Abstract
Complement plays a key role in immunity and inflammation through direct effects on immune cells or via crosstalk and regulation of other host signaling pathways. Deregulation of these finely balanced complement activities can link infection to inflammatory tissue damage. Periodontitis is a polymicrobial community-induced chronic inflammatory disease that can destroy the tooth-supporting tissues. In this review, we summarize and discuss evidence that complement is involved in the dysbiotic transformation of the periodontal microbiota and in the inflammatory process that leads to the destruction of periodontal bone. Recent insights into the mechanisms of complement involvement in periodontitis have additionally provided likely targets for therapeutic intervention against this oral disease.
Keywords: Complement, C5a receptor, periodontitis, dysbiosis, P. gingivalis
1. Introduction
Complement can be produced locally or systemically [1] and plays important roles in immunity and inflammation through direct effects on innate and adaptive immune cells or through crosstalk and regulation of other signaling pathways [2,3]. Complement activities are therefore not restricted to a linear cascade of events but involve a network of interactions with other systems to better coordinate the host response to infection or other insults. These connections of complement can enhance innate immune defense through synergy with Toll-like receptors (TLRs) [3], provide a barrier against the spread of invading bacteria by potentiating local clotting [4], and replenish the immune system through mobilization of hematopoietic stem/progenitor cells from the bone marrow [5,6]. Complement also influences the activation and differentiation of T-cell subsets [7,8].
Besides the classical group of serum proteins (C1-9), the integrated complement system includes pattern-recognition molecules, convertases and other proteases, regulators, and receptors for interactions with immune mediators [2]. The complement cascade can be triggered via distinct pathways (classical, lectin, or alternative), which converge at the third complement component (C3). The activation of the classical pathway is initiated by antigen-antibody complexes recognized by the C1q subunit of C1, whereas the lectin pathway can be triggered through interaction of a secreted pattern-recognition molecule (the mannose-binding lectin; MBL) with specific carbohydrate groups on microbial surfaces. Both the classical and lectin pathways proceed through C4 and C2 cleavage to generate the classical/lectin C3 convertase. The alternative pathway is initiated by low-level, spontaneous hydrolysis of C3 to C3[H2O], which forms the initial alternative pathway C3 convertase in the presence of factors B (fB) and D (fD). As long as there is no sufficient negative regulation (as is normally the case of microbes or other non-self surfaces), this initiation is followed by rapid propagation of the alternative pathway through an amplification loop [2,7]. The alternative pathway can also be triggered by bacterial lipopolysaccharide and lipooligosacharide via the plasma protein properdin attached to bacterial surfaces [9,10] and can potentially contribute to ≥80% of the total complement activation [11]. C3 activation by pathway-specific C3 convertases leads to the generation of effector molecules involved in (a) the recruitment and activation of inflammatory cells (e.g., the C3a and C5a anaphylatoxins that activate specific G-protein-coupled receptors, C3aR and C5aR [CD88], respectively); (b) microbial opsonization and phagocytosis (e.g., through the C3b opsonin); and (c) direct lysis of targeted pathogens (by means of the C5b-9 membrane attack complex [MAC])[2].
As alluded to above, complement is not normally activated on the surface of host cells and tissues. However, disruption of the regulatory mechanisms involved can lead to excessive complement activation, inflammation, and damage to host tissues. For instance, deficiencies, hypo-functional polymorphisms, or mutations in complement regulators have been implicated in the development of local or systemic diseases, such as age-related macular degeneration and systemic lupus erythematosus [2,12,13]. Microbial pathogens may also contribute to complement deregulation or dysfunction. In this regard, pathogens can hijack negative regulators of complement to protect themselves against host defense mechanisms, or they can degrade regulatory molecules that protect host tissues or cells [14–16]. Moreover, pathogens can exploit complement receptors to proactively promote their survival and persistence in the host [17]. These mechanisms can contribute to defective bacterial clearance and enhanced inflammation, thereby promoting infection-driven inflammatory diseases.
Periodontitis is an inflammatory disease in which complement appears to form a major link between infection and inflammation [18]. Chronic periodontal inflammation is initiated and perpetuated by a dysbiotic microbiota and may lead to tooth loss as a result of the destruction of the supporting alveolar bone [19,20]. This is a highly prevalent disease, affecting nearly half of U.S. adults [21]. In its most severe form, which affects 8.5% of U.S. adults [21], periodontitis can influence systemic health and increase the risk for atherosclerosis, diabetes, and possibly rheumatoid arthritis [22–25].
In this review, we summarize and discuss the published evidence supporting a crucial role for complement in the initiation and progression of periodontitis (Table 1). Specifically, excessive activation of complement or subversion of its normal functions contributes to the breakdown of host-microbe homeostasis in the periodontal tissue (periodontium), thereby precipitating inflammatory disease. This evidence comes from both clinical observations and experimental animal studies.
Table 1.
Clinical and experimental evidence for complement involvement in periodontitis
| Observations | Refs. |
|---|---|
| Complement components and cleavage products covering the whole complement cascade are detected in chronically inflamed gingiva and in GCF of patients; undetected or at lower levels in healthy control samples. | [45–47, 100– 105] |
| Induction of experimental gingival inflammation in humans causes progressive elevation of complement cleavage fragments correlating with increased clinical inflammatory parameters. | [108] |
| C3 conversion to C3c in GCF increases with increasing periodontal pocket depth but decreases dramatically after periodontal therapy | [110, 127] |
| Possible reduced expression of complement negative regulators in periodontitis: (a) Reduced expression of CD59 in the gingiva of periodontitis patients compared to healthy controls; (b) P. gingivalis causes shedding and degradation of membrane-anchored CD46. | [75, 103] |
| Single nucleotide polymorphism of C5 (rs17611) significantly more prevalent in periodontitis patients than in healthy controls. | [112] |
| Periodontal bacteria (e.g., P. gingivalis, T. forsythia, T. denticola, P. intermedia. A. actinomycetemcomitans) possess mechanisms for complement manipulation; evasion of antimicrobial effects and promotion of inflammatory responses. | [44, 50, 51, 53, 57–61, 71, 84, 128]. |
| C5aR-deficient mice are resistant to dysbiosis and periodontitis. | [84, 88] |
| C5aR antagonist reverses dysbiosis and prevents or halts the progression of mouse periodontitis. | [86, 88] |
2. Role of the microbiota in periodontitis
In order to better understand the interplay between bacteria and complement in periodontitis, it is instructive to first consider the role of bacteria in the pathogenesis of this chronic oral inflammatory disease. Until fairly recently, the identities of the organisms associated with periodontal disease or health were restricted to those that could be cultured in the laboratory. Cultural characterization of the periodontal microbiota in the late 1970s and early 1980s revealed dramatic compositional changes in disease as compared to health [26–31]. One way to interpret these findings was that the disease-associated microbiota contained novel pathogenic species that were either absent or barely detectable in health. On the basis of cluster analysis and association with different disease severities, oral bacteria were historically divided into six groups of different potential pathogenicity, commonly referred to by their color-coded designations [32]. Foremost among these groups was the so-called “red complex” a group of three, Gram-negative anaerobic species, including Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia, the detection of which was strongly associated with each other and with diseased sites [32]. Also associated with periodontal lesions, the “orange” complex comprised species members of the genus Prevotella, Fusobacterium, and Campylobacter, as well as Streptococcus constellatus and Eubacterium nodatum. The other four complexes (“blue”, “yellow”, “green”, and “purple”) primarily consisted of early colonizers of the tooth surfaces [32].
