Abstract
Objective
Characterization of HIV-1 sequences in newly infected individuals is important for elucidating the mechanisms of viral sexual transmission. We report the identification of transmitted/founder viruses in eight pairs of HIV-1 sexually-infected patients enrolled at the time of primary infection (“recipients”) and their transmitting partners (“donors”).
Methods
Using a single genome-amplification approach, we compared quasispecies in donors and recipients on the basis of 316 and 376 C2V5 env sequences amplified from plasma viral RNA and PBMC-associated DNA, respectively.
Results
Both DNA and RNA sequences indicated very homogeneous viral populations in all recipients, suggesting transmission of a single variant, even in cases of recent sexually transmitted infections (STIs) in donors (n = 2) or recipients (n = 3). In all pairs, the transmitted/founder virus was derived from an infrequent variant population within the blood of the donor. The donor variant sequences most closely related to the recipient sequences were found in plasma samples in 3/8 cases and/or in PBMC samples in 6/8 cases. Although donors were exclusively (n = 4) or predominantly (n = 4) infected by CCR5-tropic (R5) strains, two recipients were infected with highly homogeneous CXCR4/dual-mixed-tropic (X4/DM) viral populations, identified in both DNA and RNA. The proportion of X4/DM quasispecies in donors was higher in cases of X4/DM than R5 HIV transmission (16.7–22.0% versus 0–2.6%), suggesting that X4/DM transmission may be associated with a threshold population of X4/DM circulating quasispecies in donors.
Conclusions
These suggest that a severe genetic bottleneck occurs during subtype B HIV-1 heterosexual and homosexual transmission. Sexually-transmitted/founder virus cannot be directly predicted by analysis of the donor’s quasispecies in plasma and/or PBMC. Additional studies are required to fully understand the traits that confer the capacity to transmit and establish infection, and determine the role of concomitant STIs in mitigating the genetic bottleneck in mucosal HIV transmission.
Introduction
The transmission of human immunodeficiency virus type 1 (HIV-1) and the establishment of a productive infection are complex biological processes, and the details of the mechanisms remain to be elucidated. Initial studies of sexually acquired HIV-1 infection suggested that viral populations in the acute phase are generally highly homogeneous genetically, in contrast to the more heterogeneous viral populations found in chronic infections [1]–[6]. These findings thus suggested that HIV-1 infection is associated with a transmission “bottleneck”. However, more recent studies have reported heterogeneous virus populations shortly after infection in African female sex workers [7]–[10] and in American men who have sex with men (MSM) [11]. These observation suggest that the routes and circumstances of infection may affect the complexity of the transmitted virus [12].
The differing findings concerning the complexity of viruses during the acute and early phases of HIV-1 infection probably result from a combination of factors, including differences in the experimental designs and the methodologies used. A common approach has been to identify subjects within the first months following infection and to derive HIV sequences by bulk or near-limiting-dilution PCR amplification of proviral DNA or plasma RNA followed by cloning, sequencing and phylogenetic analyses [1], [2], [7], [10], [12]. In 2008, Keele et al. devised a novel strategy for a more precise molecular identification and enumeration of transmitted HIV-1 genomes [13]. This method, SGA-direct amplicon sequencing, was recently applied to clinical cohorts of acutely infected individuals [13]–[17], and the findings indicated that approximately 80% of heterosexual subjects and 60% of MSM are productively infected by a single viral genome. Since most of these studies only characterized the transmitted viral population in recipients, little information was available about its relationship with virus circulating in the donor. Also, few of these studies compared the viral populations identified by analysis of both RNA and DNA samples from donor/recipient pairs.
We report a study of eight transmission pairs, each of them including a sexually-infected patient enrolled into the French ANRS PRIMO cohort at the time of primary HIV-1 infection (PHI) (“recipient”) and his/her HIV-1-infected sexually-transmitting partner (“donor”). SGA-direct amplicon sequencing in plasma RNA and PBMC-derived DNA samples was used to compare C2V5 env gene sequences of the quasispecies in the donors and recipients.
Patients and Methods
Ethics Statement
The Ethics Committee of Cochin Hospital approved the study, and all the patients gave their written informed consent.
The ANRS PRIMO Cohort
The patients were defined as having PHI from a western blot (WB) profile compatible with ongoing seroconversion (incomplete WB with absence of antibodies to pol proteins) (94% of the patients), detectable plasma HIV RNA with a negative or weakly reactive ELISA (2%), or an interval of less than 6 months between a negative and a positive ELISA result (4%) [18]. The date of infection was estimated as the date of symptom onset minus 15 days or, in asymptomatic patients, the date of incomplete WB minus 1 month, or the midpoint between a negative and a positive ELISA result. Patients were enrolled if HIV infection was estimated to have occurred less than 3 months previously. All patients were antiretroviral (ART)-naïve when enrolled into the cohort. At enrolment, blood samples were collected for immunological and virological studies. Participants completed standardized questionnaires describing HIV-acquisition risk group and sexual behavior (including number and characteristics of sexual intercourses before diagnosis of PHI and history of sexually transmitted infections (STI)). Serological screening for syphilis (Treponema pallidum hemagglutination assay (TPHA) and Venereal Diseases Research Laboratory test (VDRL)), hepatitis B (HBV) and hepatitis C viruses (HCV) was performed at enrolment. In cases of positive HBV or HCV screening results, HBV DNA and HCV RNA were quantified using the COBAS®Ampliprep/COBAS®TaqMan®HBV v.2.0 assay (Roche, Meylan, France) and the Abbott RealTime HCV® assay (Abbott, Rungis, France), respectively.
This study involved patients enrolled during PHI in the PRIMO cohort, who were able to identify their partner likely to be the source of their HIV infection, and agreed to participate in the substudy. They were then asked to propose to their partner to have a questionnaire and a blood sample in the month following enrolment of the recipient. Samples from donors were collected and systematically screened for syphilis, HBV and HCV at the time of collection. Between March 1998 and October 2008, 17 donor/recipients pairs were enrolled in our cohort. We herein study 8 out of these pairs, from which plasma and PBMC samples from donors were available.
Laboratory Methods
HIV-RNA was quantified with the Cobas Taqman HIV-1 v1.5 assay (Roche Diagnostics, Meylan, France) as recommended by the manufacturers (threshold of detection of 20 copies/ml). Cell associated HIV-1 DNA in whole blood samples was quantified using the real-time HIV-1 DNA assay (Biocentric, Bandol, France) with a detection limit of 5 copies/PCR.
Drug resistance was evaluated by amplifying and sequencing the HIV-1 reverse transcriptase (RT) and protease genes in plasma HIV-RNA samples obtained at enrolment [19]. Resistance to nucleoside RT inhibitors, non-nucleoside RT inhibitors and protease inhibitors was defined according to the 2012 ANRS HIV-1 genotypic resistance interpretation algorithm (www.hivfrenchresistance.org).
HIV-1 subtype was determined by phylogenetic analysis of the RT and V3 env sequences, based on sequence comparisons with previously reported representatives of group M including the reference sequences of subtypes and sub-subtypes, and all the CRF sequences available in the HIV database or genbank (up to CRF54_01B) (http://www.hiv.lanl.gov).
Viral RNA Extraction and cDNA Synthesis
Viral RNA was extracted from each sample using a home-made protocol: the patient plasma sample (500 µl) was centrifuged at 16,000 rpm for 1 hour at +4°C. The supernatant was removed, and 600 µl of HCV LYS v2.0 (from the Amplicor® HCV Specimen Preparation Kit, Roche Diagnostics, Mannheim, Germany) was added, followed by an incubation during 10 minutes at room temperature. A 600-µl of volume of isopropanolol was added and RNA was precipitated at 14,300 rpm for 15 minutes at +20°C. The supernatant was removed and the RNA pellet was rinsed with 1,000 µl of 70% ethanol. The tube was centrifuged at 14,300 rpm for 5 minutes at +20°C. The supernatant was removed and the RNA pellet was resuspended in 30 µl of water. RNA was immediately subjected to first strand cDNA synthesis by the Titan One Tube RT-PCR System® and the Protector Rnase Inhibitor® (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Each first strand synthesis reaction included 0.4 µM of the antisense primer ED12 (see sequence below). In some experiments, a different antisense primer, Env8, was used (see sequence below). The reactions were incubated at 50°C for 30 minutes.
