Abstract
The tethering of platelets on the injured vessel surface mediated by glycoprotein Ibα (GPIbα) - Von Willebrand factor (vWF) bonds, as well as the interaction between flowing platelets and adherent platelets, are two key events that take place immediately following blood vessel injury. This early-stage platelet deposition and accumulation triggers the initiation of hemostasis, a self-defensive mechanism to prevent the body from excessive blood loss. To understand and predict this complex process, one must integrate experimentally determined information on the mechanics and biochemical kinetics of participating receptors over very small time frames (1–1000 µs) and length scales (10–100 nm), to collective phenomena occurring over seconds and tens of microns. In the present study, a unique three dimensional multiscale computational model, platelet adhesive dynamics (PAD), was applied to elucidate the unique physics of (i) a non-spherical, disk-shaped platelet interacting and tethering onto the damaged vessel wall followed by (ii) collisional interactions between a flowing platelet with a downstream adherent platelet. By analyzing numerous simulations under different physiological conditions, we conclude that the platelet’s unique spheroid-shape provides heterogeneous, orientation-dependent translocation (rolling) behavior which enhances cell-wall interactions. We also conclude that platelet-platelet near field interactions are critical for cell-cell communication during the initiation of microthrombi. The PAD model described here helps to identify the physical factors that control the initial stages of platelet capture during this process.
Keywords: Adhesion, Multiscale modeling, Platelet, Receptors, Shear flow, Von Willebrand factor
1. Introduction
One of the key events in early stage hemostasis – a self-defensive mechanism to prevent excess blood loss forming a clot to cover the disturbed vessel surface – is the initial tethering of circulating platelets onto the exposed subendothelial surface at the site of blood vessel injury. Followed by translocation of platelets linear wall-shear driven by blood flow, such platelet-vessel interactions under moderate to high shear rate (>500 s−1) are mediated through bonding between the α subunit of GPIb receptors on the platelet surface the A1 domain of subendothelial collagen-bound multimeric plasma glycoprotein Von Willebrand factor (vWF). 1, 2 The translocation of a platelet on a subendothelial surface through the GPIbα-vWF-A1 bond prolongs the duration of platelet-vessel contact to gain enough signals from receptor-ligand bonding as well as surrounding soluble platelet activation factors to trigger platelet activation 3, 4. This leads to the formation of other receptor-ligand bonds such as integrin α2β1 with subendothelial collagen to support platelet firm adhesion 5 and αIIbβ3 with its same kind on the surface of other platelets through fibrinogen (also other molecules such as vWF) to cause platelet aggregation 6. GPIbα-vWF-A1 bonds have been demonstrated to exhibit selectin-like, slip-bond kinetics that include fast intrinsic association and dissociation rates, force dependence of dissociation rate, and a requirement of a threshold shear stress for adhesion to occur 7–9. Recently, a catch-bond sub-regime that spans the lower range of bond force has also been revealed and the detailed bond kinetics for the transition of catch-slip sub-regimes – a modified quasi-first order dissociation reaction – has been established 10, 11. Qualitative defects on GPIbα and vWF, which result in platelet-type Von Willebrand Disease (vWD) and 2B-type vWD respectively 12, alter the kinetics of the GPIbα-vWF-A1 bond and lead to disordered bleeding phenotypes 9, 13–15. Specifically, for platelet-type vWD, the mutant GPIbα receptor stabilizes the interaction with inactive circulating vWF, depleting the vWF pool in plasma, causing insufficient binding of vWF to subendothelial surfaces during injury leading to hemorrhage 16.
Once the translocating platelet becomes activated and fully adherent – mediated by platelet-collagen receptors (e.g. integrin α2β1) – dramatic changes take place including: exocytosis of α-granules and dense granules (leading to the release of ADP, thrombin and other activating factors), change in shape from discoid to spherical, extension of filopodia, formation of thromboxane A2 (TXA2), and activation of other surface receptors (e.g. integrin αIIbβ3) that mediate platelet aggregation. Recent studies have shown that there are three distinct mechanisms that intitiate platelet aggregation at various shear rates 17: i) a single-stage aggregation is predominantly mediated by the interaction of fibrinogen with integrin αIIbβ3 (shear rate < 1000 s−1); ii) a distinct two-stage aggregation process with the initial-stage aggregations dependent on membrane tethering GPIbα, integrin αIIbβ3, and a later stage associated with soluble agonists such as ADP (1000 s−1 < shear rate < 10000 s−1); iii) platelet aggregation begins independently of integrin αIIbβ3 or platelet activation and is exclusively mediated by the vWF-GPIbα interaction and form a platelet plug (shear rate > 10000 s−1). The third mechanism has been described in further detail as a shear-gradient dependent aggregation where a platelet plug can form under mechanical shear-gradient caused by sharp changes in local vessel geometry (stenosis, etc) in the absence of vessel injury 18.
Though critical, few attempts were made to elucidate the physics of the platelet aggregation, and the majority of them utilize multiscale simulation methods which contain parameters and assumptions that cannot be easily determined by experiments. This greatly hampers the fidelity of the simulation work. One explanation is the difficulty of constructing a precise platelet aggregation model a priori. Accurate predictions based on the simplifying assumptions during early thrombus formation (coarse-grained methods) must first be made. A free-flowing two-platelet aggregate mediated by GPIbα-vWF-A1 bonding at physiologically high shear rate (>10000 s−1) was studied 19, but a more typical scenario to focus on is that of a flowing platelet interacting with a captured and immobilized platelet on the injured surface. The present article describes a multiscale computational study focusing on specific biophysical details including the transient formation of GPIbα-vWF-A1 bonds, which mediate the translocating motion of ellipsoid-shaped platelets on an injured surface, as well as the hydrodynamic interaction between an upstream flowing platelet and a downstream, adherent platelet. Based on the well-developed Platelet Adhesive Dynamics (PAD) numerical method, we explore these fundamental hemostasis/thrombosis procedures in depth and provide necessary results for future coarse-grained simulation study.
2. Platelet Adhesive Dynamics (PAD)
The PAD model is a multiscale three-dimensional (3-D) numerical simulation method that consists of two distinct yet interrelated components: (1) the 3-D hydrodynamic motion of multiple solid-body spheroid-shaped particles (platelets) under simple shear flow near a stationary planar surface (vessel wall), and the (2) adhesive dynamics of receptors on the platelet membrane binding to their ligands. The adhesive dynamics component utilizes the Monte Carlo method to determine each bond formation/breakage event based on specific receptor-ligand binding kinetics. It calculates the bond forces, which yield the net force and torque acting on each platelet particle. These net forces and torques will then be introduced into the hydrodynamic equations to solve the mobility problem. The PAD model has been successfully used to characterize flowing platelet convection and the high shear-induced behavior of aggregates. 19, 20
2.1 The hydrodynamic calculation
This problem involves the creeping motion of one/two rigid oblate spheroid particles of 96 or 384 QUAD9 surface elements (96 for all of the translocation simulations, and 384 for all of the collision simulations) with an aspect ratio of 0.25 in a semi-infinite three-dimensional region filled with a fluid of density of 1.0 g/cm3 bounded by an infinite flat plane at z = 0 21.
The Reynolds number of this system is calculated as NRE = γρa2 / μ < O(10−2) < 1, where the wall shear rate, γ< 10,000s−1, is within physiological range, the particle radius, a, is 1 µm, the density of blood plasma, ρ, is 1.0239 g/cm3, and the viscosity of blood plasma, μ, is 1.0 cP. The flow is within the Stokes regime. The Stokes equation for linear shear ambient flow is:
| (1) |
where u is the velocity, p is the pressure, μ is the viscosity of the fluid, γ is the shear rate, and z is the horizontal distance from the center of the particle to the flat wall.