With the advent of culture-independent, molecular-based methods of bacterial identification and enumeration, such as 16S rDNA amplification and high-throughput sequencing, our understanding of the bacterial composition of the periodontal region has changed [33–35]. The in-depth study of thousands of plaque samples derived from a variety of clinical periodontal conditions has demonstrated a more heterogeneous and diverse periodontal microbiota than previously thought [33–35]. In addition to the consensus periodontal pathogens, P. gingivalis, T. forsythia, and T. denticola, newly recognized non- or poorly cultivable organisms that increase in number in diseased sites include the Gram-positive Filifactor alocis and species in the genera Prevotella, Megasphaera, Selenomonas, and Desulfobulbus [33,34,36–39]. Many of these newly recognized organisms show as good a correlation (or better) with disease as does the classical red complex [37,39,40]. It is now increasingly recognized that chronic periodontitis is not a bacterial infection in the classical sense, i.e., caused by a single or a limited number of pathogens. Rather, periodontal disease is the result of a polymicrobial community-induced perturbation of host homeostasis in susceptible individuals [20,41]. Bacterial constituents of these communities often exhibit synergistic interactions that can enhance colonization, persistence, or virulence, and some bacteria may be involved in the breakdown of periodontal homeostasis, whereas other may trigger destructive inflammation once homeostasis is disrupted [20].
3. Mechanisms used by periodontal bacteria for protection against complement
The space between the free gingiva and the tooth surfaces, known as the gingival crevice, constitutes a niche for periodontitis-associated microbial communities [41]. The gingival crevice is bathed with an inflammatory exudate, termed gingival crevicular fluid (GCF) [42]. In a clinically healthy periodontium, in which the tooth-associated biofilm is usually confined to the gingival margin, the GCF represents a slow-flowing transudate of plasma proteins. However, if the biofilm is left undisturbed for 2–4 days, the biofilm enters the crevice by proliferation and spreading or by relocation of dislodged bacteria. The host response is therefore escalated and manifested by increased flow of GCF (in part due to increased vascular permeability of the subepithelial blood vessels) and chemotactic recruitment of inflammatory cells, mostly neutrophils [43]. Under inflammatory conditions, the GCF contains complement at up to 70% to 80% of its concentration in serum, although certain components can be found at higher levels in GCF reflecting local production of complement [44–47]. Therefore, the periodontal bacteria constantly encounter complement and, in cases of chronic periodontitis, it is reasonable to think that the bacteria have evolved mechanisms to resist its antimicrobial actions.
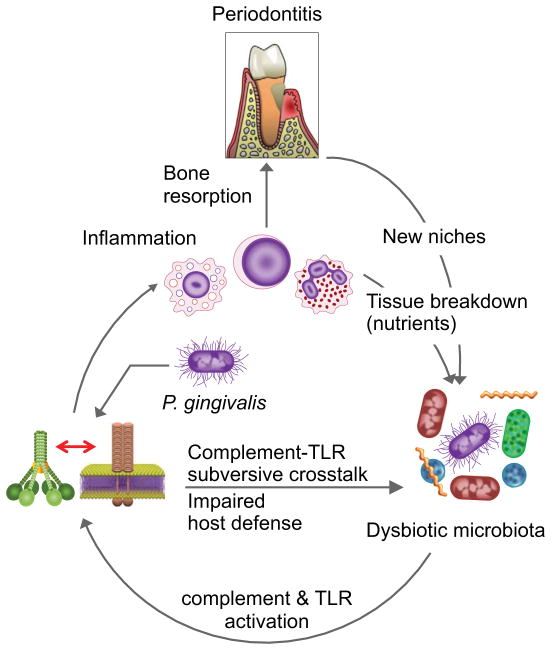
Studies with several periodontal bacteria, such as P. gingivalis, Prevotella intermidia, T. forsythia, and T. denticola, indicate that they interact with complement in complex ways that include both inhibitory and stimulatory effects [15,16,48,49]. This seemingly contradictory microbial behavior is probably due to the dynamics of survival tactics of periodontal bacteria: on the one hand trying to evade immune clearance, and on the other to stimulate inflammation and the flow of GCF as a source of nutrients [16] (Fig. 1).
Fig. 1. Subversion of host immuno-inflammatory surveillance and destructive inflammation in periodontitis.
Colonization by P. gingivalis impairs innate immunity by subverting complement-TLR crosstalk, leading to increased numbers of periodontal bacteria and therefore enhanced inflammation through the activation of synergistic complement and TLR pathways. The inflammatory environment is favorable to further bacterial growth, since the gingival inflammatory exudate is a rich source of nutrients (e.g., degraded host proteins and hemin, a source of essential iron). These environmental changes, moreover, can alter the composition of the oral microbiota, favoring those bacteria (e.g., proteolytic and asaccharolytic organisms) that can better exploit these environmental changes. These changes result in even higher inflammation and bone resorption, leading to increased niche space (deeper periodontal pockets) for the bacteria, thereby perpetuating a vicious cycle of periodontal tissue destruction. Adapted from Hajishengallis et al (ref. 92).
P. gingivalis expresses Arg- and Lys-specific cysteine proteinases known as gingipains, which can degrade C3, thereby potentially inhibiting complement activation regardless of the initiation pathway involved [44]. All three gingipains can degrade C3, although the Arg-specific enzymes (HRgpA and RgpB) are more potent in this regard than the Lys-specific gingipain (Kgp) [44]. As a consequence, the deposition of opsonins or the C5b-9 MAC on the pathogen surface is suppressed, unless the activity of the gingipains is inhibited by chemical or genetic means [50,51]. Consistent with these findings, P. gingivalis displays exquisite resistance to the lytic action of complement in vitro [44,50]. Unexpectedly, however, Arg- and Lys-gingipain deletion mutants maintained resistance to killing in 20% normal human serum (a similar viability to that seen in heat-inactivated serum) despite increased deposition of complement fragments or complexes (C3d and C5b-9 MAC) [50]. This finding suggested that the mechanism of serum resistance is largely independent of the Arg- and Lys-gingipains and has, in fact been attributed to the presence of a surface anionic polysaccharide (APS; now known as A-LPS, i.e., LPS with APS repeating units [52]), the lack of which renders P. gingivalis extremely sensitive to complement killing in 20% normal serum [50]. Taken together, these data raise the following question: What is the biological significance of gingipain-dependent C3 degradation if P. gingivalis is inherently resistant to the lytic action of complement? Since the inhibitory mechanisms of P. gingivalis against complement activation are “leaky” [44], it is possible that the A-LPS-dependent resistance represents a reinforcing mechanism to provide maximal protection against complement. In addition, or alternatively, P. gingivalis may have evolved the capacity to inactivate complement to protect susceptible species occupying the same subgingival niche (this could be one of the mechanisms for the keystone pathogen status of P. gingivalis; see below). In this regard, incubation of human serum with purified gingipains or whole cells of P. gingivalis has been shown to cause a drastic decrease in the bactericidal activity of the serum against susceptible organisms [44].
