC class I allele involves a combination of several different approaches. Typically, sequencing of naturally occurring ligands combined with identification of optimal T cell epitopes leads to a preliminary motif that can be further refined with peptide-binding assays using purified MHC (Van Bleek and Nathenson 1990; Falk et al. 1991; Kast et al. 1994; Kubo et al. 1994; Rammensee et al. 1995; Sidney et al. 2000). A comprehensive analysis requires knowledge of secondary interactions to best identify the peptides that will be bound in vivo (Ruppert et al. 1993; Kondo et al. 1995; Sidney et al. 1996). Once a peptide-binding motif is determined for a particular MHC class I allele, candidate epitopes can be efficiently identified and then tested for immunogenicity utilizing a rapid screening method, such as IFN-γ ELISPOT using PBMC from SIV-infected macaques. Those epitopes that elicit a response can be utilized to manufacture fluorescently labeled tetrameric peptide-MHC complexes (Altman et al. 1996; Kuroda et al. 1998; Ogg et al. 1998). These tetramers serve as reagents for immune monitoring in future experiments utilizing animals with defined MHC class I alleles. Additionally, the regions of the virus that contain these epitopes can be monitored for viral evolution and escape, and also be evaluated as candidates for inclusion in future vaccines. Of the 6 characterized alleles, 2 (B*008:01 and B*017:01 often need to be excluded from vaccine studies due to correlation with spontaneous SIV control. Thus characterization of additional Mamu MHC I alleles is needed to add to the cohort of MHC I characterized Rhesus macaques for use in vaccine studies.
Mamu-A1*007:01 (formerly known as A*07 before being renamed by the Comparative MHC Nomenclature Committee) has been determined by polymerase chain reaction-sequence-specific-priming (PCR-SSP) to be present in 5.27% (55 of 1165) of the macaques at the Wisconsin National Primate Research Center (WNPRC) colony, 10.6% at the Oregon National Primate Research Center (8 of 75), and total frequency of 5.08% of all rhesus macaques tested. This allele is present in five of 27 SIVmac239-infected elite controller (EC) animals in our cohort. The contribution of Mamu-A1*007:01 to viral control is not known. We sought to define the peptide-binding motif of Mamu-A1*007:01 and establish the epitopic breadth in SIV-infected Mamu-A1*007:01-positive macaques.
The Mamu-A1*007:01 group of alleles in Indian rhesus macaques contains three known subtypes (:01, :02, and :03) (IPD Database, http://www.ebi.ac.uk/cgi-bin/ipd/mhc/get_nomenclature.cgi?Mamu-A1). Additionally, Otting et al. (2007) described a related allele (A1*007:04) in Chinese rhesus macaques and Genbank lists yet another Chinese allele (A1*007:06, accession number EU334697). Recently, Naruse et al. (2010) and Otting el al. (2011) described two more related alleles in Burmese rhesus macaques (subtypes :02-like and :05, respectively). The amino acid differences between the various MHC Class I subtypes might affect peptide binding (Supplemental Figure 1). With the exception of a single animal, all of the animals that made Mamu-A1*007:01-restricted CTL responses in this study had subtype :01. Rh2001 appears to be of mixed origin, and has the A1*007:02-like allele subtype.
We defined the Mamu-A1*007:01-binding motif, generated potential SIV-derived peptide binders, and tested them in Mamu-A1*007:01-positive macaques that had either been vaccinated with SIVmac239 sequences or challenged with SIVmac239. We found six novel epitopes from Pol, Vpr, Vif, and Gag that bind Mamu-A1*007:01. Two of these were previously used in studies detailing the kinetics of epitope presentation: Pol782-789 YL8 and Vpr45-55 NL11 (Sacha et al. 2007; Sacha et al. 2010). CTL responses against four of these epitopes selected for viral variants consistent with immunologic escape. The other two epitopes were derived from highly conserved regions of SIV; we did not detect viral variation despite robust responses against these regions. This result is intriguing as it indicates these regions might be under functional or structural constraints. Responses directed against these regions might be particularly effective at controlling SIV replication. Together, our data support a potential role for the Mamu-A1*007 allele in the immune response against SIV.
Methods and materials
Animals, viruses, and vaccinations
The Indian rhesus macaques (Macaca mulatta) used in this study were from the Wisconsin National Primate Research Center colony. We identified them as Mamu-A1*007+ by PCR-SSP using conditions as previously described (Kaizu et al. 2007). The primers used to amplify Mamu-A1*007 are as follows: Forward= (GGG TGC GGC GGA GCA GA), and Reverse= (CAC GAT GGG AAT GGT GGA CTC). The animals were cared for in accordance with the regulations and guidelines of the University of Wisconsin Institutional Animal Care and Use Committee.
See Supplemental Table 1 for a list of animals, vaccinations and infecting viruses.
Peptide synthesis and positional scanning combinatorial library
Peptides for screening were purchased as crude or purified material from Genscript (Piscataway, NJ, USA), A and A Labs (San Diego, CA, USA), the Biotechnology Center at the University of Wisconsin-Madison, or were provided by the NIH AIDS Research and Reference Reagent Program (Germantown, MD, USA). They were synthesized using standard tertiary butyloxycarbonyl or fluorenylmethoxycarbonyl solid phase methods (Ruppert et al. 1993). Peptides synthesized for use as radiolabeled ligands were synthesized by A and A Labs and purified to >95% homogeneity by reversed-phase HPLC. Purity of these peptides was determined using analytical reversed-phase HPLC and amino acid analysis, sequencing, and/or mass spectrometry. Peptides were radiolabeled with the chloramine T method (Sidney et al. 2001). Lyophilized peptides were resuspended at 4-20 mg/ml in 100% DMSO, then diluted to required concentrations in PBS + 0.05% (v/v) Nonidet P-40 (Fluka Biochemika, Buchs, Switzerland). SIV peptides were derived from the SIVmac239 sequence, GenBank accession no. M33262 (Kestler et al. 1990).
The positional scanning combinatorial library (PSCL) was synthesized as previously described (Pinilla et al. 1992; Sidney et al. 2008a). Each pool in the library contains 9-mer peptides with one fixed residue at a single position. With each of the 20 naturally occurring residues represented at each position along the 9-mer backbone, the entire library consisted of 180 peptide mixtures.
Mamu A1*007:01 peptide-binding motif from endogenous ligands
We generated soluble Mamu-A1*007:01 by deletion of the transmembrane and cytosolic domains. The construct was transfected into 721.221 cell lines and stable, secreting, clones were selected for expansion into a bioreactor. Approximately 35mg of sMamu-A1*007:01 was produced and purified using antibody affinity chromatography. Peptide ligands were eluted using an acid-boil, and separated from the complex using a 3kDa cut-off filter. Ten percent of the peptide pool was subjected to Edman degradation sequencing to determine a peptide-binding motif. The remainder of the peptides was resolved into approximately 40 fractions using reverse-phase HPLC. HPLC fractions were then analyzed on an ESI QTOF mass spectrometer to generate MS ion maps (AB SCIEX, Foster City, CA, USA). Abundant ions were then fragmented and analyzed using MASCOT to determine sequence (Matrix Science, London, U.K.).
MHC purification and peptide binding assays
MHC class I purification was performed using affinity chromatography as previously described (Sidney et al. 2001; Loffredo et al. 2009). Mamu-A1*007:01 molecules were purified from cell lysates of stable membrane-bound MHC class I 721.221 transfectants using the anti-HLA class I (A, B, and C) antibody W6/32 (Barnstable et al. 1978). Protein purity, concentration, and depletion efficiency steps were monitored by SDS-PAGE.
Quantitative assays for the ability of peptide to bind to detergent-solubilized MHC class I molecules were based on the competitive inhibition of binding of a high-affinity radiolabeled standard probe peptide and performed as detailed previously (Sidney et al. 2001; Loffredo et al. 2009). Peptides were tested at six different concentrations covering a 100,000-fold dose range in three or more independent assays. The radio-labeled peptide used for the A1*007:01 assays was the SIV Pol782-789 8-mer peptide YHSNVKEL (IC50 5.8 nM). For each test, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was calculated. Under the conditions used, where [radiolabeled probe]<[MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values (Cheng and Prusoff 1973; Gulukota et al. 1997). In each experiment, a titration of the unlabeled version of the radiolabeled probe was tested as a positive control for inhibition.
Bioinformatic analysis
Analysis of the PSCL data was performed as described previously (Sidney et al. 2008a; Loffredo et al. 2009). Briefly, IC50 nM values for each mixture were standardized as a ratio to the geometric mean IC50 nM value of the entire set of 180 mixtures, and then normalized at each position so that the value associated with optimal binding at each position corresponds to 1. For each position, an average (geometric) relative binding affinity (ARB) was calculated, and then the ratio of the ARB for the entire library to the ARB for each position was derived. We have denominated this ratio, which describes the factor by which the normalized geometric average binding affinity associated with all 20 residues at a specified position differs from that of the average affinity of the entire library, as the specificity factor (SF). As calculated, positions with the highest specificity will have the highest SF value. Primary anchor positions were then defined as those with an SF ≥2.4. This criterion identifies positions where the majority of residues are associated with significant decreases in binding capacity. Preferred residues at the primary anchor positions have been defined as those with ARB values within 5-fold of the optimal residue, and tolerated residues as those with an ARB in the 5-25-fold range of the optimal residue.
Secondary anchor designations are based on the standard deviation (SD) of residue specific values at each position. Dominant secondary anchor positions were defined as those where the SD was >3 and the SF <2.4, as well as positions associated with an SD >2 if the SF is between 1.5 and 2.4. Weak secondary anchors have been defined as positions associated with a SD between 2.5-3, with an SF <1.5, or an SF in the 1.5-2.4 range with an SD <2.
To identify predicted binders, all possible 9-mer peptides in SIV sequences were scored using the matrix values derived from the PSCL analysis as described previously (Sidney et al. 2007; Sidney et al. 2008a; Loffredo et al. 2009). The final score for each peptide represents the product of the corresponding matrix values for each peptide residue-position pair. Peptides of 8, 10, or 11 residues in length were also selected using the combinatorial library as described previously (Sidney et al. 2007). Briefly, 8-mers were selected by scoring residues 1-7 with the corresponding residues in the 9-mer matrix, and the C-terminus was scored using the corresponding 9-mer C-terminal value. Similarly, for 10- and 11-mer peptides, residues 1-8 were scored using the corresponding residues of the 9-mer matrix, and the C-terminus with the 9-mer C-terminal residues. Peptides scoring amongst the top 1.5% (n = 50) for each size were selected.
