Abstract
The biological picture of cancer is rapidly advancing from disease models built from phenomenological descriptions to network models derived from systems biology, which can capture the evolving pathophysiology of the disease at the molecular level. The translation of this (still academic) picture into a clinically relevant framework can be enabling for the war on cancer, but it is a scientific and technological challenge. In this review, we discuss emerging in vitro diagnostic technologies and therapeutic approaches that are being developed to handle this challenge. Our discussion of in vitro diagnostics is guided by the theme of making large numbers of measurements accurately, sensitively, and at very low cost. We discuss diagnostic approaches based on microfluidics and nanotechnology. We then review the current state of the art of nanoparticle-based therapeutics that have reached the clinic. Our goal is to identify nanotherapeutic strategies that are designed to increase efficacy while simultaneously minimizing the toxic side effects commonly associated with cancer chemotherapies.
Keywords: in vitro diagnostics, cancer therapeutics, nanoparticles, quantum dots, nanowires, microfluidics
INTRODUCTION
More than 1500 Americans will die from cancer each day this year. Projections of the numbers of expected cancer diagnoses, and the numbers of expected deaths from cancer, are publicly available.1 While the rate of cancers being diagnosed has steadily increased, the normalized numbers of cancer-related deaths has remained virtually unchanged. Against this static background is emerging a new picture of cancer, which is inspiring hopes that the war on cancer may be winnable. In this review, we introduce this picture and discuss how it is driving the development of new diagnostic and therapeutic technologies. In particular, we focus on nanotechnologies and microfluidics for in vitro diagnostics and nanotechnologies for drug delivery. These technologies constitute only a few pieces, albeit critical ones, that are being brought together to win the war on cancer.
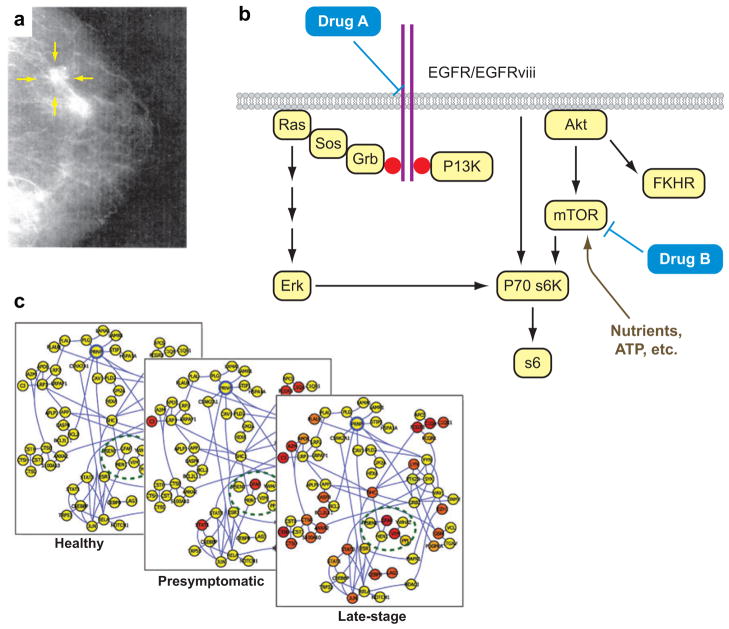
Recent advances, both conceptual and technological, are making it possible to imagine a future in which cancer is a manageable chronic ailment. Consider how cancer was viewed just a few years ago. Most pathology practices were based upon a few phenomenological measurements to assess disease (Figure 1a). An increased understanding of cancer has demonstrated that a given type of cancer (e.g. breast cancer) can be triggered by different genetic mutations, each of which can lead to a different outcome (e.g., aggressive versus nonaggressive cancer). This understanding has led to the model of cancer pathways (2) (Figure 1b). In this model, there are multiple pathways of interacting proteins, each constituting a cascade of molecular events. A given pathway, if genetically altered in specific ways, is effectively short-circuited and thus constantly activated, even in the absence of signaling molecules. Emerging cancer molecular therapeutics (3, 4) are designed against specific pathways, often targeting the genetically altered proteins. Molecular measurements, such as the identification of mRNAs (5) or pathway-associated proteins, are increasingly used to identify the altered pathway or the response of the cancer to therapy (6). Such an analysis can potentially indicate an appropriate therapy (7, 8), the progression of the cancer (9), the potential for recurrence after therapy (10), or the potential for drug resistance (8). In vivo molecular imaging is also increasingly employed as a diagnostic of drug efficacy (11).
Figure 1.
These images represent the evolving picture of medicine that is driving the development of nanotechnologies for the investigation, diagnosis, and treatment of cancer. (a) A mammogram exemplifies traditional, phenomenological, single-parameter cancer diagnostic techniques. (b) The “cancer pathways” model for understanding the differential response of certain cancers to molecular therapies. Each pathway is comprised of a cascade of interacting proteins (circled in yellow). Genetic mutation of a protein within a pathway can lead to cancer. Identifying which pathway is altered typically involves the analysis of multiple mRNAs and proteins from the cancerous tissue. If the altered pathway is correctly diagnosed, then that information can lead to the appropriate prescription of drugs. (c) A dynamic network model of disease. This model, compared to the cancer pathways model, more accurately reflects the complex interrelationships between various proteins within a biological system. It is thus a truer reflection of the molecular nature of the disease, but it also can be mined for biomarkers that can be diagnostic for the progression of the disease. Such biomarkers can potentially be harnessed for detecting disease prior to the emergence of clinical symptoms (dynamic network model images courtesy of Leroy Hood.
Pathway models are useful but limited. A pathway-based diagnosis typically requires prior knowledge that cancer is present, so it constitutes a more accurate pathology report but not an early detection strategy. Another drawback is that pathway models do not account for the dynamic evolution of cancer, and they underestimate the degree of interconnectivity among the various genes and proteins. Finally, pathway models assume that a given cancer is homogeneous, which is almost certainly incorrect.
