Abstract
Background
Previous studies have revealed that traditional real-time quantitative PCR (qPCR) underestimates adeno-associated virus (AAV) titer. Because the inverted terminal repeat (ITR) exists in all AAV vectors, the only remaining element from the wild genome could form special configurations to interfere with qPCR titration. To solve this problem, a modified and universal qPCR method was tested and established.
Material/Methods
In this work, there was a great variation in titration of ssAAV2-EGFP (Enhanced Green Fluorescence Protein) and scAAV2-EGFP genome by traditional qPCR. For ssAAV2-EGFP, the highest titer was found by using the targeting EGFP primers and the lowest titer was measured by those targeting bovine growth hormone polyA element (pBGH) primers.
Results
Experimental data were reverse for ssAAV2-EGFP and scAAV2-EGFP. Here we report an improved and universal SmaI qPCR method, based on cleaving all ITRs in AAV2 genome by SmaI with several advantages: (1) impact of all ITRs in ssAAV2 and scAAV2 was dismissed; (2) titers increased remarkably, up to 7-fold, especially for scAAV2; (3) the variation of titers was reduced when different primers were applied. A similar phenomenon was also observed in other ssAAV2 and scAAV2 products when the range of titration was at 3×107 to 7×109 V.G/μl in this study.
Conclusions
This modified qPCR strategy can increase rAAV’ titer and reduce titration variance, possibly become a universal method for titrating AAV vectors.
Keywords: SmaI, treatment, adeno-associated virus, titration, qPCR
Background
Recombinant adeno-associated virus (AAV) vector has been exploited in 86 clinical trials so far, with most of them under clinical Phase I and II trials and 8 under Phase III trial (http://www.abedia.com/wiley/search.php). The safe and long-term effective expression of AAV has been confirmed by Phase I trails [1, 2]. Currently, several Phase I and II trails are at the stage of evaluating pharmacokinetics and therapeutic efficacy. To better evaluate the safety and efficacy of AAV-based gene medicine, a precise and standard method for AAV vector titration is a prerequisite. Firstly, dose-dependence of AAV vector-derived transgene expression and therapeutic efficacy necessitate precise quantification of AAV vectors. For example, a linear relationship between AAV genomic copy and expression of á1-antitrypsin (AAT) was demonstrated by a Phase II clinical trail [3]. Secondly, the dose-dependent potential adverse effects of AAV-based gene medicine require medication accuracy. For instance, previous clinical trails have shown that antigen-specific memory CD8+ T cells, antibodies, and interferon-γ production were induced in some patients [4,5] and lower doses of AAV vectors could reduce these immune responses. These adverse effects were proposed to be induced by pre-exposure to wild-type AAV, but not associated with any transgene expression [3,6].
Quantitative PCR (qPCR) has been frequently used for quantification of AAV vector for both preclinical and clinical trails [7]. The advantages of qPCR make it an attractive method for AAV titrating: it is easy to process, and it is rapid, cheap, broad range, accurate, and frequently used. However, great variation in AAV titrating is associated with traditional qPCR, especially for self-complementary AAV (scAAV).
Previous reports have shown that traditional qPCR underestimated 5- to 10-fold relative to the titers by dot-blot and optical density measurement [8,9]. The main reason for this problem was that the AAV genome contains two inverted terminal repeat (ITR) sequences. They might form palindrome structures at each ITR or between two ITRs. In addition, scAAV vector genomes could form another dsDNA hairpin molecule, consisting of 2 monomer single-stranded genomes connected by a mutated ITR due to lack of terminal resolution site (TRS) [10] (Figure 1). All these structures would interfere with annealing of qPCR primers to AAV DNA templates, leading to inefficient binding and underestimation of AAV vectors. As a result, AAV genome was subjected to SpeI endonuclease digestion prior to qPCR to avoid this problem [8]. Nevertheless, it still could not be used in all AAV titration. In this study, we report a reliable and feasible qPCR method to measure all AAV titers.