IFN-γ ELISPOT assay
We performed ELISPOT assays as previously described (Wilson et al. 2009). Briefly, PBMC were isolated from EDTA-anticoagulated blood using Ficoll-Paque PLUS (GE Healthcare Systems, Uppsala, Sweden) and density centrifugation. 1 × 105 PBMC or 2 × 104 cultured CD8+ T cells were used per well in precoated ELISpotPLUS kits (Mabtech USA, Mariemont, OH, USA) according to the manufacturer’s instructions. All tests were performed in duplicate or triplicate using individual peptides at 10uM or serial dilutions 10uM-100pM. The positive control, Con A (Sigma-Aldrich, St. Louis, MO, USA), was used at a final concentration of 5 μg/ml. The negative control wells were devoid of any stimulation. The 96-well plates were incubated for 12-18 h at 37° C in 5% CO2.
Wells were imaged and counted with an AID EliSpot reader version 4.0 (AID, Strassberg, Germany) and analyzed as described previously (Wilson et al. 2009). Background levels were subtracted from each well, and assay results are shown as SFC per 1 × 106 PBMC. Responses were considered positive if the mean of the number of SFC was more than 50 spots per million cells and significance was determined using a one-tailed t-test where alpha = 0.05, and the null hypothesis (Ho): background level ≥ treatment level.
Intracellular cytokine Staining (ICS) Assay
SIV-specific CD8+ T-cell lines were used in TNF-α and IFN-γ ICS assays as previously described (Vogel et al. 2002). Briefly, each test contained 2 × 105 CD8+ T cells and 0.5 × 105 BLCLs or single-allele transfectants. As a positive control, phorbol myristate acetate (1 μg/ml) with ionomycin (2 μg/ml; Sigma-Aldrich) was used. Peptides were used at a concentration of 5 μM or in serial 10-fold dilutions from 5 μM to 500 pM. Approximately 2 × 105 lymphocyte-gated events were acquired on a BD LSR II (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo software 9.1 (TreeStar, Ashland, OR, USA). All values were normalized by subtracting the background level staining (negative control of CD8+ T-cell lines in media without stimulation).
Defining epitopes using peptide-specific CD8+ T cell lines
We cultured CD8+ T cell lines by incubating 5 × 106 freshly isolated PBMC with the minimal optimal peptide predicted to bind to Mamu-A1*007:01 for one to three hours before washing the cells twice. They were then incubated at 37° C with 5% CO2 in R-15 (RPMI 1640 supplemented with 15% fetal calf serum, L-glutamine, and Antibiotic/Antimycotic solution (Hyclone, Logan, UT, USA for all components)) plus 10ng of recombinant human interleukin-7 (R&D Systems, Minneapolis, MN, USA) per ml for 48 hours. The R-15 was then supplemented with 100 Units of interleukin-2 (NIH AIDS Research and Reference Reagent Program) per ml for the length of the culture. Every 7-14 days, the culture was restimulated by the addition of peptide-pulsed, gamma-irradiated autologous B-lymphoblastoid cell lines.
After several weeks in culture, we tested the lines for specificity by IFN-γ ELISPOT or by IFN-γ/TNF-α intracellular cytokine staining (ICS) assays with Mamu-A1*007:01 transfectants to verify that immunogenic peptides were restricted by Mamu-A1*007:01.
MHC class I transfectants
MHC class I sequences were cloned into expression vectors from cDNA libraries as described previously. (Loffredo et al. 2007a; Burwitz et al. 2009) Transient expression of cloned MHC class I cDNA was achieved by electroporation of plasmid DNA into the MHC class I deficient human B-cell line 721.221 (DeMars et al. 1985). Briefly, 5μg of plasmid DNA was added to 5 × 106 721.221 cells in 100μl Nucleofector™ Solution C and electroporated using program G-16 on a Nucleofector I device (Amaxa, Köln, Germany). Transfectants were used 48-72 hours post-electroporation. The stable Mamu-A1*007:01 transfectant was created as previously described (Loffredo et al. 2007a), except for the use of the Nucleofector I device according to the manufacturer’s protocols.
MHC class I surface expression on stable and transient MHC class I transfectants was measured by W6/32 antibody surface staining. Immortalized homologous macaque B-cell lines and 721.221 cells were used as positive and negative controls, respectively. Approximately 1 × 105 lymphocyte-gated events were acquired on a BD LSR II (BD Biosciences) and analyzed using FlowJo version 9.1 (Treestar).
Sequencing of plasma viral RNA (vRNA)
We extracted viral RNA from plasma using the QIAGEN MinElute kit (Qiagen, Valencia, CA, USA) or by a guanidine thiocyanate extraction as previously described (O’Connor et al. 2004; Friedrich et al. 2007). We used the QIAGEN One Step RT-PCR kit to amplify overlapping regions ~300-800 nucleotides in length that targeted parts of the SIVmac239 open reading frames (ORFs) Gag, Pol, Vif, Vpr, and Env. The RT-PCR conditions for all amplicons were as follows: 50° C for 30 min; 95° C for 15 min; 45 cycles of 94° C for 30 s, 53° C for 1 min and 72° C for 150 s; and 68° C for 20 min. Cycling ramp rates were 2° C per second. Unused primers and nucleotides were removed from the cDNAs using ExoSAP-IT° (USB Corporation).
Both strands of each amplicon were sequenced on a 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA) using DYEnamic ET Terminators (GE Healthcare) and their respective RT-PCR primers. The sequencing cycling conditions for all amplicons were as follows: 30 cycles of 95° C for 20 s, 50° C for 15 s, and 60° C for 1 min. Sequences were assembled using CodonCode Aligner version 3.7.1 (CodonCode Corporation, Deadham, MA, USA). DNA sequences were conceptually translated and aligned to wild type SIVmac239 in MacVector 11.1.1 trial version (MacVector,Inc, Cary, NC, USA).
Ultra-deep Pyrosequencing of Mamu-A1*007:01-restricted epitopes
Ultra-deep pyrosequencing of the Mamu-A1*007:01-restricted CD8+ T cell epitopes was conducted as previously described (Bimber et al. 2009). Briefly, cell-free plasma was obtained from EDTA anticoagulated whole blood using Ficoll-Paque PLUS (GE Healthcare) and density centrifugation. Plasma was frozen at −80° C until the time of viral RNA extraction. Viral RNA in the plasma was isolated using the QIAamp MinElute Virus Spin Kit (Qiagen) according to the manufacturer’s instructions. Viral RNA was reverse transcribed and amplified using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen, Carlsbad, CA) and MID-tagged primers (454-Life Sciences, Branford, CT) spanning Pol amino acid residues 271-375, 511-602, and 722-825, as well as Vif 46-215 and Vpr 43-102. Amplicons were pooled at equimolar ratios and the final library was diluted to 1 × 106 copies/μl. Emulsion PCR and subsequent bead enrichment was performed at 1.5 copies/bead using the GS FLX Titanium Lib-A emPCR kit per manufacturers instructions (Roche, Indianapolis, IN). 5 × 105 enriched DNA beads were sequenced on a single GS Junior picotiter plate.
Results
Mapping novel epitopes in a vaccinated animal that controls viral replication
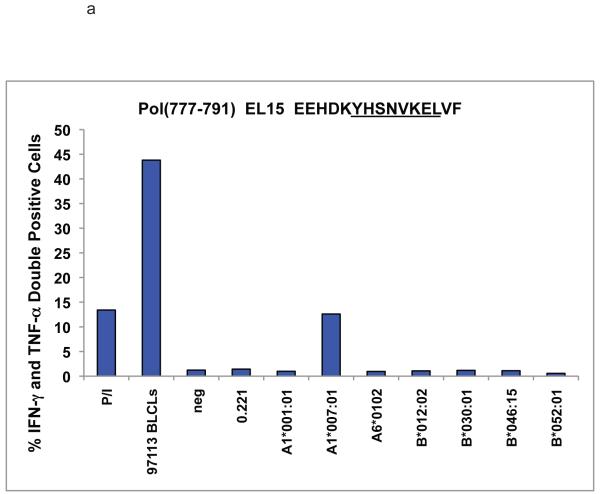
We vaccinated animal r97113 with DNA followed by Adenovirus 5 (Ad5) constructs that coded for full length SIV-Gag, -Tat, -Nef, and –Rev (Wilson et al. 2006). This animal had a peak of virus replication of 1.4 × 107 viral RNA copy eq/ml of plasma five days after the eighth low-dose intra-rectal challenge with SIVmac239 (300 TCID50 per challenge). The animal controlled viral replication for over 100 weeks before viral breakthrough occurred. Since the animal was Mamu-B*008:01-negative and Mamu–B*017:01-negative, we wanted to determine which immune responses were contributing to control of the virus.
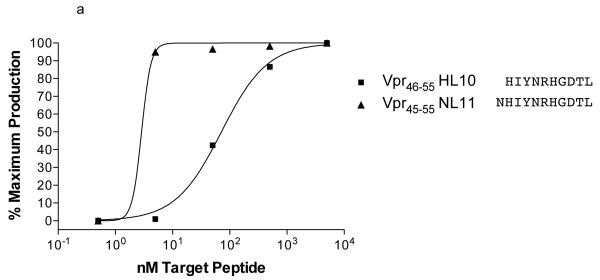
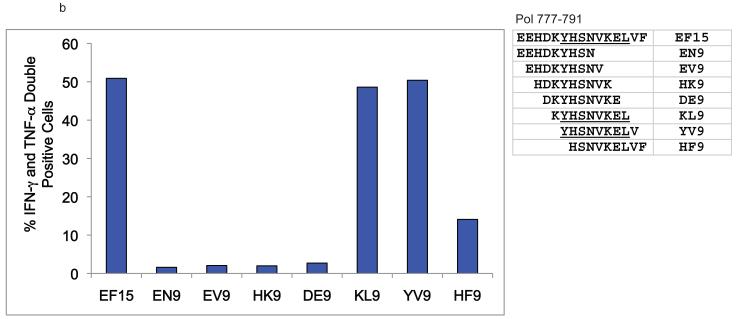
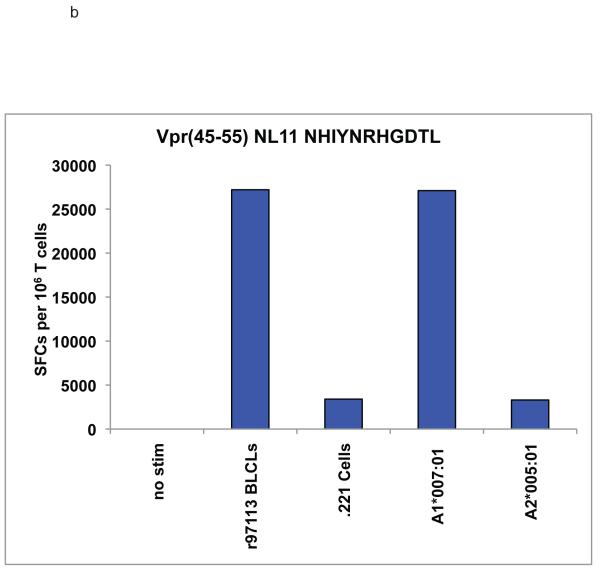
We noted novel immune responses in two pools of ten 15-mer peptides. The first pool contained peptides spanning residues 757-807 in SIVmac239 Pol, while the second spanned residues 1-55 in Vpr. In both cases, the responses were deconvoluted by testing each individual 15-mer within the pool to determine the location of the novel epitope. We then used IFN-γ ELISPOT or ICS to define the minimal optimal epitopes by testing the overlapping 8-, 9-, 10-, and 11-mer peptides contained within the reactive 15-mer (Figure 1a-c). Note that for a typical ELISPOT or ICS the peptide concentration is 10 μM,. (Figure 1b) the peptide concentration is 10 μM, whereas in Figure 1a and c, the peptide concentration is titrated down to less than 1 nM. At 10 μM (104 nM), responses are all in a maximal range. Based on a cDNA library of the animal’s MHC, we were able to narrow down the potential restricting class I alleles. R97113 had 12 different MHC class I alleles according to the cDNA library. Therefore, we first used a panel of MHC typed B-lymphocyte cell lines (BLCLs) to narrow down which alleles were likely to be involved, and to eliminate those that were not for each peptide. Then we individually transfected seven remaining different alleles into 721.221 cells, which do not express endogenous classical MHC class I molecules on the cell surface, resulting in a panel of seven different transfectants, each expressing a single MHC class I allele. We used autologous BLCL as a positive control, 721.221 cells as a negative control. As a result of our pre-screening with BLCL panels, we determined that Pol782-789 YL8 could potentially be bound by Mamu-A1*001:01, -A1*007:01, -A*15, -B*012:02, -B*030:01, -B*046 or -B*052:01 (Figure 2a), and we used this panel of transfectants as antigen presenting cells by pulsing them with the reactive 15-mer peptide containing Pol782-789 YL8. Similarly, we determined that Vpr45-55 NL11 could potentially be bound by Mamu-A1*007:01 or –A2*05:01, and we pulsed these transfectants with the reactive 15-mer peptide containing Vpr45-55 NL11 (Figure 2b). We used the cell line generated from r97113 in each case as a positive control. We were able to determine which allele restricted these two epitopes, and in both cases, the allele was Mamu-A1*007:01 (Figure 2). We subsequently determined these two immune responses to be the immunodominant Mamu-A1*007:01-restricted epitopes. We use the term immunodominant here to mean that these two epitopes are presented in most Mamu-A1*007:01-positive rhesus macaques infected with SIV, or vaccinated with these regions of Pol or Nef.