The complex nature of P. gingivalis-complement interactions is also suggested by the observation that all three gingipains (HRgpA, RgpB, and Kgp) can activate the C1 complex in serum at relatively low concentrations, resulting in the deposition of C1q on inert surfaces or on the bacteria themselves [44]. It is reasonable to speculate that P. gingivalis activates complement when present in low numbers, while attempting to establish infection. This complement activation is unlikely to eliminate P. gingivalis (since it is inherently resistant to complement-mediated lysis), but the resulting local inflammatory response may provide P. gingivalis with precious nutrients, such as GCF-derived hemin, a source of essential iron. At later stages of infection, when the concentration of proteases is high enough to destroy C3 and inhibit complement activation, P. gingivalis may promote the survival of the entire biofilm community by helping bystander bacteria evade complement killing.
Certain strains of Prevotella intermedia express and secrete interpain A (InpA), a streptopain-like cysteine protease, which can also degrade C3 and contribute to resistance against the antibacterial activity of complement [53]. Interestingly, P. intermedia shares an important feature with P. gingivalis. At low concentrations, interpain, like the gingipains, is able to activate the C1 complex in serum, causing deposition of C1q on bacterial surfaces [53]. Moreover, the interpain of P. intermedia acts in synergy with the gingipains of P. gingivalis in inactivating complement in vitro [53]. Such synergy may be of relevance for the in vivo pathogenicity of mixed biofilms, given that the two organisms can co-aggregate [54,55]. Such synergy may occur even if the two species do not intimately interact in subgingival biofilms. Since the gingipains and the interpain are secreted proteases, they can diffuse, reach, and protect bystander bacterial species, which would otherwise be eliminated by the complement bactericidal activity.
T. forsythia, another important species associated with periodontitis [56], also possess mechanisms for escaping complement. T. forsythia expresses karilysin, a metalloproteinase that mediates resistance to killing by human complement by acting at different stages of the complement cascade: Specifically, karilysin inhibits the classical and lectin pathways by degrading MBL, ficolin-2, ficolin-3, and C4, whereas it blocks the terminal pathway by degrading C5 [57]. Karilysin synergizes with interpain and gingipains in these subversive functions, suggesting that P. gingivalis, P. intermedia, and T. forsythia together can better protect susceptible bystander bacterial species and promote the survival of the entire biofilm [57].
In addition to the degradation of key complement components, other evasion mechanisms depend upon the ability of pathogens to hijack and employ circulating complement regulators [14,49]. In this regard, P. gingivalis uses its HRgpA to capture the circulating C4b-binding protein (C4BP) on the bacterial cell surface, thereby acquiring the ability to negatively regulate the classical pathway C3 convertase [58]. Aggregatibacter actinomycetemcomitans uses its outer membrane protein 100 to bind the alternative pathway inhibitor factor H and acquire resistance to complement killing in serum [59]. P. intermedia binds the serine protease factor I (FI), a major inhibitor of complement that degrades C3b and C4b in the presence of cofactors such as C4BP and factor H [60]. Importantly, P. intermedia also binds the cofactors factor H and C4BP leading, respectively, to increased degradation of C4b and C3b [60].
T. denticola expresses a 11.4-kDa cell surface lipoprotein that can bind factor H [61]. Strikingly, however, once full-length factor H becomes associated with T. denticola, the organism uses its serine protease dentilisin to generate a 50-kDa factor H fragment that remains attached to the bacterial surface [61]. This seems paradoxical, since dentilisin appears to counteract the action of the factor H-binding protein. An interesting question therefore is whether the attached fragment retains useful complement inhibitory activity to protect T. denticola against complement. Alternatively, if dentilisin does inactivate factor H, this protease may deregulate complement and promote local inflammation.
The T. denticola dentilisin can also hydrolyze the α-chain of C3 and generate iC3b [62], consistent with an early study demonstrating opsonization of T. denticola with iC3b [63]. Intriguingly, iC3b-mediated phagocytosis is often associated with weak killing mechanisms or even immunosuppressive signaling [64–67] and may be exploited by certain pathogens. Indeed, P. gingivalis, Mycobacterium tuberculosis, Bordetella pertussis, and HIV-1 promote their intracellular survival by exploiting complement receptor-3-mediated entry, either by direct interaction with the receptor or upon opsonization with iC3b [14,17,68–70].
4. Periodontal bacteria proactively utilize complement to cause inflammation
As alluded to earlier, periodontal bacteria can proactively activate complement using virulence factors (gingipains or interpain at relatively low concentrations) that directly interact with the C1 complex. Although high concentrations of P. gingivalis gingipains or P. intermedia interpain inhibit the complement cascade, the interpain can directly release anaphylatoxin C3a, while gingipains can release C5a [44,53,71]. Both enzymes preferentially attack the α-chains of C3 and C5 [44,53,72]. Since the anaphylatoxins are potent mediators of inflammatory responses [2,73], their local generation at sites heavily populated with bacteria can contribute to the inflammatory destruction of the periodontium and the generation of tissue breakdown nutrients for the bacteria (Fig. 1). Mechanistically, local complement activation can promote periodontal inflammation through C5a-induced vasodilation, increased vascular permeability, the flow of inflammatory exudate, and chemotactic recruitment of inflammatory cells, especially neutrophils [16,74]. Moreover, C5a can be also exploited by the bacteria for the immune subversion of leukocytes (see below).
Membrane-anchored negative regulators of complement, such as CD46, CD55, and CD59, protect host cells from unwarranted complement attack [2,12]. In interacting with oral epithelial cells, P. gingivalis causes the shedding of membrane-anchored CD46, which as a soluble protein can lead to IL-8 secretion from the epithelial cells or become degraded by the Kgp gingipain of the bacterium [75]. Regardless of whether shed CD46 can further promote inflammatory responses, its removal from the epithelial cell surface can render a cell susceptible to the destructive effects of complement activation. It should be noted, however, that under inflammatory conditions, oral epithelial cells upregulate the expression of CD46, CD55, and CD59 [76], presumably to prevent cell lysis by excessive complement activation and/or replenish the loss of these membrane-bound regulators as a result of attack by bacterial proteases.