Proviral DNA Extraction
The QIAamp DNA Mini Kit® (Qiagen SA, Courtaboeuf, France) was used according to the manufacturer's instructions to extract total DNA from PBMCs from samples collected at inclusion.
Single Genome Amplification (SGA)
C2V5 env amplicons were obtained from plasma cDNA or genomic DNA using nested PCR amplification by SGA [20]. Input genomic DNA or plasma cDNA was diluted to a limiting concentration such that approximately one-third or less of all second-round reactions produced a positive env amplicon. At this dilution, approximately 90% of the reactions will have originated from a single virus genome in the reaction [21]. Taq DNA Polymerase® (Roche Applied Science, Mannheim, Germany) was used according to the manufacturer's instructions for nested PCR amplifications. C2V5 amplification (627-bp fragment) was performed using the following primer combinations: first round: sense primer ED3 (5'-TTAGGCATCTCCTATGGCAGGAAGAAGCGG-3'), antisense primer ED12 (5'-AGTGCTTCCTGCTGCTCCCAAGAACCCAAG-3'); second round: sense primer ES7 (5'-GTGAATTCCTGTTAAATGGCAGTCTAGC-3'; nucleotide position relative to HXB2 genome start: 6994->7021), antisense primer ES8 (5'-GTGAATTCCACTTCTCCAATTGTCCCTCA-3′; nucleotide position relative to HXB2 genome start: 7676->7648). In cases of misamplification, different primers were used (334-bp fragment): first round: sense primer Env31 (5'-CAGTACAATGTACACATGG-3'), antisense primer Env8 (5'-ATGGGAGGGGCATACATTG-3'); second round: sense primer Env7 (5'-AATGGCAGTCTAGCAGAAG-3'; nucleotide position relative to HXB2 genome start: 7008->7026), antisense primer ED33 (5'-TTACAGTAGAAAAATTCCCCTC-3'; nucleotide position relative to HXB2 genome start: 7381->7360). PCR were performed in a reaction volume of 50 µl with cycling parameters as previously published [22]. The amplified products were purified using a QIAquick PCR Purification Kit® (Qiagen SA, Courtaboeuf, France).
DNA Sequencing
Nucleotide sequences were determined by direct sequencing according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). Electrophoresis and data collection were performed on an ABI 3130 Genetic Analyser sequencer® (Applied Biosystems). Individual sequence fragments for each amplicon were assembled and edited using Sequence Navigator software [23]. All chromatograms were inspected for sites of mixed bases (double peaks), which would be evidence of priming from more than one template or the introduction of errors in early cycles of PCR. Any sequence with evidence of double peaks was excluded from the subsequent analyses.
Nucleotide sequences have been submitted to GenBank (accession numbers [KF142735-KF143426].
Sequence Alignments
Clustal X was used for sequence alignments [24]. Phylogenetic interrelationships among viral sequences were estimated using Neighbor-Joining trees [25], and maximum likelihood methods with BioEdit and MEGA4 integrated molecular evolutionary genetic analysis software [26], [27]. The reliability of the tree topology was estimated from 1,000 bootstraps replicates.
Hypermutated Samples
Enrichment for APOBEC3G/F mutations violates the assumption of a constant mutation rate across positions, as the editing performed by these enzymes are base- and context-sensitive. Enrichment for mutations with APOBEC3G/F signatures was assessed using Hypermut 2.0 (www.hiv.lanl.gov). Sequences that yielded a p-value of 0.05 or lower were considered significantly hypermutated and excluded from subsequent analyses.
Nucleotide Sequence Diversity Analysis
Pairwise sequence similarities were calculated with DNADIST (http://cmgm.stanford.edu/phylip/dnadist.html) using the Kimura two-parameter algorithm [28]. Each set of sequences was then inspected using the Highlighter v2.2.1 tool (www.hiv.lanl.gov).
Estimation of the Date of Infection of the Donors
The date of the donors' HIV-1 infection was unknown. Two methods were used to estimate the date. First, a serum sample was sent to the French National Reference Center for HIV and tested for recent infection by EIA-RI [29], [30]; this single indirect enzyme-linked immunosorbent assay quantifies antibodies for TM (gp41) and V3 peptides and has been validated as being able to discriminate recently infected individuals from those with long-lasting infection.
Second, we used a probabilistic modeling approach. For each data set, the env sequences were aligned using MAFFT (L-INS-i option) [31]. Sites with more than 75% gaps were removed, and we estimated the dates of infection of chronic patients as the date of the most recent common ancestor (MRCA) of his sequences, calculated using the Bayesian, MCMC-based program BEAST v1.7 [32]. We assumed a GTR+I+Γ4 substitution model and a strict molecular clock with a fixed substitution rate of 6×10−3 substitutions per site and per year (a standard value for env, see, for example, [33]). We used a Bayesian skyride tree prior as a coalescent demographic model with time-aware smoothing [34]. MCMC simulations were run for 2×108 chain steps with sub-sampling every 2×105. Convergence of the chains and results were inspected using Tracer v1.5. ESS values were larger than 200 for all parameters and all data sets, except for two parameters (prior and posterior) with the MRT data set. We also tested a lognormal relaxed molecular clock, but obtained poor results as nearly identical sequences were separated by large divergence times, most notably among recipient sequences. As a consequence, the infection date of the recipient was close to the infection date of the donor, thus contradicting clinical evidences.
Viral Tropism Determination
The SVMGeno2pheno algorithm (available at: http://coreceptor.bioinf.mpi-sb.mpg.de/cgi-bin/coreceptor.pl) with a 10% false positive rate was used to determine HIV-1 co-receptor usage by each virus.
Results
Study Subjects
SGA-direct sequencing was used to identify and enumerate transmitted/founder env sequences in eight patients with PHI, who reported sexual exposure as their primary HIV-1 risk behavior and who denied injection drug use (Table 1). These eight patients included three women and one man infected through heterosexual exposure and four MSM. All were infected with subtype B strains without resistance to the three main classes of antiretrovirals. One patient (recipient#2) was coinfected with HBV and HCV and three others (recipients#1, 3 and 7) reported histories of STI in the 6 months preceding HIV diagnosis. At the time of the study, four of the subjects were ELISA+/WB indeterminate, one was ELISA+/WB+/p31- and three were ELISA+/WB+/p31+.
Table 1. Characteritics of the 8 recipients.
| Recipient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Date of sample collection | 2/12/2002 | 3/25/1998 | 01/02/1999 | 6/26/2000 | 3/1/1999 | 4/18/2007 | 6/16/1999 | 10/24/2008 |
| Estimated time since infection (days) | 33 | 37 | 37 | 40 | 43 | 85 | 32 | 38 |
| HIV-1 RNA (log10 copies/mL) | 5.93 | 4.70 | 4.44 | 5.52 | 6.07 | 4.01 | 4.83 | 4.62 |
| HIV-1 DNA (log10 copies/10e6 PBMC) | 3.25 | 3.12 | 3.15 | 2.86 | 3.64 | 2.76 | 3.84 | ND |
| CD4 cell count (cells/mm3) | 520 | 734 | 368 | 681 | 381 | 635 | 558 | 523 |
| Viral subtype | B | B | B | B | B | B | B | B |
| Viral resistance | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Age (years) | 38 | 28 | 36 | 21 | 28 | 39 | 29 | 19 |
| Gender (male/female) | M | M | F | M | F | M | M | F |
| Risk group | Het | MSM | Het | MSM | Het | MSM | MSM | Het |
| Country of birth | France | Chile | France | France | Algeria | France | France | France |
| Living place | France | France | France(West Indies) | France (West Indies) | France | France | France | France (West Indies) |
| Number of regular sexual partners * | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| Number of casual sexual partners * | 0 | 15 | 0 | 0 | 0 | 5 | 3 | 0 |
| Serological syphilis testing | – | – | – | – | – | – | – | – |
| HBV testing | ||||||||
| HBsAg | – | – | – | – | – | – | – | – |
| HBsAb | – | + | – | – | – | + | – | – |
| HBcAb | – | + | – | – | – | – | – | – |
| HBV DNA (log10 IU/mL) | n.d. | Und. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| HCV testing | ||||||||
| HCV Ab | – | + | – | – | – | – | – | – |
| HCV RNA (log10 IU/mL) | n.d. | 1.7 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Prior sexually transmitted infections § | Condyloma | No | Vaginal candidiasis | No | No | No | Condyloma | No |
In the 6 months preceding PHI diagnosis.