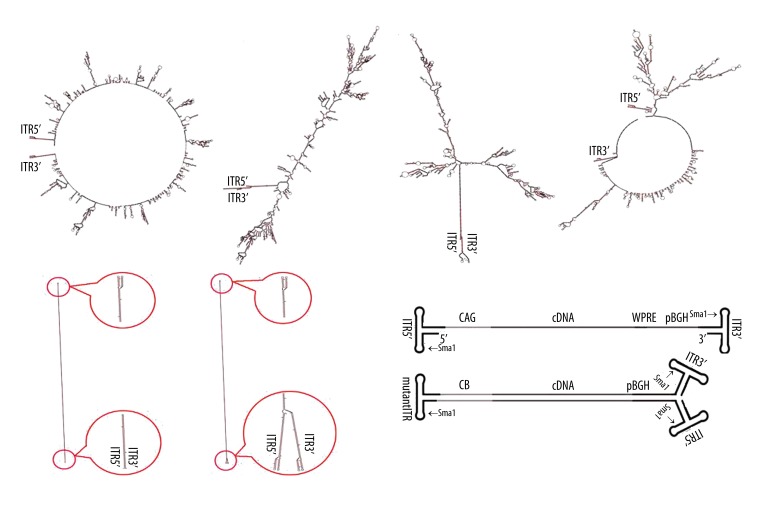
Figure 1.
The putative molecular structures of AAV genome and diagram of rAAV vector genome. (A, B) shows the main putative molecular structures of wild AAV genome. (C, D) shows the main putative molecular structures of ssAAV2-EGFP genome. (E, F) shows the putative molecular structures of scAAV2-EGFP genome. (G) Diagram of ssAAV (up) and scAAV (down) vector genome, with the different elements and restriction endonuclease (SmaI) site indicated.
Material and Methods
AAV vector construction and production
The structures of conventional ssAAV2/2-EGFP and ssAAV2/2-KS were published earlier [11–13]. The single-stand AAV genome plasmids contain cytomegalovirus enhancer/chicken β-actin promoter (CAG), transgene, a woodchuck hepatitis B virus posttranscriptional regulatory element (WPRE), and bovine growth hormone (BGH) polyA element inserted between the 2 terminal ITRs. The scAAV2 genome plasmids include CB promoter, transgene, and pBGH inserted between a mutant ITR and an intact ITR. Both scAAV2/2-TRAIL and scAAV2/2-Kallistatin (ssAAV2-KS) were constructed from scAAV2/2-EGFP.
AAV production and purification
ssAAV and scAAV were prepared using modified methods as described previously [14,15]. Briefly, 3 plasmids – AAV vector plasmids, helper plasmid (H22), and adenovirus helper plasmid (pFd6) – were transfected into HEK293 cells using calcium phosphate method. Following transfection, AAV viruses released from HEK293 cells by sonification and viruses in the medium were precipitated using ammonium sulfate. Then they were resuspended, treated by Benzonase (MERCK) at 37° for 1 h and precipitated by PEG-Na. CsCl gradient centrifugation was performed. The fractions pooled from the CsCl gradient were dialyzed against 3 changes of sterile 1×PBSS (PBS+5% Sorbitol) for at least 1 or 3 h, respectively, at 4°.
Bioinformatic analysis of AAV genome folding form
The putative ssAAV and scAAV genome folding form and hybridization prediction were investigated (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form). The online software is available for the prediction of the secondary structure of single-stranded nucleic acids [16]. The free energies used are from the laboratory of SantaLucia Jr [17]. All sequences of wild AAV, ssAAV, and scAAV genome were used for prediction and the default parameters.
AAV genome purification and endonuclease SmaI treatment
Twenty micro-liters of each AAV sample were mixed with 2 μl of DNase I buffer and 1 μl of DNase I (Qiagen) and incubated at 25° for 1 h. The virus samples were lysed by addition of 180 μl of deionized water and then 200 μl of 2× proteinase buffer (20 mM Tris, 20 mM EDTA, pH 8.0, 1% SDS) and 100 μg protease, and then kept at 37° for 1 h. The viral DNA was purified by phenol/chloroform extraction [18]. Ten micro-liters of each sample were incubated with 1 μl of SmaI (Fermentas) in a 20 μl endonuclease reaction volume at 37° for 3 h. The cleaved product was denatured at 95° for 10 min.
