SUMMARY
The transmembrane protein CD33 is a sialic acid-binding immunoglobulin-like lectin that regulates innate immunity but has no known functions in the brain. We have previously shown that the CD33 gene is a risk factor for Alzheimer’s disease (AD). Here, we observed increased expression of CD33 in microglial cells in AD brain. The minor allele of the CD33 SNP rs3865444, which confers protection against AD, was associated with reductions in both CD33 expression and insoluble amyloid beta 42 (Aβ42) levels in AD brain. Furthermore, the numbers of CD33-immunoreactive microglia were positively correlated with insoluble Aβ42 levels and plaque burden in AD brain. CD33 inhibited uptake and clearance of Aβ42 in microglial cell cultures. Finally, brain levels of insoluble Aβ42 as well as amyloid plaque burden were markedly reduced in APPSwe/PS1ΔE9/CD33−/− mice. Therefore, CD33 inactivation mitigates Aβ pathology and CD33 inhibition could represent a novel therapy for AD.
INTRODUCTION
Alzheimer’s disease (AD) is the most prevalent neurodegenerative disease and the leading cause of dementia among the elderly. The mechanisms underlying the onset and progression of neurodegeneration and cognitive decline are incompletely understood. A major breakthrough in our understanding of AD was the identification of gene mutations associated with rare familial AD (FAD) cases. Autosomal dominant mutations in the amyloid beta (A4) precursor protein (APP) and presenilin 1 and 2 (PSEN1/2) genes greatly accelerate the rate of cognitive decline leading to early-onset dementia (Bertram et al., 2010; Tanzi, 2013). The vast majority of AD cases, however, are late-onset (LOAD) forms, which lack an obvious Mendelian inheritance pattern. LOAD has a strong genetic component and is probably caused by a combination of multiple risk alleles, each with modest and partially penetrant effects, and environmental factors (Bertram et al., 2010). Although apolipoprotein E ε4 (APOE ε4) remained for a long time the only confirmed genetic risk factor for LOAD, it accounts for only 10%–20% of the LOAD risk, suggesting the existence of additional risk factors (Liu et al., 2013). Recently, genome-wide association studies (GWAS) performed on extended cohorts (thousands of individuals) led to the identification of additional confirmed genetic risk factors for AD: CD33 (Bertram et al., 2008; Hollingworth et al., 2011; Naj et al., 2011), CLU, BIN1, PICALM, CR1, CD2AP, EPHA1, ABCA7, MS4A4A/MS4A6E (Harold et al., 2009; Hollingworth et al., 2011; Lambert et al., 2009; Naj et al., 2011; Seshadri et al., 2010), and TREM2 (Guerreiro et al., 2013; Jonsson et al., 2013). Understanding the molecular and cellular activities of these novel genes, as well as their functional interactions, should greatly advance our understanding of AD.
The deposition of amyloid beta (Aβ)-containing plaques is a pathological hallmark of both FAD and LOAD. Aβ results from the amyloidogenic processing of APP, which is cleaved by the sequential action of β-secretase/BACE1 and γ-secretase/Presenilin (Querfurth and LaFerla, 2010). In FAD, both APP and PSEN1/2 mutations lead to enhanced amyloidogenic processing of APP and enhanced production of the toxic Aβ42 species (Querfurth and LaFerla, 2010). Less is known about the mechanisms of Aβ formation, self-assembly, and clearance in LOAD. Interestingly, several genes linked to LOAD have been shown to impact Aβ generation, aggregation, or clearance (Bertram et al., 2010), suggesting that Aβ dysregulation is a central pathogenic mechanism in LOAD. A widely accepted model of AD pathogenesis is the “amyloid hypothesis,” whereby increased production and self-assembly of Aβ toxic species initiates a series of progressive changes that ultimately lead to neurodegeneration (Hardy and Selkoe, 2002; Hardy and Higgins, 1992; Tanzi and Bertram, 2005). In this hypothesis, persistent Aβ proteotoxic stress triggers the hyperphosphorylation and aggregation of the microtubule-associated protein tau, leading to neurofibrillary tangles, another pathological hallmark of AD (Tanzi and Bertram, 2005). Therefore, a better understanding of the mechanisms that regulate the generation and deposition, as well as clearance, of Aβ might improve the therapeutic approaches in AD.
Two SNPs in the CD33, rs3826656 (Bertram et al., 2008) and rs3865444 (Hollingworth et al., 2011; Naj et al., 2011), have been associated with LOAD. The 67 kDa type 1 transmembrane protein CD33 (Siglec-3) is a member of the sialic acid-binding immunoglobulin-like lectins (Siglecs) and is expressed in immune and hematopoietic cells. The Siglecs recognize sialic acid residues of glycoproteins and glycolipids, have one or more immunoreceptor tyrosine-based inhibition motif (ITIM), and mediate cell-cell interactions that inhibit or restrict immune responses (Crocker et al., 2012; Pillai et al., 2012). CD33 activity has been implicated in several processes, such as adhesion processes in immune or malignant cells, endocytosis, inhibition of cytokine release by monocytes, and immune cell growth (Crocker et al., 2007; von Gunten and Bochner, 2008). To date, no functions have been described for CD33 in the brain.
Here, we show that CD33 is expressed in microglial cells in the human brain. CD33 protein levels as well as the number of CD33-positive microglia are increased in AD brains relative to age-matched controls. Conversely, we show that the minor allele of the CD33 SNP rs3865444, which protects against AD, leads to reductions in both CD33 microglial expression and levels of insoluble Aβ42 in AD brain. Furthermore, the numbers of CD33-immunoreactive microglia positively correlate with insoluble Aβ42 levels and the amyloid plaque burden in AD cases. Using cultured primary and BV2 microglial cells, we show that CD33 is both required and sufficient to inhibit the microglial uptake of Aβ42, thus impairing Aβ42 clearance. Finally, APPSwe/PS1ΔE9 transgenic mice in which the CD33 gene was knocked out exhibited a marked reduction of insoluble Aβ42 levels and Aβ plaque burden, indicating that CD33 promotes the Aβ42 pathology in vivo. Collectively, these results suggest that CD33 activity in microglia promotes Aβ42 pathology in AD. They also raise the possibility that the loss of microglial-degradative capacity of Aβ in AD could be reversed therapeutically by inhibition of CD33 activity.
RESULTS
Increased CD33 Expression in AD
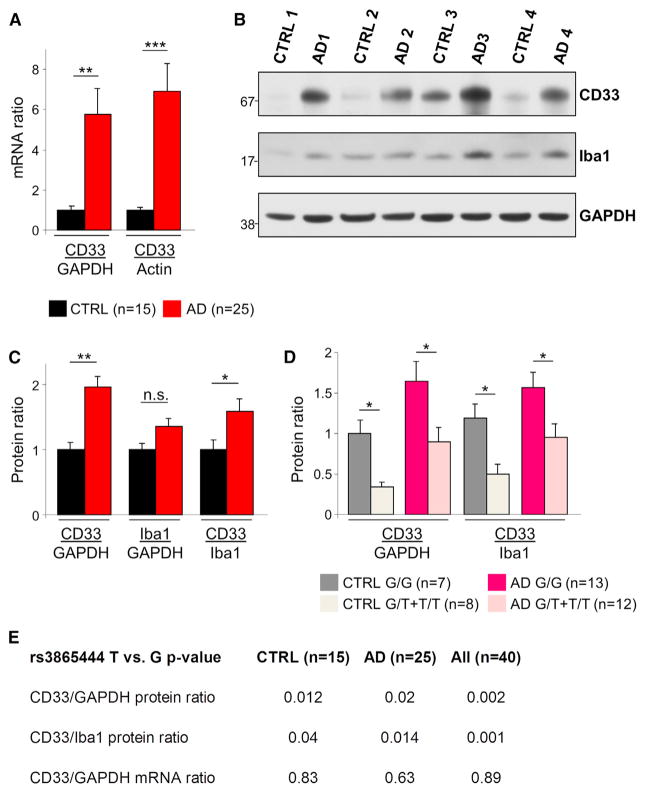
To assess the role of CD33 in AD pathology, we initially assessed the expression of CD33 in postmortem brain samples from 25 AD patients and 15 age-matched nondemented controls (cohort characteristics in Table S1, available online). To investigate the relationship between CD33 mRNA levels and AD, we performed quantitative real-time PCR on total mRNA extracted from frozen cortical samples. This revealed a 5-fold increase in CD33 mRNA levels in AD cases relative to controls (Figure 1A, p < 0.01, Student’s t test). Normalization of CD33 mRNA levels using GAPDH and β-Actin mRNAs led to similar results (Figure 1A). Next, we asked whether CD33 protein levels are increased in the frontal cortex in AD. Western blotting using a CD33-specific antibody (Hoyer et al., 2008; Rollins-Raval and Roth, 2012) revealed a 2-fold increase in CD33 protein levels in AD samples relative to controls (Figures 1B and 1C, p < 0.01, Student’s t test). This significant increase was also observed when normalizing CD33 levels to the levels of the microglial marker Iba1 (Figure 1C, p < 0.05, Student’s t test), indicating that this difference is not explained by an increased number of microglia in AD. Using stereology-based quantitative methods, we have previously observed that the total number of Iba1-positive microglia is not significantly different between AD and nondemented subjects (A.S.-P., T. Gomez-Isla, J.H. Growdon, M.P. Frosch, and B.T.H., unpublished data). Thus, CD33 mRNA and protein levels are increased in AD brain.