The completed double layer-boundary integral equation method (CDL-BIEM) 22, a boundary elements solution technique to solve the integral representation of the Stokes equation, was used to solve Eq. 1. It is based on the integral representation of the Stokes equation:
| (2) |
Here, Gij is the singularity solution due to a point force on the bounding surface S, with x acting in the j direction of the fluid. The factor n(x) is the unit normal vector at x and points out from the particle surface into the surrounding fluid, and σ is the stress tensor given by:
| (3) |
where I is the identity matrix and the superscript t denotes the transpose operator. The integrals on the RHS and LHS of Eq. 2 are known as the single-layer and double-layer potentials, respectively. Both of the expressions are carried out over the bounding surfaces, including the surface of all the particles, but not the infinite plane. The singularity solutions in half-space are used to account for the infinite wall effect 43. Eq. 2 is termed a boundary integral equation (BIE) where the velocity u(X) is evaluated at the particle surface. It has been shown that the BIE containing the single-layer potential is sufficient to represent the Stokes disturbance flow problem for particles undergoing rigid body motion. However, this integral equation takes the form of a Fredholm integral equation of the first kind, which for a mobility problem is generally ill-posed and becomes numerically unstable for more refined boundary element meshes 22. An alternative method solely involves the double-layer potential. Because the double-layer potential does not exert any net force or torque, Power and Miranda 23 incorporated an unknown density (corresponding to the velocity ui(x)), which results in a net force and torque that coincides with the behavior at infinity for Stokes flow (i.e. velocity decays as R−1) into the double-layer integrals. The summarized solution method for Eq. 2 was described by Phan-Thien et al. 43, with the final form of the boundary integral equation shown as
| (4) |
where φj is the unknown surface density of the double-layer distribution, К represents the double-layer surface integral operator, φ(p,l) are the orthonormalized null solutions corresponding to the translational (l=1,2,3) and rotational (l=4,5,6) motions of particles p, and the angled brackets represent the inner product . P takes on values from 1 to N (total particles in the system), ψ(p) (ζ) are the orthonormalized eigenvectors of the operator К with the value for ζ ∈ S(p), but 0 for ζ ∉ S(p). On the RHS of Eq. 4, bj (ζ) is calculated as
| (5) |
where F(α) and T(α) are the force and torque acting on cell α at the center of mass xc(α) of the particle, and uj∞ is any ambient fluid velocity that is a valid solution of the Stokes equation.
These equations are solved by Newton iteration for φ. Once the double-layer density, φ, is determined, the surface velocity field can be obtained as follows:
| (6) |
By taking the inner product of Eq. 6 and φ(n,m) (n = 1,…,N; m = 1~6), the rigid body motion of a particle n can be extracted.
2.2 Bond kinetics and bond force
The GPIbα receptor density on the platelet surface is about 1500 molec/µm2 24. When the circulating platelet approaches the site of vessel injury, GPIbα-vWF-A1 bonds can be formed between the platelet surface receptor GPIbα and the subendothelial-bound vWF at the injury site and are treated as linear springs throughout the simulation model. The Monte Carlo method is used to determine each individual GPIbα-vWF-A1 bond formation/dissociation event using the probability formulations Pf (probability of bond formation) and Pr (probability of bond breakage) described by Hammer and Apte 25:
| (7) |
where kf and kr are given in s−1 units and Δt is the simulation time step 10−7 s.
The mathematical expression for the rate of bond formation was derived from Bell’s expression 26 of the equilibrium constant for cell-cell bond bridging. The dependence of bond formation rate kf on the deviation bond length |xb−lb| and slip velocity between two bonding molecules takes the similar form described by Mody et al. 19 and is shown in Eq. 8,
| (8) |
where vs is the slip velocity determined by the rotational and translational velocities as well as the location of the GPIbα receptor on the platelet surface, xb is the distance spanning the endpoint of the GPIbα receptor on the platelet surface and the vWF-A1 binding site on the vessel wall, kBT is the product of Boltzmann’s constant and temperature. In our translocation assay, we assume abundant vWF ligand on the injured vessel wall surface, thus xb here is simply the z-coordinates minus the surface roughness. The physical meaning and value of the other parameters in Eq. 8 can be found in Table 1.
Table.1.
Values of bond formation kinetic parameters
The reverse rate kr was evaluated using the Bell model 27 for the force-dependent dissociation rate of weak non-covalent bonds in previous studies 19, because the GPIbα-vWF-A1 bond demonstrates selectin-like binding kinetics: (1) short-lived tethering events and (2) a fast intrinsic dissociation rate constant 7. This model involves pure slip-bond kinetics stated as following:
| (9) |
The value for the intrinsic dissociation rate k0r and the reactive compliance γ were determined from optical tweezer studies 28 with k0r = 5.47 s−1, γ = 0.71 nm.
The GPIbα-vWF-A1 bond dissociation based on the Bell model is widely accepted and applied. However, recent in-vitro experiments suggest a new catch-bond/slip-bond force-dependent interchanging mechanism for the GPIα-vWF-A1 bond 11. This explains the results from studies demonstrating the unusual platelet translocation pattern that an increase in shear stress lowers the average translocation velocity of platelets in a flowing chamber assay below 20 dyn/cm2 of shear stress 10, 29. These results suggest that two GPIbα-vWF-A1 states exist in the system during platelet rolling. More specifically, two structural arrangements of the vWF A1 domain have been revealed, with one in a native, folded state (NG) which exhibits catch-bond kinetics while another more stretched conformation of an intermediate, open state (IG) that exhibits slip-bond kinetics 11. The resulting reverse rate, kr, is a quasi-first-order dissociation rate, which consists of a slip-bond dissociation component and a catch-bond dissociation component, shown as:
| (10) |
where Φ represents the population ratio of binding vWF molecules between the IG state and NG state and is dependent on the bond force f as follows:
| (11) |
This two-state, quasi-first-order dissociation model was first applied to the P-selectin: PSGL-1 interaction 30 and simulations of quasi-first-order dissociation have successfully predicted the nano-to-micro scale dynamics of P-selectin detachment from leukocyte interfaces 31. Each IG/NG state has its own force-dependent dissociation rate based on the classic Bell Model:
| (12) |
| (13) |
The physical significance and values for all of the parameters in Eq. 10~12 can be found in Table 2. The bond force f in Eq. 9–12 is expressed as σ(xb− lb).
Table.2.
Values of bond dissociation kinetic parameters
| Parameter (unit) | Definition | Value | Reference | |
|---|---|---|---|---|
|
|
Unstressed equilibrium constant for NG – IG states | 0.4 | 11 | |
| γ’ (nm) | Force compliance for two states ratio | 0.45 | 11 | |
|
|
Intrinsic dissociation constant for NG state | 4.9 | 11 | |
|
|
Intrinsic dissociation constant for IG state | 1.84 | 11 | |
| yN (nm) | Force compliance of dissociation for NG state | −0.23 | 11 | |
| yI (nm) | Force compliance of dissociation for IG state | 0.039 | 11 |
3. Platelet translocation (rolling)
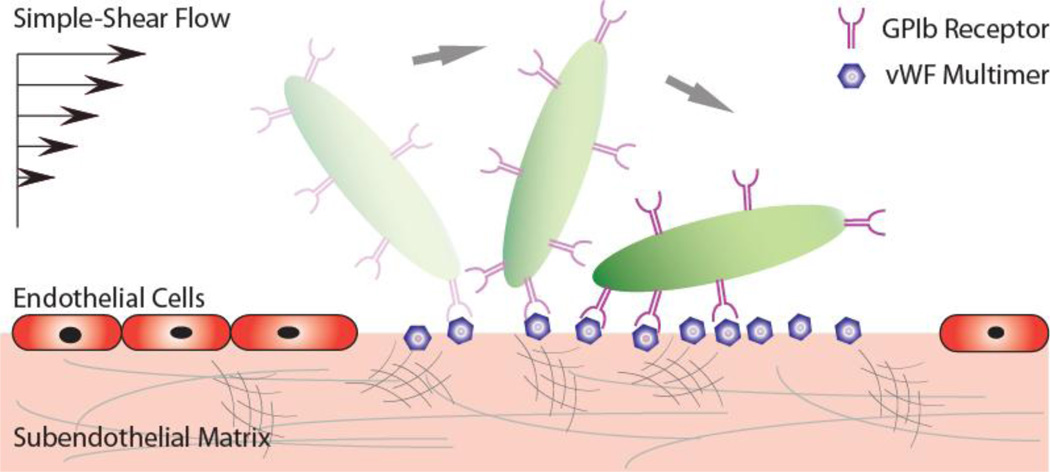
The translocating motion of a platelet at the site of injury is mediated by the frequent formation and dissociation of GPIbα-vWF-A1 bonds at the interface of damaged endothelium and the platelet surface under simple shear flow (Fig. 1). This interaction reduces the platelet translational velocity by orders of magnitude, providing an adequate time window for the platelet to become activated and fully adherent to the exposed subendothelial matrix before extravastion at the injury site.