Although CD46 was originally appreciated for its role in preventing complement deposition on host cells and tissues, recent work has ascribed additional regulatory functions to CD46, including the ability to regulate T-helper type 1 immune responses [77]. This may explain why a number of microbial pathogens exploit CD46 to promote their fitness [78]. Fusobacterium nucleatum, a periodontal bacterium that forms a co-aggregating bridge between early and late colonizers of the dental biofilm and may occur in deep periodontal pockets along with P. gingivalis [79,80], has been shown to bind CD46 on oral epithelial cells [76]. This interaction upregulates IL-6, IL-8, and matrix metalloproteinase (MMP)-9 at the mRNA and protein level and has been interpreted as a mechanism that can contribute to host tissue destruction in periodontitis [76].
Taken together, these data suggest that periodontal bacteria have evolved mechanisms to proactively regulate inflammation to their own advantage, since host tissue destruction can provide them with nutrients and new niches (as a result of deeper periodontal pockets) (Fig. 1). However, this inflammatory destruction of host tissue could be a genuine advantage only if, at the same time, the periodontal bacteria have evolved to endure inflammation. This concept is discussed below.
5. Exploitation of complement as a mechanism to cause dysbiosis and periodontitis
As discussed above, P. gingivalis proactively generates C5a. All three gingipains (especially HRgpA and RgpB) act in a C5 convertase-like manner and generate biologically active C5a through limited degradation of C5 [44,72]. When C5 is oxidized by hydroxyl radicals (as may occur in a local inflammatory environment with neutrophil-released oxidants), the gingipains generate elevated C5a activity [81]. Furthermore, P. gingivalis HRgpA and RgpB activate prothrombin to form thrombin [82] which, in turn, generates biologically active C5a by acting as a C5 convertase [83]. C5a can also be generated in vivo by P. gingivalis, but not by an isogenic mutant lacking all three gingipain genes [84].
C5a can potentially play a key role in host defense against infection [85]. However, not only is P. gingivalis survival not impaired by C5a, but it is enhanced. Mechanistically, P. gingivalis-generated C5a activates C5aR and stimulates Gαi-dependent intracellular Ca2+ signaling, which synergistically enhances an otherwise weak cAMP response by P. gingivalis-induced TLR2 activation alone [71]. The resulting crosstalk sustains high production of cAMP and leads to the activation of the cAMP-dependent protein kinase A, which inactivates the glycogen synthase kinase-3β. This, in turn, impairs the nitric oxide-dependent killing of P. gingivalis [71]. This C5aR-TLR2 subversive crosstalk undermines the killing function of macrophages, although it is yet to be shown whether a similar mechanism protects P. gingivalis against neutrophils.
The P. gingivalis-induced C5aR-TLR2 crosstalk also inhibits TLR2-induced IL-12p70, but it upregulates inflammatory and bone-resorptive cytokines such as IL-1β, IL-6, IL-17, and TNF [84,86]. This inhibition enables P. gingivalis to escape IL-12p70-dependent immune clearance in vivo and to cause inflammatory bone loss in a mouse model of periodontitis [84,86]. Because the C5aR-TLR2 crosstalk inhibits only a subset of TLR2 signaling events, C5aR is considered to be a “modulatory” receptor for TLRs, as opposed to “TLR inhibitory receptors” such as IL-10 receptor or TGF-β receptor, which inhibit most, if not all, inflammatory responses [87]. Therefore, by generating C5a to locally regulate C5aR, P. gingivalis can not only promote its survival but also contribute to destructive inflammation. The induction of totally immunosuppressive signaling would not be a safe option for P. gingivalis because P. gingivalis is a strictly asaccharolytic organism, and its survival and growth depends crucially on inflammatory tissue breakdown products (degraded proteins, as well as hemin for essential iron) [16].
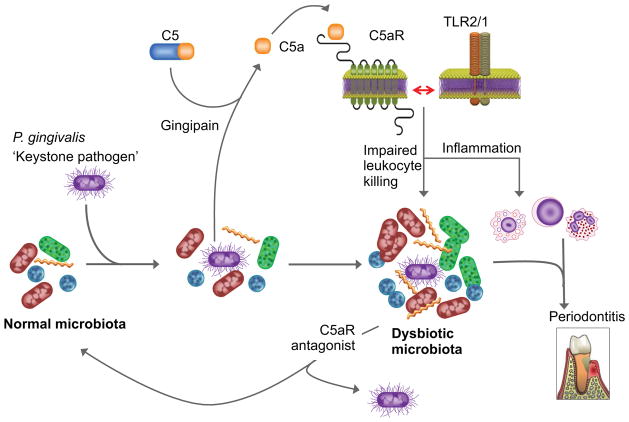
Intriguingly, the manipulation of the periodontal C5aR response by P. gingivalis benefits the entire microbial community, which becomes dysbiotic (altered composition and increased total counts) and causes inflammatory periodontal bone loss [88] (Fig. 2). Strikingly, P. gingivalis causes these community-wide effects while present at low colonization levels (<0.01% of the total bacterial counts) in the periodontal tissue. Consistent with a C5aR requirement for disease pathogenesis, P. gingivalis fails to alter the microbiota in C5aR-deficient mice, which are also protected against periodontal inflammation and bone loss [88]. These data suggest that dysbiosis is required for inflammatory bone loss. Furthermore, the essential participation of the periodontal microbiota at large in the disease pathogenesis was confirmed by the inability of P. gingivalis to cause bone loss by itself, i.e., in germ-free mice, despite colonizing this host [88].
Fig. 2. P. gingivalis-induced dysbiosis in periodontitis.
P. gingivalis exploits C5aR signaling and inhibits the host defense, leading to an altered composition and increased numbers of the periodontal microbiota which, in turn, cause complement-dependent periodontal inflammation and bone loss. This community-wide effect can occur at low colonization levels of P. gingivalis and requires its Arg-specific gingipain activity, acting in a C5 convertase-like manner and thus cleaving C5 and generating high levels of C5a locally. C5a-induced activation of C5aR triggers inflammation and is also crucially involved in subversive crosstalk with the TLR2/1 complex that impairs macrophage killing. The ability of P. gingivalis to orchestrate inflammatory disease by remodeling a symbiotic microbiota into a dysbiotic state, while being a minor constituent of this community, qualifies it as a keystone pathogen. This pathologic process is reversible, since C5aR blockade promotes the clearance of P. gingivalis and reverses its dysbiotic effects. Adapted from Hajishengallis et al (ref. 92).
Mice deficient in TLR2 are also resistant to inflammatory periodontal bone loss [84,89]. The fact that mice deficient either in C5aR or TLR2 are highly and similarly resistant to periodontitis is consistent with the notion that a crosstalk between the two receptors is synergistically involved in the disease process (indeed, as discussed above, C5aR-TLR2 crosstalk acts in synergy with inflammation and mediates immune subversion by P. gingivalis [71,84,86]). This in turn suggests that blockade of just one of the two receptors may be sufficient to inhibit the development of periodontitis. Indeed, local intragingival treatment of mice with a C5aR antagonist reverses P. gingivalis-induced dysbiosis [88] and prevents or halts the progression of periodontitis, depending on whether the antagonist is given in a preventive or therapeutic setting [86].