Figure 1. Increased CD33 Expression in AD.
(A) Quantitative real-time PCR analysis of CD33 expression in the frontal cortex reveals increased CD33 mRNA levels in AD relative to control subjects; CD33 mRNA levels have been normalized to GAPDH or β-Actin mRNA.
(B) Western blotting detection of CD33 in the frontal cortex reveals a marked upregulation in AD cases relative to age-matched controls. A less pronounced increased expression is also seen for Iba1. GAPDH served as loading control.
(C) The CD33/GAPDH and CD33/Iba1 protein ratios are increased in AD patients relative to age-matched controls.
(D) The levels of CD33 protein are decreased in carriers of the protective minor (T) allele of the CD33 SNP rs3865444. Bar graph shows CD33 protein levels in individuals from the indicated groups (CTRL or AD) and genotypes (G/G versus G/T or T/T). For (A)–(D), *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test. Data are represented as mean ± SEM.
(E) Analysis based on general linear regression model reveals that the protective minor (T) allele of rs3865444 is associated with decreased CD33 protein, but not mRNA levels, in both controls and AD cases (p < 0.05 was considered statistically significant). See also Figure S1 and Table S1.
The rs3865444 SNP in the CD33 gene was previously reported to confer protection against AD (Hollingworth et al., 2011; Naj et al., 2011). Thus, we investigated whether CD33 expression differed in carriers of the major (G) allele, relative to the carriers of the minor protective (T) allele. All subjects were genotyped by the Taqman assay using sequence-specific primers to differentiate between the alleles (Shen et al., 2009). We found that the minor protective (T) allele is not associated with changes in CD33 mRNA levels (Figure 1E; p > 0.05, general linear regression model). We used different sets of primers to amplify different regions of the CD33 mRNA, with similar results (Figures S1A and S1B). However, remarkably, carriers of the minor (T) allele had significantly reduced CD33 protein levels (normalized to GAPDH or Iba1 protein levels), in both AD and control groups (Figure 1D; p < 0.05, Student’s t test and Figure S1C). We also found that the protective (T) allele is associated with decreased CD33 protein levels (normalized to GAPDH or Iba1 protein levels) in both control and AD groups (Figure 1E, p < 0.05, general linear regression model). Thus, although the rs3865444 SNP is located on chromosome 19 at the 51,727,962 base pair (bp) position, upstream of the 5′ UTR of the CD33 gene (51,728,335-51,743,274 bp, forward strand) (Hollingworth et al., 2011; Naj et al., 2011), it does not affect CD33 mRNA stability but somehow influences mRNA translation or protein stability. One possibility is that the rs3865444 SNP is in linkage disequilibrium with functional variant(s) located in the coding region. The observations of increased CD33 expression in the AD brain and the decreased CD33 protein levels in the carriers of the protective allele of the CD33 SNP rs3865444 strongly suggest a role for CD33 in AD pathogenesis.
Microglial Localization of CD33 and Relationship to AD
CD33 has previously been shown to be expressed in cells of the immune and hematopoietic cell systems (Crocker et al., 2007). Microglial cells are responsible for the immune surveillance of the brain and regulate critical processes relevant to AD pathology, including the uptake and clearance of Aβ (Aguzzi et al., 2013; Prinz et al., 2011). Thus, we next asked whether CD33 is expressed in microglial cells. Immunolabeling of control and AD frontal cortex sections, using a CD33-specific antibody (Hoyer et al., 2008; Rollins-Raval and Roth, 2012) and the microglial marker Iba1 revealed a good colocalization between the two proteins (Figures 2A and 2C). CD33 was also expressed in neurons (Figures S2D and S2E) but not in astrocytes, oligodendrocytes, or endothelial cells (Figures S2G–S2I).
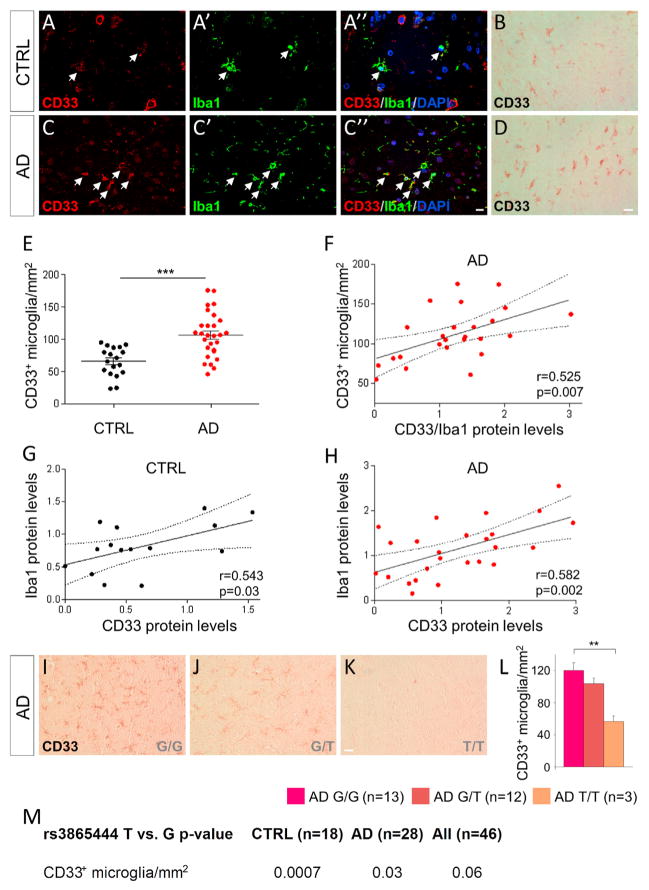
Figure 2. Increased Number of CD33-Immunoreactive Microglia in AD.
(A–A″ and C–C″) Fluorescent immunolabeling reveals colocalization between CD33 (red) and the microglial marker Iba1 (green) in the frontal cortex of CTRL (A–A″) and AD (C–C″) subjects.
(B and D) CD33 labeling using diaminobenzidine reveals numerous microglial cells that are positive for CD33.
(E) Stereology-based quantification reveals increased numbers of CD33-positive microglial cells in the frontal cortex of AD cases (n = 28) relative to controls (n = 18), ***p < 0.001, Student’s t test.
(F) CD33 protein levels normalized to Iba1 protein levels positively correlate with the numbers of CD33-immunoreactive microglia in the AD cases (n = 25; r = 0.525; p = 0.007, Pearson’s correlation test).
(G and H) CD33 protein levels correlate with the levels of the microglial marker Iba1, both in controls (G; n = 15; r = 0.543; p = 0.03, Pearson’s correlation test) and AD (H; n = 25; r = 0.582; p = 0.002, Pearson’s correlation test) cases.
(I–K) Carriers of the protective minor (T) allele of rs386544 exhibit decreased numbers of CD33-positive microglia. Shown are representative pictures from the frontal cortex of a G/G carrier (I), G/T carrier (J), and T/T AD carrier (K), stained for CD33.
(L) Stereology-based quantifications reveal reduced numbers of CD33-positive microglia in carriers of two protective (T) alleles of rs3865444 (**p < 0.01 T/T carriers versus G/G carriers, one-way Kruskal-Wallis ANOVA, Dunn’s test). Data are represented as mean ± SEM. Scale bars represent 25 μm.
(M) The protective (T) allele of rs3865444 is associated with decreased numbers of CD33-immunoreactive microglia in both controls and AD cases (general linear regression model, p < 0.05 was considered statistically significant). See also Figure S2.
Next, we asked whether the number of CD33-immunoreactive cells differed between AD and control brains. Sections immunolabeled for CD33 and stained with diaminobenzidine (DAB) were subjected to stereology-based quantifications, using previously described protocols (Serrano-Pozo et al., 2011). The total numbers of CD33-positive cells increased by 48.9% in the AD frontal cortex relative to age-matched controls (Figure S2A, n = 28 AD cases and 18 controls, p < 0.001, Student’s t test). We then identified the CD33-positive microglia using morphological criteria (Figures 2B and 2D). Stereology-based quantification of the numbers of CD33-immunoreactive microglia revealed a marked increase in the AD frontal cortex relative to age-matched controls (Figure 2E, n = 28 AD cases and 18 controls, p < 0.001, Student’s t test). We also asked whether the number of CD33-positive neurons differed between AD and control subjects. We found no significant difference between the numbers of CD33-positive neurons in AD and controls (Figures S2B and S2C). The levels of CD33 protein normalized to Iba1 protein levels positively correlated with the numbers of CD33-immunoreactive microglial cells, as expected (Figure 2F, p = 0.007, Pearson’s correlation test). To validate these findings, we assessed the relationship between the levels of CD33 protein and those of the microglial marker Iba1 using western blotting and frontal cortex protein extracts. We found a positive correlation between CD33 and Iba1 levels, both in control (Figure 2G, p = 0.03, Pearson’s correlation test) and AD (Figure 2H, p = 0.002, Pearson’s correlation test) cases.