Preparation of qPCR standards
The plasmids containing the ssAAV insert were purified using the QIAGEN Plasmid Maxi kit. The purified ssAAV and scAAV plasmids were linearized by restriction digestion. The subsequent treatment was the same as for AAV genome purification.
AAV titration by qPCR
Quantitative PCR (qPCR) analysis was performed using the Applied Biosystems StepOne Plus Real-Time PCR system. PCR reactions were performed in 20 μl of final volume using the 2×SYBR Green qPCR mix (ZOMANBIO, China) supplemented with 100 nM sense primer, 100 nM antisense primer, and 2 μl of twice serially diluted template DNA (either plasmid standard or extracted sample DNA, either SmaI-treated or not) according to the manufacturer’s instructions. Each sample and negative control was run in 6 replicates of 20 μl of reactions in alternate rows of a 96-well optical plate. The PCR profile contained an initial denaturation step at 95° for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and annealing or extension at 60°C for 1 min, followed by a melt curve stage. Data analysis was performed using the Applied Biosystems StepOne software v2.1.
Statistical analysis
Data are expressed as mean ±SD. The titers (Figures 2–5) were subjected to single- factor analysis of variance (ANOVA) using the SPSS 19.0 statistical software. P<0.05 and P<0.01 was regarded as significant and highly significance, respectively. Every experiment was performed 3 times (n=3) independently.
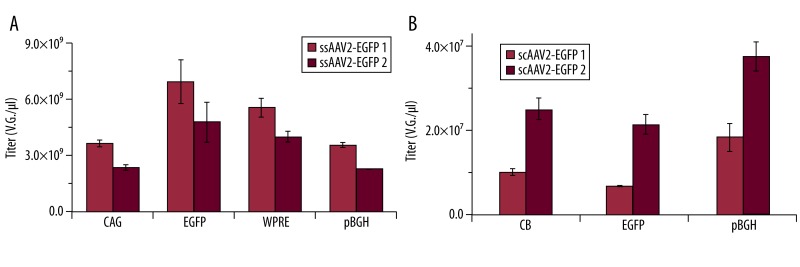
Figure 2.
There was a titration variance using different primers to target different elements of ssAAV2-EGFP or scAAV2-EGFP. (A) Titration of ssAAV2-EGFP using primers targeting CAG, EGFP, WPRE, and pBGH. (B) Titration of ssAAV2-EGFP using primers targeting CB, EGFP, and pBGH; n=6.
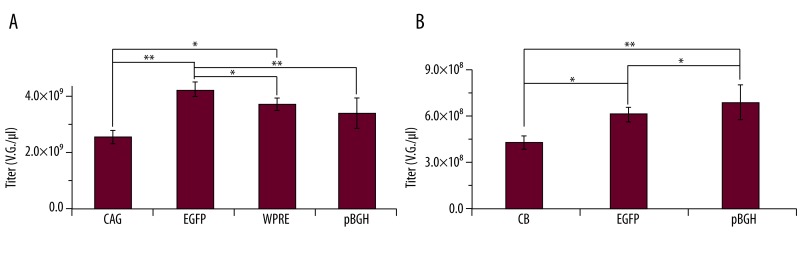
Figure 5.
Titration of ssAAV2-EGFP or scAAV2-EGFP by SmaI qPCR with all the different primers. (A) The titers of ssAAV2-EGFP using CAG, EGFP, WPRE and pBGH primers. (B) The titers of scAAV2-EGFP using CB, EGFP and pBGH primers. ** P<0.01; * P<0.05, n=6.
Results
Predicted secondary structure of AAV genome and design of qPCR primers
The secondary structures of wild AAV, ssAAV, and scAAV genome were predicted by Mfold online software using the default constraint information. The putative secondary structures of ssAAV genome containing 2 main configurations were similar to those of wild AAV. For the former, the 2 end ITRs were complementary with each other. For the latter, the 2 ITRs formed hairpins. The putative structures of the scAAV genome also had 2 main forms: one was a completely complementary configuration with a hairpin present in the middle of the genome (the mutant ITR) and the other form had some differences in the 2 end ITR domains that formed hairpins (Figure 1). These structures of AAV genome might impair the AAV titration by qPCR, resulting in great variation in AAV genome titration.