Network models of disease and disease progression (Figure 1c) are emerging out of systems biology procedures (12), which generally involve deep transcriptome analyses (13), occasionally coupled with focused proteomic investigations (14), all integrated together using computational methods (15). Network models can illustrate how the onset and progression of disease are reflected in the form of differentially expressed genes and their associated protein networks. Current network models, though unwieldy, are beginning to lend molecular-level insight into the pathophysiology of disease progression. This, in turn, has implications for cancer clinical care, including the potential for achieving a more informative diagnosis. This increased information content arises from at least three concepts. First, the proteomic and genomic databases may be comparatively mined to generate a list of candidate biomarkers that are detectable in body fluids. Blood, for example, is a powerful window into health and disease, but it is a noisy environment, containing >104 proteins that span a concentration range of >109. The ability to identify organ-specific, secreted proteins in blood is an example of a powerful strategy for extracting signal from such an environment (16). Second, if the regulatory networks associated with the relevant proteins are identified, then measurements of those proteins can be directly correlated with the developing pathophysiology of the disease. Third, the best network models will soon be dynamic models, and thus the extracted molecular signatures of disease are identified against a time-averaged background. Ultimately, dynamic network models may allow the detection of disease prior to the development of clinical symptoms, thus paving the way for prophylactic therapies.
This evolving picture of cancer holds promise for changing both diagnostics and therapeutics. For diagnostics, this scenario permits the asking of many more clinically relevant questions, but it places new demands on both measurement and computational technologies. Information will eventually become the commodity of value, implying that quantitative, sensitive, and multiparameter diagnostic measurements must be accomplished inexpensively, and the results must be rapidly integrated to produce a simple and yet accurate diagnostic conclusion. “Multiparameter” measurements consider genes, proteins, and cells. “Inexpensive” measurements are rapid and routine to execute, requiring small amounts of tissue and minimal sample handling. It is here that nanotechnologies, new chemical methods, and microfluidics are emerging as powerful tools.
The ability to detect cancer early almost always correlates with the ability to cure the disease, typically with combinations of surgery, radiation therapy, and chemotherapy. Emerging molecular therapeutics have shown promise against very specific classes of tumors (17, 18), but typically the cancer is kept at bay for only 1–2 years before returning in a drug-resistant form (19, 20). The more traditional chemotherapies are, as a rule, more effective against broad patient populations, but they are also accompanied by side effects ranging from hair loss to cardiac arrest. New nanotechnologies for drug encapsulation and delivery are being developed to increase the accuracy of drug delivery to target, while also minimizing the exposure of noncancerous tissues and reducing toxicity.
EMERGING IN VITRO DIAGNOSTIC TECHNOLOGIES
In vitro cancer diagnostics will increasingly mean the measurement of large panels of biomolecules (mRNAs and proteins) from ever-smaller samples. Samples may consist of body fluids, tissues, cells sorted from resected or biopsied tumors, circulating tumor cells (21), etc. To focus our discussion on the rapidly expanding fields of microfluidics (22) and nanotechnologies for in vitro cancer diagnostics, we take a lesson from the semiconductor industry. Integrated circuit manufacture has advanced so that the cost of producing a transistor is a fraction of a penny. This has required high-throughput manufacturing protocols that integrate hundreds of processing steps and many different materials. The analogy to the transistor is the measurement of some biomolecule, such as a protein. For clinical diagnostics to keep pace with the evolving picture of human disease, such measurements must cost a few pennies or less each. Our discussion highlights both the progress and the challenges associated with integrating chemistry, biology, device fabrication, etc., into a seamless and highly parallel manufacturing process for enabling inexpensive biological measurements.
Antibody reagents constitute a severe roadblock standing in the way of making biological measurements inexpensive. Antibodies are expensive and unstable, and they don’t always have a high affinity or a high selectivity for their cognate proteins. A description of alternative protein capture agents (e.g., peptides, aptamers, biligands) is not presented here, but we would be remiss to ignore this issue.
Protein Assays
A common diagnostic technique for measuring proteins is the enzyme-linked immunosorbent assay (ELISA) (23). ELISA variations are often referred to as sandwich assays (24)—the biomolecule to be detected is sandwiched between a surface- bound primary (1°) antibody and a fluorophore-labeled secondary (2°) antibody. The binding event of the 2° antibody is optically detected. ELISA assays are typically carried out in multiwell plates, with one type of protein detected per well. The well is incubated first with the 1° antibody and then with the sample and the 2° antibody. This protocol takes a few hours, but this long timescale is not intrinsic (see below). With a 1° antibody that has 10−9 M affinity, an ELISA assay can detect proteins present in the few-picoM concentration range.
The limitations of ELISA are multifold. First, ELISA is a single-protein detection method (although extendable to multiple wells). Second, the concentration range over which a given biomolecule may be quantitated is ~102, limited by the minimal detectable signal over background and the tendency of fluorophores to photobleach. Third, the need for two antibodies per biomolecule is nontrivial. Significant work has gone into improving sandwich assays (25–27). Improvements have included the introduction of amplification using electroless Ag deposition on Au nanoparticle-labeled 2° antibodies (28), or using nanoparticle-loaded DNA or Raman bio-barcodes (29). These nanotechnology-based amplification strategies can (when high-affinity antibodies are available) push the detection threshold into the 100-attoM (10−16 M) range , although they also add noise into the measurement. Nevertheless, such high sensitivity will almost certainly have diagnostic value.