We next asked whether the minor (T) allele of the CD33 SNP rs3865444 is associated with changes in the number of CD33-positive microglia. We found that carriers of the protective (T) allele had lower numbers of CD33-positive microglial cells (Figures 2I–2K); this effect was dose dependent, i.e., carriers of two (T) alleles exhibited a dramatic reduction of CD33-positive microglia numbers relative to carriers of one (T) allele or carriers of the major (G) allele (Figure 2L, p < 0.01 T/T versus G/G carriers, one-way Kruskal-Wallis ANOVA, Dunn’s test). We found that the minor protective (T) allele is associated with decreased CD33-immunoreactive microglia numbers in both the control and AD groups (Figure 2M, p < 0.001 and p < 0.05, respectively, general linear regression model). Therefore, the numbers of CD33-positive microglia are increased in AD cases and are reduced in carriers of two protective (T) alleles, suggesting that CD33 activity in microglia might impact the etiology and/or pathogenesis of AD.
CD33 Microglial Expression Correlates with Amyloid Beta Pathology in AD
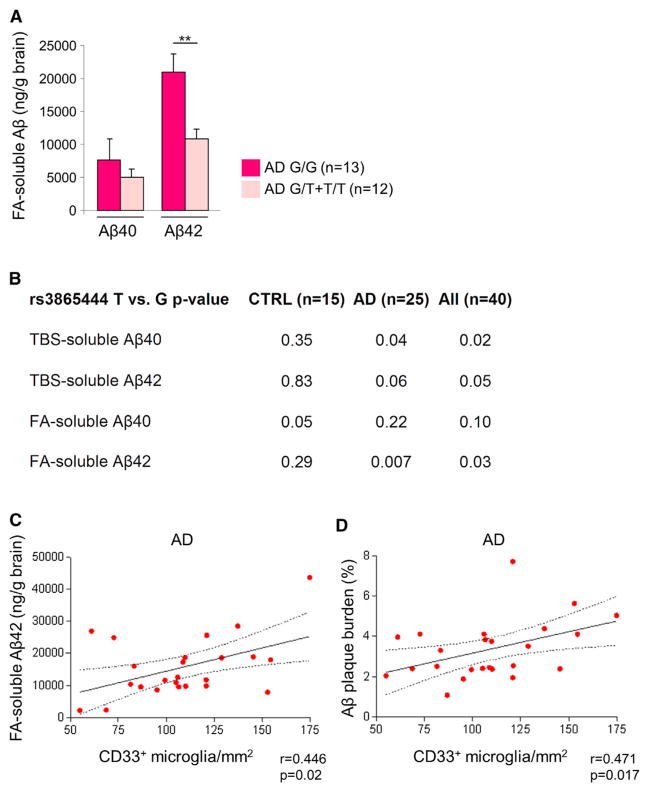
Increased production and deposition of aggregation-prone Aβ species are hallmarks of AD pathology (Selkoe, 2012; Tanzi and Bertram, 2005). We investigated whether Aβ levels were different in carriers of the major (G) allele of the CD33 SNP rs3865444 in comparison to the carriers of the protective (T) allele. We generated Tris-buffered saline (TBS)-soluble and formic acid (FA)-soluble fractions from the frontal cortex tissue of controls and AD cases (Wang et al., 2011), and we used these fractions for Aβ ELISA experiments. Remarkably, we found that the carriers of the minor (T) allele had significantly reduced FA-soluble Aβ42 levels but not FA-soluble Aβ40 in comparison to the carriers of the major (G) allele in AD (Figure 3A, p < 0.01, Student’s t test). We also observed that the minor protective (T) allele is associated with decreased levels of both TBS-soluble Aβ40 and FA-soluble Aβ42 in AD cases (Figure 3B, p < 0.05 and p < 0.01, respectively, general linear regression model).
Figure 3. CD33 Microglial Expression Positively Correlates with Formic Acid-Soluble Aβ42 Levels and Amyloid Plaque Burden in AD.
(A) Formic acid (FA)-soluble Aβ42 levels are decreased in carriers of the rs3865444 minor (T) allele. ELISA analysis of Aβ40 and Aβ42 in FA-soluble fractions isolated from the frontal cortex of AD cases of the indicated genotypes (**p < 0.01, Student’s t test). Data are represented as mean ± SEM.
(B) The protective rs3865444 (T) allele is associated with decreased levels of both FA-soluble Aβ42 and TBS-soluble Aβ40 in AD cases (n = 25 AD cases, n = 15 controls, general linear regression model, p < 0.05 was considered statistically significant).
(C) The numbers of CD33-immunoreactive microglia positively correlate with the FA-soluble Aβ42 levels in AD cases (n = 25; r = 0.446; p = 0.02, Spearman’s correlation test).
(D) The numbers of CD33-positive microglia positively correlate with amyloid plaque burden in AD brain (n = 25; r = 0.471; p = 0.017; Spearman’s correlation test). See also Figure S3.
Microglial cells regulate Aβ levels in the brain by a process of uptake and degradation, which plays a key role in AD pathogenesis (Aguzzi et al., 2013; Prinz et al., 2011). To explore the relationship between CD33 microglial expression and amyloid pathology, we labeled the AD frontal cortex with Thioflavin S (to detect amyloid plaques) and antibodies directed against CD33 and the microglial marker Iba1. This revealed a broad distribution of CD33-positive microglia throughout the AD cortex together with an enrichment of CD33-positive microglia around amyloid plaques (Figure S3). We then explored the possibility that increased CD33 microglial expression in the aging brain promotes Aβ pathology by preventing the efficient Aβ clearance. We found that the numbers of CD33-immunoreactive microglia positively correlated with the levels of FA-soluble Aβ42 in AD brain (Figure 3C, p = 0.02, Spearman’s correlation test). We then asked whether Aβ plaque burden in AD cases correlates with the numbers of CD33-positive microglia. We estimated the Aβ plaque burden in the frontal cortex of AD subjects as the proportion of area of full-width cortex occupied by Aβ-immunoreactive deposits in sections immunostained with an antibody directed against Aβ (residues 3–7, 10D5) (Serrano-Pozo et al., 2011). Remarkably, we found a positive correlation between amyloid plaque burden and numbers of CD33-immunoreactive microglial cells in AD cases (Figure 3D, p = 0.017, Spearman’s correlation test). These results suggest that increased microglial expression of CD33 prevents Aβ clearance and strongly implicate microglial CD33 function in Aβ pathology in AD brain.
CD33 Inactivation Promotes the Uptake of Amyloid Beta by Microglia
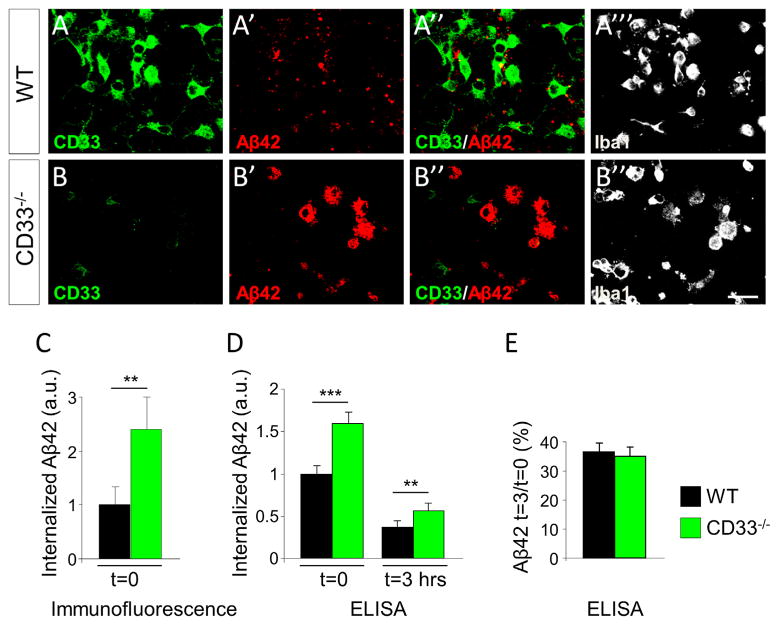
Our genetic, biochemical, and histopathological data strongly suggest that the activity of CD33 in microglia impacts the accumulation of Aβ in the brain. To test for a causal relationship between CD33 activity and Aβ pathology in AD, we employed an in vitro assay of Aβ uptake and clearance, using primary microglia isolated from mice with a constitutive inactivation of the CD33 gene and littermate wild-type (WT) controls. CD33 inactivation in mice does not lead to obvious developmental, histological, and behavioral abnormalities, and CD33−/− mice breed normally (Brinkman-Van der Linden et al., 2003). We established a mixed glial (microglia/astrocyte) primary culture using the forebrain of WT and CD33−/− postnatal day 1 (P1) pups as source of cells and subsequently enriched for microglial cells (Choi et al., 2008; Gorlovoy et al., 2009); the microglial cultures contained more than 93% microglial (Iba1-positive) cells. We did not detect any differences in proliferation, growth, and morphological parameters between WT and CD33−/− microglia (data not shown). We stained these cells with a CD33 antibody that we developed in our laboratory (Figure S4).