Urethritis, rectitis, genital herpes infection, vulvo-vaginal candidosis, condyloma and/or syphilis.
PBMC = peripheral blood mononuclear cells; Het = heterosexual; MSM = man having sex with men; HBV = hepatits B virus; HBsAg = HBV surface antigen; HBsAb = HBV surface antibodies; HBcAb = HBV core antibodies; HCV = hepatitis C virus; n.d. = not done; und = undetectable.
Blood samples from the donors were obtained concomitantly (donor#4), 14 (donors#1-3 and 6-7) or 30 days (donors#5 and 8) after the enrolment of their respective recently infected partner (Table 2). Envelope sequences in these samples were analyzed: the EIA-RI test suggested that the donors had been infected less than 6 months previously in two cases (donors#3 and 7), and that the other six patients had long-lasting infections. These findings were confirmed by our modeling approach: donors#3 and 7 were estimated to have been infected 0.46 and 0.20 years, respectively, prior to collection of the blood sample and the remaining donors were estimated to have been infected between 2.66 and 11.17 years previously (Table 2). Histories of STI were not available for donors. However, the microbiological screening for coinfections was positive in five patients; the pathogens identified were syphilis (n = 2, donors #4 and 6), HBV (n = 4, #2, 3, 4 and 5) and HCV (n = 1, #5). There was no evidence of transmission of these infections to their partners.
Table 2. Characteritics of the 8 donors.
| Donor | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| Date of sample collection (compared with that of the recipient) | 2 weeks later | 2 weekslater | 2 weekslater | at the sametime | 1 monthlater | 2 weekslater | 2 weekslater | 1 monthlater |
| Date of diagnosis of HIV infection (compared with that of the recipient) | 12 years before | 1 week before | <2 months before | 1 week later | 1 year before | 11 years before | <6 months before | n.a. |
| Time of infection (estimated with a EIA-RI test) (months) | >6 | >6 | <6 | >6 | >6 | >6 | <6 | >6 |
| Time of infection (estimated using a mathematical model) (years) (mean, range) | 3.85(2.95–4.86) | 2.66(2.06–3.36) | 0,46(0.32–0.62) | 9.41(7.51–11.39) | 11.17(8.66–14.03) | 4.14(3.05–5.41) | 0.20(0.09–0.31) | 7.18(5.39–8.86) |
| HIV-1 RNA (log10 copies/mL) | 5.89 | 4.75 | 5.22 | 4.20 | 4.40 | 5.80 | 5.10 | 4.60 |
| HIV-1 DNA (log10 copies/10e6 PBMC) | 3.53 | 3.64 | 3.03 | 3.73 | 2.81 | 3.90 | 3.33 | 3.91 |
| CD4 cell count (cells/mm3) | 216 | 503 | n.a. | 200–500 | n.a. | 200–500 | 800 | n.a. |
| Viral subtype | B | B | B | B | B | B | B | B |
| Viral resistance | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Age (years) | 40 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Gender (male/female) | F | M | M | M | M | M | M | M |
| Risk group | Het | MSM | Het | MSM | Het | MSM | MSM | Het |
| Country of birth | n.a. | Egypt | France | n.a. | n.a. | France | France | France |
| Serological syphilis testing | – | – | – | + | – | + | – | – |
| HBV testing | ||||||||
| HBsAg | – | – | – | + | – | – | – | – |
| HBsAb | – | + | + | – | – | + | + | – |
| HBcAb | – | + | + | + | + | – | – | – |
| HBV DNA (log10 IU/mL) | n.d. | Und. | Und. | 1.3 | Und. | n.d. | n.d. | n.d. |
| HCV testing | ||||||||
| HCV Ab | – | – | – | – | + | – | – | – |
| HCV RNA (log10 IU/mL) | n.d. | n.d. | n.d. | n.d. | 5.9 | n.d. | n.d. | n.d. |
Results based on the declaration of the recipient.
EIA-RI test = enzyme-linked immunosorbent assay for recent infection [29], PBMC = peripheral blood mononuclear cells; Het = heterosexual; MSM = man having sex with men; HBV = hepatits B virus; HBsAg = HBV surface antigen; HBsAb = HBV surface antibodies; HBcAb = HBV core antibodies; HCV = hepatitis C virus; n.d. = not done; n.a. = data not available; und = undetectable.
HIV-1 env Diversity Analysis
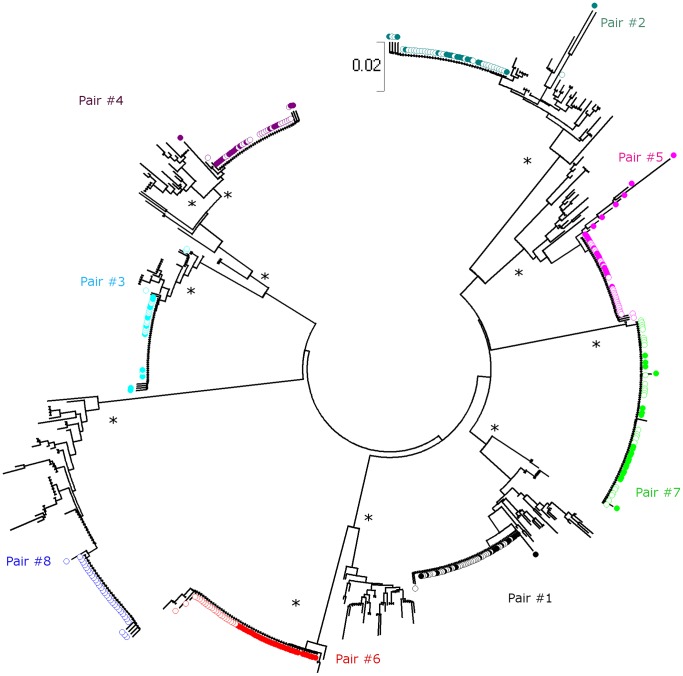
Totals of 316 and 376 C2V5 env sequences were obtained from plasma vRNA and PBMC-associated DNA, respectively (median of 21 RNA sequences per subject; range 12–33; and 22.5 DNA sequences per subject; range 14–39). DNA env sequences could not be obtained from recipient#8 because appropriate samples were not available. A composite Neighbor-Joining phylogenetic tree was generated (Figure 1): the viral sequences formed eight distinct donor/recipient-pair-specific monophyletic lineages, each with strong statistical support (bootstrap values >98%), indicating that neither cross-contamination from other samples nor related transmission networks have occurred. Sequences from PBMC DNA and plasma RNA were distributed throughout the branch patterns of each donor and recipient, suggesting that these two sources were not compartmentalized. In all transmission pairs, recipient env sequences were highly homogeneous forming a distinct monophyletic subcluster within the tree of donor sequences. An example of the trees of donor/recipient pairs is given in Figure 2 (pair#1).
Figure 1. Evolutionary relationships between the HIV-1 env genes in the eight donor/recipient pairs.
The evolutionary history was inferred using the Neighbor-Joining method [38]. The optimal tree with the sum of branch length = 2.01912678 is shown. The tree is drawn to scale, with branch length in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [39] and the unit is the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+noncoding. All positions containing gaps and missing data were eliminated from the dataset. There were a total of 230 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [40]. For each recipient, viruses isolated from PBMC-derived DNA (•) and plasma RNA (○) are represented, with a different color for each donor/recipient pair. Asterisks indicate branches with bootstrap values greater than 98%.