The ability of P. gingivalis to orchestrate inflammatory disease through community-wide effects, while being a quantitatively minor constituent of the periodontal microbiota, has prompted its characterization as a keystone pathogen, in analogy to the crucial role of a keystone that holds an entire arch together [88,90,91]. It is also plausible that keystone pathogens underlie the pathogenesis of several other polymicrobial inflammatory diseases (e.g., inflammatory bowel disease), a concept that would lead to a better understanding of their etiology and to targeted diagnostic and treatment modalities [92].
Although established in the mouse model, the keystone pathogen concept in periodontitis is consistent with observations in other animal models and in humans: In rabbits, P. gingivalis causes a shift to a more anaerobic flora and an overall increase in the bacterial load of the dental biofilm [93]. In non-human primates, in which P. gingivalis is a natural inhabitant of the periodontal biofilm, a gingipain-based vaccine causes a reduction both in Pg numbers and the total subgingival bacterial load [94], suggesting that the presence of P. gingivalis benefits the entire biofilm. The keystone pathogen concept is also consistent with P. gingivalis being a quantitatively minor constituent of human periodontitis-associated biofilms [26,40,95], despite its high prevalence and association with progressive bone loss in periodontal patients [96,97]. Importantly, the mouse model is similar to the situation in humans in terms of innate defense status, including complement and neutrophil transit [98], and, moreover, a significant increase in the total microbial load is also observed in human periodontitis as compared to health [99].
6. Potential for complement-targeted therapeutics in periodontitis
The data obtained from animal models suggest that complement inhibition may have therapeutic potential in human periodontitis, where complement involvement is supported by clinical and histological observations (Table 1). Complement components and cleavage products covering the whole complement cascade have been detected in chronically inflamed gingiva, although they are undetected or at lower levels in healthy control samples [45,47,100–104]. The gingival crevicular fluid (GCF) from periodontitis patients displays complement-dependent hemolytic activity, suggesting the presence of a functional complement system (C1–C9) in gingival inflammatory exudates [100,104]. Moreover, the GCF of patients contains activated complement fragments at higher concentrations than in the GCF from healthy individuals [46,105–108]. Importantly, induction of experimental gingival inflammation in human volunteers causes a progressive elevation of complement cleavage products that is correlated with increased clinical indices of inflammation [108]. C3 is among the top 5% of genes that are most strongly downregulated following periodontal therapy [109]. Moreover, C3 conversion to C3c in GCF decreases dramatically after periodontal therapy [110]. Interestingly, a single nucleotide polymorphism of C5 (rs17611), which is associated with increased serum C5 levels and susceptibility to liver fibrosis (a complement-associated disease) [111], has been shown to be more prevalent in periodontitis patients than in healthy controls [112]. An immunohistochemical study showed weaker expression of CD59 in the gingiva of periodontitis patients than in healthy controls, suggesting reduced protection of diseased tissues against possible tissue damage by autologous MAC formation [103].
The involvement of complement in human periodontitis has recently prompted intervention studies in preclinical models, on the basis of mechanistic studies implicating C5aR in disease pathogenesis. When used preventively, local intragingival injection of a C5aR antagonist (C5aRA; PMX-53) in mice completely inhibits P. gingivalis-induced bone loss [86]. Mice receiving this treatment elicit minimal inflammatory responses (TNF, IL-1β, and IL-17), like those of sham-infected mice. C5aRA is able to protect against inflammatory bone loss even when it is administered 2 weeks after the onset of experimental periodontitis [86]. This is important, since periodontal treatment is more likely to be implemented in a therapeutic than a preventive mode. Nevertheless, preventive intervention may be appropriate for individuals at high risk for periodontitis, such as cigarette smokers, diabetic patients, or individuals with systemic diseases affecting neutrophil function [113].
Since periodontitis fundamentally represents a disruption of host-microbe homeostasis [20,41] and is associated with multiple etiologies and disease modifiers [19,25,114,115], P. gingivalis is not likely to be involved in all cases of periodontitis. C5aRA is able to inhibit periodontal bone loss in a mouse model in which periodontitis is induced independently of P. gingivalis, specifically the “ligature-induced periodontitis model,” In this model, a silk ligature is placed around the molar teeth, resulting in massive local bacterial accumulation and rapid induction of severe bone loss in specific-pathogen-free (but not germ-free) animals [116]. Mice locally treated with C5aRA at the ligated sites display significantly decreased gingival inflammation and about 50% less bone loss when compared to controls [86]. In a ligature-induced periodontitis study in rats, a PMX53 analog, PMX205, was administered in the drinking water prior to and during the disease, and it inhibited bone loss by <20% when compared to controls [117]. These differences in efficacy might be due to the different modes of administration used, although the different animal species used might have been another contributory factor. Modes of local administration, which restrict the action of C5aRA to the periodontal tissue, are likely to be safer, since systemic inhibition of complement may predispose to increased susceptibility to microbial infections.
7. Conclusions
In summary, complement is a target of immune subversion that leads to the dysbiotic transformation of the microbiota, which in turn causes complement-dependent destructive inflammation (Fig. 1). Specifically, P. gingivalis exploits C5aR to impair antimicrobial (but not proinflammatory) leukocyte responses, leading to uncontrolled growth and altered composition of the periodontal microbiota at large, which precipitates periodontitis [71,88,91,92] (Fig. 2). A better understanding of the role of the various complement pathways in periodontitis may identify additional targets for additional treatment options. For instance, blocking complement activity upstream of C5aR may be more beneficial if additional pathways contribute to the disease. This notion is supported by unpublished studies from our group: In ligature-induced periodontitis studies, C3-deficient mice or non-human primates locally treated with the C3 inhibitor compstatin [123–126] are protected against periodontal inflammation and bone loss.
The high prevalence of periodontitis (>47% of US adults, with 8.5% presenting with severe periodontitis) [21], its economic burden [118,119], and the fact that many periodontitis cases are refractory to conventional therapy [120,121] underscore the importance of implementing innovative and cost-effective therapeutic interventions. Although these efforts currently involve preclinical models, the availability of complement-specific drugs that have already undergone successful safety trials for other diseases [122–126] indicates a potential for rapid translation to the clinic.
Highlights.