Fig. 1.
Schematic depicting the translocational motion of a platelet at a site of injury (removed endothelial layer) via GPIbα-vWF-A1 bonds under simple-shear near-wall blood flow.
In-vitro platelet translocation experiments provide a useful technique to study the GPIbα-vWF-A1 bond kinetics and have been successfully applied to explain mechanisms of bleeding disorders resulting from various vWF pathologies 10, 15. Our simulation model utilizes recovers the platelet translocation behavior and serves as a potentially powerful predictive tool that elucidates mechanisms of disease through altered bond kinetics, platelet shape or surface receptor count.
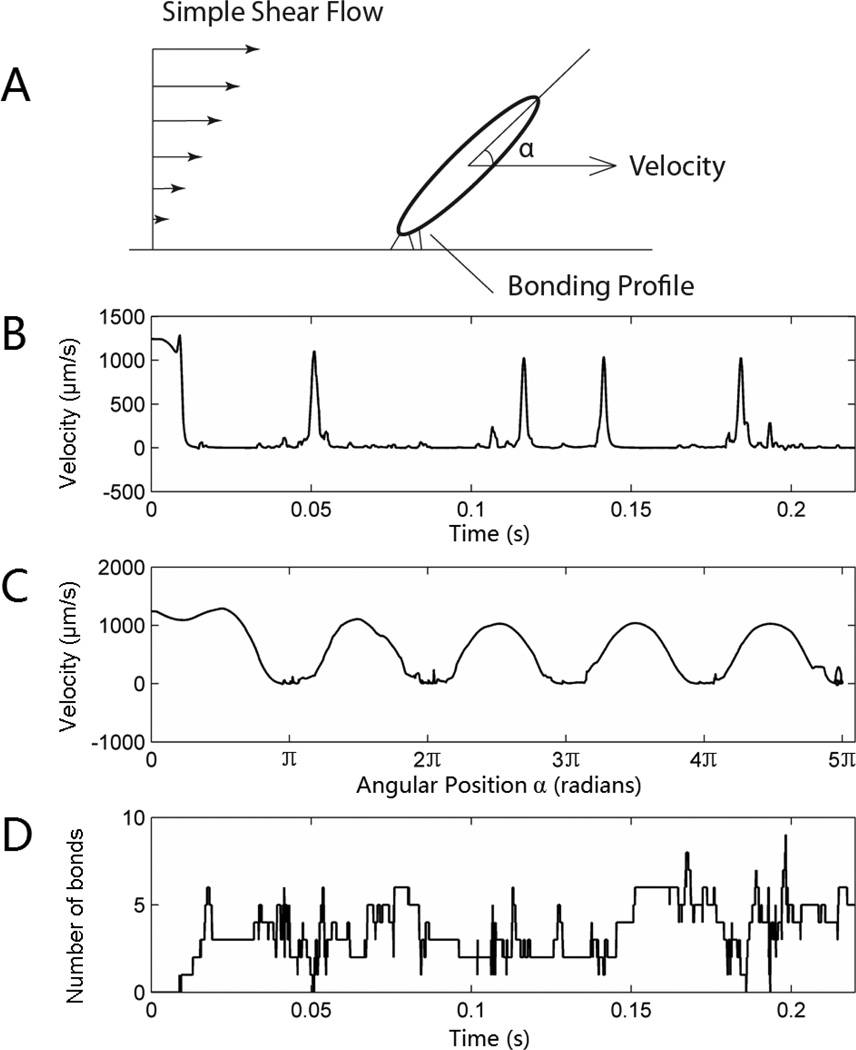
The translocation of an ellipsoid-shaped platelet (Fig. 2) was demonstrated using traditional slip-bond dissociation kinetics (Bell Model). The intrinsic cross-linking formation rate constant, k0f,2−D, was tested over a range of values (from 0.001 to 10 s−2/µm) under varying physiological shear rates (100~ 2000s−1) and then determined by its translocation (larger k0f,2−D resulted in fully adherent status and flipping motion was never observed in the simulation time range of 1 s, while smaller k0f,2−D resulted in undetectable tethering events, data not shown). The determined k0f,2−D was 0.1 s−2/µm. The rolling motion was composed of a fast flipping period (sharp peaks in Fig. 2B) where the velocity of the platelet’s center of mass reached the flow velocity, followed by a long residence period where the lever arm of the hydrodynamic force acting on the platelet was small, thus increasing the difficulty of breaking existing GPIbα-vWF-A1 bonds. The duration of residence time for each flipping cycle may be different, and depends on the number of existing bonds in the system as well as the uncertainty of a specific bond lifespan due to the dissociation probability distribution, but the velocity profile was quite homogeneous relative to the angular position (Fig. 2C), indicating that the rapid flipping motion is mainly determined by the hydrodynamic environment, rather than the bonding profile. For spherical cells such as a leukocyte rolling on an endothelial layer, both the homogeneity of the velocity profile with the angular position and the heterogeneity of translocation velocity versus real-time were not obvious 32, suggesting there are hidden functions of the platelet’s ellipsoid shape. Interestingly, the number of total bonds present in the system oscillated with no obvious accordance to either time or angular position (Fig. 2D), which deviates from the intuition that more bonds may be formed during the residence period (long axis parallel to the wall surface) due to the larger contact area rather than during the fast flipping period. One possible explanation is the near-zero slip velocity reached by the transiently resting platelet, which reduces the likelihood of new bond formation. Conversely, when the platelet rapidly rotates through the “standing” position the significant increase in relative slip velocity increases the probability of new bond formation despite the reduced contact area.
Fig. 2.
Translocation motion of a simulated platelet under simple shear of 1250 s−1 (A). The velocity is plotted versus time (B) and relative angular position (C). The bottom graph (D) shows the instantaneous number of bonds. Note: this simulation is based on a slip-bond-kinetic dissociation.