Microfluidics Chips
Glass (30), elastomeric (31), or multilayer integrated elastomeric (32, 33) microfluidics platforms provide the framework for most of the emerging technologies, and they enable cost reductions both in the consumption of reagents and, under certain conditions, in the time required to perform an assay. The binding kinetics of microfluidics-entrained surface immunoassays has been modeled (34, 35). Zimmerman et al. found two kinetic limits. Under low flow velocity, the surface-bound antigen is able to exploit a large fraction of the analyte, but with slow, diffusion-limited capture kinetics. Higher flow velocities (of order 1 mm·s−1) and small active areas (150 nm2) provide a limit at which the immunoassay binding kinetics reflects the analyte/antigen kon and koff binding constants:
| 1 |
with a characteristic binding time, τ, can be expressed as
| 2 |
Under the (common) conditions in which konC ≪ koff, τ ≈ 1/koff. Here, θt is the surface density of bound analyte at time t, θmax is the maximum surface density of molecules possible, and C is the target protein concentration. Over fairly broad protein concentration ranges (10−15–10−10 M), the time to detection can be fast (minutes) for an analyte/antigen binding affinity in the nanomolar range. Thus, the slow rate associated with developing an ELISA immunoassay can be improved through microfluidics design. This potential time savings represents a key reduction in the cost of measurements.
Chips for Blood and Tissue Handling
Serum proteomics involves separating the cells from the plasma using centrifugation, followed by detection of proteins using mass spectrometry, Western blot, or sandwich assays. Centrifugation requires significant handling and sample volumes, and so is expensive. Various methods for the on-chip separation of biological materials have been advanced, including dielectrophoresis (36), microfiltration (37), acoustic forces (38), and lateral displacement (39), to name a few. Yang et al. (40) recently reported on a microfluidics design for the separation of plasma from whole blood. They took advantage of the Zweifach-Fung effect, in which a microfluidic channel is split into low-resistance and high-resistance channels. In an optimized design, the blood cells pass into the channel that has the higher flow rate, with ~20% of the plasma flowing into the low-flow-rate channel. They reported a plasma selectivity with respect to blood hematocrit level of almost 100% regardless of the inlet hematocrit. This technology is remarkably efficient, has no moving parts, is composed only of plastic and glass, and can handle very small amounts of blood.
The culturing and handling of tissue and the sorting of cells are important procedures for cancer research, drug screening, cancer immunotherapy (41), and in vitro diagnostics. On-chip techniques for cell culture (42) and sorting are being developed by a number of groups (43). Cell-sorting chips rely largely on microfluidics variations of fluorescence-activated cell sorting (FACS) (44, 45), dielectrophoresis (4), or the antibody array–based technique of panning (47, 48). As of this writing, we have not identified chip-based automated tissue processors, such as would be necessary to separate and sort specific cancer cells, immune cells, etc., starting from a solid tumor.
A major challenge associated with these blood/tissuehandling chips is to find methods to integrate them with measurement assays. Integration of disparate technologies is often considered an applied engineering problem, and is ignored in the academic labs where many of the individual components are invented. However, for the applications discussed herein, integration issues constitute relatively uncharted science.
Label-Free Measurement Techniques
Surface plasmon resonance (SPR) (49), nanowire (50), nanotube (51, 52), and nanocantilever (53, 54) biomolecular sensors are all classified as label-free measurement technologies, meaning that the binding of the target biomolecule to its surface-bound capture agent is directly detected. Furthermore, under flowing sample conditions, kon, koff, and/or analyte concentration (see Equations 1 and 2) can be directly measured (55, 56) from the dynamic sensing response. SPR is a commercial product that is rapidly being improved (57),2 but the nanotechnologies have distinct advantages. These include direct electrical readout of the signal [which, for the case of cantilevers, requires piezoresistive nanocantilevers (59)], increased sensitivity (60) and dynamic range, the ability to detect small molecule-binding events, and a large degree of substrate independence (61). Although nanowire and nanotube sensors have demonstrated the ability to sense single (62) or small panels (63) of cancer serum biomarkers, they are limited for protein sensing in biological media (e.g., an electrolyte) by Debye screening (64). Nanocantilevers and nanowire technologies are immature technologies and, despite promising demonstrations, it is not clear if they will emerge as useful cancer diagnostic tools. Nevertheless, the impressive sensitivity, dynamic range, and batch processability of these devices, coupled with emerging and novel applications (59), imply that they will become significant measurement tools.
Tissue Analysis
Immunohistochemical staining and related methods (65) represent in situ protein assays that are important for obtaining a molecular diagnosis of cancer from resected tumors. Emerging nanotechnology variants of this method have included the use of semiconductor quantum dots as the antibody fluorescent labels (66) for the staining of breast cancer tissues. The relevant advantages (67) arise from the robust fluorescence properties and the sharp and size-tunable emission spectra of quantum dots, which permit increased multiplexing.
Multiparameter Measurements
Cancer diagnostic platforms will ultimately integrate multiplexed assays of cells, mRNAs, and proteins. However, the surface chemistries that are required for different classes of biomolecules, are often not compatible with one another nor with device fabrication procedures. Antibodies are immobilized onto aldehyde, epoxy, maleimide, or hydrophobic solid supports (68–71). Variables such as pH, ionic strength, hydration, etc., must be controlled to prevent protein denaturation. The best surface for reducing nonspecific binding of cells while maintaining full antibody functionality is acrylamide (72), which is incompatible with DNA. DNA microarrays are electrostatically absorbed (via spotting) onto amine surfaces. One option for detecting DNA, proteins, and cells on the same platform is to utilize differentially functionalized surfaces (or beads, such as are used by certain Illumina® systems for the codetection of DNA oligomers and proteins). For microfluidics-based assays, this adds significant manufacturing complexity and cost. In addition, not all of the above-described surface chemistries are stable to the processing steps associated with microfluidics fabrication. A second alternative recently demonstrated (48) is to utilize the technique of DNA Encoded Antibody Libraries (DEAL) for multiplexed gene and protein detection and cell sorting. This provides one common surface chemistry (spotted DNA arrays) that is fully compatible with microfluidics manufacture. Furthermore, an optimized microfluidics-based DEAL assay can be significantly faster and more sensitive than conventional immunoassays; it can cover a broader dynamic range; and it is far superior to panning as a multiplexed cell-sorting technique.