Based on the different structures of the AAV genome, we designed different qPCR primers to target the different elements in the ssAAV2-EGFP and scAAV2-EGFP genomes (Table 1). All primers were designed for annealing at 60°. Each primer was performed for qualitative PCR analysis using gradient PCR, annealing at 55° to 65° (gradient). It was found that 60° was the best annealing temperature for qPCR (date not shown).
Table 1.
The primers used for qPCR in the present study.
| Vector | Targeted element | Primer sequence (5’→ 3’) |
|---|---|---|
| ssAAV2-EGFP | CAG | Sense primer: CTGACCGCGTTAATCCCACA |
| Antisense primer: ACAAGCCGTGATTAAACCAAGA | ||
|
|
||
| EGFP | Sense primer: CACCCACGTGACCACCCTTAC | |
| Antisense primer: GGATGTTGCAGTCCTCCCTG | ||
|
| ||
| WPRE | Sense primer: TTGGATGCTCGCCTGGGTTG | |
| Antisense primer: AGGAAGGTCCGCTGGATCGA | ||
|
| ||
| pBGH | Sense primer: CATATAAAATGAGGAAATTGC ATCGCA | |
| Antisense primer: TCAGAACCCATAGAGCCC ACCG | ||
|
| ||
| scAAV2-EGFP | CB | Sense primer: AGTTTAGTCTTTTTGTCTTTTATTTCAGGTCCCG |
| Antisense primer: GCAGCTTTTAGAGCAGAAGTAACACTTCCGTAC | ||
|
|
||
| EGFP | Sense primer: ACAAGCAGGAGAACAGCATCAAGGT | |
| Antisense primer: GTCTTTGCTCTGGGCGGAATG | ||
|
| ||
| pBGH | Sense primer: CGTGGCTTCCTTGACCGTGG | |
| Antisense primer: GAATAGAGTCACACCTACCCAGACAATG | ||
There was great titration variation by traditional qPCR using primers to target different elements of AAV genome
After determination of specificity and annealing temperature of all primers by gradient PCR, qPCR was performed to compare the titration variance. For ssAAV2-EGFP, those primers targeted at EGFP reached the highest titer, followed by WPRE. Those targeting CAG and pBGH primers reached the lowest titer. The ratio of the highest and the lowest titer was about 2.
The titration bias also occurred for titration of scAAV. Among of them, the highest titer was found for pBGH primers, followed by CB primers, and EGFP primers were the lowest. The ratios between the highest and the lowest were 2.02 and 2.61 for 2 groups of samples. To further confirm that this is common phenomenon, 2 groups of AAV from different production dates were also analyzed for each titration.
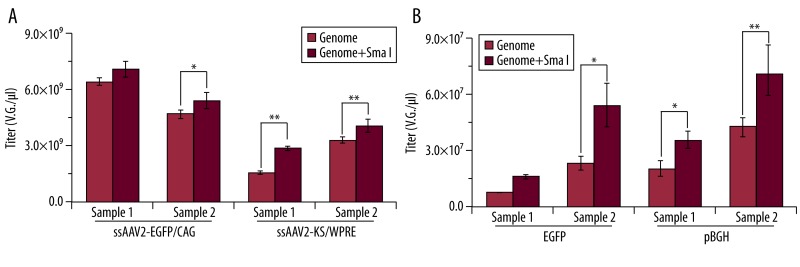
Higher titers were measured by qPCR of SmaI digested AAV genome than by traditional qPCR
To test if incubation with SmaI prior to qPCR (SmaI qPCR) could reduce the effect of ITR’s special configurations in ssAAV2 or scAAV genome by qPCR titration, we examined ssAAV2-EGFP first, and primers targeting CAG were selected due to CAG existence in all of these vectors. The results of SmaI qPCR revealed that titers of ssAAV2-EGFP increased with primers targeting CAG of ssAAV2-EGFP genome (Figure 3A). Titers detected also increased when the primers targeting WPRE of ssAAV2-KS were applied (Figure 3A). Our study also revealed that it was unnecessary to further purify genome after SmaI digestion (data not shown).