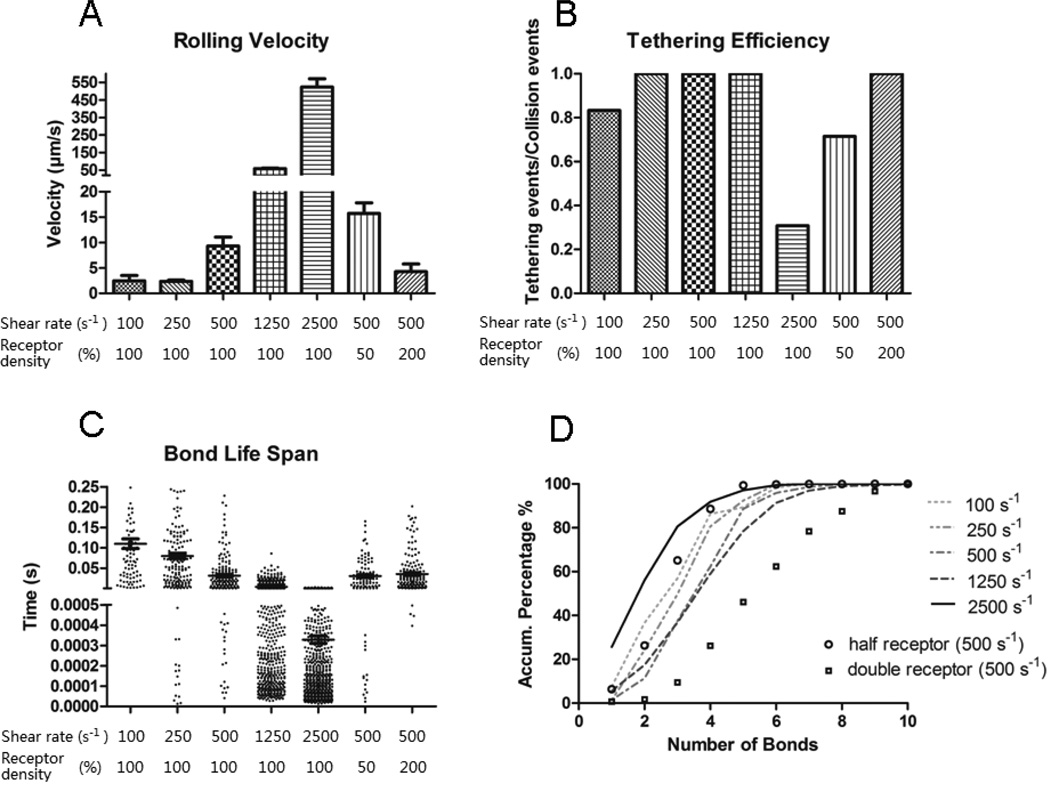
The translocational velocity (Fig. 3A) was maintained at steady state at a lower range shear rate (100~250 s−1). In this regime, one would expect that increasing the shear rate would increase the platelet-wall slip velocity and resulted in a larger number of GPIbα-vWF-A1 bonds formed. However, the additional number of bonds drags the platelet backwards and reduces its translocation velocity, which in turn would result in reduced bond formation rate. Such dynamic fluctuations caused by this self-stabilizing negative feedback mechanism made rolling platelets reach a steady translocation velocity of about 2.5 µm/s. At shear rates greater than 500 s−1, the steady translocation velocity increases in a superlinear manner with increasing shear rate.
Fig. 3.
Simulation results of translocation velocity (A), tethering efficiency (B), bond life span (C) and the probability distribution of number of bonds (D) are shown at different shear rates. These simulations were based on slip bond kinetics. In A, the main bars indicate the mean rolling velocity and error bar shows the standard error. In C the longer line section shows the average bond lifetime with the two shorter lines indicating the standard error. Each simulation was repeated 10 times. In (A), the difference between the rolling velocity for 100s−1 and 250s−1 conditions is not significant (P = 0.88 from unpaired two-tailed t-test). For the comparison between 500s−1 and the double receptor 500s−1 conditions, P = 0.102. For any other two combinations, P < 0.05. In (C), the bond life span at the 100s−1, 250s−1, 500s−1 or 2500s−1 condition is significantly different than any other condition (P < 0.05).
The tethering efficiency (Fig. 3B) was measured by dividing the total number of tethering events with the total number of collision events. The results showed a similar pattern to the in-vitro bead-tethering assays 15 that the tethering efficiency reached maximum over a specific range of shear stress and deviation from that “optimal” tethering would result in a lower tethering efficiency. The initial increase in tethering efficiency was mainly caused by the increased slip velocity between the platelet and wall surface. Furthermore, the drop at the higher end of shear rate was most likely caused by an increase in the hydrodynamic force acting on the bonds, which reduces the bond life span and makes tethering events reversible (Fig. 3C).
The number of bonds in the system showed a similar pattern to tethering efficiency and displayed an optimal range of shear rate (around 1250s−1) which maximized the total number of bonds formed (Fig. 3D). This phenomenon resulted from the combined effects of the increased rate of bond formation and the decreased bond lifespan with incremental increases in shear rate.
The receptor density on the cell surface is a critical factor determining the platelet translocation pattern. Previous simulation studies dealing with leukocytes showed that increased receptor counts will reduce the rolling velocity and vice versa 33. In our study, such sensitivity tests (groups with half or double the surface receptor density) were also performed. Reducing the total number of GPIbα receptors on the platelet surface by half increased the translocation velocity by 60%, lowered the tethering efficiency, and reduced average number of bonds as well as bond lifespan, while doubling the surface receptor density decreased the translocation velocity by 50%, increased the number of bonds and extended the bond lifetime (Fig. 3). These results indicate that the ellipsoid-particle translocation system behaves similar to a spherical-particle translocation system in the sense that the mobility of the particle (cell) has an inverse relation to the receptor density on particle (cell) surface.
Though slip-bond dissociation kinetics (based on the Bell Model) have been widely accepted, recently a catch-bond regime in the lower shear rate range (<1000 s−1) was discovered and is said to show similar binding kinetics to selectin-ligand bonds 10, 29 based on both in-vitro platelet and microsphere rolling assays. It is interesting that previous work – employing platelets, mammalian cells engineered to express the GPIbα receptor on the cell surface 9 or spherical beads – did not display catch-bond kinetics in the lower shear rate regime. Since multiscale simulation models are prone to experimental updates 34, this new types of receptor-ligand bonding kinetics and reproduces expected results based on each mechanism, which is validated by in-vitro experimental data.
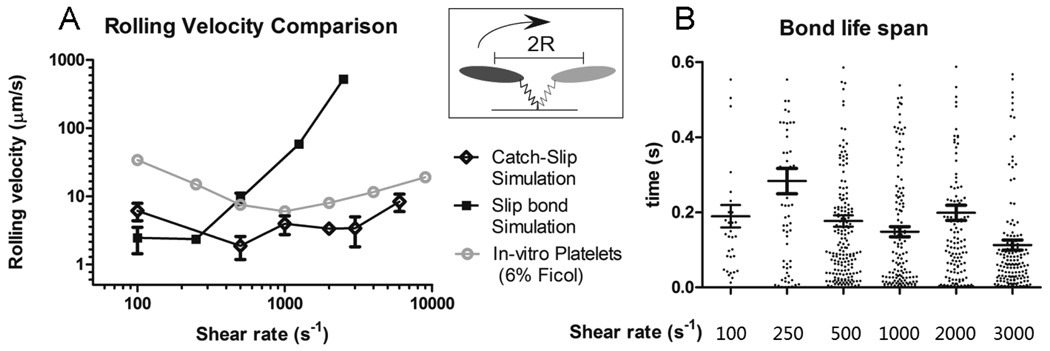
The intrinsic cross-linking formation rate constant, k0f,2−D, was re-determined using a similar method (indicated above) with a value of 0.05 s−2/µm. The translocation velocity for the catch-slip combination kinetics is shown in Fig. 4. Both the traditional slip-bond model (Bell Model) and the catch-slip combination model were plotted and compared with the experimental results of in-vitro platelet rolling assays 10. The catch-slip combination model indeed generated a similar parabolic relationship between the translocation velocity and increasing shear rate. However, over the low shear rate range (100 – 500 s−1), the translocation velocity differs by several orders of magnitude compared to experimental data. In order to explain this deviation, we examined the distribution of bond lifetimes under various bond forces (data from Auton, et al, 2010) 11. It was shown that within the catch-bond regime (bond force < 20pN), the shortest lifetime for the GPIbα-vWF-A1 bond is around 0.075 s where there is no net stress acting on the bond (bond force ~ 0pN). By assuming that there is no “jumping” period at any given time, i.e., there is at least one GPIbα-vWF-A1 bond existing at any moment, the fastest possible translocation velocity of the rolling platelet would happen in the scenario shown in the upright box of Fig. 4A, where there was only one bond formed at the long axis end of the translocating platelet: Vmax = 2*R/0.075s = 26.7µm/s. (R is the major axis of the ellipsoid-shaped platelet.) On average, we would expect at least a 50% drop in the maximum possible translocation velocity due to the multiple bonds formed as well as the random surface distribution of the GPIbα receptors (bonding profile).