EMERGING NANOPARTICLE THERAPEUTICS
Informative diagnoses of the future will exploit new advances in nanotechnology in order to provide in vitro molecular measurements of pathophysiology from body fluids such as blood. These advanced diagnostic methods will provide information that will allow the design of new intervention strategies, provided appropriate therapeutics are available. Nanotechnology is playing a role in providing new types of therapeutics for cancer. These nanotherapeutics have the potential to provide effective therapies with minimal side effects. Most cancer patients die from drug-resistant, metastatic disease. Thus, the ultimate goal for cancer therapies would be the ability to treat this stage as well as any of those leading up to it. It is hoped that as diagnostic methods improve, treament can be initiated at earlier and earlier stages of disease progression. However, in the most general sense, it would be advantageous to develop therapies that could be employed at all stages of cancer because of the enormous resources that are required to bring a new therapeutic to market.
Targeted nanoparticles have the potential to provide therapies not achievable with any other drug modalities. By tuning the size and surface properties of the nanoparticle, manipulation of the pharmacokinetics (PK) from a systemic administration is achievable. Nanoparticles should be larger than ~10 nm to avoid single-pass renal clearance and not be positively charged to any great extent (minimizing nonspecific interactions with proteins and cells) in order to allow these PK manipulations. The particles can be tuned to provide long or short circulation times, and with careful control of size and surface properties, they can be directed to specific cell types within target organs (e.g., hepatocytes versus Kupffer cells in the liver). Other types of therapeutics, such as molecular conjugates (e.g., antibody-drug conjugates), can also meet these minimum specifications, but targeted nanoparticles are distinguished from all other therapeutic entities by at least four features:
The nanoparticle can carry a very large “payload.” For example, a 70-nm nanoparticle can contain ~2000 siRNA drug molecules (73) whereas antibody conjugates have <10 (74). The nanoparticle payloads are located within the particle and do not participate in the control over PK and biodistribution. In molecular conjugates, by contrast, the type and number of therapeutic entities conjugated to the targeting ligand (e.g., an antibody) significantly modify the overall properties of the conjugate.
Nanoparticles are sufficiently large to contain multiple targeting ligands that can provide multivalent binding to cell surface receptors (75). Nanoparticles have two parameters for tuning the binding to target cells: (a) the affinity of the targeting moiety and (b) the densities of the targeting moiety. The multivalency effects can lead to very high “effective” affinities when arrangements of low-affinity ligands are used (75–77). Thus, the repertoire of molecules that can be used as targeting agents is greatly expanded, since many low-affinity ligands can be installed on nanoparticles to create higher affinity via multivalent binding to cell surface receptors.
Nanoparticles are sufficiently large to accommodate multiple types of drug molecules. Numerous therapeutic interventions can be simultaneously applied with a nanoparticle in a controlled manner.
Nanoparticles bypass multidrug resistance mechanisms that involve cell surface protein pumps, e.g., glycoprotein P, because they enter cells via endocytosis.
These properties provide the opportunity to create therapeutic strategies not possible with non-nanoparticle drugs. A controlled combination of these features can minimize side effects while enhancing drug efficacy, and offers the potential to treat drug-resistant disease if the resistance is from cell surface pumps. Clinical results are emerging that suggest nanoparticle therapeutics will lead to new methods of treatment for cancer.
Therapeutics that are now classified as nanoparticles have existed for some time. Table 1 lists nano-scaled systems for systemic therapy and their latest stage of development. Liposomes carrying chemotherapeutic small-molecule drugs have been approved since the mid-1990s. Liposomes (~100 nm and larger) can give extended circulation times if they are stabilized (see Doxil® in Table 2) but do not provide intracellular delivery of drug molecules (78). Thus, they are not effective against disease that is resistant to cell surface pumps. Additionally, they provide no control for the time of drug release. Their use is mainly in solubilizing drugs and extending circulation times to favor higher tumor uptake of drugs. Albumin-based nanoparticles were approved by the US Food and Drug Administration in 2005 (79), but are not nanoparticle therapeutics, in that they dissolve upon administration into the circulatory system (note PK parameters for Abraxane versus Taxol® in Table 2). Nanocrystals of drug molecules are also approved for oral administration; however, these nanoparticles never reach the bloodstream. These first approved nanoparticle formulations prove that nanoparticle-based therapeutics can safely be administered to patients and can enhance the safety and efficacy of other drug molecules. However, newer nanoparticle systems have great advantages over these early nanoparticle products.
Table 1.
Nano-scaled systems for systemic therapy
| Platform | Latest stage of development | Examples |
|---|---|---|
| liposomes | FDA approved | DaunoXome, Doxil® |
| albumin-based particles | FDA approved | Abraxane |
| nanocrystals | FDA approved | Rapamune (oral), Emend (oral) |
| polymeric micelles | clinical trials | Genexol-PM, SP1049C, NK911, NK012 |
| polymer-based particles | clinical trials | XYOTAX, IT-101, CT-2106, AP5346 |
| dendrimers | preclinical | polyamidoamine (PAMAM) |
| inorganic or other solid particles | preclinical | carbon nanotubes, silica particles, gold particles |
Table 2.