Human periodontitis is associated with increased complement activation
Periodontal bacteria evade and subvert complement
Experimental periodontitis is mediated by complement-dependent dysbiosis
Antagonistic blockade of C5a receptor protects against experimental periodontitis
Acknowledgments
The authors are supported by grants from the U.S. National Institutes of Health, DE015254, DE021580, and DE021685 (GH) and AI003040, AI068730, AI072106, AI097805, EY020633, GM097747 (JDL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li K, Sacks SH, Zhou W. The relative importance of local and systemic complement production in ischaemia, transplantation and other pathologies. Mol Immunol. 2007;44:3866–74. doi: 10.1016/j.molimm.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010;31:154–63. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markiewski MM, Nilsson B, Ekdahl KN, Mollnes TE, Lambris JD. Complement and coagulation: strangers or partners in crime? Trends Immunol. 2007;28:184–92. doi: 10.1016/j.it.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Jalili A, Shirvaikar N, Marquez-Curtis L, Qiu Y, Korol C, Lee H, et al. Fifth complement cascade protein (C5) cleavage fragments disrupt the SDF-1/CXCR4 axis: Further evidence that innate immunity orchestrates the mobilization of hematopoietic stem/progenitor cells. Exp Hematol. 2010;38:321–32. doi: 10.1016/j.exphem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HM, Wysoczynski M, Liu R, Shin DM, Kucia M, Botto M, et al. Mobilization studies in complement-deficient mice reveal that optimal AMD3100 mobilization of hematopoietic stem cells depends on complement cascade activation by AMD3100-stimulated granulocytes. Leukemia. 2009;24:573–82. doi: 10.1038/leu.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 8.Friec GL, Kemper C. Complement: coming full circle. Arch Immunol Ther Exp (Warsz) 2009;57:393–407. doi: 10.1007/s00005-009-0047-4. [DOI] [PubMed] [Google Scholar]
- 9.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179:2600–8. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 10.Kimura Y, Miwa T, Zhou L, Song W-C. Activator-specific requirement of properdin in the initiation and amplification of the alternative pathway complement. Blood. 2008;111:732–40. doi: 10.1182/blood-2007-05-089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harboe M, Mollnes TE. The alternative complement pathway revisited. J Cell Mol Med. 2008;12:1074–84. doi: 10.1111/j.1582-4934.2008.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–40. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 13.Ricklin D, Lambris JD. Complement in Immune and Inflammatory Disorders: Pathophysiological Mechanisms. J Immunol. 2013;190 doi: 10.4049/jimmunol.1203487. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–42. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krauss JL, Potempa J, Lambris JD, Hajishengallis G. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol 2000. 2010;52:141–62. doi: 10.1111/j.1600-0757.2009.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajishengallis G. Complement and periodontitis. Biochem Pharmacol. 2010;80:1992–2001. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–20. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 20.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The Polymicrobial Synergy and Dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–19. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–20. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 22.Genco RJ, Van Dyke TE. Prevention: Reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010;7:479–80. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- 23.Tonetti MS, D’Aiuto F, Nibali L, Donald A, Storry C, Parkar M, et al. Treatment of periodontitis and endothelial function. N Engl J Med. 2007;356:911–20. doi: 10.1056/NEJMoa063186. [DOI] [PubMed] [Google Scholar]
- 24.Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6:727–30. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 25.Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol. 2011;7:738–48. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- 26.Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1982;38:1137–48. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Socransky SS. Microbiology of periodontal disease -- present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 28.Tanner ACR, Haffer C, Bratthall GT, Visconti RA, Socransky SS. Study of the Bacteria Associated with Advancing Periodontitis in Man. Journal of Clinical Periodontology. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 29.Slots J. Microflora in the healthy gingival sulcus in man. Scand J Dent Res. 1977;85:247–54. doi: 10.1111/j.1600-0722.1977.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 30.Slots J. The predominant cultivable microflora of advanced periodontitis. Scand J Dent Res. 1977;85:114–21. doi: 10.1111/j.1600-0722.1977.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 31.Moore WE, Holdeman LV, Cato EP, Smibert RM, Burmeister JA, Ranney RR. Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect Immun. 1983;42:510–5. doi: 10.1128/iai.42.2.510-515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 33.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffen AL, Beall CJ, Firestone ND, Gross EL, Difranco JM, Hardman JH, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10:302–6. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 37.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–55. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–73. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–90. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 42.Lamster IB, Ahlo JK. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann N Y Acad Sci. 2007;1098:216–29. doi: 10.1196/annals.1384.027. [DOI] [PubMed] [Google Scholar]
- 43.Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontol 2000. 2003;31:55–76. doi: 10.1034/j.1600-0757.2003.03105.x. [DOI] [PubMed] [Google Scholar]
- 44.Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–50. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 45.Schenkein HA, Genco RJ. Complement cleavage products in inflammatory exudates from patients with periodontal diseases. J Immunol. 1978;120:1796. [Google Scholar]
- 46.Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. II. Evidence for cleavage of complement components C3, C3 proactivator (factor B) and C4 in gingival fluid. J Periodontol. 1977;48:778–84. doi: 10.1902/jop.1977.48.12.778. [DOI] [PubMed] [Google Scholar]
- 47.Lally ET, McArthur WP, Baehni PC. Biosynthesis of complement components in chronically inflamed gingiva. J Periodontal Res. 1982;17:257–62. doi: 10.1111/j.1600-0765.1982.tb01152.x. [DOI] [PubMed] [Google Scholar]
- 48.Slaney JM, Curtis MA. Mechanisms of evasion of complement by Porphyromonas gingivalis. Front Biosci. 2008;13:188–96. doi: 10.2741/2669. [DOI] [PubMed] [Google Scholar]
- 49.Blom AM, Hallstrom T, Riesbeck K. Complement evasion strategies of pathogens-acquisition of inhibitors and beyond. Mol Immunol. 2009;46:2808–17. doi: 10.1016/j.molimm.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 2006;74:5352–61. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schenkein HA. Complement factor D-like activity of Porphyromonas gingivalis W83. Oral Microbiol Immunol. 1991;6:216–20. doi: 10.1111/j.1399-302x.1991.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 52.Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, et al. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol. 2005;58:847–63. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- 53.Potempa M, Potempa J, Kantyka T, Nguyen KA, Wawrzonek K, Manandhar SP, et al. Interpain A, a cysteine proteinase from Prevotella intermedia, inhibits complement by degrading complement factor C3. PLoS Pathog. 2009;5:e1000316. doi: 10.1371/journal.ppat.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamaguch A, Nakayama K, Ohyama T, Watanabe T, Okamoto M, Baba H. Coaggregation of Porphyromonas gingivalis and Prevotella intermedia. Microbiol Immunol. 2001;45:649–56. doi: 10.1111/j.1348-0421.2001.tb01298.x. [DOI] [PubMed] [Google Scholar]