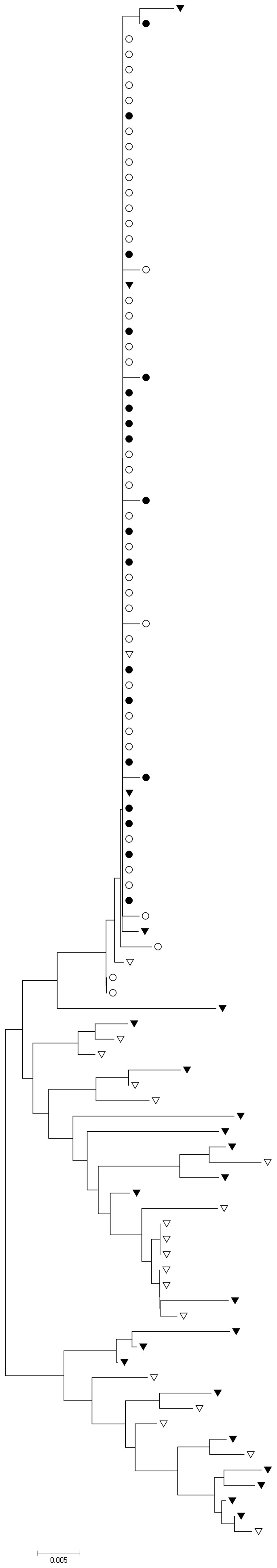
Figure 2. Evolutionary relationships between the HIV-1 env genes in donor/recipient pair #1.
The evolutionary history was inferred using the Neighbor-Joining method [38]. The optimal tree with the sum of branch length = 0.28572580 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [39] and the unit is the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 500 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [40]. For the recipient, viruses from PBMC-derived DNA (•) and plasma RNA (○) are represented. For the donor, strains from PBMC-derived DNA (▾) and plasma RNA (∇) are represented.
In 6/8 cases, the donor sequences were highly heterogeneous (mean within-patient env diversity = 4.03% (range 2.70–5.93)), consistent with the long-lasting infection of their host. By contrast, the env diversities were below 1% in donors infected less than 6 months ago (mean 0.51% and 0.14% in donors #3 and 7, respectively). Maximum within-patient env diversities for the eight recipients ranged from 0.04% to 0.34% for DNA sequences (mean 0.18%) and from 0.11% to 0.33% for RNA sequences (mean 0.17%) (Table 3). Mean within-patient env diversities were similar in cases of heterosexual and homosexual transmission (0.26% versus 0.14%, and 0.18% versus 0.13% for DNA and RNA sequences, respectively). All nucleotide diversity values were <0.75%, consistent either with single variant transmission or with transmission of only closely related viruses [13], [15]–[17], [35].
Table 3. Diversity and tropism analyses of V3 env sequences from 8 HIV-1 infected donor/recipients pairs.
| Donor/recipient pair | Origin of sequences | No. Of SGA env sequences | APOBEC- mediated hypermutation | Nucleotide sequence diversity (%) | No. of CXCR4/DM-tropic virus | |||
| Mean | Range | n | % | |||||
| 1 | Recipient | DNA | 20a/21b | Yes | 0.18 | 0.00–0.51 | 0/20 | 0.0% |
| RNA | 39/39 | No | 0.11 | 0.00–0.49 | 0/39 | 0.0% | ||
| Donor | DNA | 22/23 | Yes | 3.83 | 0.17–6.10 | 0/22 | 0.0% | |
| RNA | 19/19 | No | 3.23 | 0.00–5.31 | 0/19 | 0.0% | ||
| 2 | Recipient | DNA | 15/16 | Yes | 0.60 | 0.00–2.31 | 0/15 | 0.0% |
| RNA | 29/29 | No | 0.33 | 0.00–2.59 | 0/29 | 0.0% | ||
| Donor | DNA | 18/19 | Yes | 2.88 | 0.33–4.86 | 1/18 | 5.6% | |
| RNA | 21/21 | No | 2.07 | 0.00–3.29 | 0/21 | 0.0% | ||
| 3 | Recipient | DNA | 12/12 | No | 0.09 | 0.00–0.53 | 0/12 | 0.0% |
| RNA | 14/14 | No | 0.11 | 0.00–0.47 | 0/14 | 0.0% | ||
| Donor | DNA | 23/23 | No | 0.38 | 0.00–0.99 | 0/23 | 0.0% | |
| RNA | 25/25 | No | 0.63 | 0.00–1.30 | 1/25 | 4.0% | ||
| 4 | Recipient | DNA | 19/19 | No | 0.34 | 0.00–1.37 | 0/19 | 0.0% |
| RNA | 21/21 | No | 0.17 | 0.00–0.80 | 0/21 | 0.0% | ||
| Donor | DNA | 16/16 | No | 7.55 | 0.00–12.77 | 0/16 | 0.0% | |
| RNA | 23/23 | No | 4.80 | 0.00–10.46 | 0/23 | 0.0% | ||
| 5 | Recipient | DNA | 19/23 | Yes | 0.34 | 0.00–1.60 | 19/19 | 100.0% |
| RNA | 25/25 | No | 0.21 | 0.00–0.82 | 25/25 | 100.0% | ||
| Donor | DNA | 15/15 | No | 7.30 | 0.00–12.89 | 1/15 | 6.7% | |
| RNA | 15/15 | No | 5.32 | 0.00–12.61 | 4/15 | 26.7% | ||
| 6 | Recipient | DNA | 33/33 | No | 0.04 | 0.00–0.32 | 0/33 | 0.0% |
| RNA | 22/22 | No | 0.16 | 0.00–0.86 | 0/22 | 0.0% | ||
| Donor | DNA | 24/25 | Yes | 4.85 | 0.00–10.37 | 0/24 | 0.0% | |
| RNA | 21/21 | No | 1.73 | 0.00–3.50 | 0/21 | 0.0% | ||
| 7 | Recipient | DNA | 20/20 | No | 0.04 | 0.00–0.46 | 0/20 | 0.0% |
| RNA | 26/26 | No | 0.14 | 0.00–1.71 | 0/26 | 0.0% | ||
| Donor | DNA | 23/23 | No | 0.16 | 0.00–0.89 | 0/23 | 0.0% | |
| RNA | 20/20 | No | 0.11 | 0.00–1.15 | 0/20 | 0.0% | ||
| 8 | Recipient | RNA | 31/31 | No | 0.18 | 0.00–0.88 | 31/31 | 100.0% |
| Donor | DNA | 25/28 | Yes | 4.01 | 0.00–6.80 | 4/25 | 16.0% | |
| RNA | 25/25 | No | 2.80 | 0.00–40.15 | 7/25 | 28.0% | ||
a = initial sequence set.
b = total number of sequence analyzed after exclusion of APOBEC-mediated hypermutations.
Within-patient env diversities for the eight donors ranged from 0.16% to 7.55% (mean 3.92%) for DNA sequences, and from 0.11% to 5.32% (mean 2.44%) for RNA sequences. Again, viral diversity in both DNA (0.38%, 0.16%) and RNA sequences (0.63%, 0.11%) was low in donors#3 and 7, consistent with recent infection. The viral diversity in the six other donors was significantly higher in DNA than in RNA sequences.
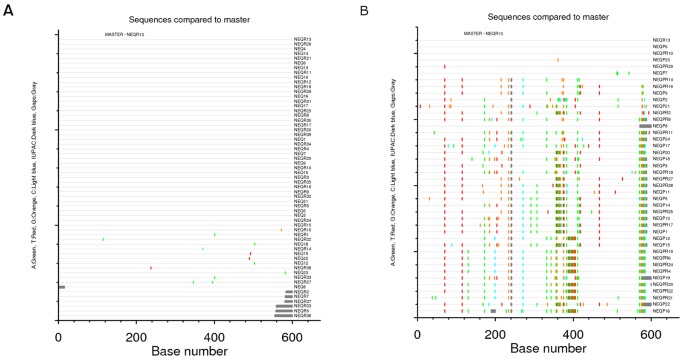
To evaluate the extent of sequence diversity in recipients, env sequences from the recipients were examined using the Highlighter tool, which allows comparison of each recipient env sequence to a reference recipient sequence and graphically depicts all nucleotide differences between the two. An example of the output of this tool for one recipient (recipient#1) is given in Figure 3A; it shows a remarkable degree of homogeneity of the env sequences in this recipient, whose infection was estimated to have occurred 33 days prior to sample collection. Approximately 80% (47/59) of the sequences are identical despite each being amplified from a unique viral genome. Similar proportions of identical sequences were observed in sequences obtained from both plasma (32/39) and PBMC (15/20). Compared to the reference amplicon, approximately 17% of the sequences exhibited a single nucleotide change, and these changes were randomly dispersed over the C2V5 region; 2% of the sequences carried two nucleotide changes. Similar results were obtained by Highlighter analyses of the seven other recipients, consistent with single-variant transmission in all cases.