Microglia derived from CD33 −/− mice were not immunoreactive to the CD33 antibody, as expected (Figures 4A and 4B). The enriched microglial cultures were incubated with Aβ42 for 3 hr, which allows efficient Aβ42 uptake. Subsequently, the Aβ42 was washed out and the cells were incubated for an additional 3 hr, to allow for Aβ42 degradation (Jiang et al., 2008; Mandrekar et al., 2009). To visualize the process of Aβ42 uptake, we used fluorescently labeled Aβ42 (Lee et al., 2012). Visually, we noted a strong increase in Aβ42 levels in CD33−/− microglia relative to WT microglia (Figures 4A′ and 4B′). Quantification of the fluorescent Aβ42 signal revealed a significant increase in Aβ42 uptake in CD33−/− relative to WT cells (Figure 4C, p < 0.01, Student’s t test). The process of Aβ42 degradation cannot be assessed by imaging with fluorescently labeled Aβ42, because microglial cells degrade Aβ42 but they do not completely degrade the attached fluorophore (Mandrekar et al., 2009). To validate the Aβ42 uptake findings, and investigate whether CD33 inactivation impacts Aβ42 degradation as well, we measured Aβ42 levels by ELISA on extracts prepared from cultures incubated with unlabeled Aβ42 (Figures 4D and 4E). We found that CD33−/− microglia contained increased Aβ42 levels after 3 hr of incubation (Figure 4D), confirming our imaging findings. However, after 3 hr of Aβ42 washout, we found similar rates of Aβ42 degradation in both CD33−/− and WT microglia (Figure 4E). Together, these results suggest that CD33 directly impacts Aβ42 uptake, but not Aβ42 degradation, in microglial cells.
Figure 4. CD33 Inactivation Leads to Increased Microglial Uptake of Aβ42.
(A–A‴ and B–B‴) Primary microglial cell cultures were incubated with fluorescently labeled Aβ42 for 3 hr and then subjected to immunofluorescent labeling for CD33 (A and B) and Iba1 (A‴ and B‴). Microglial cultures derived from CD33−/− mice, at postnatal day 1, exhibited a markedly increased Aβ42 uptake (compare A′ and B′). Scale bar represents 25 μM.
(C) Quantification of Aβ42 average signal intensity in individual cells reveals a strong increase in Aβ42 signal in CD33−/− cells relative to WT cells (at least 30 cells were scored per genotype, **p < 0.01, Student’s t test).
(D and E) Microglial cell cultures were treated with unlabeled Aβ42 and incubated for 3 hr. After incubation, cells were either collected for ELISA analysis (t = 0) or washed and incubated for an additional 3 hr in Aβ-free medium, followed by ELISA analysis (t = 3 hr). (D) CD33−/− microglia exhibit increased Aβ42 uptake levels relative to WT microglia (t = 0) (results were derived from four independent experiments; ***p < 0.001, Student’s t test). (E) Similar rates of Aβ42 degradation in CD33−/− and WT microglial cells incubated in the absence of Aβ42 for an additional 3 hr (the percentage of remaining Aβ42 was the ratio of Aβ42 remaining at t = 3 hr to the total amount of Aβ42 internalized at t = 0). Data are represented as mean ± SEM. See also Figure S4.
Increased CD33 Levels Inhibit Microglial Uptake of Amyloid Beta
We next assessed whether increasing CD33 levels impairs Aβ42 uptake by microglial cells. For this purpose, we employed the BV2 microglial cell line, which was previously found to efficiently take up and degrade exogenously added Aβ42 (Jiang et al., 2008; Mandrekar et al., 2009). Cells transfected with a WT-CD33 construct displayed a decreased uptake of fluorescently labeled Aβ42 relative to cells transfected with an empty construct (Figures 5A′, 5B′, and 5E, p < 0.01, one-way ANOVA, Tukey’s test). These results were confirmed by ELISA quantifications (Figure 5F, p < 0.05, one-way ANOVA, Tukey’s test). Furthermore, ELISA also revealed that Aβ42 was degraded at similar rates by cells transfected with WT-CD33 or an empty vector (Figure 5G). Similar differences were observed when a GFP construct was used as a control (Figures 5E–5G). Thus, increasing CD33 levels is sufficient to inhibit Aβ42 uptake, but not degradation, by microglial cells.
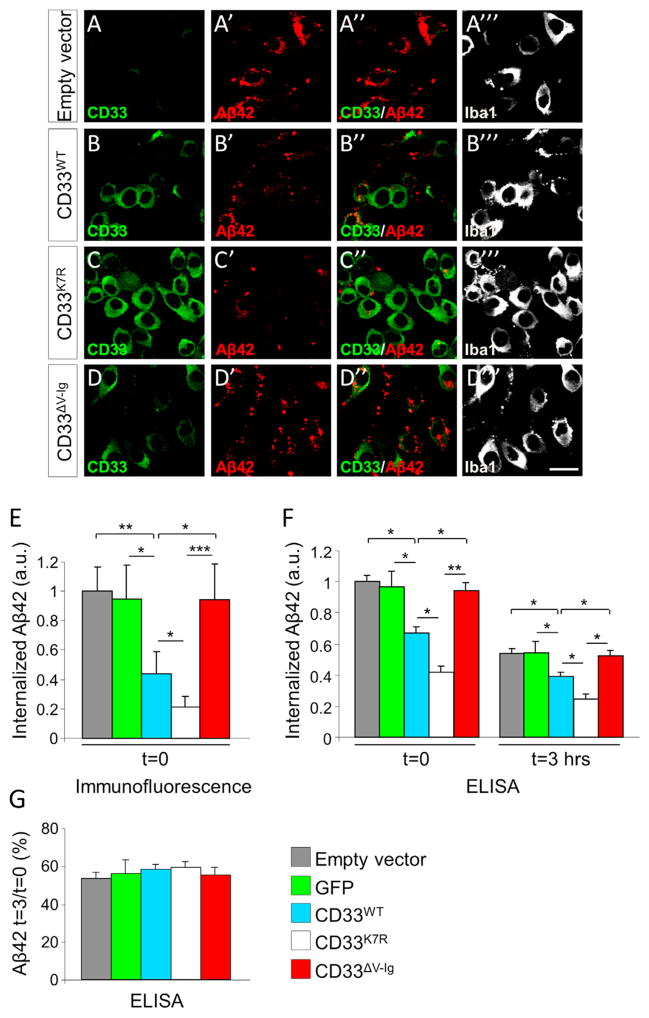
Figure 5. CD33 Inhibits Microglial Uptake of Aβ42.
(A–D‴) BV2 microglial cells were incubated with fluorescently labeled Aβ42 for 3 hr and then subjected to immunofluorescent labeling for CD33 (A–D) and Iba1 (A‴–D‴). Overexpression of WT-CD33 decreases the amount of internalized Aβ42 (compare A′ and B′). Expression of an ubiquitylation-defective CD33 variant (K7R-CD33) further decreases the amount of internalized Aβ42 (compare B′ and C′). A CD33 variant lacking the sialic acid-binding V-type immunoglobulin-like extracellular domain (ΔV-Ig-CD33) no longer inhibits Aβ42 uptake (compare B′, C′, with D′). Scale bar represents 25 μM.
(E) Quantification of Aβ42 signal intensity in individual cells transfected with the indicated constructs or with an empty vector; at least 30 cells were scored per condition (*p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA, Tukey’s test).
(F and G) BV2 cells were treated with unlabeled Aβ42 and incubated for 3 hr. After incubation, cells were either collected for ELISA analysis (t = 0) or washed and incubated for an additional 3 hr in Aβ-free medium, followed by ELISA analysis (t = 3 hr). (F) WT-CD33 inhibits microglial uptake of Aβ42. ELISA quantifications of Aβ42 levels in BV2 cells transfected with the indicated constructs or with empty vector (results were obtained from four independent experiments; *p < 0.05, **p < 0.01, one-way ANOVA, Tukey’s test). (G) CD33 overexpression does not affect the rate of Aβ42 degradation by BV2 cells (the percentage of remaining Aβ42 was the ratio of Aβ42 remaining at t = 3 hr to the total amount of Aβ42 internalized at t = 0). Both empty vector and a GFP vector served as controls. Data are represented as mean ± SEM.