- 55.Kamaguchi A, Ohyama T, Sakai E, Nakamura R, Watanabe T, Baba H, et al. Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia. Microbiology. 2003;149:1257–64. doi: 10.1099/mic.0.25997-0. [DOI] [PubMed] [Google Scholar]
- 56.Sharma A. Virulence mechanisms of Tannerella forsythia. Periodontol 2000. 2010;54:106–16. doi: 10.1111/j.1600-0757.2009.00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jusko M, Potempa J, Karim AY, Ksiazek M, Riesbeck K, Garred P, et al. A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J Immunol. 2012;188:2338–49. doi: 10.4049/jimmunol.1101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, et al. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537–44. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asakawa R, Komatsuzawa H, Kawai T, Yamada S, Goncalves RB, Izumi S, et al. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2003;50:1125–39. doi: 10.1046/j.1365-2958.2003.03748.x. [DOI] [PubMed] [Google Scholar]
- 60.Malm S, Jusko M, Eick S, Potempa J, Riesbeck K, Blom AM. Acquisition of complement inhibitor serine protease factor I and its cofactors C4b-binding protein and factor H by Prevotella intermedia. PLoS One. 2012;7:e34852. doi: 10.1371/journal.pone.0034852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDowell JV, Huang B, Fenno JC, Marconi RT. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun. 2009;77:1417–25. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki T, Miyamoto M, Yamada S, Okuda K, Ishihara K. Surface protease of Treponema denticola hydrolyzes C3 and influences function of polymorphonuclear leukocytes. Microbes Infect. 2006;8:1758–63. doi: 10.1016/j.micinf.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 63.Schenkein HA, Berry CR. Activation of complement by Treponema denticola. J Dent Res. 1991;70:107–10. doi: 10.1177/00220345910700020201. [DOI] [PubMed] [Google Scholar]
- 64.Mevorach D, Mascarenhas JO, Gershov D, Elkon KB. Complement-dependent clearance of apoptotic cells by human macrophages. J Exp Med. 1998;188:2313–20. doi: 10.1084/jem.188.12.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21:643–53. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 66.Wright SD, Silverstein SC. Receptors for C3b and C3bi promote phagocytosis but not the release of toxic oxygen from human phagocytes. J Exp Med. 1983;158:2016–23. doi: 10.1084/jem.158.6.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berton G, Laudanna C, Sorio C, Rossi F. Generation of signals activating neutrophil functions by leukocyte integrins: LFA-1 and gp150/95, but not CR3, are able to stimulate the respiratory burst of human neutrophils. J Cell Biol. 1992;116:1007–17. doi: 10.1083/jcb.116.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ernst JD. Macrophage receptors for Mycobacterium tuberculosis. Infect. Immun. 1998;66:1277–81. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hellwig SM, van Oirschot HF, Hazenbos WL, van Spriel AB, Mooi FR, van De Winkel JG. Targeting to Fcγ receptors, but not CR3 (CD11b/CD18), increases clearance of Bordetella pertussis. J. Infect. Dis. 2001;183:871–9. doi: 10.1086/319266. [DOI] [PubMed] [Google Scholar]
- 70.Hajishengallis G, McIntosh ML, Nishiyama S-i, Yoshimura F. Mechanism and implications of CXCR4-mediated integrin activation by Porphyromonas gingivalis. Mol Oral Microbiol. 2012;28 doi: 10.1111/omi.12021. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 1992;267:18902–7. [PubMed] [Google Scholar]
- 73.Klos A, Tenner AJ, Johswich KO, Ager RR, Reis ES, Kohl J. The role of the anaphylatoxins in health and disease. Mol Immunol. 2009;46:2753–66. doi: 10.1016/j.molimm.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snyderman R. Role for endotoxin and complement in periodontal tissue destruction. J Dent Res. 1972;51:356–61. doi: 10.1177/00220345720510022201. [DOI] [PubMed] [Google Scholar]
- 75.Mahtout H, Chandad F, Rojo JM, Grenier D. Porphyromonas gingivalis mediates the shedding and proteolysis of complement regulatory protein CD46 expressed by oral epithelial cells. Oral Microbiol Immunol. 2009;24:396–400. doi: 10.1111/j.1399-302X.2009.00532.x. [DOI] [PubMed] [Google Scholar]
- 76.Mahtout H, Curt S, Chandad F, Rouabhia M, Grenier D. Effect of periodontopathogen lipopolysaccharides and proinflammatory cytokines on CD46, CD55, and CD59 gene/protein expression by oral epithelial cells. FEMS Immunol Med Microbiol. 2011;62:295–303. doi: 10.1111/j.1574-695X.2011.00813.x. [DOI] [PubMed] [Google Scholar]
- 77.Cardone J, Le Friec G, Vantourout P, Roberts A, Fuchs A, Jackson I, et al. Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nat Immunol. 2010;11:862–71. doi: 10.1038/ni.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cardone J, Le Friec G, Kemper C. CD46 in innate and adaptive immunity: an update. Clin Exp Immunol. 2011;164:301–11. doi: 10.1111/j.1365-2249.2011.04400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 80.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–87. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 81.Discipio RG, Daffern PJ, Kawahara M, Pike R, Travis J, Hugli TE, et al. Cleavage of human complement component C5 by cysteine proteinases from Porphyromonas (Bacteroides) gingivalis Prior oxidation of C5 augments proteinase digestion of C5. Immunology. 1996;87:660–7. doi: 10.1046/j.1365-2567.1996.478594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Imamura T, Banbula A, Pereira PJ, Travis J, Potempa J. Activation of human prothrombin by arginine-specific cysteine proteinases (Gingipains R) from Porphyromonas gingivalis. J Biol Chem. 2001;276:18984–91. doi: 10.1074/jbc.M006760200. [DOI] [PubMed] [Google Scholar]
- 83.Huber-Lang M, Sarma JV, Zetoune FS, Rittirsch D, Neff TA, McGuire SR, et al. Generation of C5a in the absence of C3: a new complement activation pathway. Nat Med. 2006;12:682–7. doi: 10.1038/nm1419. [DOI] [PubMed] [Google Scholar]
- 84.Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, et al. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol. 2011;186:869–77. doi: 10.4049/jimmunol.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo RF, Ward PA. Role of C5a in inflammatory responses. Annu Rev Immunol. 2005;23:821–52. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- 86.Abe T, Hosur KB, Hajishengallis E, Reis ES, Ricklin D, Lambris JD, et al. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J Immunol. 2012;189:5442–8. doi: 10.4049/jimmunol.1202339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kagan JC. “Complementing” toll signaling. Sci Signal. 2010;3:pe15. doi: 10.1126/scisignal.3120pe15. [DOI] [PubMed] [Google Scholar]
- 88.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: Activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- 90.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91:816–20. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Honda K. Porphyromonas gingivalis sinks teeth into the oral microbiota and periodontal disease. Cell Host Microbe. 2011;10:423–5. doi: 10.1016/j.chom.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 92.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–25. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hasturk H, Kantarci A, Goguet-Surmenian E, Blackwood A, Andry C, Serhan CN, et al. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J Immunol. 2007;179:7021–9. doi: 10.4049/jimmunol.179.10.7021. [DOI] [PubMed] [Google Scholar]