Figure 1. Fine mapping of Vpr45-55 NL11 and Pol782-789 YL8 epitopes in T cell lines derived from a vaccinated animal that controls viremia.
a. Overlapping peptides were presented by autologous BLCL to a Vpr45-55 NL11-specific T cell line in a series of 10-fold dilutions in a functional avidity ELISPOT assay. The 11-mer elicits nearly 100% maximal IFN-γ secretion out to the fourth dilution. b. Breakdown of an individual Pol 15-mer peptide into overlapping 9-mer peptides in an ICS assay. We stained for both IFN-γ and TNF-α and gated on CD8+ cells. Peptide is present at high concentration in the ICS assay (10 μM) c. Overlapping peptides were presented by autologous BLCL to a Pol782-789 YL8-specific T cell line in a series of 10-fold dilutions in a functional avidity ICS assay. Normalized frequencies of IFN-γ positive CD8+ cells are shown.
Figure 2. Mamu-A1*007:01 restricts both the Pol782-789 YL8 and Vpr45-55 NL11 epitopes.
721.221 cells, which do not express endogenous classical MHC class I molecules, were either stably or transiently transfected with the alleles shown. By W6/32 staining, MHC expression level was between 2% and 24%, at least 50% of cells had receptor on the surface. The transfectants were pulsed with either the 15-mer peptides used to raise the specific CD8+ T cell lines, no peptide (data not shown), or an A1*002:01-restricted irrelevant peptide. Peptide-pulsed cells were used in either ICS or ELISPOT assays with peptide-specific CD8+ T cell lines specific for a) a Pol-derived 15-mer peptide containing YL8 or b) a Vpr-derived 15-mer peptide containing NL11. Both cell lines were cultured from animal r97113; autologous BLCLs from this animal were used to check the specificity of the cell line. Peptide-pulsed 721.221 cells were used as a negative control. P/I refers to PMA and ionomycin, which are used as a positive control for inducing cytokine secretion.
Endogenous Mamu-A1*007:01 peptides
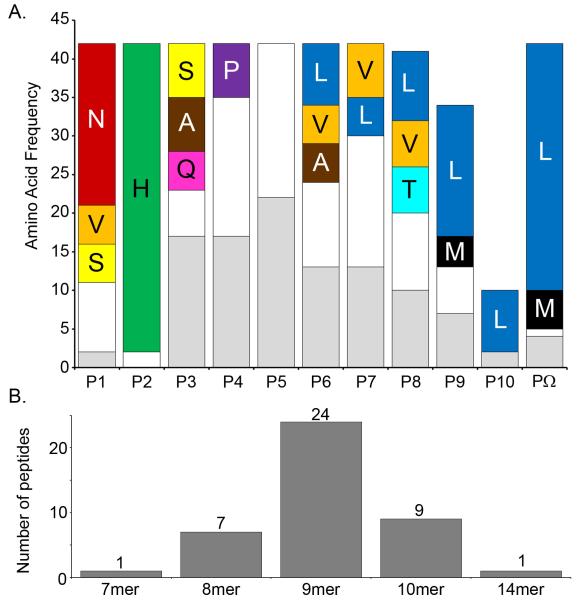
As an initial step to characterize the Mamu-A1*007:01 binding specificity, we eluted and sequenced by mass spectroscopy a total of 42 peptides from Mamu-A1*007:01 (Supplemental Table 2). In addition, Edman degradation sequencing of the pooled peptides eluted from Mamu- A1*007:01 provided a motif consistent with the mass spectrometric sequencing of individual peptides (data not shown). The majority of the individual peptides sequenced by mass spectroscopy have a histidine at the P2 anchor residue with a strong preference to be preceded by an asparagine at P1 (Figure 3). The C-terminal anchor was dominated by leucine, although a few peptides have a methionine residue at this position (Figure 3). When compared to HLA peptide binding motifs, these ligands are most similar to those bound by HLA alleles in the B*27 (Sidney et al. 2008) or B*39 supertypes (Lund et al. 2004). Alleles in these supertypes are characterized by specificity for ligands with positively charged residues in position 2 and hydrophobic residues at the C-terminus. Interestingly, amongst the alleles associated with the B*27 and B*39 supertypes are B*2705 and B*38:01, both of which have been linked with control of HIV viremia (Hendel et al. 1999; Kaslow et al. 2001; Carrington and O’Brien 2003; Gao et al. 2005; Altfeld et al. 2006; Deeks and Walker 2007; Salgado et al. 2010). The panel of sequenced ligands was synthesized and tested for their capacity to bind to purified Mamu- A1*007:01 molecules. All but 2 of the peptides (95%) bound with affinities of 500 nM or better, and half (22/42) bound with affinities of 50 nM or better (Supplemental Table 2). Taken together, these data suggest a putative binding motif for Mamu- A1*007:01 , and validate the binding assay utilized.
Figure 3. Summary of sequencing results for eluted endogenous ligands.
a. Single letter abbreviations for amino acids are used. The percent? frequency of specific amino acid residues at each position for eluted ligands is shown. We observed strong preferences at P2 for histidine and the C-terminus for leucine, indicating these are the binding anchors. P1 also prefers asparagine, although subsequent analysis indicated it to be a weak secondary anchor. b. Frequency of the various lengths of peptides eluted from A1*007:01.
Determination of a quantitative Mamu-A1*007:01 peptide-binding motif using a positional scanning combinatorial peptide library
To complement the endogenous ligand sequencing analysis and derive a more detailed quantitative motif for Mamu- A1*007:01, we next tested the capacity of a positional scanning combinatorial library (PSCL) to bind purified Mamu- A1*007:01 molecules. The binding capacity (IC50 nM) for each mixture was determined, and then normalized, as described in the Materials and Methods, to generate the resulting Mamu- A1*007:01 matrix shown in Table 1.
Table 1.
SIV-derived peptides binding Mamu-A1*007:01 with high affinity
| Protein | Amino acid positions |
Length | Sequence | IC50 (nM) | # of Animals Responding |
|---|---|---|---|---|---|
| Vpr | 45-55 | 11 | NHIYNRHGDTL | 0.20 | 5 |
| Env | 764-772 | 9 | WQIEYIHFL | 3.2 | |
| Pol | 782-789 | 8 | YHSNVKEL | 5.8 | 6 |
| Pol | 782-791 | 10 | YHSNVKELVF | 6.2 | 5 |
| Vif | 22-29 | 8 | WHSLIKYL | 10 | |
| Pol | 279-288 | 10 | WRMLIDFREL | 10 | |
| Vif | 145-153 | 9 | VPSLQYLAL | 13 | 2 |
| Pol | 567-575 | 9 | THTNGVRLL | 13 | 1 |
| Env | 795-802 | 8 | YQILQPIL | 24 | |
| Pol | 332-340 | 9 | FRQYTAFTL | 39 | |
| Pol | 817-825 | 9 | IHGQANSDL | 65 | 1 |
| Env | 230-240 | 11 | FRYCAPPGYAL | 101 | 1 |
| Vpx | 38-46 | 9 | NHLPRELIF | 144 | |
| Env | 184-192 | 9 | NETWYSADL | 147 | |
| Pol | 333-340 | 8 | RQYTAFTL | 151 | |
| Gag | 42-50 | 9 | DRFGLAESL | 177 | 1 |
| Env | 35-42 | 8 | WRNATIPL | 200 | |
| Vif | 65-75 | 11 | SHLEVQGYWHL | 209 | 1 |
| Env | 769-776 | 8 | IHFLIRQL | 223 | |
| Vpr | 33-43 | 11 | KHFDPRLLTAL | 227 | |
| Pol | 368-376 | 9 | FQYTMRHVL | 234 | |
| Pol | 760-769 | 10 | SQGIRQVLFL | 242 | 2 |
| Env | 562-569 | 8 | VQQQQQLL | 254 | |
| Env | 735-743 | 9 | THIQQDPAL | 306 | |
| Env | 792-802 | 11 | SRVYQILQPIL | 310 | |
| Pol | 104-114 | 11 | DRGFAAPQFSL | 322 | |
| Gag | 42-51 | 10 | DRFGLAESLL | 325 | 1 |
| Pol | 105-114 | 10 | RGFAAPQFSL | 359 | |
| Env | 782-790 | 9 | WLFSNCRTL | 379 | |
| Gag | 195-202 | 8 | DHQAAMQI | 380 | |
| Pol | 289-298 | 10 | NRVTQDFTEV | 447 | |
| Gag | 195-203 | 9 | DHQAAMQII | 473 | |
| Vpr | 71-78 | 8 | MHFRGGCI | 498 |
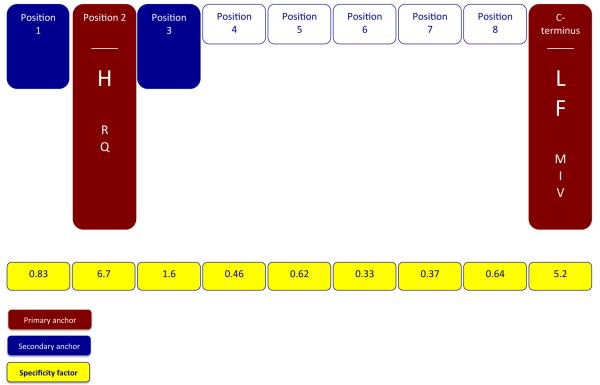
Next, for each position, an average (geometric) relative binding affinity (ARB) was calculated, and then the ration of the ARB for the entire library to the ARB for each position was derived (Table 2). This ratio, which we have denominated as the specificity factor (SF), represents the factor by which the normalized geometric average binding affinity associated with all 20 residues at a specified position differs from that of the average affinity of the entire library. As calculated, positions with the highest specificity will have the highest SF value. For Mamu- A1*007:01, the highest SF values were noted for position 2 and the C-terminus, with SF of 6.7 and 5.2 respectively; SF for all other positions were found to be <1.6. On this basis, and following previously established criteria (Sidney et al. 2008a), P2 and the C-terminus can thus be identified as the main anchor positions for Mamu-A1*007:01 binding.