Comparison of pharmacokinetics (human) of small-molecule drugs to nanoparticle therapeutics
| Name | Formulationa | Diameter (nm) | t1/2 (h) | CL (ml/min·kg) | Reference |
|---|---|---|---|---|---|
| DOX | 0.9% NaCl | — | 0.8 | 14.4 | 80 |
| SP1049C | pluronic micelle/DOX | 22–27 | 2.4 | 12.6 | 80 |
| NK911 | PEG-Asp micelle/DOX | 40 | 2.8 | 6.7 | 80 |
| Doxil® | PEG-liposome/DOX | 80–90 | 84.0 | 0.02 | 80 |
| Taxol® | Cremophor® EL | — | 21.8 (20.5) | 3.9 (9.2) | 79, 80 |
| Genexol | PEG-PLA micelle/paclitaxel | 20–50 | 11.0 | 4.8 | 80 |
| Abraxane | albumin/paclitaxel | 120b | 21.6 | 6.5 | 79 |
| XYOTAX | PG/paclitaxel | ? | 70–120 | 0.07–0.12 | 82 |
| LE-SN38 | liposome/SN-38 | ? | 7–58 | 3.5–13.6 | 83 |
| CT-2106 | PG/CPT | ? | 65–99 | 0.44 | 84 |
| IT-101 | CD polymer/CPT | 30–40 | 38 | 0.03 | 85 |
Abbreviations: DOX, doxorubucin; PEG-PLA, block copolymer of polyethylene glycol-poly(L-lactic acid); PG, polyglutamic acid; CPT, camptothecin; CD polymer, cyclodextrin-containing polymer.
Dissolves upon exposure to blood.
Table 2 compares some nanoparticle-based therapeutics to the drug molecules that they are carrying. The types of particles include liposomes, polymer micelles, and polymer-based nanoparticles. For each case, e.g., DOX versus SP1049C, NK911 and Doxil®, the nanoparticle alters the PK properties of the drug molecule. The listed circulation half-lives are difficult to compare because different models are used for their determination. Clearance rate (the volume of blood/plasma cleared of the drug per time; lower clearance rates indicate higher circulation times) is a common and available PK parameter, and it is a better indicator of circulation differences among these therapeutics. Dramatically reduced clearance rates have been obtained with nanoparticles, e.g., Doxil®, XYOTAX, CT-2106, and IT-101. These nanoparticles can provide longer circulation times that allow them to adequately interrogate the body for the presence of tumors if in fact they extravasate into tumors. Small particles like polymeric micelles (<100 nm) have been shown to accumulate more readily in tumors than the larger liposomes (80). Additionally, movement of a particle throughout a tumor is also size-dependent. It is speculated that nanoparticles between 10 and 100 nm in diameter will be optimal for tumor penetration. Therefore, careful control of size will be important to the PK, biodistribution, tumor accumulation, and tumor penetration. Some of the nanoparticles that are now in clinical testing also have mechanisms to control the release of the drug. These mechanisms rely on cleavage of a chemical bond between the particle and the drug by (a) hydrolysis, (b) enzymes that are located within and outside of cells (e.g., esterases), or (c) enzymes that are located only within cells (e.g., cathepsin B). This feature is designed into the structures of polymer micelle-based (NK911 and NK012) and polymer-based (XYTOTAX, CT-2106, and IT-101) nanoparticles. Finally, some of the newer nanoparticles (e.g., IT-101) enter tumor cells and are thus effective against tumors that are resistant to the drug via surface pump mechanisms. Results from clinical trials with the newer nanoparticle-based experimental therapeutics are confirming that common side effects of the drug molecules used can be altered or reduced. Vastly improved side-effect profiles are emerging from these nanoparticle treatments.
The nanoparticles listed in Table 2 reach tumors by passive targeting, meaning they accumulate in tumors because the leaky vasculature of tumors allows the nanoparticles to extravasate while normal vasculature does not (this property partially accounts for the difference in biodistribution between nanoparticles and drug molecules). Active targeting via the inclusion of a targeting ligand on the nanoparticles is envisioned to provide the most effective therapy. Table 3 lists the very few ligand-targeted therapeutics that are either approved or in the clinic. PK-2 can be considered the first ligand-targeted nanoparticle to reach the clinic. The galactose ligand was used to target the asialoglycoprotein receptor (ASGPR) that is expressed on hepatocytes in hopes that it was still highly expressed on primary liver cancer cells. However, because the ASGPR is expressed on healthy hepatocytes, the targeted nanoparticles accumulated in normal liver as well as in the tumor. As of mid-2007, MBP-426 and SGT-53 are the only targeted nanoparticle in the clinic. Clinical trials of CALAA-01 are planned to begin in late 2007. All three of these nanoparticles (liposomal delivery of a small-molecule chemotherapeutic, liposomal delivery of plasmid DNA, and polymer delivery of siRNA) use the human protein transferrin to target the transferrin receptors on cancer cells. Transferrin receptor is known to be upregulated in many types of cancer. A targeted nanoparticle can be very multifunctional [e.g., CALAA-01 is a targeted nanoparticle that has high drug (siRNA) pay-load per targeting ligand, proven multivalent binding to cancer cell surfaces, and an active drug (siRNA) release mechanism that is triggered upon intracellular localization (73, 81)], and it is expected that these new nanoparticles should perform in superior ways to the older, less functional nanoparticles.
Table 3.