Fig. 4.
Simulation results of translocation velocity (A) and bond life span (B) based on catch-slip combination kinetics. In A, the main bars indicate the mean translocation velocity and error bar shows the standard error. The in-vitro experimental data is from Coburn et al 29. In B, the longer line section shows the average bond lifetime with the two shorter lines indicating the standard error.
Another reason for such deviation may come from the fact that the governing bond association equation (Eq. 8) may not truly represent the most bond-forming kinetics. Unlike dissociation rate, which can be more easily determined by tethering assays or atomic force microscopy, there is a lack of straightforward experimental methods to determine the association rate under flow condition. This may result in overestimates of the bond association rate in the lower predicted shear rate region, overpredicting the number of bonds in the system and a slower translocation velocity. Nevertheless, the translocation velocity from the simulations, though disparate from the experimental data, makes reasonable predictions based on this formula.
Fig. 4B shows the duration of bonds under different shear rates. It is interesting to note that the average bond lifespan does not show an obvious parabolic pattern, which would reverse the trend of translocation velocity at different shear rates. One may argue that shorter bond life span would directly relate to faster rolling velocity. However, this statement may not always hold. There is an additional parameter, the average number of bonds existing between the rolling cell and the vessel wall, which also contributes to the translocation velocity. Reducing the bond lifespan will immediately increase the translocation velocity of the cell, however, because the bond formation rate is dependent on the 2D slip velocity between the bond-forming region on the cell and the vessel wall (Eq. 8), faster translocation velocities cause bonds to form faster, though not necessarily increase the average number of bonds existing at one moment since the bonds break faster as well. The quantitative change of the average number of bonds existing in the rolling event after a reduction in bond lifespan is thus a result of balancing the faster bond formation rate and faster bond breakage rate and can be predicted only by simulations or by calculation if a complete analytical model of the cell rolling existed. If the average number of bonds existing in the system truely increases, the distance that a cell will move in response to a single bond breakage event will be smaller, which attenuates the propelling effect of faster translocation velocity caused by shorter bond duration.
4. Two Platelet Hydrodynamic Interaction—with one adherent platelet on the vessel wall
The fast expansion of a platelet plug to cover the exposed sub-endothelial surface cannot be fulfilled if thrombus development is purely reliant on platelet/vessel wall interactions. In fact, an adherent platelet provides additional anchoring mechanisms to recruit flowing platelets to the site of injury to facilitate the process of thrombosis, which are strongly associated with the characteristics of platelet collisions including: collision frequency, contact duration during a collision, and the extent of surface area on each interacting platelet that enters the particle-particle reactive zone. The contact duration and contact area can be combined to generate a metric – the time integral of contact area – that can be used to assess the probability of platelet-platelet adhesion during the hydrodynamic encounter between the upstream flowing platelet and the downstream immobilized platelet. Hundreds of collisions were simulated with different initial configurations of relative distances between the particle centroids in the y- (parallel to the wall plane, perpendicular to the direction of flow) and z-direction (perpendicular to the wall plane). Significant trends between the relative position of the two platelets to the duration and quality of their interactions were found, which serve as indicators of growth potential of the “micro-thrombi”. The detailed hydrodynamic and geometrical information during two-platelet interactions are also summarized.
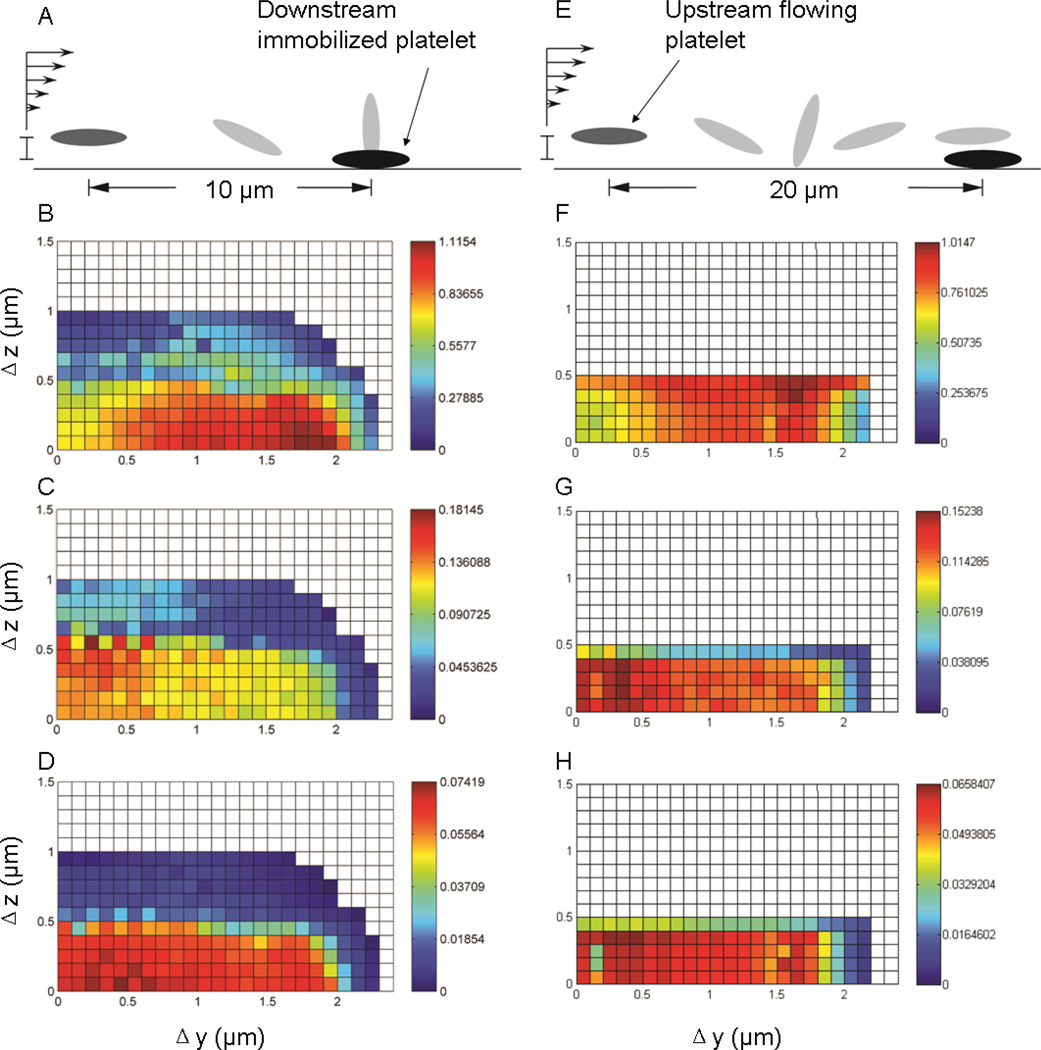
Collision simulations were initiated with two different initial distances (either 10µm or 20µm in the direction of flow) between the centroids of the two platelets, which would, in the absence of flow disturbances from the downstream adherent platelet, result in two limiting orientations (“perpendicular” orientation and “parallel” orientation) for the flowing platelet (see Fig.5A, E). A “collision” was defined as any event in which the distance between the two platelet surfaces became less than the maximum reactive distance (260 nm), an approximation of the length of the trimolecular bond integrin αIIbβ3 – vWF - integrin αIIbβ3 which is comparable of the length of GPIb - vWF- GPIbα trimolecular bond as used in our previous studies [Mody 2008] (integrin αIIbβ3 is comparable in size to GPIb, with the former slightly smaller than 30nm 35 and the latter slightly larger than 30nm 36). Threshold distance as the length of integrin αIIbβ3 - Fibrinogen- integrin αIIbβ3 bond, which supports the platelet- platelet firm adhesion, is because 1) It has been shown that vWF can support platelet adhesion via interaction with activated integrin αIIbβ3 37 and 2) the single fibrinogen molecule spans only about 50nm 38 which is much smaller than the average size of vWF multimers; this gives the likelihood that an integrin αIIbβ3 – vWF - integrin αIIbβ3 bond has a greater chance to establish