Table 2.
PSCL-derived matrix describing 9-mer binding to Mamu-A1*007:01
| Position |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| A | 0.061 | - | 0.35 | 0.21 | 0.14 | 1.0 | 0.14 | 1.0 | 0.016 |
| C | 0.22 | - | 0.019 | 0.16 | 0.087 | 0.22 | 0.21 | 0.14 | - |
| D | 0.14 | - | 0.094 | 0.13 | 0.048 | 0.15 | 0.066 | 0.10 | - |
| E | 0.023 | - | - | 0.024 | 0.044 | 0.12 | 0.038 | 0.018 | - |
| F | 0.095 | - | 1.0 | 1.0 | 0.42 | 0.28 | 0.72 | 0.21 | 0.51 |
| G | 0.063 | - | 0.042 | 0.33 | 0.18 | 0.24 | 0.17 | 0.12 | - |
| H | 0.16 | 1.0 | 0.059 | 0.27 | 0.18 | 0.22 | 0.32 | 0.039 | - |
| I | 0.12 | 0.015 | 0.097 | 0.15 | 0.25 | 0.56 | 0.70 | 0.29 | 0.075 |
| K | 0.023 | - | 0.026 | 0.095 | 0.061 | 0.10 | 0.14 | 0.078 | - |
| L | 0.057 | 0.011 | 0.025 | 0.13 | 0.098 | 0.023 | 0.52 | 0.25 | 1.0 |
| M | 0.21 | 0.022 | 0.077 | 0.97 | 0.092 | 0.62 | 0.30 | 0.13 | 0.14 |
| N | 1.0 | - | 0.061 | 0.21 | 0.11 | 0.69 | 0.16 | 0.083 | - |
| P | 0.015 | - | 0.015 | 0.39 | 0.12 | 0.65 | 0.22 | 0.11 | - |
| Q | 0.077 | 0.12 | 0.050 | 0.26 | 0.15 | 0.51 | 0.27 | 0.15 | - |
| R | 0.13 | 0.14 | - | 0.22 | 0.11 | 0.25 | 0.22 | 0.17 | - |
| S | 0.30 | 0.012 | 0.12 | 0.21 | 0.23 | 0.94 | 0.25 | 0.22 | - |
| T | 0.081 | - | 0.13 | 0.13 | 0.14 | 0.39 | 0.22 | 0.35 | 0.014 |
| V | 0.22 | - | 0.11 | 0.16 | 0.23 | 0.28 | 0.42 | 0.67 | 0.063 |
| W | 0.28 | - | 0.025 | 0.16 | 1.0 | 0.13 | 0.32 | 0.047 | 0.021 |
| Y | 0.20 | - | 0.16 | 0.17 | 0.34 | 0.26 | 1.0 | 0.13 | - |
|
| |||||||||
| Geomean | 0.11 | 0.014 | 0.059 | 0.20 | 0.15 | 0.28 | 0.25 | 0.14 | 0.017 |
| SD | 2.7 | 3.9 | 3.2 | 2.2 | 2.1 | 2.5 | 2.2 | 2.5 | 4.9 |
| SF | 0.83 | 6.7 | 1.6 | 0.46 | 0.62 | 0.33 | 0.37 | 0.64 | 5.2 |
In agreement with the pool sequencing derived motif, at position 2 the positively charged residue histidine was the most preferred. The positively charged residue arginine, and the polar residue glutamine were also tolerated, with normalized average relative binding (ARB) values of 0.14 and 0.12, respectively. All other residues at position 2 were associated with ARB values <0.04, corresponding to a 25-fold or more decrease in binding affinity, compared to histidine.
At the C-terminus, the hydrophobic aliphatic residue leucine was identified as most preferred, followed by the hydrophobic aromatic residue phenylalanine, with an ARB value of 0.51. The hydrophobic residues methionine, isoleucine, and valine were tolerated, with ARBs of 0.14, 0.075 and 0.063, respectively. All other residues at the C-terminus were associated with binding capacities more than 25-fold lower than the optimal residue, leucine.
Influences on binding at other positions tended to be minor, reflected by low SF values in the 0.33 to 1.6 range. Defining secondary anchors on the basis of standard deviations (SD), following the rubric previously defined (Sidney et al. 2008a; Loffredo et al. 2009), only position 3 is identified as a dominant secondary anchor. Here, the aromatic residue phenylalanine is the most preferred. Small residues alanine, serine, threonine, and valine, and the aromatic residue tyrosine, are all tolerated, with ARBs in the 0.11 to 0.35 range, representing 3- to 10-fold reductions in binding compared to phenylalanine.
In contrast to the pool sequencing data, the PSCL analysis did not identify position 1 as either a primary or dominant secondary anchor position, although asparagine was identified as the optimal residue. With an SD of 2.7, P1 would appear to function as a weak secondary anchor. To examine this further, we also measured the Mamu-A1*007:01 binding capacity of a panel of single amino acid analogs of the Human Ribosomal protein L9 39-48 peptide (sequence NHINVELSL), which was identified above as an endogenously bound Mamu-A1*007:01 ligand (IC50 13 nM, Supplemental Table 2). This panel (data not shown) similarly identified position 2 and the C-terminus as the primary anchors, with histidine as the preferred residue in position 2 and leucine and phenylalanine as preferred at the C-terminus. While asparagine was again identified as the optimal residue in position 1, this position does not appear to function as a primary anchor, although it may function as an important secondary anchor.
In summary, we used the PSCL to define a detailed motif for Mamu-A1*007:01. This motif is characterized by primary anchor specificity for the positively charged residue histidine in position 2, where arginine and glutamine are also tolerated. A second primary anchor is found at the C-terminus, where a preference for the hydrophobic/aromatic residues leucine and phenylalanine was identified, and methionine, isoleucine, and valine were tolerated. Positions 1 and 3 were identified as the most prominent secondary anchors. This motif is in agreement with analyses based on sequencing of eluted ligands, as well as analysis of the binding capacity of a panel of single amino acid substitution analogs of a high affinity Mamu-A1*007:01 binder. A summary cartoon depicting the Mamu-A1*007:01 motif is shown in Figure 4.
Figure 4. The binding motif for Mamu A1*007:01.
As determined using eluted ligands, a PSCL binding analysis, and a panel of single amino acid substitution analogs, the motif is summarized to show the main (red fill) and secondary (blue fill) anchor positions. The most preferred residues at the primary anchor positions are indicated by enlarged font. Tolerated anchor residues are also shown. SF values for each position as determined in the PSCL binding analysis, reflecting the relative overall contribution of each position to peptide binding are also shown (yellow fill).
Identification of SIVmac239-derived Mamu-A1*007:01-binding peptides
To identify SIV-derived Mamu-A1*007:01 restricted T cell epitopes, we first sought to identify a panel of high affinity Mamu-A1*007:01 binding peptides. PSCL matrices have proven to be an efficient means to identify MHC class I binding peptides (Udaka et al. 1995; Stryhn et al. 1996; Udaka et al. 2000; Lauemoller et al. 2001; Peters et al. 2006; Sidney et al. 2007; Sidney et al. 2008a; Loffredo et al. 2009). Thus, for this purpose, we utilized the PSCL-based matrix shown in Table 2 to score all 9-mer peptides in the SIVmac239 proteome. As before (Sidney et al. 2007; Sidney et al. 2008a; Loffredo et al. 2009), we also adapted the matrix to score peptides of 8, 10 or 11 residues in length. For each size, the peptides scoring in the top 1.5% range (n=50), a selection threshold shown to be associated with the vast majority of epitopes (Moutaftsi et al. 2006; Assarsson et al. 2007; Kotturi et al. 2007; Loffredo et al. 2009), were selected for analysis. All 200 peptides were synthesized and tested for their capacity to bind purified Mamu-A1*007:01. The binding affinities of these peptides, as well as several additional other SIV-derived peptides tested in the course of the present study, are provided in Supplemental Table 3; all of the binding data will also be submitted to the IEDB (www.iedb.org). As in previous studies, the cutoff for defining a peptide as a candidate binder is an IC50 value ≤500nM. This is based on previous work in humans and macaques that have correlated these values to T cell recognition in vivo (Sette et al. 1994a; Sette et al. 1994b; Allen et al. 1998).
In total, 33 of the 200 peptides (17%) bound Mamu-A1*007:01 with an affinity of 500 nM or better (Table 1). Ten of these peptides bound very strongly (IC50 ≤50nM). Binders were identified from six different SIV proteins, with the greatest numbers in Env and Pol (10 and 12, respectively). Interestingly, no binders were identified in the Nef, Tat, or Rev proteins.
Several binders, including those with very high affinity (IC50 <50 nM), were identified for each size examined. However, as expected from the analysis of endogenous ligands, 9-mers were the best overall, both in terms of the percentage of binders (12/50, 24%) and average affinity of the selected peptides (2520 nM). After 9-mers, 8-mers were the next best (9 binders, average affinity 4861 nM), followed by 10-mers (6 binders, 6915 nM), and then 11-mers (6 binders, 9906 nM).