We next transfected cells with a mutant version of CD33 in which seven lysine residues from the intracellular C-terminal domain were mutated to arginine (CD33K7R). This prevents CD33 ubiquitylation and subsequent internalization from the plasma membrane (Walter et al., 2008). We confirmed that the CD33K7R protein displayed enhanced cell surface expression of CD33 (Figure 5C) in comparison to the CD33WT protein (Figure 5B). Expression of the CD33K7R protein led to a further inhibition of Aβ42 uptake (Figures 5C′ and 5E, p < 0.05 CD33K7R versus CD33WT, one-way ANOVA, Tukey’s test and Figure 5F) but did not impair subsequent Aβ42 degradation (Figure 5G), further indicating that CD33 inhibits Aβ42 uptake by microglial cells.
CD33 and the related Siglecs perform their biological functions by interacting with sialic acids, which are attached to the outer membrane of cells and can mediate cis- or trans-cellular interactions (Paulson et al., 2012). To determine whether the interaction between CD33 and sialic acids is involved in Aβ42 uptake, we employed a CD33 mutant construct in which the sialic acid-binding V-type Immunoglobulin-like (V-Ig) domain was deleted (Pérez-Oliva et al., 2011). The mutant CD33ΔV-Ig protein is present at the plasma membrane in BV2 cells and is expressed at levels similar to CD33WT (Figures 5B and 5D and Pérez-Oliva et al., 2011). Remarkably, inhibition of Aβ42 uptake by CD33 was completely abolished in cells expressing the CD33ΔV-Ig protein; Aβ42 levels were similar to those in cells expressing empty vector or GFP (Figures 5E and 5F), indicating that sialic acid binding is required for CD33 to mediate Aβ42 uptake. Collectively, these experiments indicate that CD33 modulates microglial uptake of Aβ42. Specifically, lower levels of cell surface CD33 enhance Aβ42 internalization, while higher levels impair this process. Moreover, interaction of CD33 with sialic acids is necessary to mediate these effects.
CD33 Activity Promotes Amyloid Beta Pathology In Vivo
The above in vitro experiments, together with our genetic, biochemical, and histopathological observations, strongly implicate CD33 activity in Aβ pathology in AD. To provide direct in vivo evidence that CD33 activity potentiates Aβ pathology in vivo, we used a model of AD, the APPSwe/PS1ΔE9 mouse (subsequently referred to as APP/PS1), in which we inactivated CD33 function. APP/PS1 mice exhibit elevated Aβ (including Aβ42) production and develop amyloid plaques (Jankowsky et al., 2004). APP PS1 mice lacking CD33 (APP/PS1/CD33−/−) were born at Mendelian ratios and exhibited no gross anatomical defects (data not shown). We isolated the cortex from 4-month-old APP/PS1 and APP/PS1/CD33−/− mice and their littermate controls and derived detergent-soluble fractions that we used for subsequent western blotting as well as TBS-soluble and FA-soluble fractions (Wang et al., 2011) that we used for Aβ ELISA experiments. Using western blotting we found, as expected, increased and similar levels of full-length APP and APP C-terminal fragments (APP-CTF) in both APP/PS1 and APP/PS1/CD33−/− mice as compared to controls (Figures 6A and 6B). BACE1 levels also did not differ across genotypes (data not shown).
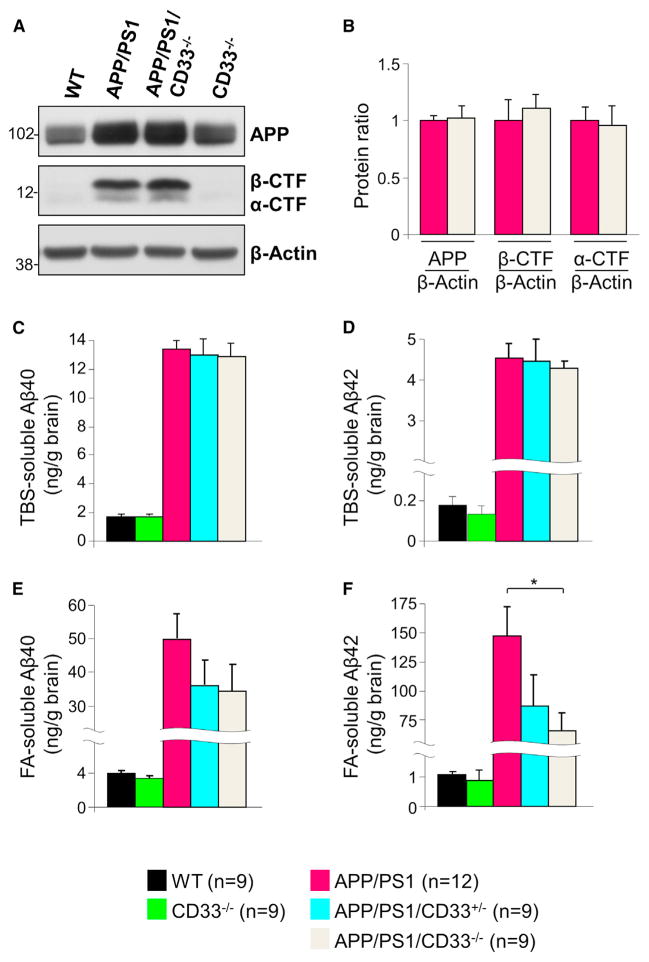
Figure 6. CD33 Deletion Decreases Formic Acid-Soluble Aβ42 Levels in APP/PS1 Mice.
(A) Western blotting analysis of cortical extracts reveals increased levels of APP and APP C-terminal fragments (CTFs) in 4-month-old APP/PS1 mice in comparison to controls. However, APP and APP-CTFs levels are similar in APP/PS1 and APP/PS1/CD33−/− mice. β-Actin served as loading control.
(B) Quantification of APP, α-CTF, and β-CTF levels in APP/PS1 and APP/PS1/CD33−/− mice (n = 9 male mice were analyzed per group).
(C–F) ELISA analysis of Aβ40 (C and E) and Aβ42 (D and F) in TBS-soluble (C and D) or formic acid (FA)-soluble (E and F) fractions isolated from the cortex of 4-month-old male mice of the indicated genotypes. APP/PS1 mice exhibit increased levels of Aβ40 and Aβ42 relative to WT and CD33−/− mice, as expected. No differences in Aβ40 levels and TBS-soluble Aβ42 levels were seen in APP/PS1 and APP/PS1/CD33−/− mice. However, the FA-soluble Aβ42 levels were markedly decreased in APP/PS1/CD33−/− relative to APP/PS1 mice (n = 9–12 mice were analyzed per group, *p < 0.05, one-way Kruskal-Wallis ANOVA, Dunn’s test). Data are represented as mean ± SEM. See also Figure S5.
Next, we asked whether knockout of CD33 affects Aβ levels in the APP/PS1 mice. As expected, we found that Aβ40 and Aβ42 levels (measured by ELISA) in the TBS-soluble fraction were markedly increased in APP/PS1 mice relative to WT and CD33−/− mice (Figures 6C and 6D). Furthermore, the levels of Aβ40 and Aβ42 in the TBS-soluble fraction were similar in the APP/PS1, APP/PS1/CD33+/−, and APP/PS1/CD33−/− mice (Figures 6C and 6D). Remarkably, however, the levels of Aβ42 were significantly decreased in the APP/PS1/CD33−/− FA-soluble fraction of mice relative to APP/PS1 mice (Figure 6F, p < 0.05 APP/PS1/CD33−/− versus APP/PS1 mice, one-way Kruskal-Wallis ANOVA, Dunn’s test). This effect was not due to increased numbers of microglial cells or astrocytes in APP/PS1/CD33−/− mice (Figure S5) or alterations in the levels of either inducible nitric oxide synthase or microglia-derived cytokines, e.g., IL-1β and TNFα (data not shown). These data suggest that CD33-deficient microglia possess increased Aβ42 uptake/clearance capacity in vivo and that the CD33-mediated effect on microglial clearance of Aβ42 is cell autonomous.
Finally, we investigated whether CD33 deletion impacts the process of Aβ deposition in the APP/PS1 brain. Aβ deposition is obvious by 6 months in APP/PS1 mice when both compact and diffuse Aβ-containing plaques can be observed in the fore-brain (Jankowsky et al., 2004). We stained coronal sections from 6- to 7-month-old control, CD33−/−, APP/PS1, and APP/PS1/CD33−/− mice with an antibody directed against Aβ (residues 1–5, 3D6) that recognizes both compact and diffuse Aβ plaques (Reilly et al., 2003). The 3D6 antibody did not label any plaques in control and CD33−/− brains (data not shown). Analysis of the APP/PS1 brains revealed numerous Aβ plaques, both in the cortex and the hippocampus (Figures 7A and 7C). Sections spanning the cortex or the hippocampus were subjected to quantification of amyloid plaque burden. Remarkably, the Aβ plaque burden was robustly decreased in the APP/PS1/CD33−/− brains relative to the APP/PS1 brains (Figures 7B and 7D). The quantification revealed a 37.2% and 33.5% reduction of Aβ plaque burden in the APP/PS1/CD33−/− cortex and hippocampus, respectively (Figures 7E and 7F; n = 9–11 animals/group, p < 0.01 and p < 0.05, APP/PS1/CD33−/− versus APP/PS1 cortex and hippocampus, respectively, Student’s t test). Therefore, CD33 promotes Aβ deposition and plaque formation in vivo.