Figure 3. Comparative Highlighter analyses of env diversity in a donor-recipient HIV-1 transmission pair.
(A) Recipient#1 shows evidence of infection with a single virus. (B) Donor#1 was the chronically infected partner of recipient#1. The same reference amplicon, a V3 RNA sequence from recipient plasma, was used to depict the viral diversity in both individuals.
Relationships between Recipient and Donor Quasispecies
The infection in each of the eight recipients appeared to have been established from a single donor variant. We sought to determine the frequency of this variant or closely related variants within the donor quasispecies and to examine whether such variants predominated in plasma- or PBMC-derived viruses in the donor. The number of nucleotide differences between each donor virus sequence and the consensus recipient virus sequence were calculated for each of the eight transmission pairs. In all cases, at least one donor variant was identified that differed by fewer than seven nucleotides from the consensus recipient variant (Table 4). The donor variants most closely related to the recipient sequences were found in both plasma (3/8) and PBMC (6/8) samples, such that it was not possible to establish whether the infecting virus was specifically derived from a single compartment. In the donor/recipient pairs #3 and 7, the viral quasispecies were largely homogeneous in both donors and recipients. In the other pairs, fewer than 20% of the donor sequences contained fewer than 10 nucleotide differences to the recipient consensus sequence. This is illustrated in Figure 3B, depicting the extent of sequence diversity in the donor#1 using the Highlighter tool, using the same reference amplicon as shown in Figure 3A (an env RNA sequence isolated from recipient plasma). These findings suggest than the virus establishing the new infection is derived from an infrequent circulating variant population in the donor.
Table 4. Relationship of donor variants to recipient consensus sequence.
| Recipient | Number of donor variants analyzed | Blood compartment of originof nearest donor variant | Number of nucleotide differencescompared to nearest donor variant | Number of donor sequences<10 nucleotides different | ||
| in plasmacompartment | in PBMCcompartment | |||||
| 1 | 41 | Plasma | 0 | 8 | 5 | 12% |
| 2 | 39 | PBMC | 11 | 2 | 3 | 8% |
| 3 | 48 | PBMC | 5 | 4 | 48 | 100% |
| 4 | 39 | Plasma | 6 | 13 | 5 | 13% |
| 5 | 30 | PBMC | 4 | 3 | 5 | 17% |
| 6 | 45 | PBMC | 58 | 6 | 5 | 11% |
| 7 | 43 | PBMC or Plasma | 1 | 1 | 43 | 100% |
| 8 | 50 | PBMC | 16 | 2 | 7 | 14% |
PBMC = peripheral blood mononuclear cells.
Characterization of Coreceptor Tropism of Transmitted/founder Viruses
HIV-coreceptor usage determination using SVMGeno2pheno10% showed that all donors were predominantly infected with CCR5-tropic strains (“R5-strains”). However, there was a minority of CXCR4/dual-mixed (“X4/DM” strains) viruses among DNA and/or RNA sequences in 4/8 donors; these strains represented 2.1% (donor#3), 2.6% (#2), 16.7% (#5) and 22.0% (#8) of the viral quasispecies.
Again, the viral quasispecies identified in the eight recipients were highly homogeneous and displayed exclusive CCR5-tropism in 6/8 cases and exclusive X4/DM-tropism in 2 cases (recipients#5 and 8). The two patients harboring X4/DM viruses were women infected through heterosexual contact each with a donor harboring a minority of X4/DM-tropic strains (6.7% of DNA and 26.7% of RNA sequences for donor#5; 16.0% of DNA and 28.0% of RNA sequences for donor#8). The proportion of X4/DM quasispecies in donors was higher in cases of X4/DM than R5 HIV transmission (16.7–22.0% versus 0–2.6%).
Discussion
We report an analysis of eight transmission pairs, each including a sexually infected patient enrolled into the French ANRS PRIMO cohort at the time of primary HIV-1 infection and his/her donor. This provided a unique opportunity to compare epidemiologically linked virus populations in donors and recipients very close to the time of virus transmission, contrary to many previous studies which only characterized the transmitted viral populations in recipients without available virological data in donors.
Our study was based on the novel SGA-based approach involving more precise methods than those used in many previous studies for estimating the multiplicity of HIV-1 infection in recently infected individuals [3], [5], [7], [8], [10]–[12], [36], [37]. We found that the viral populations, as assessed from both DNA and RNA sequences, were highly homogeneous in each of the eight sexually PHI patients. Our findings are consistent with recent studies, based on similar methods, which suggest that there is a severe genetic bottleneck associated with sexual transmission of HIV-1 [13], [14], [16], [17]. The observed highly homogeneous viral population in PBMC is consistent with a single variant being transmitted, which then massively fuels the cellular reservoir, rather than multiple variants being transmitted rapidly followed by virus population homogenization in the recipient [37].
We herein show that the donor variant most closely related to the strain establishing infection was infrequently found within the quasispecies present in donor blood. This result suggests that the recipient’s infection is not due to the preferential transmission of a strain overrepresented among circulating quasispecies in donor. There are several possible explanations for this finding. First, the viral transmission could result from a stochastic process of a donor’s variant whatever its frequency among blood quasispecies. Second, the severe genetic bottleneck which occurs during sexual transmission, involves properties of the “mucosal barrier” and/or selection of blood variants with properties favoring transmission, as recently suggested [16]. In both hypotheses, we cannot exclude that compartmentalization between blood and genital viral subpopulations may contribute to the selection of the transmitted/founder strain: at the time of transmission, the predominant strains may be different in the blood and genital compartments. Boeras et al recently analyzed viral envelope sequences in the blood and genital fluids of eight transmission pairs and found that the viruses establishing infection were in most cases more closely related to blood-derived variants than to the variants prevalent in the genital compartment [38]. However, a recent study in the SIV macaque model suggested that SIV sequences are intermixed between the blood and the semen at the time of peak virus replication, but that SIV replication evolves to compartmentalization in the male genital tract after peak viremia resolves [39]. Similarly, Redd et al working with Ugandan HIV-1-discordant couples showed preferential transmission of ancestral as opposed to contemporary strains circulating in the donor [40]. The authors suggested that transmitted strains may be sequestered in a long-lived reservoir during the early stage of infection, such as latently infected cells of the genital tract and persist at a low level in blood and are potentially preferentially selected for subsequent transmission.
In 2010, a retrospective comparison of multivariant HIV-1 transmission among patient cohorts using new SGA-based determinations concluded that the multiplicity infection is higher in MSM than in heterosexuals (38% versus 19%) [17], consistent with previous studies using less precise methods [36]. We report an homogeneous viral population in each of eight sexually infected patients enrolled near the time of PHI, including four male-to-male, three male-to-female and one female-to-male transmissions. The small number of MSM donors included in our study and the fact than one of them (donor#7) harbored a homogeneous viral population may explain the absence of differences between MSM and heterosexuals. However, our findings are in line with a much larger study of clonal env sequences obtained from 145 patients at the time of PHI, which suggested that homosexual (versus heterosexual) transmission mode did not predict transmission of more heterogeneous founder virus populations [41].