Defining six novel epitopes restricted by Mamu-A1*007:01
Next, we examined whether any of the 33 SIV-derived Mamu-A1*007:01 binders would be recognized in SIVmac239-infected macaques. We used freshly isolated or frozen PBMC from 15 SIV-infected and two vaccinated but uninfected, Mamu-A1*007:01-positive rhesus macaques in IFN-γ ELISPOT assays. In total, the PBMC from these animals secreted IFN-γ in response to 11 of the 33 binders (Table 1). The magnitude of the responses detected in the ELISPOT assays ranged from 50 to 930 SFCs per 106 PBMC. Of the 11 peptides recognized, the number of responding animals ranged from one to six per epitope. Animals r98030 and r97113 responded to the broadest repertoire of binders (6 and 5, respectively). The highest frequency response was also in r97113, at 930 SFCs per million cells to Vpr45-55 NL11. This epitope was recognized in five animals, making it the second most-frequently targeted after Pol782-789 YL8.
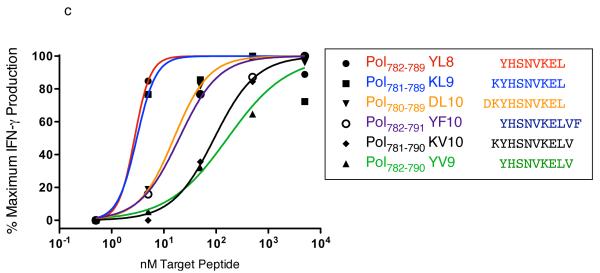
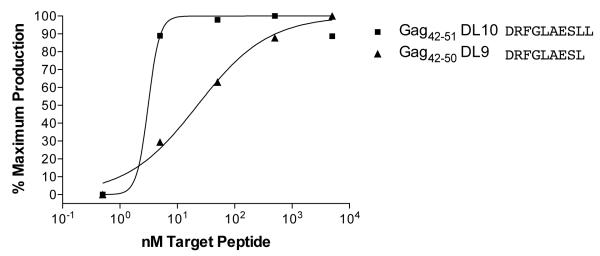
Of the 11 binders that elicited IFN-γ in ELISPOT, there are two pairs of overlapping peptides that most likely represent a single epitope. Pol782-791 YF10 contains the minimal optimal Pol782-789YL8, while Gag42-51 DL10 contains the smaller binder Gag42-50 DL9. We performed functional avidity assays with a minimum of five 10-fold dilutions of these peptides and determined in both cases the shorter peptide to be the minimal optimal (Figure 1 and Figure 5). Additionally, we were unable to confirm restriction by Mamu-A1*007:01 for three of the peptides that elicited IFN-γ in the ELISPOT assays. Env230-240 FL11, Pol817-825 IL9, and Vif65-75 SL11 each elicited a response by a single animal (range=70-220 SFC per 1 × 106), but we were unable to generate epitope-specific CTL lines with which to confirm the restriction by Mamu-A1*007:01 for the first two responses. For Vif65-75 SL11, the CTL line that we generated did not secrete IFN-γ to the SL11 peptide in the context of Mamu-A1*007:01; we would hypothesize that this epitope may be restricted by a different MHC class I allele. Thus, we have defined six novel Mamu-A1*007:01-restricted epitopes (Table 3).
Figure 5. Functional Avidity Assay for Gag42-50 DL9 epitope.
As in Figure. 1, we performed 10-fold peptide dilutions and presented the peptides to a specific CTL line. The results concur with the binding affinities as predicted by the binding studies.
Table 3.
Mamu-A1*007:01-restricted epitopes in SIVmac239
| Epitope | Protein | Length | Position | IC50 (nM) |
|---|---|---|---|---|
| NHIYNRHGDTL | Vpr | 11 | 45-55 | 0.2 |
| YHSNVKEL | Pol | 8 | 782-789 | 5.8 |
| SQGIRQVLFL | Pol | 10 | 760-769 | 242 |
| DRFGLAESL | Gag | 9 | 42-50 | 177 |
| VPSLQYLAL | Vif | 9 | 145-153 | 13 |
| THTNGVRLL | Pol | 9 | 567-575 | 13 |
As a negative control, we tested all 33 peptides in two SIVmac239-infected Mamu-A1*007:01-negative rhesus macaques (r95061 and rhAJ11). No reactivity was observed against any of the peptides (data not shown). Two uninfected Mamu-A1*007:01-positive macaques were also screened in ELISPOT assays to test for cross-reactivity with other non-SIV epitopes. None of the peptides elicited a response from either animal (data not shown).
Mamu-A1*007:01-restricted CD8+ T cells select for viral variation in multiple SIVmac239 epitopes
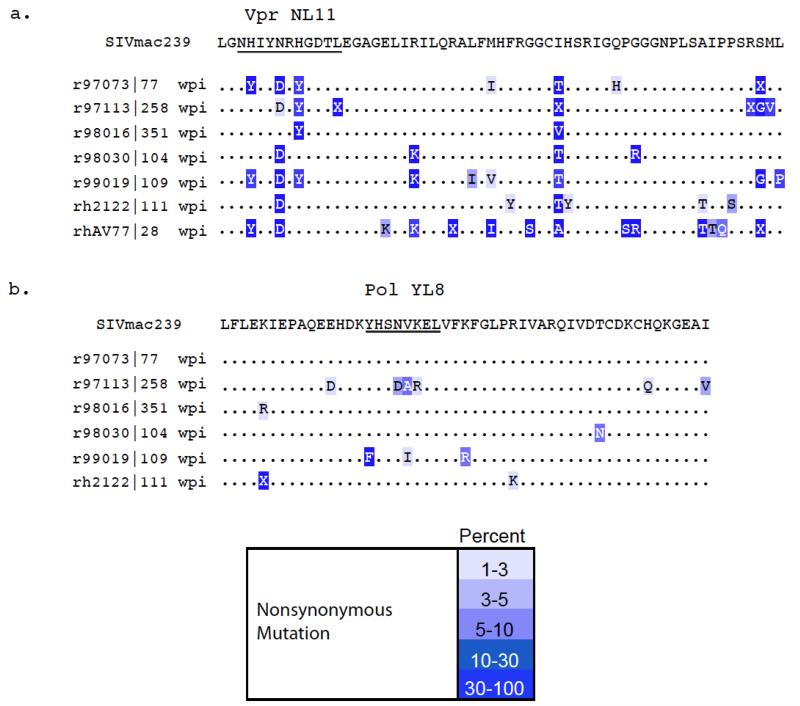
We isolated virus from plasma of chronically infected Mamu-A1*007:01-positive macaques to look for evidence that these novel CD8+ T cell responses were capable of selecting for viral variation. We sequenced open reading frames from the SIV proteins Gag, Pol, Vif, and Vpr in the regions where we identified epitopes that bind to Mamu-A1*007:01. We compared the amino acid sequences of the novel epitope regions of the virus from the A1*007:01-positive macaques to those from time-of-death sequences previously obtained from 35 Mamu-A1*007:01-negative macaques infected with SIVmac239 (O’Connor et al. 2004). Amino acid substitutions existed in four of the six epitopes predicted to bind to Mamu-A1*007:01. Two of these are shown in Figure 6. Vpr45-55 NL11 had variation at positions two (histidine to tyrosine), five (asparagine to aspartic acid), seven (histidine to tyrosine), and eleven (leucine to isoleucine) (Figure 6a). In total, virus from nine of the twelve Mamu-A1*007:01-positive animals had variation in at least one position within this epitope by Sanger sequencing, while virus from seven of seven animals had variation in a minimum of 15 percent of sequencing reads across the epitope by ultradeep pyrosequencing. By comparison, only one of the 35 Mamu-A1*007:01-negative animals had any variation within the epitope: virus from r95003 contained the same P5 asparagine to aspartic acid seen in five of the Mamu-A1*007:01-positive animals (O’Connor et al. 2004). Thus, we detected viral variation at five positions within the epitope, including both primary binding anchor positions, indicating a CD8-mediated selective pressure in Mamu-A1*007:01-positive animals.
Figure 6. Ultradeep pyrosequencing results across the NL11 and YL8 epitopes in Mamu-A1*007:01-positive animals.
a. Vpr45-55 NL11 and b. Pol782-789 YL8 epitopes. The Vpr epitope has broad and consistent variation at multiple positions within the epitope demonstrating escape in binding anchor positions and TCR-contact residues. The Pol epitope has some variation; however, none of the substitutions has become the dominant circulating species, even after six years of replication. This may indicate that this region is under functional constraints.
In the Pol782-789 YL8 epitope, we could detect variation in the virus from two animals. Animal r97113 had substitutions in three consecutive positions (four through six) of the epitope (Figure 6b). Using ultradeep pyrosequencing of this region, we noticed that the P4 asparagine to aspartic acid change occurred in 67 of 1361 sequence reads (4.9%), while the P5 valine to alanine change occurred in 68 of 1361 reads (5%). The P6 lysine to arginine substitution was only present in 1 percent of reads. The virus we sequenced from this animal was from 258 weeks post infection, and while there are three changes within this epitope, they are all present at low frequencies overall within the circulating virus population. The other animal from which we detected viral variation was r99019. Its virus had a P1 tyrosine to phenylalanine change present in 43 percent of circulating virus. Additionally, this animal’s virus had a P5 valine to isoleucine change present in only 2 percent of the circulating virus at 109 weeks post-infection. None of the 35 Mamu-A1*007:01-negative animals had any variation within this region of the virus by Sanger sequencing (data not shown). We were able to consistently detect CTL responses to this epitope in both r97113 and r99019, but even after two years of virus replication under this selective pressure, the amino acid substitutions that we could detect were all minority species within the animals. Taken together, these data may indicate that this region of the virus is under functional constraints.
Additionally, we detected a single mutation in two other epitopes. Gag42-50 DL9 in five chronically infected Mamu-A1*007:01-positive macaques contained a single position eight substitution in animal rh2161 from serine to glycine by conventional sequencing. In the Pol760-769 SL10 epitope, one of the six animals that we investigated with ultradeep pyrosequencing had variation in its virus. Animal r97073 had a position seven valine to isoleucine switch at a frequency of 24.9 percent. Neither of these regions showed any variation in the 35 Mamu-A1*007:01-negative animals at time of death (data not shown).
While the number of variants is not overwhelming, we are hampered by the low frequency of Mamu-A1*007:01 positive animals in the population, limiting the number of examples we are able to observe. Additionally, we have shown that sequence variation in the epitope has a strong impact on binding affinity to Mamu-A1*007:01. (Table 4). The effect of variation on binding affinity, along with the complete lack of variation in these regions in Mamu-A1*007:01 negative animals indicates that, while these mutations are limited in number, they are indeed CTL driven.
Table 4.