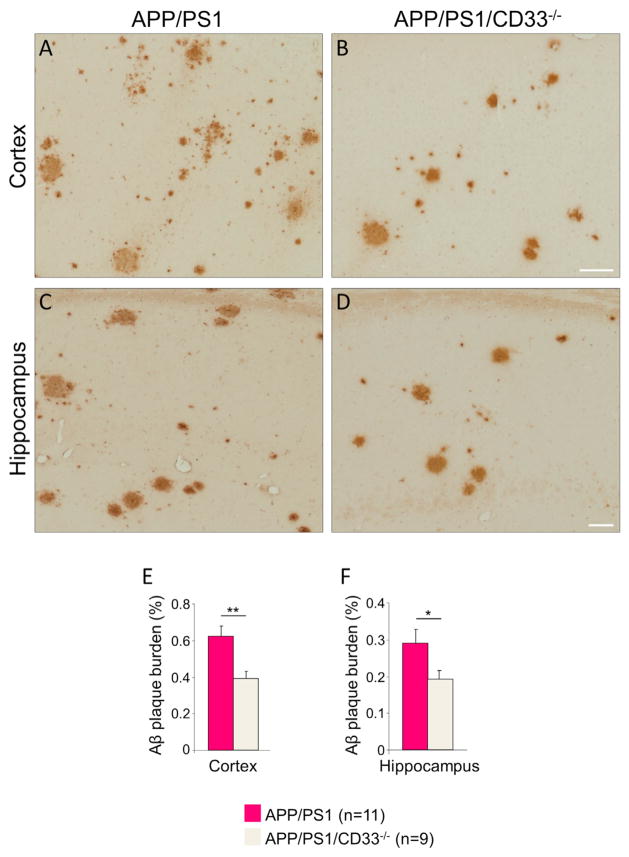
Figure 7. CD33 Deletion Mitigates Aβ Plaque Pathology in APP/PS1 Mice.
(A–D) Photomicrographs of selected cortical (A and B) and hippocampal (C and D) fields from 6- to 7-month-old APP/PS1 (A and C) and APP/PS1/CD33−/− (B and D) brains were stained with the anti-Aβ antibody 3D6 to reveal Aβ plaques. The Aβ plaque burden is decreased in APP/PS1/CD33−/− brains relative to APP/PS1 brains (compare B with A and D with C).
(E and F) Quantification of amyloid plaque burden in the cortex (E) and hippocampus (F) of 6- to 7-month-old APP/PS1 and APP/PS1/CD33−/− brains (n = 9–11 male mice were analyzed per group, *p < 0.05, **p < 0.01, Student’s t test). Data are represented as mean ± SEM. Scale bar represents 50 μm.
DISCUSSION
Despite great strides toward the development of an effective therapy, no treatment is currently able to prevent, delay, or stop AD progression. This is due in part to our incomplete understanding of AD pathogenesis. While important advances have been made toward a better understanding of FAD, comparatively little is known about LOAD. Great advances in DNA sequencing technologies coupled with the investigation of large human cohorts have provided an unprecedented analytic power and have led to the identification of multiple risk factors for LOAD, thus opening new research avenues that might advance our understanding of this devastating disease.
We and others have found that the CD33 gene is associated with risk for LOAD (Bertram et al., 2008; Hollingworth et al., 2011; Naj et al., 2011). CD33 is a member of the Siglec family of lectins that binds sialic acid residues leading to cis (same CD33-expressing cell) or trans (adjacent cells) interactions that regulate several aspects of innate immunity (Crocker et al., 2007; Paulson et al., 2012). To date, the role of CD33 in the brain has remained unknown. To bridge this gap, and to understand how CD33 misregulation contributes to LOAD pathogenesis, we performed a comprehensive analysis of CD33 expression and mechanism of action, using a combination of human and mouse genetics, biochemistry, neuropathology, and in vitro modeling. Based on our findings, we propose that increased CD33 activity in microglial cells inhibits Aβ clearance in LOAD; thus, CD33 inhibition may represent a novel means for preventing and treating AD.
CD33 Expression Is Increased in AD
We found that the levels of CD33 protein are increased in AD. This increase was paralleled by an increase in the number of CD33-immunoreactive microglia. Moreover, the ratio CD33/Iba1 is also increased in AD, suggesting that individual microglia display increased CD33 levels in AD. Thus, AD cases are characterized by both an increase in the number of CD33-immunoreactive microglia and an increase in CD33 levels in these CD33-positive cells. Moreover, the fact that the levels of CD33 mRNA were also strongly increased in AD suggests a potential upregulation of CD33 transcription in microglial cells; alternatively, the CD33 mRNA might exhibit increased stability in microglia in the AD brain. It will be important to determine whether CD33 is part of a broader transcriptional program that operates in microglial cells in the aging brain and to identify the upstream activators of such a program.
To further explore the relationship between CD33 expression and AD, we assessed CD33 expression in carriers of the minor (T) allele of the CD33 SNP rs3865444, which protects against AD. We found that the minor (T) allele is associated with reduced CD33 protein levels, both in the AD and control subjects (Figure 1E). Interestingly, the levels of CD33 mRNA were not reduced in the carriers of this allele. The rs3865444 SNP is located upstream of the 5′ UTR of the CD33 gene (Hollingworth et al., 2011; Naj et al., 2011). One possibility is that the rs3865444 SNP is in linkage disequilibrium with functional variant(s) located in the coding region. This, in turn, could influence mRNA translation without affecting mRNA stability. Recently, increased CD33 mRNA levels were independently shown to be associated with AD; CD33 mRNA levels correlated with Iba1 mRNA levels and CD33 mRNA expression normalized to Iba1 expression correlated with disease status and Clinical Dementia Rating scores (Karch et al., 2012). The increased CD33 (mRNA and protein) levels in the AD brain and decreased CD33 protein levels in the carriers of the protective allele suggest that increased CD33 expression plays a direct role in the etiology and/or pathogenesis of AD.
CD33 Is a Modifier of Amyloid Beta Pathology
Several genes associated with LOAD have been shown to be critically involved in the control of Aβ homeostasis. ApoE binds Aβ and influences its oligomerization (Hashimoto et al., 2012). We recently found that lipidated apoE, in particular its pathogenic variant apoE4, increases the oligomerization of Aβ; as a consequence, APOE ε4/ε4 AD brains displayed higher levels of Aβ oligomers relative to APOE ε3/ε3 brains (Hashimoto et al., 2012). The LOAD-associated gene Clusterin (CLU) encodes an extracellular chaperone apolipoprotein J (apoJ) that interacts with Aβ prefibrillar structures and inhibits Aβ aggregation (Yerbury et al., 2007). ApoJ also facilitates the transport of plasma-derived Aβ across the blood-brain barrier (BBB) and Aβ42 clearance at the BBB in mice (Bell et al., 2007). Interestingly, apoJ cooperates with apoE to suppress Aβ levels and deposition in vivo (DeMattos et al., 2004).
Besides the control of Aβ self-assembly, LOAD-associated genes are able to control the trafficking of APP and thus influence the formation of Aβ. A genetic screen in yeast revealed that several LOAD genes (including PICALM and BIN1) were modifiers of Aβ toxicity and suggested that these genes regulate the endocytic trafficking of APP (Treusch et al., 2011). Moreover, PICALM promotes APP internalization, endocytic trafficking, and Aβ generation in neurons in vitro (Xiao et al., 2012). The ABCA7 LOAD risk gene also regulates APP trafficking, by stimulating cholesterol efflux, which decreases the levels of APP at the plasma membrane and Aβ generation (Chan et al., 2008). Therefore, targeting the intracellular trafficking of APP in neurons might represent a novel therapeutic approach in AD.
Our results identify CD33 as a modifier of Aβ pathology in vivo. We observed an association between the protective minor (T) allele of the CD33 SNP rs3865444 and decreased FA-soluble Aβ42 levels in AD as well as a positive correlation between CD33 microglial expression and FA-soluble Aβ42 levels and amyloid plaque burden in the AD cortex (Figure 3). Moreover, CD33 levels directly impact microglial uptake of Aβ42 (Figures 4 and 5) and modulate the accumulation of FA-soluble Aβ42 and Aβ plaque burden in APP/PS1 transgenic mice (Figures 6 and 7). Based on these observations, we propose that increased activity of CD33 in microglial cells contributes to the etiology and/or pathogenesis of AD by preventing Aβ uptake and thus potentiating its toxicity.