Ligand-targeted therapeutic agents
| Name | Targeting agent | Therapeutic agent | Status | Reference |
|---|---|---|---|---|
| Mylotarg (antibody-drug) | humanized antibody anti-CD33 | calicheamicin | FDA approved | 86 |
| Ontak (fusion protein) | IL-2 | diphtheria toxin fragment | FDA approved | 87 |
| Zevalin (radioimmunotherapy) | mouse antibody anti-CD20 | 90Y | FDA approved | 88 |
| Bexxar (radioimmunotherapy) | mouse antibody anti-CD20 | 131I | FDA approved | 88 |
| PK-2 (polymer-drug) | galactose | DOX | Phase I (stopped) | 89 |
| MBP-426 (liposome-drug) | transferrin | oxaliplatin | Phase I | 90 |
| SGT-53 liposome-plasmid DNA | antibody fragment to transferrin receptor | plasmid DNA with p53 gene | Phase I | 91 |
| CALAA-01 (polymer-siRNA) | transferrin | siRNA | Phase I (planned for 2007) | 81 |
Increasingly sophisticated nanoparticles are reaching the clinic, with trial results already inspiring enthusiasm for this type of therapeutic modality. This is only the beginning. The nanoparticle provides the ability to design and tune properties in ways not possible with other types of therapeutics. Thus, as more clinical data become available, the nanoparticle approach will become better and better as the optimal properties will be elucidated from previous experiences in humans. The better side-effect profiles enabled by the newer nanoparticles are already improving the quality of life for patients, and there is hope for even more improvement in the future. It is not unreasonable to predict the design of nanoparticle therapeutics sufficiently nontoxic for prophylactic use. Such nanotherapeutics would provide a powerful companion to very early in vitro diagnostic detection.
Footnotes
Statistics related to cancer deaths, cases diagnosed, etc., for years 1997–2007 are available from the American Cancer Society at http://www.cancer.org/docroot/stt/stt0.asp.
For example, Lumera sells an SPR product for evaluating ~103 protein-protein interactions simultaneously (see http://www.lumera.com/Bioscience/Products/ProteomicProcessor.php).
DISCLOSURE STATEMENT
Dr. Davis is a consultant and has stock in Calando Pharmaceuticals and Insert Therapeutics.
Dr. Heath is on the board of Homestead Clinical Corp., which has obtained licensing rights to the DEAL technology.
LITERATURE CITED
- 1.Deleted in proof
- 2.Weinberg R. The Biology of Cancer. New York: Garland Sci; 2006. [Google Scholar]
- 3.Htoy S. New additions to the oncology arsenal: Assessing their place in therapy. Formulary. 2001;36:838–52. [Google Scholar]
- 4.Lombardo LJ, Lee FY, Chen P, et al. Discovery of N-(2-chloro-6-methylphenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–61. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 5.Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 6.Hughes T, Branford S. Molecular monitoring of chronic myeloid leukemia. Semin Hemat. 2003;40:62–68. doi: 10.1053/shem.2003.50044. [DOI] [PubMed] [Google Scholar]
- 7.Price ND, Trent J, El-Naggar AK, et al. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leimyosarcomas. PNAS. 2007;104:3414–19. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang F, Reeves K, Han X, et al. Identification of candidate molecular markers predicting sensitivity in solid tumors to Dasatinib: rationale for patient selection. Cancer Res. 2007;67:2226–38. doi: 10.1158/0008-5472.CAN-06-3633. [DOI] [PubMed] [Google Scholar]
- 9.Radich JP, Dai HY, Mao M, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. PNAS USA. 2006;103:2794–99. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 11.Waldherr C, Mellinghoff IK, Tran C, et al. Monitoring antiproliferative responses to kinase inhibitor therapy in mice with 3′-deoxy-3′-F-18-fluorothymidine PET. J Nucl Med. 2005;46:114–20. [PubMed] [Google Scholar]
- 12.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman & Hall/CRC; 2007. [Google Scholar]
- 13.Lin B, White JT, Lu W, et al. Evidence for the presence of disease-perturbed networks in prostate cancer cells by genomic and proteomic analyses: a systems approach to disease. Cancer Res. 2005;65:3081–91. doi: 10.1158/0008-5472.CAN-04-3218. [DOI] [PubMed] [Google Scholar]
- 14.Shiio Y, Suh KS, Lee H, et al. Quantitative proteomic analysis of Myc-induced apoptosis—a direct role for Myc induction of the mitochondrial chloride ion channel, mtCLIC/CLIC4. J Biol Chem. 2006;281:2750–56. doi: 10.1074/jbc.M509349200. [DOI] [PubMed] [Google Scholar]
- 15.Hwang D, Rust AG, Ramsey JM, et al. A data integration metholodology for systems biology. Proc Natl Acad Sci USA. 2005;102:17296–301. doi: 10.1073/pnas.0508647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hood L, Heath JR, Phelps ME, et al. After the genome project: systems biology and new technologies enable predictive and preventive medicine. Science. 2004;306:640–43. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 17.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–97. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 18.Druker BJ. Molecularly targeted therapy: Have the floodgates opened? Oncologist. 2004;9:357–60. doi: 10.1634/theoncologist.9-4-357. [DOI] [PubMed] [Google Scholar]
- 19.Wadleigh M, DeAngelo DJ, Griffin JD, et al. After chronic myelogenous leukemia: tyrosine kinase inhibitors in other hematologic malignancies. Blood. 2005;105:22–30. doi: 10.1182/blood-2003-11-3896. [DOI] [PubMed] [Google Scholar]
- 20.Carter TA, Wodicka LM, Shah NP, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci USA. 2005;102:1011–16. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 22.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 23.Engvall E, Perlmann PO. Enzyme-linked immunosorbent assay, ELISA. 3 Quantitation of specific antibodies by enzyme-labeled anti-immunoglobulin in antigen-coated tubes. J Immunol. 1972;109:129–35. [PubMed] [Google Scholar]
- 24.Weller TH, Coons AH. Fluorescent antibody studies with agents of varicella and herpes zoster propagated in vitro. Proc Soc Exp Biol. 1954;86:789–94. doi: 10.3181/00379727-86-21235. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Cheng X, Richter M, et al. A sensitive and high-throughput assay to detect low-abundance proteins in serum. Nat Med. 2006;12:473–77. doi: 10.1038/nm1378. [DOI] [PubMed] [Google Scholar]