Figure 3.
Comparison of titers by traditional qPCR or qPCR after digestion with SmaI (A) showed qPCR titration of ssAAV2-EGFP using primers targeting CAG and ssAAV2-KS using primers targeting WPRE; (B) showed qPCR titration of scAAV2-EGFP using primers targeting EGFP and pBGH. ** P<0.01; * P< 0.05, n=6.
The results of the traditional qPCR showed that the lowest titer for the EGFP primers and the highest for the pBGH (Figure 2B). For titration of scAAV2-EGFP, both primers targeting EGFP and pBGH were selected for this investigation. The titration of scAAV2-EGFP increased remarkably by SmaI qPCR using either the EGFP or pBGH primers (Figure 3B). The ratios of the titers by SmaI qPCR and traditional qPCR with EGFP primers were 2.26 and 2.61, respectively, and the ratios were 1.79 and 1.83, respectively, for pBGH primers.
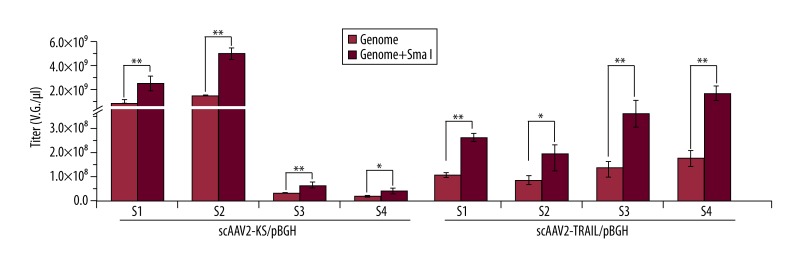
To determine if the SmaI qPCR could increase titers of other scAAV2, scAAV2-KS and scAAV2-TRAIL were examined (n=6). The titers were also elevated by SmaI qPCR for pBGH primers when compared with those analyzed by traditional qPCR (Figure 4). The ratio increased about 1.96~3.89. There was an approximately 7-fold increase in ratio with these primers, analyzed by SmaI qPCR, when compared with those by traditional qPCR. The titer range of scAAV2 was 3×107~5×109 V.G./μl.
Figure 4.
Comparison of titers of scrAAV2-KS and scrAAV2-TRAIL by traditional qPCR or qPCR after vectors were digested with SmaI digestion. S1, S2, S3, and S4 represent sample1, 2, 3, and 4, respectively. ** P<0.01; * P< 0.05, n=6.
SmaI qPCR reduced the titration variation using different primers
Titration of ssAAV2-EGFP or scAAV2- EGFP was performed with all the different primers by SmaI qPCR. The ratios of titer using CAG, EGFP, WPRE, and pBGH primers were 1:1.5:1.2:1.1, respectively, for ssAAV2- EGFP (Figure 5A), and titers were similar using either CAG, WPRE, or pBGH primers. The highest ratio (1.5) was also lower for those analyzed by SmaI qPCR than by traditional qPCR (1.95 and 2.09) (Figure 2A).
The ratio of titer using CB, EGFP, and pBGH primers was 1:1.5:1.7, respectively, for scAAV2-EGFP (Figure 5A). The highest ratio (1.7) was also lower for those analyzed by SmaI qPCR than by traditional qPCR (2.6 and 2.0) (Figure 2B).
Discussion
qPCR is a popular method for titration of AAV and is chosen as one of the characterization assays of AAV2 reference standard material [20]. This method depends on accurate measurements of packaged vector genome, which is key mediator of therapeutic efficacy [21]. Nevertheless, titers of ssAAV2 and scAAV2 were varied by traditional qPCR when the primers targeting different elements were used. The present study further confirms that there is great variation by traditional qPCR with different primers. However, the titers analyzed by SmaI qPCR were consistently higher than those by traditional qPCR, suggesting that the titers by traditional qPCR might be underestimated.