Taken together, the multiple lines of evidence implicating multiple LOAD risk genes in the control of Aβ production, clearance, and deposition provide additional support for the amyloid hypothesis of AD by demonstrating that failure of multiple systems that ensure Aβ homeostasis is associated with an increased risk for developing AD. These findings also suggest immense therapeutic opportunities, since the identification of drug targets that are critically involved in control of Aβ pathogenicity by the novel LOAD risk genes might provide novel strategies targeting the earliest stages of cognitive decline, well before the occurrence of overt neurodegeneration.
CD33 Inhibits the Microglial Uptake of Amyloid Beta
Based on our finding that CD33 microglial expression is elevated in AD but decreased in carriers of the protective minor (T) allele of the CD33 SNP rs3865444, we hypothesized that the activity of CD33 in microglia promotes AD pathogenesis. To test this hypothesis and to unravel the molecular underpinnings of CD33 action in microglial cells, we investigated whether CD33 is involved in the process of Aβ clearance by microglia. Using an assay of microglial uptake and clearance optimized for Aβ42 (Jiang et al., 2008), we found that mouse primary microglial cells lacking CD33 expression exhibit an increased uptake of Aβ42 relative to WT cells (Figures 4A–4D). Interestingly, CD33-deficient and WT cells degraded Aβ42 at a similar rate (Figure 4E). In CD33-deficient cells, however, Aβ42 clearance was accelerated, due to the increased overall uptake. To further explore the involvement of CD33 in Aβ42 uptake by microglial cells, we employed a well-characterized microglial cell line (BV2) that effectively internalizes and degrades added Aβ42 (Mandrekar et al., 2009). We found that microglia overexpressing WT CD33 were markedly impaired in their capacity to internalize Aβ42 but degraded the internalized Aβ42 at a similar rate to cells transfected with an empty plasmid or a GFP plasmid (Figures 5E–5G). This finding was also validated by the transfection with an ubiquitylation-defective CD33 mutant (CD33K7R), which exhibits enhanced cell surface expression of CD33 (Figure 5C and Walter et al., 2008) and further exacerbates the inhibition of Aβ42 uptake (Figures 5E and 5F). Thus, CD33 directly modulates uptake, but not degradation, of Aβ42 by microglial cells.
CD33 and the related Siglecs perform their biological functions by interacting with sialic acids, which are attached to the outer membrane of cells, and can mediate cis- or trans-cellular interactions (Crocker et al., 2007). To explore the requirement of sialic acid binding in the process of CD33-mediated microglial uptake of Aβ42, we transfected microglial BV2 cells with a CD33 mutant in which the sialic acid-binding V-type immunoglobulin-like domain was removed. The mutant CD33ΔV-Ig protein is present at the plasma membrane in BV2 cells and is expressed at levels similar to CD33WT (Figures 5B and 5D and Pérez-Oliva et al., 2011). Cells expressing the CD33ΔV-Ig protein were no longer impaired in their capacity to uptake Aβ42, indicating that sialic acid binding is necessary for the ability of CD33 to inhibit Aβ42 uptake. Collectively, these experiments indicate that CD33 directly modulates microglial uptake of Aβ42, via interaction with sialic acids.
Microglial cells have been suggested to play critical roles as mediators of Aβ clearance in the brain. A previous study (Grathwohl et al., 2009) challenged this view by showing that ablation of microglial cells (using drug-induced microglial toxicity) did not alleviate Aβ pathology in two AD mouse models, within a 4 week time window after the ablation. These results await confirmation in a setting of prolonged microglial ablation (several months to years). It also remains to be determined whether an acute removal of microglial cells has a specific effect on the dynamics of monomeric and oligomeric as opposed to fibrillar Aβ.
Mounting evidence implicates genes associated with risk for LOAD in the process of microglial clearance of Aβ42. For example, apoE promotes the clearance of Aβ in the brain, partly through its capacity to enhance Aβ uptake and degradation by microglia (Jiang et al., 2008; Lee et al., 2012). Interestingly, another LOAD risk gene, ApoJ/Clusterin (CLU) cooperates with APOE to suppress Aβ levels and deposition in PDAPP transgenic mice (DeMattos et al., 2004). A rare variant of the TREM2 gene has been recently shown to confer increased risk for LOAD (Guerreiro et al., 2013; Jonsson et al., 2013). TREM2 is an innate immune receptor, similar to CD33, and is expressed in a subset of myeloid cells including microglia (Klesney-Tait et al., 2006). Remarkably, TREM2 is upregulated in amyloid plaque-associated microglia in aging APP23 transgenic mice (Frank et al., 2008) and in CRND8 transgenic mice (expressing mutant KM670/671NL and V717F APP) (Guerreiro et al., 2013) and promotes the phagocytic clearance of amyloid proteins (Melchior et al., 2010). Interestingly, TREM2 interacts with its ligand TREM2-L, expressed by apoptotic neurons, and mediates removal of dying neuronal cells by microglia (Hsieh et al., 2009).
Our results suggest that CD33 represents a regulator of microglial clearance of Aβ and a target for the treatment and prevention of AD. It is interesting to note that therapies targeting CD33 have already been developed in acute myeloid leukemia (AML), due to its high membrane expression in myeloid cells (Jandus et al., 2011; O’Reilly and Paulson, 2009). Naked humanized anti-CD33 and calicheamicin-conjugated humanized murine anti-CD33 antibodies have been developed and tested in several phase III clinical trials of AML, with mixed results (Jandus et al., 2011; Jurcic, 2012; Ricart, 2011). This suggests that the development of a CD33 antibody that is able to cross the BBB is feasible in principle. A chimeric antibody in which the CD33 antibody is fused to a monoclonal antibody against the human insulin receptor could facilitate the receptor-mediated passage of the chimera across the BBB (O’Reilly and Paulson, 2009). An alternative approach is the development of small compounds, e.g., sialic acid-based antagonists that target CD33 specifically and inhibit its function. Furthermore, a better understanding of CD33 action in microglial cells should lead to the identification of critical cellular targets that link the activity of CD33 in microglia to the process of Aβ recognition and uptake and will hopefully lead to the development of novel therapeutics for the prevention and treatment of AD.
EXPERIMENTAL PROCEDURES
Please see the Supplemental Experimental Procedures for detailed methods on SNP genotyping, RNA extraction and real-time PCR, immunohistochemistry and stereology, assessment of Aβ plaque burden in mice, ELISA, western blot analysis, generation of an anti-mouse CD33 antibody, primary microglia isolation, cell culture and transfection, Aβ uptake, and degradation assays.
Brain Specimens
Formalin-fixed, paraffin-embedded 8-μm-thick sections, as well as frozen tissue specimens from the fontal cortex of 25 patients with AD and 15 age-matched nondemented control subjects were obtained from the Massachusetts Alzheimer’s Disease Research Center Brain Bank. All the study subjects or their next of kin gave written informed consent for the brain donation, and the Massachusetts General Hospital Institutional Review Board approved the study protocol. The demographic characteristics of both groups are shown in Table S1. All patients with AD fulfilled the National Institute of Neurological and Communicative Disorders and Stroke-Alzheimer’s Disease and Related Disorders Associations criteria for probable AD and the National Institute on Aging-Reagan Institute criteria for high likelihood of AD.
Animals
APPSwe/PS1ΔE9 transgenic mice (referred to as APP/PS1) (Jankowsky et al., 2004) and constitutive CD33 knockout mice (Brinkman-Van der Linden et al., 2003) were obtained from The Jackson Laboratory (catalog 005864 and 006942, respectively). Both mouse strains are on the C57Bl/6 background. All mice were housed under standard conditions with free access to food and water. All animal experiments were performed in accordance with national guidelines (National Institutes of Health) and approved by Massachusetts General Hospital and McLaughlin Institute Institutional Animal Care and Use Committees.
Statistical Analysis
A general linear regression model adjusting for appropriate covariates was used to test for allelic association between the rs3865444 SNP and quantitative traits, as implemented in PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink) (Purcell et al., 2007). To identify covariates that maximize the regression model’s predictive ability and predict the quantitative traits for the human samples, we performed a stepwise regression procedure using age, gender, disease status, postmortem interval, and presence of APOE ε4 allele. The stepwise regression analysis was performed using the R (v2.10.0) software package (R Development Core Team, 2009). Statistics and correlations of different quantitative traits were performed using the GraphPad Prism software, version 5.0 (GraphPad). The normality of quantitative trait data sets was tested with the D’Agostino-Pearson omnibus test. Student’s t test and Pearson’s correlation test were performed for normally distributed data sets and Mann-Whitney U and Spearman’s correlation tests otherwise. Multiple group analyses were performed by one-way ANOVA followed by Tukey’s post hoc test or by one-way Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
Supplementary Material
Acknowledgments
We thank Ronald B. Walter (Fred Hutchinson Cancer Research Center, Seattle, WA, USA) for the CD33 plasmids and Linda Van Eldik (University of Kentucky, Lexington, KY, USA) for the BV2 microglial cell line. We thank Karlotta Fitch for assistance with tissue sectioning. We thank Lars Bertram, Zhongcong Xie, Jaehong Suh, Can Zhang, and Doo Kim for stimulating discussions. This work was supported by grants from the National Institutes of Health (5R37MH060009 and 5P01AG15379 to R.E.T. and AG08487 and P50AG05134 to B.T.H.), and Cure Alzheimer’s Fund. A.G. is supported by a fellowship from the Deutsche Forschungsgemeinschaft (Germany). B.T.H. is on the Scientific Advisory Board of Neurophage, which was not involved in this study.