- 94.Page RC, Lantz MS, Darveau R, Jeffcoat M, Mancl L, Houston L, et al. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol Immunol. 2007;22:162–8. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 95.Doungudomdacha S, Rawlinson A, Douglas CW. Enumeration of Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in subgingival plaque samples by a quantitative-competitive PCR method. J Med Microbiol. 2000;49:861–74. doi: 10.1099/0022-1317-49-10-861. [DOI] [PubMed] [Google Scholar]
- 96.Chaves ES, Jeffcoat MK, Ryerson CC, Snyder B. Persistent bacterial colonization of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in periodontitis and its association with alveolar bone loss after 6 months of therapy. J Clin Periodontol. 2000;27:897–903. doi: 10.1034/j.1600-051x.2000.027012897.x. [DOI] [PubMed] [Google Scholar]
- 97.Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–39. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 98.Page RC, Schroeder HE. Periodontitis in man and other animals- A comparative review. Karger; 1982. [Google Scholar]
- 99.Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 100.Courts FJ, Boackle RJ, Fudenberg HH, Silverman MS. Detection of functional complement components in gingival crevicular fluid from humans with periodontal diseases. J Dent Res. 1977;56:327–31. doi: 10.1177/00220345770560032001. [DOI] [PubMed] [Google Scholar]
- 101.Toto PD, Lin L, Gargiulo A. Identification of C3a, IgG, IgM in inflamed human gingiva. J Dent Res. 1978;57:696. doi: 10.1177/00220345780570050501. [DOI] [PubMed] [Google Scholar]
- 102.Nikolopoulou-Papaconstantinou AA, Johannessen AC, Kristoffersen T. Deposits of immunoglobulins, complement, and immune complexes in inflamed human gingiva. Acta Odontol Scand. 1987;45:187–93. doi: 10.3109/00016358709098858. [DOI] [PubMed] [Google Scholar]
- 103.Rautemaa R, Meri S. Protection of gingival epithelium against complement-mediated damage by strong expression of the membrane attack complex inhibitor protectin (CD59) J Dent Res. 1996;75:568–74. doi: 10.1177/00220345960750010901. [DOI] [PubMed] [Google Scholar]
- 104.Boackle RJ. The interaction of salivary secretions with the human complement system--a model for the study of host defense systems on inflamed mucosal surfaces. Crit Rev Oral Biol Med. 1991;2:355–67. doi: 10.1177/10454411910020030401. [DOI] [PubMed] [Google Scholar]
- 105.Attstrom R, Laurel AB, Lahsson U, Sjoholm A. Complement factors in gingival crevice material from healthy and inflamed gingiva in humans. J Periodont Res. 1975;10:19–27. doi: 10.1111/j.1600-0765.1975.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 106.Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. I. Quantitative study of immunoglobulins, complement components, and other plasma proteins. J Periodontol. 1977;48:772–7. doi: 10.1902/jop.1977.48.12.772. [DOI] [PubMed] [Google Scholar]
- 107.Challacombe SJ, Shirlaw PJ. Immunology of diseases of the oral cavity. In: Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, Mayer L, editors. Mucosal Immunology. Elsevier Academic Press; 2005. pp. 1517–46. [Google Scholar]
- 108.Patters MR, Niekrash CE, Lang NP. Assessment of complement cleavage in gingival fluid during experimental gingivitis in man. J Clin Periodontol. 1989;16:33–7. doi: 10.1111/j.1600-051x.1989.tb01609.x. [DOI] [PubMed] [Google Scholar]
- 109.Beikler T, Peters U, Prior K, Eisenacher M, Flemmig TF. Gene expression in periodontal tissues following treatment. BMC Med Genomics. 2008;1:30. doi: 10.1186/1755-8794-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Niekrash CE, Patters MR. Simultaneous assessment of complement components C3, C4, and B and their cleavage products in human gingival fluid. II. Longitudinal changes during periodontal therapy. J Periodontal Res. 1985;20:268–75. doi: 10.1111/j.1600-0765.1985.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 111.Hillebrandt S, Wasmuth HE, Weiskirchen R, Hellerbrand C, Keppeler H, Werth A, et al. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat Genet. 2005;37:835–43. doi: 10.1038/ng1599. [DOI] [PubMed] [Google Scholar]
- 112.Chai L, Song Y-Q, Zee K-Y, Leung WK. Single nucleotide polymorphisms of complement component 5 and periodontitis. J Periodont Res. 2010;45:301–8. doi: 10.1111/j.1600-0765.2009.01234.x. [DOI] [PubMed] [Google Scholar]
- 113.Heitz-Mayfield LJ. Disease progression: identification of high-risk groups and individuals for periodontitis. J Clin Periodontol. 2005;32 (Suppl 6):196–209. doi: 10.1111/j.1600-051X.2005.00803.x. [DOI] [PubMed] [Google Scholar]
- 114.Hajishengallis G. Too old to fight? Aging and its toll on innate immunity. Mol Oral Microbiol. 2010;25:25–37. doi: 10.1111/j.2041-1014.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kornman KS. Interleukin 1 genetics, inflammatory mechanisms, and nutrigenetic opportunities to modulate diseases of aging. Am J Clin Nutr. 2006;83:475S–83S. doi: 10.1093/ajcn/83.2.475S. [DOI] [PubMed] [Google Scholar]
- 116.Graves DT, Fine D, Teng YT, Van Dyke TE, Hajishengallis G. The use of rodent models to investigate host-bacteria interactions related to periodontal diseases. J Clin Periodontol. 2008;35:89–105. doi: 10.1111/j.1600-051X.2007.01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Breivik T, Gundersen Y, Gjermo P, Taylor SM, Woodruff TM, Opstad PK. Oral treatment with complement factor C5a receptor (CD88) antagonists inhibits experimental periodontitis in rats. J Periodontal Res. 2011;46:643–7. doi: 10.1111/j.1600-0765.2011.01383.x. [DOI] [PubMed] [Google Scholar]
- 118.Brown LJ, Johns BA, Wall TP. The economics of periodontal diseases. Periodontol 2000. 2002;29:223–34. doi: 10.1034/j.1600-0757.2002.290111.x. [DOI] [PubMed] [Google Scholar]
- 119.Beikler T, Flemmig TF. Oral biofilm-associated diseases: trends and implications for quality of life, systemic health and expenditures. Periodontol 2000. 2011;55:87–103. doi: 10.1111/j.1600-0757.2010.00360.x. [DOI] [PubMed] [Google Scholar]
- 120.Armitage GC. Classifying periodontal diseases--a long-standing dilemma. Periodontol 2000. 2002;30:9–23. doi: 10.1034/j.1600-0757.2002.03002.x. [DOI] [PubMed] [Google Scholar]
- 121.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wagner E, Frank MM. Therapeutic potential of complement modulation. Nat Rev Drug Discov. 2010;9:43–56. doi: 10.1038/nrd3011. [DOI] [PubMed] [Google Scholar]
- 123.Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat Biotechnol. 2007;25:1265–75. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leslie M. Immunology. The new view of complement. Science. 2012;337:1034–7. doi: 10.1126/science.337.6098.1034. [DOI] [PubMed] [Google Scholar]
- 125.Ricklin D, Lambris JD. Progress and trends in complement therapeutics. Adv Exp Med Biol. 2013;734:1–22. doi: 10.1007/978-1-4614-4118-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ricklin D, Lambris JD. Complement in Immune and Inflammatory Disorders: Therapeutic Interventions. J Immunol. 2013;190 doi: 10.4049/jimmunol.1203200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Niekrash CE, Patters MR. Assessment of complement cleavage in gingival fluid in humans with and without periodontal disease. J Periodontal Res. 1986;21:233–42. doi: 10.1111/j.1600-0765.1986.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 128.Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–45. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]