One study introduced a qPCR method using primers targeting ITR of the AAV2 genome [19] but the primers could only be used in the TaqMan probe qPCR system but not in the SYBR Green qPCR system. It would be more convenient and cheaper to use the SYBR Green qPCR system.
Although ITR is the only wild element remaining in all of the ssAAV genome, its main secondary configurations are similar to that of the wild AAV. The complementary mutant ITR and 2 ending wild ITRs of scAAV might induce the sequence to form complementary configurations. This means that ITRs may be responsible for all special configurations. We therefore propose that special secondary configurations of wild-type ITR in 2 endings of the ssAAV genome might interfere with primer annealing during traditional qPCR and lead to underestimation. Both CAG promoter and pBGH of the ssAAV2 genome might interact with ITR. As a consequence, the configurations may affect the binding of primers, leading to varied template formation.
If the EGFP element is far away from the ITR in ssAAV, the influence would be weaker, resulting in a highest titer reading for EGFP primers. Although the lowest reading was found with pBGH primers, it may be that pBGH is much closer to the 2 ending ITRs. Nevertheless, the effect of complementary structure was greater than the 2 ending ITRs in scAAV2-EGFP, leading to the highest titers by using pBGH primers. Interestingly, titers obtained by using CB primer that was nearest to the mutant ITR were higher than those with EGFP primers. This phenomenon was similar to that found in a previous report [8]. It was assumed that the sequence of EGFP in the scAAV genome might form a special configuration that could also play an important role. The exact reasons for this require further investigation.
The AAV2 genome is the first and most common used in gene therapy. AAV2 is also a well characterized serotype. Therefore, it should become the first AAV reference standard material [20]. ITR is essential for AAV genome replication, second-strand synthesis, and genome packaging [22], with only a wild AAV2 genome element remaining in the AAV2-derived vector. As a result, variance exists in the titer of AAV2 in all investigations by traditional qPCR.
A previous report described digestion of scAAV DNA prior to qPCR by SpeI or PstI endonuclease existing in the middle of scAAV2 genome could prevent the mutant ITR and complementary configuration in scAAV, and the titers measured were similar to dot-blot [9]. Nevertheless, the endonuclease positions might not exist in other scAAV2 genomes. Furthermore, after digestion, titers of vectors did not increase with the primers designed from near the 2 ending ITRs. The results of the present study show that after SmaI digestion, titers of vectors increased remarkably when using pBGH primers also designed from near 2 ending ITRs. All of these findings clearly indicated that configurations of the mutant ITR and also 2 ending ITRs could impair titration of qPCR, suggesting that endonuclease that did not exist within ITR could only eliminate complementary structure, but not the 2 ending ITR configurations. SmaI qPCR could overcome this hindrance.
Moreover, SmaI qPCR was suitable not only for ssAAV2 or scAAV2-EGFP but also other rAAV2, including other gene cDNA (Figure 4). In addition, the range of the detecting titer was very wide, from 3×107 to 7×109 V.G./μl in the present study. The SmaI qPCR method not only increased the titers, but also reduced the variation found in traditional qPCR for ssAAV2 and scAAV2 titration. Because the pBGH element was commonly used in almost all of scAAV2, it therefore would be better to choose these targeting pBGH primers in SmaI qPCR for scAAV2 titration.
Conclusions
In summary, special configurations of ITR could affect titers of all ssAAV2 and scAAV2 by traditional qPCR. Such an impact could be eliminated by incubation with SmaI prior to qPCR. Such a reliable and feasible qPCR strategy could increase titers remarkably and reduce titration variance for both AAVs, becoming a universal method to titrate all ssAAV2s and scAAV2s.