Footnotes
Supplemental Information includes five figures, one table, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2013.04.014.
References
- Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–161. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, Angata T, Reynolds SA, Powell LD, Hedrick SM, Varki A. CD33/Siglec-3 binding specificity, expression pattern, and consequences of gene deletion in mice. Mol Cell Biol. 2003;23:4199–4206. doi: 10.1128/MCB.23.12.4199-4206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Kim WS, Kwok JB, Hill AF, Cappai R, Rye KA, Garner B. ATP-binding cassette transporter A7 regulates processing of amyloid precursor protein in vitro. J Neurochem. 2008;106:793–804. doi: 10.1111/j.1471-4159.2008.05433.x. [DOI] [PubMed] [Google Scholar]
- Choi SH, Veeraraghavalu K, Lazarov O, Marler S, Ransohoff RM, Ramirez JM, Sisodia SS. Non-cell-autonomous effects of presenilin 1 variants on enrichment-mediated hippocampal progenitor cell proliferation and differentiation. Neuron. 2008;59:568–580. doi: 10.1016/j.neuron.2008.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Crocker PR, McMillan SJ, Richards HE. CD33-related siglecs as potential modulators of inflammatory responses. Ann N Y Acad Sci. 2012;1253:102–111. doi: 10.1111/j.1749-6632.2011.06449.x. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- Frank S, Burbach GJ, Bonin M, Walter M, Streit W, Bechmann I, Deller T. TREM2 is upregulated in amyloid plaque-associated microglia in aged APP23 transgenic mice. Glia. 2008;56:1438–1447. doi: 10.1002/glia.20710. [DOI] [PubMed] [Google Scholar]
- Gorlovoy P, Larionov S, Pham TT, Neumann H. Accumulation of tau induced in neurites by microglial proinflammatory mediators. FASEB J. 2009;23:2502–2513. doi: 10.1096/fj.08-123877. [DOI] [PubMed] [Google Scholar]
- Grathwohl SA, Kälin RE, Bolmont T, Prokop S, Winkelmann G, Kaeser SA, Odenthal J, Radde R, Eldh T, Gandy S, et al. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat Neurosci. 2009;12:1361–1363. doi: 10.1038/nn.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Serrano-Pozo A, Hori Y, Adams KW, Takeda S, Banerji AO, Mitani A, Joyner D, Thyssen DH, Bacskai BJ, et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid β peptide. J Neurosci. 2012;32:15181–15192. doi: 10.1523/JNEUROSCI.1542-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer JD, Grogg KL, Hanson CA, Gamez JD, Dogan A. CD33 detection by immunohistochemistry in paraffin-embedded tissues: a new antibody shows excellent specificity and sensitivity for cells of myelomonocytic lineage. Am J Clin Pathol. 2008;129:316–323. doi: 10.1309/E36008Y2H08Q1AYY. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Koike M, Spusta SC, Niemi EC, Yenari M, Nakamura MC, Seaman WE. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J Neurochem. 2009;109:1144–1156. doi: 10.1111/j.1471-4159.2009.06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandus C, Simon HU, von Gunten S. Targeting siglecs—a novel pharmacological strategy for immuno- and glycotherapy. Biochem Pharmacol. 2011;82:323–332. doi: 10.1016/j.bcp.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, Mann K, Lamb B, Willson TM, Collins JL, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcic JG. What happened to anti-CD33 therapy for acute myeloid leukemia? Curr Hematol Malig Rep. 2012;7:65–73. doi: 10.1007/s11899-011-0103-0. [DOI] [PubMed] [Google Scholar]
- Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS ONE. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol. 2006;7:1266–1273. doi: 10.1038/ni1411. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lee CY, Tse W, Smith JD, Landreth GE. Apolipoprotein E promotes β-amyloid trafficking and degradation by modulating microglial cholesterol levels. J Biol Chem. 2012;287:2032–2044. doi: 10.1074/jbc.M111.295451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:184. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar S, Jiang Q, Lee CY, Koenigsknecht-Talboo J, Holtzman DM, Landreth GE. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J Neurosci. 2009;29:4252–4262. doi: 10.1523/JNEUROSCI.5572-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior B, Garcia AE, Hsiung BK, Lo KM, Doose JM, Thrash JC, Stalder AK, Staufenbiel M, Neumann H, Carson MJ. Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer’s disease. ASN Neuro. 2010;2:e00037. doi: 10.1042/AN20100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–248. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JC, Macauley MS, Kawasaki N. Siglecs as sensors of self in innate and adaptive immune responses. Ann N Y Acad Sci. 2012;1253:37–48. doi: 10.1111/j.1749-6632.2011.06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Oliva AB, Martínez-Esparza M, Vicente-Fernández JJ, Corral-San Miguel R, García-Peñarrubia P, Hernández-Caselles T. Epitope mapping, expression and post-translational modifications of two isoforms of CD33 (CD33M and CD33m) on lymphoid and myeloid human cells. Glycobiology. 2011;21:757–770. doi: 10.1093/glycob/cwq220. [DOI] [PubMed] [Google Scholar]
- Pillai S, Netravali IA, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2009. http://www.R-project.org. [Google Scholar]
- Reilly JF, Games D, Rydel RE, Freedman S, Schenk D, Young WG, Morrison JH, Bloom FE. Amyloid deposition in the hippocampus and entorhinal cortex: quantitative analysis of a transgenic mouse model. Proc Natl Acad Sci USA. 2003;100:4837–4842. doi: 10.1073/pnas.0330745100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricart AD. Antibody-drug conjugates of calicheamicin derivative: gemtuzumab ozogamicin and inotuzumab ozogamicin. Clin Cancer Res. 2011;17:6417–6427. doi: 10.1158/1078-0432.CCR-11-0486. [DOI] [PubMed] [Google Scholar]
- Rollins-Raval MA, Roth CG. The value of immunohistochemistry for CD14, CD123, CD33, myeloperoxidase and CD68R in the diagnosis of acute and chronic myelomonocytic leukaemias. Histopathology. 2012;60:933–942. doi: 10.1111/j.1365-2559.2012.04175.x. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Preventing Alzheimer’s disease. Science. 2012;337:1488–1492. doi: 10.1126/science.1228541. [DOI] [PubMed] [Google Scholar]
- Serrano-Pozo A, Mielke ML, Gomez-Isla T, Betensky RA, Growdon JH, Frosch MP, Hyman BT. Reactive glia not only associates with plaques but also parallels tangles in Alzheimer’s disease. Am J Pathol. 2011;179:1373–1384. doi: 10.1016/j.ajpath.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen GQ, Abdullah KG, Wang QK. The TaqMan method for SNP genotyping. Methods Mol Biol. 2009;578:293–306. doi: 10.1007/978-1-60327-411-1_19. [DOI] [PubMed] [Google Scholar]
- Tanzi RE. A brief history of Alzheimer’s disease gene discovery. J Alzheimers Dis. 2013;33(Suppl 1):S5–S13. doi: 10.3233/JAD-2012-129044. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Treusch S, Hamamichi S, Goodman JL, Matlack KE, Chung CY, Baru V, Shulman JM, Parrado A, Bevis BJ, Valastyan JS, et al. Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gunten S, Bochner BS. Basic and clinical immunology of Siglecs. Ann N Y Acad Sci. 2008;1143:61–82. doi: 10.1196/annals.1443.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter RB, Häusermann P, Raden BW, Teckchandani AM, Kamikura DM, Bernstein ID, Cooper JA. Phosphorylated ITIMs enable ubiquitylation of an inhibitory cell surface receptor. Traffic. 2008;9:267–279. doi: 10.1111/j.1600-0854.2007.00682.x. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Wang X, Lu JJ, Li QX, Gao CY, Liu XH, Sun Y, Yang M, Lim Y, Evin G, et al. p75NTR regulates Abeta deposition by increasing Abeta production but inhibiting Abeta aggregation with its extracellular domain. J Neurosci. 2011;31:2292–2304. doi: 10.1523/JNEUROSCI.2733-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, Perez R, Cirrito JR, Lee JM. Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem. 2012;287:21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007;21:2312–2322. doi: 10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.