before forming the integrin αIIbβ3 - Fibrinogen- integrin αIIbβ3 bond during platelet collision. The contact area was calculated by summation of the surface patches on the flowing platelet, which had “collided” with the adherent platelet. Note that all of the simulations related to one flowing platelet colliding with the immobilized platelet on the wall were performed at a shear rate of 5000 s−1. Because of the linearity of Stokes’ flow, collision patterns under variable shear rates will be similar and can be inferred directly from these simulation results. The collision grid map showed significant differences between the two different initial configurations. The initial configuration where the flowing platelet tended to project its edge into the second platelet had not only a larger collision regime in the z-direction, but also showed approximately 10% longer contact time, 15% larger contact area and 15% larger time integral of contact area respectively (Fig.5 scale bars). Overall, the integrated contact duration was larger for platelets flowing closer to the vessel wall, such as would be created by the margination effect caused by the existence of larger blood cells such as erythrocytes. There was a sharp decrease in the time integral of contact area when the initial z-direction deviation of the two platelet centroids extended beyond a threshold distance of 0.5 µm (Fig.5 D). Interestingly, for both cases, the contact time reached a maximum when the two platelets started with a separation distance of 1.7 µm along the y axis, which differs from the expectation that gliding directly over an immobilized platelet provides the longest contact time (Fig.5 B, F). This is likely due to the fact that, when the flowing platelet glided over the immobilized platelet, the lubrication force generated by the thin fluid layer between the platelet surfaces pushed the flowing platelet further away from the vessel wall, as compared to the situation where the position of the platelet pair had a deviation in the y-direction. The resulting differences in the z-coordinates gave the gliding platelet a greater velocity, based on the bulk simple shear flow profile. On the other hand, the maximum contact area reached a peak value when the flowing platelet’s trajectory passed right above the adherent platelet, which, together with the resulting contact time, resulted in a more uniformly-distributed contact area-time integral along the y-direction (Fig. 5C, D, G, H). The comparison of this grid map to the grid map resulting from a free-flowing platelet collision 20 shows that the platelet’s unique ellipsoid shape encourages better interaction when one is tethered to the vessel wall, which to some degree, reduces the probability of flowing platelets to form homotypic aggregates under physiologically high shear stress 39, 40.
Fig. 5.
The color-scaled binary collision map. Each grid point represents an individual collision experiment with the flowing platelet starting at a specific location represented by δx, δy and δz. A–D (E–H) are results of binary collisions between a flowing platelet 10µm (20µm) upstream of a downstream immobilized platelet. The grey ellipsoids in A, E represent the non-disturbed flowing platelet trajectory path (with adjusted length scale). B, F are the platelet contact time, with the unit associated with numbers shown on the color bar as micro-seconds. C, G are the contact area for the flowing platelet (unit with numbers µm2). D, H are the time integral of contact area for the flowing platelet (µm2 ms).
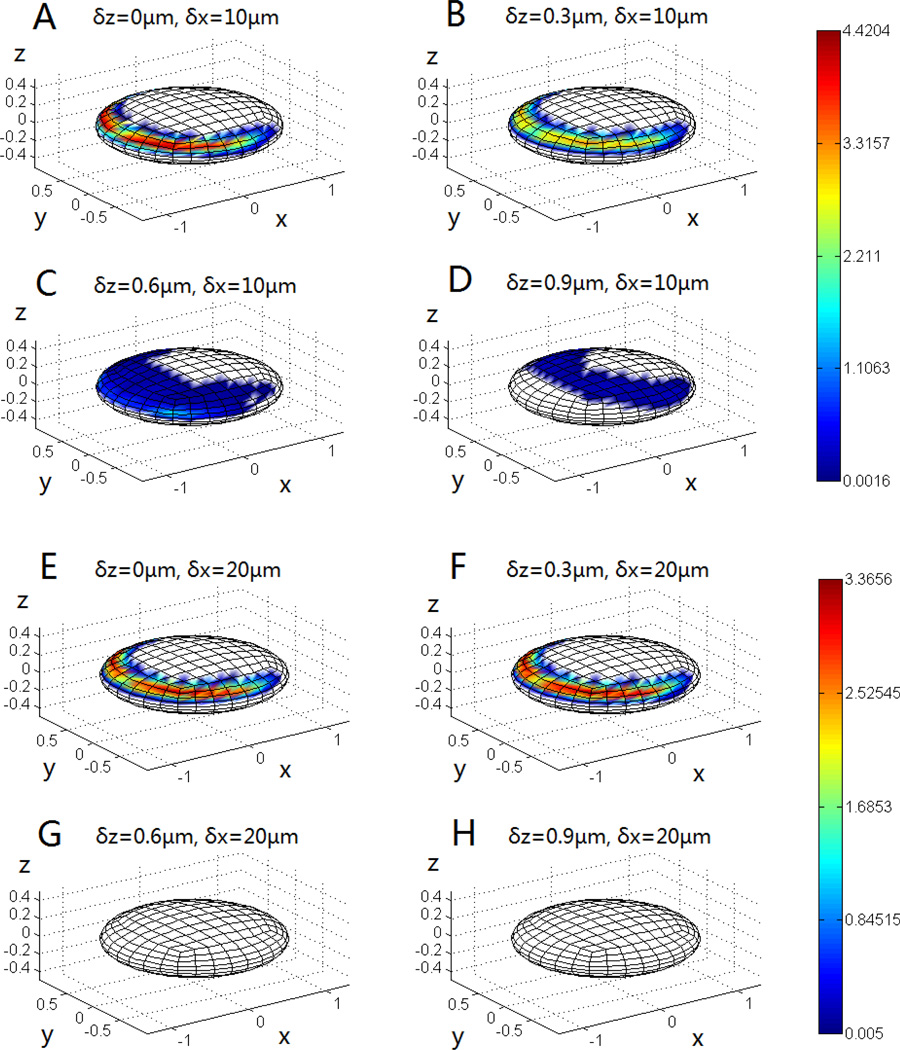
The contact zone of the immobilized platelet was examined in detail to identify the region on the immobilized platelet surface where the flowing platelet comes into contact. Fig. 6 shows the mapping of the contact area of the surface of the immobilized platelet and each sub-figure is resulted from a series of parallel collision simulations with the flowing platelet started at a specific height (z-coordinates) and position (only x-coordinates). The results match the expectation that proximity of the flowing platelet to the vessel wall leads to longer contact time. However, when the circulating platelet was very close to the vessel wall, it never contacted the top, central part of the adherent platelet. Increasing the initial height (z-coordinate) of the flowing platelet shifted the contact zone on the immobilized platelet from the upstream region towards the central top and downstream regions, but also reduced the contact quality (Fig. 6A–D). Since longer contact time resulted in higher chances of forming receptor-ligand bonds (fibrinogen, vWF mediated integrin αIIbβ3 bridging), this unique contact character reduced the probability for the flowing platelet to stack directly above the immobilized platelet. Instead, platelets tend to remain longer in the peripheral region of the immobilized platelet, approaching a side by side orientation to each other and gaining better coverage efficiency at the injury site while reducing likelihood of decreasing the effective vessel diameter and potential vessel occlusion. As described above, the collision pattern is sensitive to the x-direction separation between the two platelets, thus we also mapped out the mapping of contact area for collision series with an x-axis deviation of 20µm. Fig.6E–H confirms the results shown in Fig.5 that there is no contact when the flowing platelet is at least 0.5µm higher than the stationary platelet. The scale bars show that the overall trend of the collision quality (i.e., time and area) is better for the 10µm condition, which is consistent with the results indicated by the color map in Fig.5.