In addition, we did not evidence any case of multiple infection in patients with STI, although 3/8 recipients reported STI in the 6 months prior to PHI diagnosis and 2/8 donors tested positive for syphilis at the time of blood collection. These findings are in line with the Rieder’s study, which did not find elevated complexity of transmitted viruses in patients infected through sexual intercourse and presenting with a concomitant STI during PHI [41]. In contrast to these results, Haaland et al reported that multiple variant transmission was associated with the presence of genital inflammation or ulceration or with self-reported lower abdominal pain in 42 recently infected heterosexuals [16]; however, the authors did not find any association between multiple variant transmission and vaginal/urethral discharge, cystitis or the presence of genital inflammation or genital ulceration when analyzed as independent risk factors. Sagar et al previously suggested that the presence of genital tract infections was associated with the acquisition of multiple variants in Kenyan female sex workers [42]. However, samples from the sexual partners of these patients were not available in this study, so it is not clear whether some of these multiple infections were or were not due to successive infections rather than to concomitant transmission of multiple variants. Further studies including samples from both donors and recipients are needed to characterize the type of genital disease susceptible to increase the risk of multiple variant transmission.
The eight donors in our study were predominantly infected by R5-tropic strains, but two recipients were infected with a homogeneous X4/DM viral population, isolated in both DNA and RNA samples. Interestingly, the proportions of X4/DM viruses in the viral quasispecies in their respective donors were significantly higher than those in the other donors. There are at least two possible explanations for these findings. First, our cases of X4-tropic viral transmission may have been driven by a disproportionately higher proportion of X4 strains in the donor’s genital fluids than in the blood compartment. However, this is not consistent with previous reports, which indicate that the frequency of X4/DM quasispecies in both male [43] and female [44] genital tracts are lower than in blood plasma. Second, the transmission of X4/DM variants could result from a stochastic process. This would be inconsistent with the conclusions of numerous studies which have attempted to correlate the predominance of CCR5 strains during the acute phase of infection with a biological bottleneck inherent to the genital mucosa [45], [46]. However, no conclusive evidence has been provided to indicate that X4 viruses are less transmissible [47] and a recent study concluded that R5 and X4-infections may result from a stochastic process [48]. This conclusion is coherent with our results, which suggest that the transmission of X4 strains is associated with a threshold population of X4 quasispecies in donor plasma and PBMC samples.
That a single virus, derived from an infrequent variant of the donor quasispecies, establishes infection in patients confirms that a severe genetic bottleneck occurs during subtype B HIV-1 heterosexual and homosexual transmission. Additional studies are required to fully understand the traits that confer the capacity to transmit and establish infection, and determine the role of concomitant STIs in mitigating the genetic bottleneck in mucosal transmission. Such studies will be critical for guiding interventions aimed at preventing HIV-1 sexual transmission.
Acknowledgments
We thank all patients and their physicians for their participation in the ANRS PRIMO cohort study (http://u822.kb.inserm.fr.gate2.inist.fr/COHAD/participantsPRIMO.htm), and Francis Barin (French National Reference Center for HIV, Tours, France) and Stéphane Le Vu (Institut National de Veille Sanitaire) for technical assistance and continuous helpful discussions.
ANRS PRIMO Cohort Study Group:
- Bruno ABRAHAM, Service de Médecine interne, Centre hospitalier de Brive, Brive, France.
- Thierry ALLEGRE, Service d’Hématologie, Centre hospitalier du pays d’Aix, Centre hospitalier intercommunal Aix-Pertuis, Aix-en-Provence, France.
- Odile ANTONIOTTI, Service de Dermatologie, Centre hospitalier, Montluçon, France.
- Raymond ARMERO, Service de Dermatologie – Infectiologie, Centre hospitalier intercommunal de Fréjus – Saint Raphaël, Fréjus, France.
- Bruno AUDHUY, Service de Médecine interne, Hôpitaux Civils de Colmar, Hôpital Louis Pasteur, Colmar, France.
- Hughes AUMAITRE, Service des Maladies infectieuses, Centre hospitalier Joffre, Perpignan, France.
- Gilles BEAUCAIRE, Service des Maladies infectieuses et de Dermatologie, Centre hospitalier universitaire Abymes, Pointe à Pitre, Guadeloupe, France.
- Geneviève BECK-WIRTH, Unité fonctionnelle Déficit immunitaire, Hôpital E. Muller, Mulhouse, France.
- Jean-Luc BERGER, CHU de Reims, Hôpital Robert Debré, Reims, France.
- Louis BERNARD, Service de Médecine interne C et Maladies infectieuses, Hôpital Bretonneau, Tours, France.
- Claire BEUSCART, Service de Médecine interne et Maladies infectieuses, Centre Hospitalier de Saint Brieux, Hôpital de La Beauchée, Saint Brieuc, France.
- Loïc BODARD, Service de Médecine interne, Institut Mutualiste Jourdan-Monsouris, Paris, France.
- Olivier BOUCHAUD, Service des Maladies infectieuses et tropicales, Hôpital Avicenne, Bobigny, France.
- François BOUE, Service de Médecine interne et Immunologie clinique, Assistance Publique – Hôpitaux de Paris, Hôpital Antoine Béclère, Clamart, France.
- Jean-Paul CABANE, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Saint Antoine, Paris, France.
- Andre CABIE, CISIH, Centre hospitalo-universitaire de Fort de France, Fort de France, Martinique, France.
- Hélène CHAMPAGNE, Service de Pneumologie et Maladies infectieuses, Centre hospitalier de Valence, Valence, France.
- Christine CHENEAU, Centre de Soins de l’Infection par le VIH, Hôpitaux universitaires de Strasbourg, Le Trait d’Union, Nouvel Hôpital Civil, Strasbourg, France.
- Jean-Marie CHENNEBAULT, Service des Maladies infectieuses, Centre hospitalo-universitaire d’Angers, Angers, France.
- Antoine CHERET, Service universitaire des Maladies infectieuses et du Voyageur, Centre hospitalier de Tourcoing, Tourcoing, France.
- Bernard CHRISTIAN, Service de Médecine A, Groupement des Hôpitaux de Metz, Hôpital Notre-Dame du Bonsecours, Metz, France.
- Alexandra COMPAGNUCCI, Service de Pneumologie, Assistance Publique – Hôpitaux de Paris, Hôtel Dieu, Paris, France.
- Vincent DANELUZZI, Service de Médecine A, Centre d’Accueil et de Soins hospitaliers, Hôpital Max Fourestier, Nanterre, France.
- Yasmine DEBAB, Service des Maladies infectieuses et tropicales, Hôpital Charles Nicolle, Rouen, France.
- Pierre DELLAMONICA, Service des Maladies infectieuses et tropicales, Centre hospitalo-universitaire de Nice, Hôpital de l’Archet, Nice, France.
- Jean-François DELFRAISSY, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Bicêtre, Le Kremlin Bicêtre, France.
- Alain DEVIDAS, Service d’hématologie, Centre Hospitalier Gilles de Corbeil, Corbeil Essonnes, France.
- Georges DIAB, Service de Médecine interne, Centre hospitalier de la Vallée de l’Oise, Noyon, France.
- Jacques DOLL, Service d’Hépato-gastro-entérologie, Centre hospitalier du Chesnay, Hôpital André Mignot, Versailles, France.
- Marie-Christine DROBACHEFF-THEBAUT, Service de Dermatologie, Hôpital Saint Jacques, Besançon, France.
- Aurélie DUREL, Service de Médecine interne A, Assistance Publique – Hôpitaux de Paris, Hôpital Lariboisière, Paris, France.
- Claudine DUVIVIER, Service des Maladies infectieuses et tropicales, Assistance Publique – Hôpitaux de Paris, Centre hospitalo-universitaire Necker – Enfants malades et Institut Pasteur, Paris, France.
- Jean-Luc ESNAULT, Service Médecine Post-Urgences, Centre hospitalier départemental Les Oudairies, La Roche-sur-Yon, France.
- Lydia FABA, Service de Médecine interne, Hôpital Saint Louis, La Rochelle, France.
- Eric FROGUEL, Service des Maladies infectieuses et tropicales et Médecine interne, Centre hospitalier de Lagny, Lagny, France.
- Daniel GARIPUY, Service de Médecine interne, Hôpital Joseph Ducuing, Toulouse, France.
- Valérie GARRAIT, Service de Médecine interne, Centre hospitalier intercommunal de Créteil, Créteil, France.
- Loïck GEFFRAY, Service de Médecine interne, Hôpital Robert Bisson, Lisieux, France.
- Claire GENET, Service des Maladies infectieuses et tropicales, Centre hospitalo-universitaire Dupuytren, Limoges, France.