- 26.Schweitzer B, Wiltshire S, Lambert J, et al. Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. PNAS USA. 2000;97:10113–19. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredriksson S, Dixon W, Ji H, et al. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 2007;4:327–29. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 28.Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2003;295:1503–6. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 29.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–86. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 30.Waters LC, Jacobsen SC, Kroutchinina N, et al. Microchip device for cell lysis, multiplex PCR amplification, and electrophoretic sizing. Anal Chem. 1998;70:158–62. doi: 10.1021/ac970642d. [DOI] [PubMed] [Google Scholar]
- 31.Xia YN, Whitesides GM. Soft lithography. Angew Chem Int Ed. 1998;37:550. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 32.Unger MA, Chou HP, Thorsen T, et al. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–16. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 33.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large scale integration. Science. 2002;298:580–84. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 34.Zimmerman M, Delamarche E, Wolf M, et al. Modeling and optimization of high-sensitivity, low-volume microfluidic-based surface immunoassays. Biomed Microdev. 2005;7:99–110. doi: 10.1007/s10544-005-1587-y. [DOI] [PubMed] [Google Scholar]
- 35.Sheehan PE, Whitman LJ. Detection limits for nanoscale biosensors. Nanolett. 2005;5:803–7. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 36.Gascoyne P, Mahidol C, Ruchirawat M, et al. Microsample preparation by dielectrophoresis: isolation of malaria. Lab Chip. 2005;2:70–75. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crowley TA, Pizzoconi V. Isolation of plasma from whole blood using planar microfilters for lab-on-a-chip applications. Lab Chip. 2005;5:922–29. doi: 10.1039/b502930a. [DOI] [PubMed] [Google Scholar]
- 38.Petersson F, Nilsson A, Holm C, et al. Continuous separation of lipid particles from erythrocytes by means of laminar flow and acoustic standing wave forces. Lab Chip. 2005;5:20–22. doi: 10.1039/b405748c. [DOI] [PubMed] [Google Scholar]
- 39.Huang LR, Cox EC, Austin RH, et al. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–90. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Undar A, Zahn JD. A microfluidic device for continuous, real time blood plasma separation. Lab Chip. 2006;6:871–80. doi: 10.1039/b516401j. [DOI] [PubMed] [Google Scholar]
- 41.Chen DS, Davis MM. Cellular immunotherapy: antigen recognition is just the beginning. Springer Semin Immunopathol. 2005;27:119–24. doi: 10.1007/s00281-005-0200-z. [DOI] [PubMed] [Google Scholar]
- 42.Lee PJ, Hung PJ, Rao VM, et al. Nanoliter scale microbioreactor array for quantitative cell biology. Biotech Bioeng. 2006;94:5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- 43.Toner M, Irimia D. Blood-on-a-chip. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rosa SC, Herzenberg LA, Herzenberg LA, et al. 11 color, 13 parameter flow cytometry: identification of native T cells by phenotype, function and T cell receptor. Nat Med. 2001;7:245–48. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 45.Fu AY, Spence C, Scherer A, et al. A microfabricated fluorescence-activated cell sorter. Nat Biotech. 1999;17:1109–11. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 46.Deleted in proof
- 47.Cardoso AA, Watt SM, Batard P, et al. An improved panning technique for the selection of CD34+ human bone marrow hematopoietic cells with high recovery of early progenitors. Exp Hemat. 1995;23:407–12. [PubMed] [Google Scholar]
- 48.Bailey RC, Kwong GA, Radu CG, et al. DNA-encoded antibody libraries: a unified platform for multiplexed cell sorting and detection of genes and proteins. J Am Chem Soc. 2007;129:1959–67. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karlsson R, Falt A. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance. J Immunol Methods. 1997;200:121–33. doi: 10.1016/s0022-1759(96)00195-0. [DOI] [PubMed] [Google Scholar]
- 50.Cui Y, Wei Q, Park H, et al. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–92. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 51.Kong J, Franklin NR, Zhou C, et al. Nanotube molecular wires as chemical sensors. Science. 2000;287:622–25. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- 52.Heath JR. Label-free nanowire and nanotube biomolecular sensors for in vitro diagnosis of cancer and other diseases. In: Mirkin C, Niemeyer C, editors. Nanobiotechnology II. Weinheim, Ger: Wiley-VCH; 2007. pp. 213–28. [Google Scholar]
- 53.Backmann N, Zahnd C, Huber F, et al. A label-free immunosensor array using single-chain antibody fragments. PNAS USA. 2005;102:14587–92. doi: 10.1073/pnas.0504917102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lang HP, Hegner M, Gerber C. Cantilever array sensors for bioanalysis and diagnostics. In: Mirkin C, Niemeyer C, editors. Nanobiotechnology II. Weinheim, Ger: Wiley-VCH; 2007. [Google Scholar]
- 55.Jung LS, Campbell CT, Chinowsky TM, et al. Quantitative interpretation of the response of surface plasmon resonance sensors to adsorbed films. Langmuir. 1998;14:5636–48. [Google Scholar]
- 56.Bunimovich YL, Shin Y-S, Yeo W-S, et al. Quantitative real-time measurements of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J Am Chem Soc. 2006;128:16323–31. doi: 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Safsten P, Klakamp SL, Drake AW, et al. Screening antibody-antigen interactions in parallel using Biacore A100. Anal Biochem. 2006;353:181–90. doi: 10.1016/j.ab.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 58.Deleted in proof
- 59.Li M, Tang HX, Roukes ML. Ultra-sensitive NEMS-based cantilevers for sensing, scanned probe and very high-frequency applications. Nat Nanotech. 2007;2:114–20. doi: 10.1038/nnano.2006.208. [DOI] [PubMed] [Google Scholar]