Sequence variation in epitopes greatly decreases binding affinity for MHC.
| Peptide | Type | Sequence | Protein | Position | IC50 (nM) | Relative binding (WT) |
Fold decrease |
|---|---|---|---|---|---|---|---|
| 3408.0001 | WT | NHIYNRHGDTL | Vpr | 11 | 0.20 | 1.0 | |
| 3408.0002 | variant | NYIYDRYGDTL | Vpr | 11 | 765 | 0.00026 | 3823 |
| 3408.0003 | variant | NHIYDRYGDTV | Vpr | 11 | 230 | 0.00087 | 1151 |
| 3408.0004 | variant | NHIYDRYGDTI | Vpr | 11 | 44 | 0.0045 | 221 |
| 3408.0005 | variant | NHIYNRYGDTL | Vpr | 11 | 16 | 0.012 | 80 |
| 3408.0006 | variant | NHIYDRHGDTL | Vpr | 11 | 42 | 0.0048 | 210 |
| 3408.0007 | variant | NYIYDRHGDTL | Vpr | 11 | - | <0.000003 | >350000 |
|
| |||||||
| 3408.0008 | WT | YHSNVKEL | Pol | 782 | 5.8 | 1.0 | |
| 3408.0009 | variant | YHSDAREL | Pol | 782 | 4.5 | 1.3 | 0.77 |
| 3408.0010 | variant | FHSNIKEL | Pol | 782 | 9.9 | 0.58 | 1.7 |
|
| |||||||
| 3408.0011 | WT | DRFGLAESL | Gag | 42 | 177 | 1.0 | |
| 3408.0012 | variant | DRFGLAEGL | Gag | 42 | 4247 | 0.042 | 24 |
|
| |||||||
| 3408.0013 | WT | SQGIRQVLFL | Pol | 760 | 242 | 1.0 | |
| 3408.0014 | variant | SQGIRQILFL | Pol | 760 | - | 0.0035 | 289 |
A dash indicates IC50 >70000 nM.
Epitope IC50 values from previous synthesis, to compare with values in ms tables. 3408 values are not significantly different.
Interestingly, in two epitope regions, we did not detect viral variation of any kind. Pol567-575 TL9 and Vif145-153 VL9 showed no mutations by Sanger or ultradeep pyrosequencing (data not shown). Indeed, in at least six chronically-infected Mamu-A1*007:01-positive animals, despite a minimum of 1500 sequencing reads across both the Pol and Vif epitopes, we were unable to detect a single mutation that occurred at our minimum cutoff frequency of one percent. These regions of the virus may be under particularly strong functional constraints. Indeed, this Vif region contains a highly conserved motif both among geographically disparate strains of HIV (Stephens et al. 2001) and among lentivirus Vif proteins in general (Oberste and Gonda 1992). CTL specific for epitopes contained within such highly conserved regions may be especially effective at controlling viral replication.
Discussion
It is well established that CTL play a vital role in controlling immunodeficiency virus replication after infection. These T cells are specific for HIV/SIV derived epitopes presented in the context of specific MHC class I alleles. However, even though we have been using SIV-infected rhesus macaques for more than two decades, we still have incomplete knowledge of the immunogenetics of the macaque model when compared to humans. To date, 650 unique rhesus MHC class I alleles have been identified (IPD database, http://www.ebi.ac.uk/ipd/mhc/nhp/nomen_mamu.html), and only six alleles from Indian-origin rhesus macaques have been well-characterized with regard to their ability to bind peptides derived from SIV and the potential role they may play in control of viral replication. Many of the studied alleles were present at high frequencies, which expanded the group of animals that were useful for vaccine studies. Others have helped define animals that should be excluded from vaccine trials due to the high incidence of spontaneous viral control. Each characterized allele increases our knowledge of this animal model and broadens the pool of animals available for future study.
In this study, we demonstrated that Mamu-A1*007:01 has a binding motif with primary anchor residues at positions two and the C terminus. Leucine and phenylalanine are the preferred residues at the C-terminus although other hydrophobic amino acids can be tolerated. Histidine is the dominant residue at position 2. We also determined that position three functions as a strong secondary anchor and that position one may also have some influence on Mamu-A1*007:01-peptide binding. This overall motif is similar to the one recognized by HLA alleles classified into the B*27 and B*39 supertypes (Lund et al. 2004; Sidney et al. 2008b). Indeed, on the basis of primary sequence structure, the B and F pockets of Mamu A1*007:01, associated with the binding specificity for position 2 and the C-terminus, respectively, are identical with those of several B*27 and/or B*39 supertype alleles (Sidney et al. 2008). We have initiated studies to examine crossreactivity between HLA B*3801, which also has a prominent P2 specificity for H, and Mamu A1*007:01. B*3801 is of particular interest, as Salgado et al. (2011) have recently demonstrated that it is part of an allele combination (with HLA-Cw*1203) that may have an effect in long-term HIV non-progression. We tested Mamu-A1*007:01 binders for their capacity to bind HLA-B*3801. (Supplemental Table 4) About half of these peptides bound HLA-B*3801, including the majority of the epitopes identified in this paper. However, none of the known HLA-B*3801 restricted HIV-derived epitopes bound Mamu-A1*007:01. So, while it is apparent that these two molecules share some binding characteristics, the use of Mamu-A1*007:01-positive animals as an HLA-B*3801 surrogate would be questionable.
We identified 33 SIV-derived peptides with the capacity to bind Mamu-A1*007:01 with high affinity (IC50 ≤500nM). Using PBMC derived from SIVmac239-infected or vaccinated, uninfected Mamu-A1*007:01-positive rhesus macaques, we found that eleven of these peptides, covering nine viral protein regions, could elicit IFN-γ responses in ELISPOT assays. Two of these (Pol782-789 YL8 and Vpr45-55 NL11) were previously identified as A1*007:01-restricted epitopes (Sacha et al. 2007; Sacha et al. 2010), while four of them represent newly defined epitopes. We were unable to definitively confirm restriction of the other three epitopes by Mamu-A1*007:01.
The SIV-derived peptides utilized in the present study were selected on the basis of predicted relative affinity using a PSCL matrix. This approach has proven successful for identifying Mamu B*008:01 restricted SIV epitopes (Loffredo et al. 2009), and as a general means for predicting class I binding peptides (Sidney et al. 2007; and Sidney et al. 2008a). On the basis of previous observations (Moutaftsi et al. 2006; Kotturi et al. 2007; Loffredo et al. 2009) we selected peptides scoring in the top 1.5% range, which would be hypothesized to encompass the vast majority of Mamu A1*007:01 epitopes. It is possible that additional epitopes may be identified by systematically selecting and testing peptides scoring in lower prediction ranges. However, of the 40 peptides tested in the course of the present study that had PSCL scores in the 1.5 to 50% range, none bound Mamu-A1*007:01 with an affinity of 500 nM or better. Furthermore, and most importantly, 5 of the 6 epitopes identified herein had scores in the top 0.8% or better. Thus, it is likely that the current selection process has identified the preponderance of SIV-derived Mamu-A1*007:01 restricted T cell epitopes.
The phenylalanine discrepancy between the endogenous ligand and PSCL analyses is difficult to speculate upon, but likely reflects the lower frequency of this residue in the endogenous setting compared to L. The eluted ligand analysis will reflect the in vivo “environment” of the cell at the time of analysis, while the binding analysis reflects the in vitro potential of the molecule. Thus, it is also possible that the discrepancy may reflect differences in processing/transport efficiencies associated with the respective residues. This latter hypothesis is perhaps supported by the observation that all of the SIV epitopes identified herein bore L at the C-terminus, although it is also notable that L is present with about a 3-fold higher frequency in SIV than F, and is associated with a 2-fold higher average affinity.
Mamu-A1*007:01-restricted CD8+ T lymphocyte responses selected for viral escape. Using both Sanger sequencing and ultradeep pyrosequencing, we observed variation in at least one animal in four of the six regions. In the Vpr45-55 NL11 epitope, variation was widespread and consistent. While the Pol782-789 YL8, Gag42-50 DL9, and Pol760-769 SL10 epitope variation was less frequent, it is possible that the defined mutations in these epitopes might result in TCR escape. Interestingly, two of the A1*007:01-restricted epitope regions exhibited no variation in circulating virus, even well into the chronic phase in a number of animals. In five of the seven animals we used for the pyrosequencing analysis, the samples were from more than 100 weeks post-infection, and two of them were beyond 250 weeks post-infection. These particular conserved epitopes may be located within regions of the virus that are under severe fitness constraints. Stephens et al. (2001) demonstrated that one of these regions in Vif is highly conserved among geographically diverse isolates of HIV-1. Additionally, Oberste and Gonda (1992) described highly conserved sequence motifs in lentivirus Vif proteins. The SLQXLA motif was present in 34 of 38 of the lentiviruses they examined. This motif is conserved within SIVmac239, and the Vif145-153 VL9 epitope that Mamu-A1*007:01 binds contains this exact motif. CTL that target these highly conserved regions may be especially effective at controlling viral replication.
In summary, we have characterized, in depth, the peptide binding specificity of the rhesus macaque class I molecule Mamu A1*007:01. Our studies have led to the identification of six novel SIV-derived CD8+ T cell epitopes restricted by Mamu-A1*007:01. We have demonstrated that these CD8+ T cells are capable of selecting for viral variation. Additionally, we have described two highly conserved regions of SIVmac239 targeted by Mamu-A1*007:01-restricted CD8+ T cell responses. These responses might be useful in testing the hypothesis that CTL that target conserved regions of the virus are more effective at controlling viral replication than those that target variable regions.
Supplementary Material
Acknowledgments
We thank Gretta Borchardt and Chrystal Glidden for PCR-SSP typing and Lyle Wallace for the cDNA library work and resulting database. We also appreciate Alex Bean for assistance with immunological assays. The following reagents were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: IL-2, human (item no. 136) from Hoffman-La Roche and SIVmac239 Vpr Peptides (6449) and SIVmac239 Pol Peptides (6443) from DAIDS, NIAID. This research was supported in part by grant R24 RR015371 (D.I.W.) from the National Institutes of Health, and in part by NIH grant number P51 RR000167 to the Wisconsin National Primate Research Center, University of Wisconsin-Madison. We would also like to thank Carrie Moore, Amiyah Steen and Sandy Ngo for performing MHC purification and peptide binding studies.