- Philippe GENET, Service d’Hématologie, Centre hospitalier Victor Dupouy, Argenteuil, France.
- Laurence GERARD, Service d’Immunologie clinique, Assistance Publique – Hôpitaux de Paris, Hôpital Saint Louis, Paris, France.
- Jean-Jacques GIRARD, Service de Médecine interne, Centre hospitalier, Loches, France.
- Pierre-Marie GIRARD, Service des Maladies infectieuses et tropicales, Assistance Publique – Hôpitaux de Paris, Hôpital Saint-Antoine, Paris, France.
- Claire GODIN-COLLET, Service de Médecine C, Centre hospitalier du Chesnay, Hôpital André Mignot, Versailles, France.
- Bruno HOEN, Service des Maladies infectieuses, Hôpital Saint Jacques, Besançon, France.
- Dominique HOULBERT, Service de Médecine interne 2, Centre hospitalier d’Alençon, Alençon, France.
- Christine JACOMET, Service des Maladies infectieuses et tropicales, Centre hospitalo-universitaire de Clermont-Ferrand, Hôpital Gabriel Montpied, Clermont-Ferrand, France.
- Vincent JEANTILS, Polyclinique de Médecine, Assistance Publique – Hôpitaux de Paris, Hôpital Jean Verdier, Bondy, France.
- Kaoutar JIDAR, Service de Médecine, Centre hospitalier Marc Jacquet, Melun, France.
- Christine KATLAMA, Service des Maladies infectieuses et tropicales, Assistance Publique – Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, Paris, France.
- Elise KLEMENT, Service des Maladies infectieuses, Centre médico-chirurgical de Bligny, Briis-sous-Forges, France.
- Alain LAFEUILLADE, Service d’Infectiologie, Centre hospitalier intercommunal de Toulon – La Seyne sur Mer, Hôpital Sainte Musse, Toulon, France.
- Caroline LASCOUX, Service des Maladies infectieuses, Assistance Publique – Hôpitaux de Paris, Hôpital Saint Louis, Paris, France
- Vincent LAUNAY, Service de Médecine C, Centre hospitalier Louis Pasteur, Cherbourg, France.
- Annie LEPRETRE, Unité de Prise en Charge ambulatoire de la Maladie VIH/SIDA, Hôpital Simone Veil, Eaubonne, France.
- Yves LEVY, Service d’Immunologie clinique, Hôpital Henri Mondor, Créteil, France.
- Djamila MAKHLOUFI, Service d’Immunologie clinique, Hospices civils de Lyon, Hôpital Edouard Herriot, Lyon, France.
- Denis MALBEC, Service de Médecine interne, Centre hospitalier intercommunal Robert Ballanger, Aulnay-sous-Bois, France.
- Benoit MARTHA, Service de Médecine interne, Centre hospitalier William Morey, Chalon-sur-Saône, France.
- Thierry MAY, Service des Maladies infectieuses et tropicales, Centre hospitalo-universitaire de Nancy, Hôpitaux de Brabois, Nancy, France.
- Dominique MERRIEN, Service de Médecine interne, Centre hospitalier, Compiègne, France.
- Patrick MIAILHES, Service des Maladies infectieuses et tropicales, Hôpital de la Croix Rousse, Lyon, France.
- Christian MIODOVSKI, Paris, France.
- Jean-Michel MOLINA, Service des Maladies infectieuses et tropicales, Assistance Publique – Hôpitaux de Paris, Hôpital Saint Louis, Paris, France.
- Philippe MORLAT, Service de Médecine interne et Maladies infectieuses, Centre hospitalo-universitaire de Bordeaux, Hôpital Saint André, Bordeaux, France.
- Emmanuel MORTIER, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Louis Mourier, Colombes, France.
- Didier NEAU, Service des Maladies infectieuses A, Centre hospitalo-universitaire de Bordeaux, Hôpital Pellegrin, Bordeaux, France.
- Martine OBADIA, Service des Maladies infectieuses et tropicales, Hôpital Purpan, Toulouse, France.
- Olivier PATEY, Service de Médecine interne, Centre hospitalier intercommunal de Villeneuve Saint Georges, Villeneuve-Saint-Georges, France.
- Jean-Luc PELLEGRIN, Service de Médecine interne et Maladies infectieuses, Centre hospitalo-universitaire de Bordeaux, Hôpital Haut Lévêque, Bordeaux, France.
- Véronique PERRONNE, Service des Maladies infectieuses, Centre hospitalier François Quesnay, Mantes-la-Jolie, France.
- Patrick PHILIBERT, Consultations de Médecine interne, Hôpital Ambroise Paré, Marseille, France.
- Gilles PIALOUX, Service des Maladies infectieuses, Assistance Publique – Hôpitaux de Paris, Hôpital Tenon, Paris, France.
- Gilles PICHANCOURT, Service de Médecine interne, Onco-hématologie et Maladies infectieuses, Centre hospitalier d’Avignon – Henri Duffaut, Avignon, France.
- Lionel PIROTH, Service des Maladies infectieuses, Hôpital du Bocage, Dijon, France.
- Yves POINSIGNON, Service de Médecine interne, Centre hospitalier Bretagne Atlantique, Vannes, France.
- Isabelle POIZOT-MARTIN, Service d’hématologie, Centre d'Information et de Soins de l'Immunodéficience humaine, Hôpital Sainte Marguerite, Marseille, France.
- Thierry PRAZUCK, Service des Maladies infectieuses, Centre hospitalier régional d’Orléans, Hôpital Porte Madeleine, Orléans, France.
- Virginie PRENDKI, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Jean Verdier, Bondy, France.
- François PREVETEAU DU CLARY, Service de Dermatologie et Médecine sociale, CHU La Grave, Toulouse, France.
- Denis QUINSAT, Service de Médecine 3, Centre hospitalier d’Antibes – Juan les Pins, Antibes, France.
- François RAFFI, Service de Médecine interne et des Maladies infectieuses, Centre hospitalo-universitaire de Nantes, Hôpital Hôtel Dieu, Nantes, France.
- Alain REGNIER, Service de Médecine interne, Centre hospitalier de Vichy, Vichy, France.
- Jacques REYNES, Service des Maladies infectieuses et tropicales, Centre hospitalo-universitaire de Montpellier, Hôpital Gui de Chauliac, Montpellier, France.
- Eric ROSENTHAL, Service de Médecine interne, Centre hospitalo-universitaire de Nice, Hôpital de l’Archet, Nice, France.
- Elisabeth ROUVEIX, Service de Médecine interne, Hôpital Ambroise Paré, Boulogne, France.
- Dominique SALMON, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Cochin, Paris, France.
- Fabienne SANLAVILLE, Service de Médecine C, Hôpital de Sens, Sens, France.
- Jean-Luc SCHMIT, Service de Pathologie infectieuse, Centre hospitalo-universitaire d’Amiens, Hôpital Nord, Amiens, France.
- Anne SIMON-COUTELLIER, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, Paris, France.
- Albert SOTTO, Service des Maladies infectieuses et tropicales, Hôpital Carémeau, Nîmes, France.
- Faouzi SOUALA, Clinique des Maladies infectieuses, Hôpital Pontchaillou, Rennes, France.
- Andréas STEIN, Service des Maladies infectieuses, Hôpital de la Conception, Marseille, France.
- Françoise TIMSIT, Clinique et Biologie des Maladies sexuellement transmissibles, Assistance Publique – Hôpitaux de Paris, Hôpital Saint Louis, Paris, France.
- Pierre de TRUCHIS, Service des Maladies infectieuses et tropicales, Assistance Publique – Hôpitaux de Paris, Hôpital Raymond Poincaré, Garches, France.
- Agnès ULUDAG, Service de Médecine interne, Assistance Publique – Hôpitaux de Paris, Hôpital Beaujon, Paris, France.
- Odile VAILLANT, Service d’Hématologie, Centre hospitalier Bretagne-Sud, Lorient, France.
- Renault VERDON, Service des Maladies infectieuses, Centre hospitalo-universitaire Côte de Nacre, Caen, France.
- Annie VERLESCH-LANGLIN, Service de Dermatologie, Centre hospitalier de Valenciennes, Valenciennes, France.