Footnotes
Source of support: This study was supported by a National Natural Science Foundation of China (No.81072578) and a National High Tech 863 Grant, China
References
- 1.Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358(21):2240–48. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainbridge JW, Smith AJ, Barker SS, et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N Engl J Med. 2008;358(21):2231–39. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 3.Flotte TR, Trapnell BC, Humphries M, et al. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing alpha1-antitrypsin: interim results. Hum Gene Ther. 2011;22(10):1239–47. doi: 10.1089/hum.2011.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntosh JH, Cochrane M, Cobbold S, et al. Successful attenuation of humoral immunity to viral capsid and transgenic protein following AAV-mediated gene transfer with a non-depleting CD4 antibody and cyclosporine. Gene Ther. 2012;19(1):78–85. doi: 10.1038/gt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laredj LN, Beard P. Adeno-Associated Virus Activates an Innate Immune Response in Normal Human Cells but Not in Osteosarcoma. Cells. J Virol. 2011;85(24):13133–43. doi: 10.1128/JVI.05407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mingozzi F, High KA. Immune responses to AAV in clinical trials. Curr Gene Ther. 2011;11(4):321–30. doi: 10.2174/156652311796150354. [DOI] [PubMed] [Google Scholar]
- 7.Mayginnes JP, Reed SE, Berg HG, et al. Quantitation of encapsidated recombinant adeno-associated virus DNA in crude cell lysates and tissue culture medium by quantitative, real-time PCR. J Virol Methods. 2006;137(2):193–204. doi: 10.1016/j.jviromet.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Wright JF. New adeno-associated virus strategies to support momentum in the clinic. Hum Gene Ther. 2011;22(5):519–21. doi: 10.1089/hum.2011.4080. [DOI] [PubMed] [Google Scholar]
- 9.Fagone P, Wright JF, Nathwani AC, et al. Systemic Errors in Quantitative Polymerase Chain Reaction Titration of Self-Complementary Adeno-Associated Viral Vectors and Improved Alternative Methods. Hum Gene Ther Methods. 2012;23(1):1–7. doi: 10.1089/hgtb.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty DM. Self-complementary AAV vectors: advances and applications. Mol Ther. 2008;16(10):1648–56. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Zheng D, Liu Y, et al. Overexpression of soluble TRAIL induces apoptosis in human lung adenocarcinoma and inhibits growth of tumor xenografts in nude mice. Cancer Res. 2005;65(5):1687–92. doi: 10.1158/0008-5472.CAN-04-2749. [DOI] [PubMed] [Google Scholar]
- 12.Tse LY, Sun X, Jiang H, et al. Adeno-associated virus-mediated expression of kallistatin suppresses local and remote hepatocellular carcinomas. J Gene Med. 2008;10(5):508–17. doi: 10.1002/jgm.1180. [DOI] [PubMed] [Google Scholar]
- 13.Diao Y, Zhao XF, Lin JS, et al. Protection of the liver against CCl4-induced injury by intramuscular electrotransfer of a kallistatin-encoding plasmid. World J Gastroenterol. 2011;17(1):111–17. doi: 10.3748/wjg.v17.i1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu R, Rahimi M, Ma H, Fung P, et al. Stability of infectious recombinant adeno-associated viral vector in gene delivery. Med Sci Monit. 2005;11(9):BR305–8. [PubMed] [Google Scholar]
- 15.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc. 2006;1(3):1412–28. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95(4):1460–65. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Liu Y, Guo J, et al. Expression of an anti-DR5 single chain antibody in muscles of nude mice prevents lung cancer growth. Cancer Res. 2006;66(24):11946–53. doi: 10.1158/0008-5472.CAN-06-1227. [DOI] [PubMed] [Google Scholar]
- 19.Aurnhammer C, Haase M, Muether N, et al. Universal Real-Time PCR for the Detection and Quantification of Adeno-Associated Virus Serotype 2-Derived Inverted Terminal Repeat Sequences. Hum Gene Ther Methods. 2012;23(1):18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- 20.Lock M, McGorray S, Auricchio A, et al. Characterization of a recombinant adeno-associated virus type 2 Reference Standard Material. Hum Gene Ther. 2010;21(10):1273–85. doi: 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer JM, Smith PH, Parthasarathy S, et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol Ther. 2003;7(1):122–28. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 22.Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21(4):583–93. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]