Abbreviations used
- Ad5
Adenovirus serotype 5
- MVA
Modified Vaccinia Ankara
References
- Alexander L, Denekamp L, Czajak S, Desrosiers R. Suboptimal nucleotides in the infectious, pathogenic simian immunodeficiency virus clone SIVmac239. J Virol. 2001;75:4019–4022. doi: 10.1128/JVI.75.8.4019-4022.2001. doi:10.1128/JVI.75.8.4019-4022.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, Mothe B, Sidney J, Jing P, Dzuris J, Liebl M, Vogel T, O’Connor DH, Wang X, Wussow M, Thomson J, Altman J, Watkins D, Sette A. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738–749. doi: 10.1128/JVI.75.2.738-749.2001. doi:10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, O’Connor DH, Jing P, Dzuris J, Mothe B, Vogel T, Dunphy E, Liebl M, Emerson C, Wilson N, Kunstman K, Wang X, Allison D, Hughes A, Desrosiers R, Altman J, Wolinsky S, Sette A, Watkins D. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. doi:10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- Allen TM, Sidney J, Del Guercio MF, Glickman R, Lensmeyer G, Wiebe D, Demars R, Pauza C, Johnson R, Sette A, Watkins D. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- Altfeld M, Kalife E, Qi Y, Streeck H, Lichterfeld M, Johnston M, Burgett N, Swartz M, Yang A, Alter G, Yu XG, Meier A, Rockstroh J, Allen T, Jessen H, Rosenberg E, Carrington M, Walker B. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. doi:10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman JD, Moss P, Goulder P, Barouch D, Mcheyzer-Williams MG, Bell J, Mcmichael A, Davis M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui H, Frahm N, Brander C, Peters B, Grey H, Sette A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- Barnstable C, Bodmer W, Brown G, Galfre G, Milstein C, Williams A, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- Barouch D, Kunstman J, Glowczwskie J, Kunstman K, Egan M, Peyerl F, Santra S, Kuroda M, Schmitz J, Beaudry K, Krivulka G, Lifton M, Gorgone D, Wolinsky S, Letvin N. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J Virol. 2003;77:7367–7375. doi: 10.1128/JVI.77.13.7367-7375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Kunstman J, Kuroda M, Schmitz J, Santra S, Peyerl F, Krivulka G, Beaudry K, Lifton M, Gorgone D, Montefiori D, Lewis M, Wolinsky S, Letvin N. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 2002;415:335–339. doi: 10.1038/415335a. doi:10.1038/415335a. [DOI] [PubMed] [Google Scholar]
- Bimber BN, Burwitz B, O’Connor S, Detmer A, Gostick E, Lank S, Price D, Hughes A, O’Connor D. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J Virol. 2009;83:8247–8253. doi: 10.1128/JVI.00897-09. doi:10.1128/JVI.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Hahn B, Shaw G, Oldstone M. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz M, Peffer N, Meyers H, Nelson J, Gairin J, Hahn B, Oldstone M, Shaw G. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Boyson J, Shufflebotham C, Cadavid L, Urvater J, Knapp L, Hughes A, Watkins D. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- Burwitz B, Pendley C, Greene J, Detmer A, Lhost J, Karl J, Piaskowski S, Rudersdorf R, Wallace L, Bimber B, Loffredo J, Cox D, Bardet W, Hildebrand W, Wiseman R, O’Connor SL, O’Connor DH. Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus-specific CD8+ T cells. J Virol. 2009;83:6011–6019. doi: 10.1128/JVI.00199-09. doi:10.1128/JVI.00199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Nelson GW, Martin M, Kissner T, Vlahov D, Goedert J, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. doi:10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- Chen Z, Craiu A, Shen L, Kuroda M, Iroku U, Watkins D, Voss G, Letvin N. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J Immunol. 2000;164:6474–6479. doi: 10.4049/jimmunol.164.12.6474. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff W. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Deeks S, Walker B. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. doi:10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Demars R, Rudersdorf R, Chang C, Petersen J, Strandtmann J, Korn N, Sidwell B, Orr H. Mutations that impair a posttranscriptional step in expression of HLA-A and -B antigens. Proc Natl Acad Sci U S A. 1985;82:8183–8187. doi: 10.1073/pnas.82.23.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, O’Connor DH, Jing P, Dzuris J, Sidney J, Da Silva J, Allen T, Horton H, Venham J, Rudersdorf R, Vogel T, Pauza C, Bontrop R, Demars R, Sette A, Hughes A, Watkins D. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. doi:10.1038/15224. [DOI] [PubMed] [Google Scholar]
- Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee H. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. doi:10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Frye C, Yant L, O’Connor DH, Kriewaldt N, Benson M, Vojnov L, Dodds E, Cullen C, Rudersdorf R, Hughes A, Wilson N, Watkins D. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J Virol. 2004a;78:2581–2585. doi: 10.1128/JVI.78.5.2581-2585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Mcdermott A, Reynolds M, Piaskowski S, Fuenger S, De Souza IP, Rudersdorf R, Cullen C, Yant L, Vojnov L, Stephany J, Martin S, O’Connor DH, Wilson N, Watkins D. Consequences of cytotoxic T-lymphocyte escape: common escape mutations in simian immunodeficiency virus are poorly recognized in naive hosts. J Virol. 2004b;78:10064–10073. doi: 10.1128/JVI.78.18.10064-10073.2004. doi:10.1128/JVI.78.18.10064-10073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Valentine L, Yant L, Rakasz E, Piaskowski S, Furlott J, Weisgrau K, Burwitz B, May G, Leon E, Soma T, Napoe G, Capuano SR, Wilson N, Watkins D. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. doi:10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Bashirova A, Iversen A, Phair J, Goedert J, Buchbinder S, Hoots K, Vlahov D, Altfeld M, O’Brien SJ, Carrington M. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. doi:10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- Goulder P, Phillips R, Colbert R, Mcadam S, Ogg G, Nowak M, Giangrande P, Luzzi G, Morgan B, Edwards A, Mcmichael A, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Gulukota K, Sidney J, Sette A, Delisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. doi:10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O’Brien S, Andrieu J, Schachter F, Zagury D, Rappaport J, Winkler C, Nelson G, Zagury J. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- Jin X, Bauer D, Tuttleton S, Lewin S, Gettie A, Blanchard J, Irwin C, Safrit J, Mittler J, Weinberger L, Kostrikis L, Zhang L, Perelson A, Ho DD. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizu M, Borchardt G, Glidden C, Fisk D, Loffredo J, Watkins D, Rehrauer W. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. doi:10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah A, Goedert J, Winkler C, O’Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann D. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Kaslow R, Rivers C, Tang J, Bender T, Goepfert P, El Habib R, Weinhold K, Mulligan M. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J Virol. 2001;75:8681–8689. doi: 10.1128/JVI.75.18.8681-8689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast WM, Brandt RM, Sidney J, Drijfhout J, Kubo R, Grey H, Melief C, Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J Immunol. 1994;152:3904–3912. [PubMed] [Google Scholar]
- Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Et A. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- Klein M, Van Baalen CA, Holwerda A, Kerkhof Garde SR, Bende R, Keet I, Eeftinck-Schattenkerk JK, Osterhaus A, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo A, Sidney J, Southwood S, Del Guercio MF, Appella E, Sakamoto H, Celis E, Grey H, Chesnut RW, Kubo R, Sette A. Prominent roles of secondary anchor residues in peptide binding to HLA-A24 human class I molecules. J Immunol. 1995;155:4307–4312. [PubMed] [Google Scholar]
- Kotturi M, Peters B, Buendia-Laysa FJ, Sidney J, Oseroff C, Botten J, Grey H, Buchmeier M, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. doi:10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koup RA, Safrit J, Cao Y, Andrews C, Mcleod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo RT, Sette A, Grey H, Appella E, Sakaguchi K, Zhu N, Arnott D, Sherman N, Shabanowitz J, Michel H, Et A. Definition of specific peptide motifs for four major HLA-A alleles. J Immunol. 1994;152:3913–3924. [PubMed] [Google Scholar]
- Kuroda M, Schmitz J, Barouch D, Craiu A, Allen T, Sette A, Watkins D, Forman M, Letvin N. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauemoller S, Holm A, Hilden J, Brunak S, Holst Nissen M, Stryhn A, Ostergaard Pedersen L, Buus S. Quantitative predictions of peptide binding to MHC class I molecules using specificity matrices and anchor-stratified calibrations. Tissue Antigens. 2001;57:405–414. doi: 10.1034/j.1399-0039.2001.057005405.x. [DOI] [PubMed] [Google Scholar]
- Loffredo J, Friedrich T, Leon E, Stephany J, Rodrigues D, Spencer S, Bean A, Beal D, Burwitz B, Rudersdorf R, Wallace L, Piaskowski S, May G, Sidney J, Gostick E, Wilson N, Price D, Kallas E, Piontkivska H, Hughes A, Sette A, Watkins D. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS ONE. 2007a;2:e1152. doi: 10.1371/journal.pone.0001152. doi:10.1371/journal.pone.0001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo J, Maxwell J, Qi Y, Glidden C, Borchardt G, Soma T, Bean A, Beal D, Wilson N, Rehrauer W, Lifson J, Carrington M, Watkins D. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007b;81:8827–8832. doi: 10.1128/JVI.00895-07. doi:10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo J, Sidney J, Bean A, Beal D, Bardet W, Wahl A, Hawkins O, Piaskowski S, Wilson N, Hildebrand W, Watkins D, Sette A. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. doi:10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo J, Sidney J, Piaskowski S, Szymanski A, Furlott J, Rudersdorf R, Reed J, Peters B, Hickman-Miller HD, Bardet W, Rehrauer W, O’Connor DH, Wilson N, Hildebrand W, Sette A, Watkins D. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+ T lymphocyte responses to SIVmac239. J Immunol. 2005;175:5986–5997. doi: 10.4049/jimmunol.175.9.5986. [DOI] [PubMed] [Google Scholar]
- Loffredo J, Sidney J, Wojewoda C, Dodds E, Reynolds M, Napoe G, Mothe B, O’Connor DH, Wilson N, Watkins D, Sette A. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J Immunol. 2004;173:5064–5076. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- Lund O, Nielsen M, Kesmir C, Petersen A, Lundegaard C, Worning P, Sylvester-Hvid C, Lamberth K, Roder G, Justesen S, Buus S, Brunak S. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810. doi: 10.1007/s00251-004-0647-4. doi:10.1007/s00251-004-0647-4. [DOI] [PubMed] [Google Scholar]
- Matano T, Shibata R, Siemon C, Connors M, Lane HC, Martin M. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcneil A, Yap P, Gore S, Brettle R, Mccoll M, Wyld R, Davidson S, Weightman R, Richardson A, Robertson J. Association of HLA types A1-B8-DR3 and B27 with rapid and slow progression of HIV disease. QJM. 1996;89:177–185. doi: 10.1093/qjmed/89.3.177. [DOI] [PubMed] [Google Scholar]