- Jean-Paul VIARD, Centre de Diagnostic et de Thérapeutique, Assistance Publique – Hôpitaux de Paris, Hôtel Dieu, Paris, France.
- Daniel VITTECOQ, Service des Maladies infectieuses, Assistance Publique – Hôpitaux de Paris, Hôpital Bicêtre, Le Kremlin Bicêtre, France.
- Laurence WEISS, Service d’Immunologie clinique, Assistance Publique – Hôpitaux de Paris, Hôpital Européen Georges Pompidou, Paris, France.
- Patrick YENI, Service des Maladies infectieuses et tropicales, Assistance Publique – Hôpitaux de Paris, Hôpital Bichat, Paris, France.
Funding Statement
This work was supported by grants from the French National Agency for Research on AIDS and viral hepatitis [ANRS PRIMO CO06] and by a scholarship (P. Frange) from SIDACTION [BI22-1-01851]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of this manuscript.
References
- 1. Delwart E, Magierowska M, Royz M, Foley B, Peddada L, et al. (2002) Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. AIDS 16: 189–195. [DOI] [PubMed] [Google Scholar]
- 2. Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, et al. (2004) Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303: 2019–2022. [DOI] [PubMed] [Google Scholar]
- 3. Wolfs TF, Zwart G, Bakker M, Goudsmit J (1992) HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189: 103–110. [DOI] [PubMed] [Google Scholar]
- 4. Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, et al. (1993) Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol 67: 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu T, Mo H, Wang N, Nam DS, Cao Y, et al. (1993) Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261: 1179–1181. [DOI] [PubMed] [Google Scholar]
- 6. Zhu T, Wang N, Carr A, Nam DS, Moor-Jankowski R, et al. (1996) Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol 70: 3098–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poss M, Martin HL, Kreiss JK, Granville L, Chohan B, et al. (1995) Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol 69: 8118–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grobler J, Gray CM, Rademeyer C, Seoighe C, Ramjee G, et al. (2004) Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J Infect Dis 190: 1355–1359. [DOI] [PubMed] [Google Scholar]
- 9. Long EM, Rainwater SMJ, Lavreys L, Mandaliya K, Overbaugh J (2002) HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res Hum Retroviruses 18: 567–576. [DOI] [PubMed] [Google Scholar]
- 10. Long EM, Martin HL Jr, Kreiss JK, Rainwater SM, Lavreys L, et al. (2000) Gender differences in HIV-1 diversity at time of infection. Nat Med 6: 71–75. [DOI] [PubMed] [Google Scholar]
- 11. Ritola K, Pilcher CD, Fiscus SA, Hoffman NG, Nelson JAE, et al. (2004) Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J Virol 78: 11208–11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sagar M, Kirkegaard E, Long EM, Celum C, Buchbinder S, et al. (2004) Human immunodeficiency virus type 1 (HIV-1) diversity at time of infection is not restricted to certain risk groups or specific HIV-1 subtypes. J Virol 78: 7279–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, et al. (2008) Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U.S.A. 105: 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, et al. (2008) Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 82: 3952–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abrahams M-R, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, et al. (2009) Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol 83: 3556–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, et al. (2009) Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog 5: e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Bar KJ, Wang S, Decker JM, Chen Y, et al. (2010) High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men. PLoS Pathog 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Troude P, Chaix ML, Tran L, Deveau C, Seng R, et al. (2009) No evidence of a change in HIV-1 virulence since 1996 in France. AIDS 23: 1261–1267. [DOI] [PubMed] [Google Scholar]
- 19. Chaix ML, Descamps D, Harzic M, Schneider V, Deveau C, et al. (2003) Stable prevalence of genotypic drug resistance mutations but increase in non-B virus among patients with primary HIV-1 infection in France. AIDS 17: 2635–2643. [DOI] [PubMed] [Google Scholar]
- 20. Shriner D, Rodrigo AG, Nickle DC, Mullins JI (2004) Pervasive genomic recombination of HIV-1 in vivo. Genetics 167: 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI (1997) Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res Hum Retroviruses 13: 737–742. [DOI] [PubMed] [Google Scholar]
- 22. Frange P, Chaix ML, Raymond S, Galimand J, Deveau C, et al. (2010) Low frequency of CXCR4-using viruses in patients at the time of primary non-subtype-B HIV-1 infection. J Clin Microbiol 48: 3487–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parker SR (1997) Sequence Navigator. Multiple sequence alignment software. Methods Mol Biol 70: 145–154. [PubMed] [Google Scholar]
- 24. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 26. Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U.S.A. 101: 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 28. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 29. Barin F, Meyer L, Lancar R, Deveau C, Gharib M, et al. (2005) Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J Clin Microbiol 43: 4441–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Vu S, Meyer L, Cazein F, Pillonel J, Semaille C, et al. (2009) Performance of an immunoassay at detecting recent infection among reported HIV diagnoses. AIDS 23: 1773–1779. [DOI] [PubMed] [Google Scholar]
- 31. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bello G, Passaes CP, Guimarães ML, Lorete RS, Matos Almeida SE, et al. (2008) Origin and evolutionary history of HIV-1 subtype C in Brazil. AIDS 22: 1993–2000. [DOI] [PubMed] [Google Scholar]
- 34. Minin VN, Bloomquist EW, Suchard MA (2008) Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25: 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee HY, Giorgi EE, Keele BF, Gaschen B, Athreya GS, et al. (2009) Modeling sequence evolution in acute HIV-1 infection. J Theor Biol 261: 341–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gottlieb GS, Heath L, Nickle DC, Wong KG, Leach SE, et al. (2008) HIV-1 variation before seroconversion in men who have sex with men: analysis of acute/early HIV infection in the multicenter AIDS cohort study. J Infect Dis 197: 1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Learn GH, Muthui D, Brodie SJ, Zhu T, Diem K, et al. (2002) Virus population homogenization following acute human immunodeficiency virus type 1 infection. J Virol 76: 11953–11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boeras DI, Hraber PT, Hurlston M, Evans-Strickfaden T, Bhattacharya T, et al. (2011) Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci U.S.A. 108: E1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitney JB, Hraber PT, Luedemann C, Giorgi EE, Daniels MG, et al. (2011) Genital tract sequestration of SIV following acute infection. PLoS Pathog 7: e1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Redd AD, Collinson-Streng AN, Chatziandreou N, Mullis CE, Laeyendecker O, et al. (2012) Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis 206: 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rieder P, Joos B, Scherrer AU, Kuster H, Braun D, et al. (2011) Characterization of human immunodeficiency virus type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin Infect Dis 53: 1271–9. [DOI] [PubMed] [Google Scholar]
- 42. Sagar M, Lavreys L, Baeten JM, Richardson BA, Mandaliya K, et al. (2004) Identification of modifiable factors that affect the genetic diversity of the transmitted HIV-1 population. AIDS 18: 615–619. [DOI] [PubMed] [Google Scholar]
- 43. Pillai SK, Good B, Pond SK, Wong JK, Strain MC, et al. (2005) Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J Virol 79: 1734–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haaland RE, Sullivan ST, Evans-Strickfaden T, Lennox JL, Hart CE (2012) Female genital tract shedding of CXCR4-tropic HIV Type 1 is associated with a majority population of CXCR4-tropic HIV Type 1 in blood and declining CD4(+) cell counts. AIDS Res Hum Retroviruses 28: 1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Margolis L, Shattock R (2006) Selective transmission of CCR5-utilizing HIV-1: the “gatekeeper” problem resolved? Nat Rev Microbiol 4: 312–317. [DOI] [PubMed] [Google Scholar]
- 46. Grivel JC, Shattock RJ, Margolis LB (2011) Selective transmission of R5 HIV-1 variants: where is the gatekeeper? J Transl Med 9 (Suppl 1)S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuitemaker H, Van ’t Wout AB, Lusso P (2011) Clinical significance of HIV-1 coreceptor usage. J Transl Med 9 (Suppl 1)S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chalmet K, Dauwe K, Foquet L, Baatz F, Seguin-Devaux C, et al. (2012) Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J Infect Dis 205: 174–184. [DOI] [PubMed] [Google Scholar]