Fig. 6.
The mapping of the contact region on the immobilized platelet for different starting positions of the flowing platelet. The color is scaled to represent the contact time (ms) and the length unit of the Cartesian coordinates is µm.
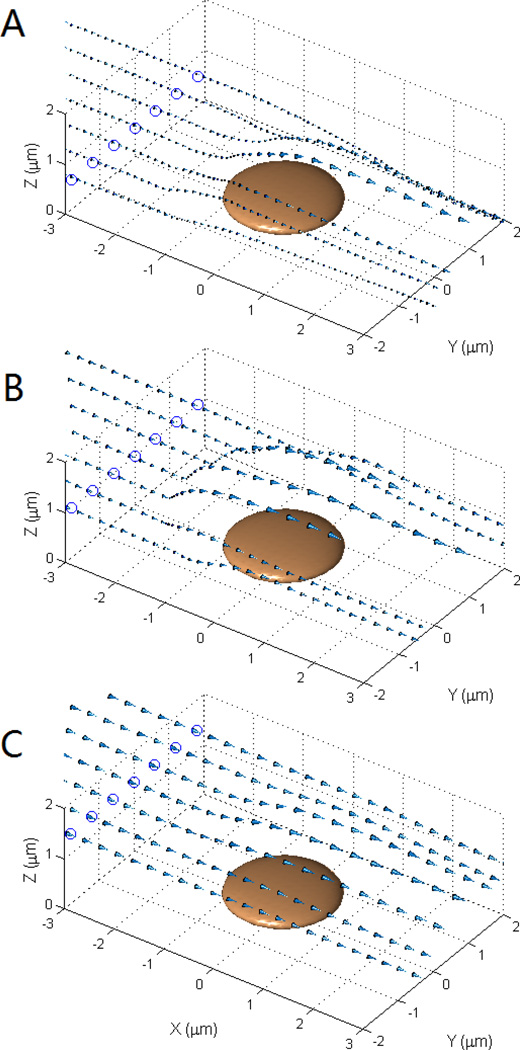
It is evident from Fig. 6 that the flowing platelet approaches the immobilized platelet mostly in the upstream region. It is interesting to consider how the flowing platelet is able to contact and deposit on the injured surface near the central and downstream regions of the immobilized platelet. Fig. 7 shows a vector plot representing the velocity of the centroid motion of the flowing platelet starting from a horizontal orientation (long axis parallel to the wall surface) 10µm upstream of the immobilized platelet. The arrows represent the relative motion of the flowing platelet for each collision event, with the circled dots showing the intercept (height) of the trajectories to the plane x = −3µm. The closer that the flowing platelet is to the vessel wall (plane z = 0), the slower the platelet convection, and importantly, the more altered the directional change of the trajectories. Fig. 7A shows that for flowing platelets that glide over the immobilized platelet, a slight deviation in the y-coordinates pushed the flowing platelet up and more towards the side (outward, or larger y-coordinates). However, when the flowing platelet starts with a y-coordinate deviation large enough to pass by the side of the adherent platelet without collision, the disturbed flow pulls the flowing platelets inward close to the downstream end of the immobilized platelet (Fig. 7A).
Fig. 7.
The velocity vector for the center of the flowing platelet when flowing past an adherent platelet. A, the flowing platelet was started at 10 µm upstream with a δz of 0.2 µm to the immobilized platelet. B, similar to A, with a δz of 0.6 µm. C, similar to B, with a δz of 1.0 µm.
5. Conclusion
We describe a multiscale model that incorporates detailed hydrodynamics as well as experimentally based receptor-ligand binding kinetics. This model is capable of revealing some of the subtle physiological processes during hemostasis, such as platelet translocation and interaction. It also demonstrates some of the mechanisms that platelets employ to efficiently form clots to seal wounds on the vessel wall, including the interaction between flowing and adherent platelets, as well as the characteristics of its translocational behavior. Importantly, the stability and versatility of our model suggests the future application of predicting bleeding phenotypes and multiple physiological states, such as a diseased status or therapeutic condition. Once additional types of platelet receptor-ligand interactions are incorporated, we will be able to simulate more comprehensive processes from platelet translocation to irreversible adherence, followed by the formation of platelet aggregates.
Acknowledgement
The authors thank John P. Lindsey for discussions and review of the manuscript. The work was funded by NIH Grant No. HL097971.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sakariassen KS, Bolhuis PA, Sixma JJ. Human blood platelet adhesion to artery subendothelium is mediated by factor VIII-Von Willebrand factor bound to the subendothelium. Nature. 1979;279(5714):636–638. doi: 10.1038/279636a0. [DOI] [PubMed] [Google Scholar]
- 2.Savage B, Saldivar E, Ruggeri ZM. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84(2):289–297. doi: 10.1016/s0092-8674(00)80983-6. [DOI] [PubMed] [Google Scholar]
- 3.Kroll MH, Harris TS, Moake JL, Handin RI, Schafer AI. von Willebrand factor binding to platelet GpIb initiates signals for platelet activation. J Clin Invest. 1991;88(5):1568–1573. doi: 10.1172/JCI115468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davi G, Patrono C. Platelet activation and atherothrombosis. N Engl J Med. 2007;357(24):2482–2494. doi: 10.1056/NEJMra071014. [DOI] [PubMed] [Google Scholar]
- 5.Jung SM, Moroi M. Activation of the platelet collagen receptor integrin alpha(2)beta(1): its mechanism and participation in the physiological functions of platelets. Trends Cardiovasc Med. 2000;10(7):285–292. doi: 10.1016/s1050-1738(01)00064-0. [DOI] [PubMed] [Google Scholar]
- 6.Bennett JS. Structure and function of the platelet integrin alphaIIbbeta3. J Clin Invest. 2005;115(12):3363–3339. doi: 10.1172/JCI26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doggett TA, Girdhar G, Lawshe A, Schmidtke DW, Laurenzi IJ, Diamond SL, Diacovo TG. Selectin-like kinetics and biomechanics promote rapid platelet adhesion in flow: the GPIb(alpha)-vWF tether bond. Biophys J. 2002;83(1):194–205. doi: 10.1016/S0006-3495(02)75161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arya M, Kolomeisky AB, Romo GM, Cruz MA, Lopez JA, Anvari B. Dynamic force spectroscopy of glycoprotein Ib-IX and von Willebrand factor. Biophys J. 2005;88(6):4391–4401. doi: 10.1529/biophysj.104.046318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar RA, Dong JF, Thaggard JA, Cruz MA, Lopez JA, McIntire LV. Kinetics of GPIbalpha-vWF-A1 tether bond under flow: effect of GPIbalpha mutations on the association and dissociation rates. Biophys J. 2003;85(6):4099–4109. doi: 10.1016/S0006-3495(03)74822-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yago T, Lou J, Wu T, Yang J, Miner JJ, Coburn L, Lopez JA, Cruz MA, Dong JF, McIntire LV, McEver RP, Zhu C. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 2008;118(9):3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auton M, Zhu C, Cruz MA. The Mechanism of VWF-Mediated Platelet GPIb alpha Binding. Biophysical Journal. 2010;99(4):1192–1201. doi: 10.1016/j.bpj.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadler JE. A revised classification of von Willebrand disease. For the Subcommittee on von Willebrand Factor of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis. Thromb Haemost. 1994;71(4):520–525. [PubMed] [Google Scholar]
- 13.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297(5584):1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 14.Dumas JJ, Kumar R, McDonagh T, Sullivan F, Stahl ML, Somers WS, Mosyak L. Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J Biol Chem. 2004;279(22):23327–23334. doi: 10.1074/jbc.M401659200. [DOI] [PubMed] [Google Scholar]