- 60.Patolsky F, Zheng G, Hayden O, et al. Electrical detection of single viruses. PNAS USA. 2004;101:14017–22. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McAlpine M, Ahmad H, Wang D, et al. Highly ordered nanowire arrays on plastic substrates for ultrasensitive flexible chemical sensors. Nat Mat. 2007;6:379. doi: 10.1038/nmat1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li C, Curreli M, Lin H, et al. Complementary detection of prostate-specific antigen using ln2O3 nanowires and carbon nanotubes. J Am Chem Soc. 2005;127:12484–85. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- 63.Zheng G, Patolsky F, Cui Y, et al. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotech. 2005;23:1294–301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 64.Israelachvili J. Intermolecular and Surface Forces. London: Academic; 1985. [Google Scholar]
- 65.Javois LC, editor. Immunocytochemical Methods and Protocols. 2 Totowa, NJ: Humana; 1999. [Google Scholar]
- 66.Yezhelyev MV, Gao X, Xing Y, et al. Emerging use of nanoparticles in diagnosis and treatment of breast cancer. Lancet Oncol. 2006;7:657–67. doi: 10.1016/S1470-2045(06)70793-8. [DOI] [PubMed] [Google Scholar]
- 67.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 68.Liu X, Wang H, Herron J, et al. Photopatterning of antibodies on biosensors. Bioconjug Chem. 2000;11:755. doi: 10.1021/bc000006d. [DOI] [PubMed] [Google Scholar]
- 69.Macbeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–63. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 70.Pal M, Moffa A, Sreekumar A, et al. Differential photophoprotein mapping in cancer cells using protein microarrays produced from 2-D liquid fractionation. Anal Chem. 2006;78:702–10. doi: 10.1021/ac0511243. [DOI] [PubMed] [Google Scholar]
- 71.Thirumalapura NR, Morton RJ, Ramachandran A, et al. Lipopolysaccharide microarrays for the detection of antibodies. J Immunol Methods. 2005;298:73–81. doi: 10.1016/j.jim.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Soen Y, Chen DS, Kraft DL, et al. Detection and characterization of cellular immune responses using peptide-MHC microarrays. PLoS Biol. 2003;1:429–38. doi: 10.1371/journal.pbio.0000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bartlett DW, Davis ME. Physicochemical and biological characterization of targeted, nucleic acid-containing nanoparticles. Bioconjug Chem. 2007;18:456–68. doi: 10.1021/bc0603539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–17. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 75.Hong S, Leroueil PR, Majoros IJ, et al. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14:107–15. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 76.Montet X, Funovics M, Montet-Abou K, et al. Multivalent effects of RGD peptides obtained by nanoparticle display. J Med Chem. 2006;49:6087–93. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 77.Carlson CB, Mowery P, Owen RM, et al. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2:119–27. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 78.Zamboni WC. Liposomal, nanoparticle, and conjugated formulation of anticancer agents. Clin Cancer Res. 2005;11:8230–34. doi: 10.1158/1078-0432.CCR-05-1895. [DOI] [PubMed] [Google Scholar]
- 79.Sparreboom A, Scripture CD, Trieu V, et al. Comparative preclinical and clinical pharmacokinetics of a Cremophor-free, nanoparticle albumin-bound paclitaxel (ABI-007) and paclitaxel formulated in Cremophor (Taxol) Clin Cancer Res. 2005;11:4136–43. doi: 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
- 80.Sutton D, Nasongkla N, Blanco E, et al. Functionalized micellar systems for cancer targeted drug delivery. Pharmaceut Res. 2007;24:1029–46. doi: 10.1007/s11095-006-9223-y. [DOI] [PubMed] [Google Scholar]
- 81.Heidel JD, Yu Z, Liu JYI, et al. Administration in nonhuman primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci USA. 2007;104:5715–21. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boddy AV, Plummer ER, Todd R, et al. A phase I and pharmacokinetic study of paclitaxel poliglumex (XYOTAX), investigating both 3-weekly and 2-weekly schedules. Clin Cancer Res. 2005;11:7834–40. doi: 10.1158/1078-0432.CCR-05-0803. [DOI] [PubMed] [Google Scholar]
- 83.Kraut EH, Fishman MN, LoRusso PM, et al. Final results of a phase I study of liposome encapsulated SN-38 (LE-SN38): safety, pharmacogenomics, pharmacokinetics, and tumor response. Poster presented at Annu. Meet. Am. Soc. Clin. Oncol., 41st; Orlando, FL. 2005. [Google Scholar]
- 84.Daud A, Garrett C, Simon GR, et al. Phase I trial of CT-2106 (polyglutamated camptothecin) administered weekly in patients with advanced solid tumor malignancies. Poster presented at Annu. Meet. Am. Soc. Clin. Oncol., 42nd; Atlanta, GA. 2006. [Google Scholar]
- 85.Yen Y, Synold T, Schluep T, et al. First-in-human phase I trial of a cyclodextrin-containing polymer-camptothecin nanoparticle in patients with various solid tumors. Presented at Annu. Meet. Am. Soc. Clin. Oncol., 43rd; Chicago, IL. 2007. [Google Scholar]
- 86.Bross P, Beitz J, et al. Approval summary: gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin Cancer Res. 2001;7:1490–96. [PubMed] [Google Scholar]
- 87.Kawakami K, Nakajima O, Morishita R, et al. Targeted anticancer immunotoxins and cytotoxic agents with direct killing moieties. Sci World J. 2006;6:781–90. doi: 10.1100/tsw.2006.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–63. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 89.Duncan R. Polymer conjugates as anticancer nanomedicines. Nat Rev Cancer. 2006;6:688–701. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 90.Safety study of MBP-426 (liposomal oxaliplatin suspension for injection) to treat advanced or metastatic solid tumors. Clinical Trials.gov. 2006 Identifier NCT00355888. http://www.clinicaltrials.gov/ct/show/NCT00355888.
- 91.2007. Safety Study of Infusion of SGT-53 to Treat Solid Tumors. Clinical Trials.gov. Identifier NCT0040613. http://www.clinicaltrials.gov/ct/show/NCT00355888.