- Metzner K, Jin X, Lee F, Gettie A, Bauer D, Di Mascio M, Perelson A, Marx P, Ho DD, Kostrikis L, Connor R. Effects of in vivo CD8(+) T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Exp Med. 2000;191:1921–1931. doi: 10.1084/jem.191.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles S, Sabbaghian M, Shupert W, Bettinotti M, Marincola F, Martino L, Hallahan C, Selig S, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. doi:10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortara L, Letourneur F, Gras-Masse H, Venet A, Guillet J, Bourgault-Villada I. Selection of virus variants and emergence of virus escape mutants after immunization with an epitope vaccine. J Virol. 1998;72:1403–1410. doi: 10.1128/jvi.72.2.1403-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, Sidney J, Dzuris J, Liebl M, Fuenger S, Watkins D, Sette A. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol. 2002;169:210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M, Peters B, Pasquetto V, Tscharke D, Sidney J, Bui H, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. doi:10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Naruse TK, Chen Z, Yanagida R, Yamashita T, Saito Y, Mori K, Akari H, Yasutomi Y, Miyazawa M, Matano T, Kimura A. Diversity of MHC class I genes in Burmese-origin rhesus macaques. Immunogenetics. 2010;62:601–611. doi: 10.1007/s00251-010-0462-z. doi:10.1007/s00251-010-0462-z. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, Allen T, Vogel T, Jing P, Desouza I, Dodds E, Dunphy E, Melsaether C, Mothe B, Yamamoto H, Horton H, Wilson N, Hughes A, Watkins D. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat Med. 2002;8:493–499. doi: 10.1038/nm0502-493. doi:10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, Mcdermott A, Krebs K, Dodds E, Miller J, Gonzalez E, Jacoby T, Yant L, Piontkivska H, Pantophlet R, Burton D, Rehrauer W, Wilson N, Hughes A, Watkins D. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J Virol. 2004;78:14012–14022. doi: 10.1128/JVI.78.24.14012-14022.2004. doi:10.1128/JVI.78.24.14012-14022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M, Gonda M. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes. 1992;6:95–102. doi: 10.1007/BF01703760. [DOI] [PubMed] [Google Scholar]
- Ogg G, Jin X, Bonhoeffer S, Dunbar P, Nowak M, Monard S, Segal J, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon D, Mcmichael A. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- Otting N, De Vos-Rouweler AJ, Heijmans C, De Groot NG, Doxiadis G, Bontrop R. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics. 2007;59:367–375. doi: 10.1007/s00251-007-0201-2. doi:10.1007/s00251-007-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, Morner A, Bontrop R. Novel major histocompatibility complex class I alleles extracted from two rhesus macaque populations. Tissue Antigens. 2011;77:79–80. doi: 10.1111/j.1399-0039.2010.01565.x. doi:10.1111/j.1399-0039.2010.01565.x. [DOI] [PubMed] [Google Scholar]
- Peters B, Bui H, Frankild S, Nielson M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, Wilson S, Sidney J, Lund O, Buus S, Sette A. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. doi:10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyerl FW, Barouch D, Yeh W, Bazick H, Kunstman J, Kunstman K, Wolinsky S, Letvin NL. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J Virol. 2003;77:12572–12578. doi: 10.1128/JVI.77.23.12572-12578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R, Rowland-Jones S, Nixon D, Gotch F, Edwards J, Ogunlesi A, Elvin J, Rothbard J, Bangham C, Rizza C, Et A. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. doi:10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- Pinilla C, Appel J, Blanc P, Houghten R. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques. 1992;13:901–905. [PubMed] [Google Scholar]
- Price DA, Goulder P, Klenerman P, Sewell A, Easterbrook P, Troop M, Bangham C, Phillips R. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci U S A. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee H, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- Reimann K, Tenner-Racz K, Racz P, Montefiori D, Yasutomi Y, Lin W, Ransil B, Letvin N. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert J, Sidney J, Celis E, Kubo R, Grey H, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- Sacha J, Buechler M, Newman L, Reed J, Wallace L, Loffredo J, Wilson N, Watkins D. Simian immunodeficiency virus-specific CD8+ T cells recognize Vpr- and Rev-derived epitopes early after infection. J Virol. 2010;84:10907–10912. doi: 10.1128/JVI.01357-10. doi:10.1128/JVI.01357-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacha JB, Chung C, Reed J, Jonas A, Bean A, Spencer S, Lee W, Vojnov L, Rudersdorf R, Friedrich T, Wilson NA, Lifson JD, Watkins DI. Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. J Virol. 2007;81:11703–11712. doi: 10.1128/JVI.00926-07. doi:10.1128/JVI.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado M, Simon A, Sanz-Minguela B, Rallon N, Lopez M, Vicario J, Benito J, Rodes B. An additive effect of protective host genetic factors correlates with HIV-non progression status. J Acquir Immune Defic Syndr. 2010 doi: 10.1097/QAI.0b013e3182036f14. 10.1097/QAI.0b013e3182036f14. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Kuroda M, Santra S, Sasseville V, Simon M, Lifton M, Racz P, Tenner-Racz K, Dalesandro M, Scallon B, Ghrayeb J, Forman M, Montefiori D, Rieber E, Letvin N, Reimann K. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, Bui H, Del Guercio MF, Alexander J, Loffredo J, Watkins D, Mothe B. Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics. 2005;57:53–68. doi: 10.1007/s00251-004-0749-z. doi:10.1007/s00251-004-0749-z. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, Del Guercio MF, Southwood S, Ruppert J, Dahlberg C, Grey H, Kubo R. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol Immunol. 1994a;31:813–822. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast W, Melief C, Oseroff C, Yuan L, Ruppert J, Sidney J, Del Guercio MF, Southwood S, Kubo R, Chesnut R, Grey H, Chisari F. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994b;153:5586–5592. [PubMed] [Google Scholar]
- Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008a;4:2. doi: 10.1186/1745-7580-4-2. doi:10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Dzuris JL, Newman M, Johnson R, Kaur A, Amitinder K, Walker C, Appella E, Mothe B, Watkins D, Sette A. Definition of the Mamu A*01 peptide binding specificity: application to the identification of wild-type and optimized ligands from simian immunodeficiency virus regulatory proteins. J Immunol. 2000;165:6387–6399. doi: 10.4049/jimmunol.165.11.6387. [DOI] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008b;9:1. doi: 10.1186/1471-2172-9-1. doi:10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, Moore C, Pencille T, Ngo S, Masterman K, Asabe S, Pinilla C, Chisari F, Sette A. Characterization of the peptide-binding specificity of the chimpanzee class I alleles A 0301 and A 0401 using a combinatorial peptide library. Immunogenetics. 2007;59:745–751. doi: 10.1007/s00251-007-0243-5. doi:10.1007/s00251-007-0243-5. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Del Guercio MF, Grey H, Chesnut RW, Kubo R, Sette A. Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules. J Immunol. 1996;157:3480–3490. [PubMed] [Google Scholar]
- Sidney J, Southwood S, Oseroff C, Del Guercio MF, Sette A, Grey H. Measurement of MHC/peptide interactions by gel filtration. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1803s31. Chapter 18:Unit 18.3. doi:10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- Soudeyns H, Paolucci S, Chappey C, Daucher M, Graziosi C, Vaccarezza M, Cohen O, Fauci A, Pantaleo G. Selective pressure exerted by immunodominant HIV-1-specific cytotoxic T lymphocyte responses during primary infection drives genetic variation restricted to the cognate epitope. Eur J Immunol. 1999;29:3629–3635. doi: 10.1002/(SICI)1521-4141(199911)29:11<3629::AID-IMMU3629>3.0.CO;2-O. doi:10.1002/(SICI)1521-4141(199911)29:11<3629::AID-IMMU3629>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Stephens E, Singh D, Pacyniak E, Mccormick C. Comparison of Vif sequences from diverse geographical isolates of HIV type 1 and SIV(cpz) identifies substitutions common to subtype C isolates and extensive variation in a proposed nuclear transport inhibition signal. AIDS Res Hum Retroviruses. 2001;17:169–177. doi: 10.1089/08892220150217256. doi:10.1089/08892220150217256. [DOI] [PubMed] [Google Scholar]
- Stryhn A, Pedersen L, Romme T, Holm C, Holm A, Buus S. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur J Immunol. 1996;26:1911–1918. doi: 10.1002/eji.1830260836. doi:10.1002/eji.1830260836. [DOI] [PubMed] [Google Scholar]
- Tang J, Tang S, Lobashevsky E, Myracle A, Fideli U, Aldrovandi G, Allen S, Musonda R, Kaslow R. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaka K, Wiesmuller K, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- Udaka K, Wiesmuller K, Kienle S, Jung G, Walden P. Decrypting the structure of major histocompatibility complex class I-restricted cytotoxic T lymphocyte epitopes with complex peptide libraries. J Exp Med. 1995;181:2097–2108. doi: 10.1084/jem.181.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urvater J, Otting N, Loehrke J, Rudersdorf R, Slukvin I, Piekarczyk M, Golos T, Hughes A, Bontrop R, Watkins D. Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol. 2000;164:1386–1398. doi: 10.4049/jimmunol.164.3.1386. [DOI] [PubMed] [Google Scholar]
- Van Baalen CA, Schutten M, Huisman R, Boers P, Gruters R, Osterhaus A. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J Virol. 1998;72:6851–6857. doi: 10.1128/jvi.72.8.6851-6857.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bleek GM, Nathenson S. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature. 1990;348:213–216. doi: 10.1038/348213a0. doi:10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- Vogel T, Friedrich T, O’Connor DH, Rehrauer W, Dodds E, Hickman H, Hildebrand W, Sidney J, Sette A, Hughes A, Horton H, Vielhuber K, Rudersdorf R, De Souza IP, Reynolds M, Allen T, Wilson N, Watkins D. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J Virol. 2002;76:11623–11636. doi: 10.1128/JVI.76.22.11623-11636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CC, Brown R, Korber B, Wilkes B, Ruhl D, Sakamoto D, Kunstman K, Luzuriaga K, Hanson I, Widmayer S, Wiznia A, Clapp S, Ammann A, Koup R, Wolinsky S, Walker B. Frequent detection of escape from cytotoxic T-lymphocyte recognition in perinatal human immunodeficiency virus (HIV) type 1 transmission: the ariel project for the prevention of transmission of HIV from mother to infant. J Virol. 1999;73:3975–3985. doi: 10.1128/jvi.73.5.3975-3985.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NA, Keele B, Reed J, Piaskowski S, Macnair C, Bett A, Liang X, Wang F, Thoryk E, Heidecker G, Citron M, Huang L, Lin J, Vitelli S, Ahn C, Kaizu M, Maness N, Reynolds M, Friedrich T, Loffredo J, Rakasz E, Erickson S, Allison D, Piatak MJ, Lifson J, Shiver J, Casimiro D, Shaw G, Hahn B, Watkins D. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–6521. doi: 10.1128/JVI.00272-09. doi:10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N, Reed J, Napoe G, Piaskowski S, Szymanski A, Furlott J, Gonzalez E, Yant L, Maness N, May G, Soma T, Reynolds M, Rakasz E, Rudersdorf R, Mcdermott A, O’Connor DH, Friedrich T, Allison D, Patki A, Picker L, Burton D, Lin J, Huang L, Patel D, Heindecker G, Fan J, Citron M, Horton M, Wang F, Liang X, Shiver J, Casimiro D, Watkins D. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J Virol. 2006;80:5875–5885. doi: 10.1128/JVI.00171-06. doi:10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinsky S, Korber B, Neumann A, Daniels M, Kunstman K, Whetsell A, Furtado M, Cao Y, Ho DD, Safrit J. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- Yant LJ, Friedrich T, Johnson R, May G, Maness N, Enz A, Lifson J, O’Connor DH, Carrington M, Watkins D. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. doi:10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutomi Y, Reimann K, Lord C, Miller M, Letvin N. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.