- 15.Doggett TA, Girdhar G, Lawshe A, Miller JL, Laurenzi IJ, Diamond SL, Diacovo TG. Alterations in the intrinsic properties of the GPIbalpha-VWF tether bond define the kinetics of the platelet-type von Willebrand disease mutation, Gly233Val. Blood. 2003;102(1):152–160. doi: 10.1182/blood-2003-01-0072. [DOI] [PubMed] [Google Scholar]
- 16.Ruggeri ZM. Type IIB von Willebrand disease: a paradox explains how von Willebrand factor works. J Thromb Haemost. 2004;2(1):2–6. doi: 10.1111/j.1538-7836.2003.00523.x. [DOI] [PubMed] [Google Scholar]
- 17.Jackson SP. The growing complexity of platelet aggregation. Blood. 2007;109(12):5087–5095. doi: 10.1182/blood-2006-12-027698. [DOI] [PubMed] [Google Scholar]
- 18.Nesbitt WS, Westein E, Tovar-Lopez FJ, Tolouei E, Mitchell A, Fu J, Carberry J, Fouras A, Jackson SP. A shear gradient-dependent platelet aggregation mechanism drives thrombus formation. Nat Med. 2009;15(6):665–673. doi: 10.1038/nm.1955. [DOI] [PubMed] [Google Scholar]
- 19.Mody NA, King MR. Platelet adhesive dynamics. Part II: high shear-induced transient aggregation via GPIbalpha-vWF-GPIbalpha bridging. Biophys J. 2008;95(5):2556–2574. doi: 10.1529/biophysj.107.128520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mody NA, King MR. Platelet adhesive dynamics. Part I: characterization of platelet hydrodynamic collisions and wall effects. Biophys J. 2008;95(5):2539–2555. doi: 10.1529/biophysj.107.127670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mody NA, King MR. Three-dimensional simulations of a platelet-shaped spheroid near a wall in shear flow. Physics of Fluids. 2005;17(11) [Google Scholar]
- 22.Kim S, Karrila SJ. Butterworth-Heinemann series in chemical engineering. Boston: Butterworth-Heinemann; 1991. Microhydrodynamics : principles and selected applications; p. 507. xxiii. [Google Scholar]
- 23.Power H, Miranda G. 2nd Kind Integral-Equation Formulation of Stokes Flows Past a Particle of Arbitrary Shape. Siam Journal on Applied Mathematics. 1987;47(4):689–698. [Google Scholar]
- 24.Reininger AJ, Heijnen HF, Schumann H, Specht HM, Schramm W, Ruggeri ZM. Mechanism of platelet adhesion to von Willebrand factor and microparticle formation under high shear stress. Blood. 2006;107(9):3537–3545. doi: 10.1182/blood-2005-02-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammer DA, Apte SM. Simulation of cell rolling and adhesion on surfaces in shear flow: general results and analysis of selectin-mediated neutrophil adhesion. Biophys J. 1992;63(1):35–57. doi: 10.1016/S0006-3495(92)81577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell GI, Dembo M, Bongrand P. Cell adhesion. Competition between nonspecific repulsion and specific bonding. Biophys J. 1984;45(6):1051–1064. doi: 10.1016/S0006-3495(84)84252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 28.Arya M, Anvari B, Romo GM, Cruz MA, Dong JF, McIntire LV, Moake JL, Lopez JA. Ultralarge multimers of von Willebrand factor form spontaneous high-strength bonds with the platelet glycoprotein Ib-IX complex: studies using optical tweezers. Blood. 2002;99(11):3971–3977. doi: 10.1182/blood-2001-11-0060. [DOI] [PubMed] [Google Scholar]
- 29.Coburn LA, Damaraju VS, Dozic S, Eskin SG, Cruz MA, McIntire LV. GPIb alpha-vWF Rolling under Shear Stress Shows Differences between Type 2B and 2M von Willebrand Disease. Biophysical Journal. 2011;100(2):304–312. doi: 10.1016/j.bpj.2010.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(31):11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King MR, Heinrich V, Evans E, Hammer DA. Nano-to-micro scale dynamics of P-selectin detachment from leukocyte interfaces. III. Numerical simulation of tethering under flow. Biophysical Journal. 2005;88(3):1676–1683. doi: 10.1529/biophysj.104.051805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King MR, Hammer DA. Multiparticle adhesive dynamics. Interactions between stably rolling cells. Biophys J. 2001;81(2):799–813. doi: 10.1016/S0006-3495(01)75742-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang KC, Hammer DA. Adhesive dynamics simulations of sialyl-Lewis(x)/E-selectin-mediated rolling in a cell-free system. Biophys J. 2000;79(4):1891–1902. doi: 10.1016/S0006-3495(00)76439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, King MR. Multiscale Modeling of Platelet Adhesion and Thrombus Growth. Ann Biomed Eng. doi: 10.1007/s10439-012-0558-8. [DOI] [PubMed] [Google Scholar]
- 35.Carrell NA, Fitzgerald LA, Steiner B, Erickson HP, Phillips DR. Structure of human platelet membrane glycoproteins IIb and IIIa as determined by electron microscopy. J Biol Chem. 1985;260(3):1743–1749. [PubMed] [Google Scholar]
- 36.Fox JE, Aggerbeck LP, Berndt MC. Structure of the glycoprotein Ib.IX complex from platelet membranes. J Biol Chem. 1988;263(10):4882–4890. [PubMed] [Google Scholar]
- 37.Naimushin YA, Mazurov AV. Von Willebrand factor can support platelet aggregation via interaction with activated GPIIb-IIIa and GPIb. Platelets. 2004;15(7):419–425. doi: 10.1080/09537100410001721333. [DOI] [PubMed] [Google Scholar]
- 38.Tunc S, Maitz MF, Steiner G, Vazquez L, Pham MT, Salzer R. In situ conformational analysis of fibrinogen adsorbed on Si surfaces. Colloids Surf B Biointerfaces. 2005;42(3–4):219–225. doi: 10.1016/j.colsurfb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Alevriadou BR, Moake JL, Turner NA, Ruggeri ZM, Folie BJ, Phillips MD, Schreiber AB, Hrinda ME, McIntire LV. Real-time analysis of shear-dependent thrombus formation and its blockade by inhibitors of von Willebrand factor binding to platelets. Blood. 1993;81(5):1263–1276. [PubMed] [Google Scholar]
- 40.Peterson DM, Stathopoulos NA, Giorgio TD, Hellums JD, Moake JL. Shear-induced platelet aggregation requires von Willebrand factor and platelet membrane glycoproteins Ib and IIb-IIIa. Blood. 1987;69(2):625–628. [PubMed] [Google Scholar]
- 41.Singh I, Shankaran H, Beauharnois ME, Xiao Z, Alexandridis P, Neelamegham S. Solution structure of human von Willebrand factor studied using small angle neutron scattering. J Biol Chem. 2006;281(50):38266–38275. doi: 10.1074/jbc.M607123200. [DOI] [PubMed] [Google Scholar]
- 42.Chtcheglova LA, Shubeita GT, Sekatskii SK, Dietler G. Force spectroscopy with a small dithering of AFM tip: a method of direct and continuous measurement of the spring constant of single molecules and molecular complexes. Biophys J. 2004;86(2):1177–1184. doi: 10.1016/S0006-3495(04)74192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phan-Thien N, Tullock D, Kim S. Completed double-layer in half-space: a boundary element method. Comput. Mech. 1992;9:121–135. [Google Scholar]