Abstract
Microglia, the resident immune cell of the central nervous system (CNS), are thought to contribute to the pathogenesis of age-related neurodegenerative disorders. It has been hypothesized that microglia undergo age-related changes in gene expression patterns that give rise to pathogenic phenotypes. We compared the gene expression profiles in microglia isolated ex vivo from the mouse retinas of ages ranging from early adulthood to late senescence. We discovered that microglial gene expression demonstrated progressive change with increasing age and involved genes that regulate microglial supportive functions and immune activation. Molecular pathways involving immune function and regulation, angiogenesis, and neurotrophin signaling demonstrated age-related change. In particular, expression levels of complement genes, C3 and CFB, previously associated with age-related macular degeneration (AMD), increased with aging, suggesting that senescent microglia may contribute to complement dysregulation during disease pathogenesis. Taken together, senescent microglia demonstrate age-related gene expression changes capable of altering their constitutive support functions and regulation of their activation status in ways relating to neuroinflammation and neurodegeneration in the CNS.
Keywords: aging, microglia, retina, microarray, gene expression, complement, activation, angiogenesis, neurotrophic factors, senescence
1. Introduction
Advancing age is strongly associated with the increasing prevalence of neurodegenerative disease of the retina, such as age-related macular degeneration, glaucoma, and diabetic retinopathy (Friedman et al. 2004a; Friedman et al. 2004b), which constitute the leading causes of low vision and legal blindness in the developed world (Congdon et al. 2003; Congdon et al. 2004). While the pathogenic mechanisms for these age-related retinal diseases remain unclear, chronic neuroinflammation resulting from the activation of the immune system features prominently (Wax & Tezel 2009; Buschini et al. 2011; Tang & Kern 2011) and appears to be causally related to disease progression. Histopathological specimens from affected humans (Yuan & Neufeld 2001; Gupta et al. 2003; Zeng et al. 2008) and from animal models of disease (Krady et al. 2005; Combadiere et al. 2007; Bosco et al. 2011) demonstrate the early involvement of retinal microglia, implicating them as an initiating source of neuroinflammatory change underlying disease pathogenesis.
The common elements in aging and microglial changes in neurodegenerative disease suggest that senescent changes in microglia may play a causal role in pathogenic neuroinflammation (Streit & Xue 2009; von Bernhardi et al. 2010). Recent studies utilizing the technique of parabiosis to create chimerism in bone-marrow derived precursors have revealed that microglia indeed have long tenures in the course of an animal’s regular life span in the undiseased CNS (Ajami et al. 2007; Mildner et al. 2007). The resulting low turnover rate of microglia in situ indicates their susceptibility to senescence-related changes, which can influence the aging CNS milieu in potentially pathogenic ways.
There is accumulating evidence that microglia can exhibit phenotypic changes with advancing organismal age. Microglia have a unique phenotype in the uninjured CNS by virtue of their highly ramified morphology and rapidly and continuously moving processes, which allow their constant contact with neighboring neurons and glia (Davalos et al. 2005; Nimmerjahn et al. 2005; Lee et al. 2008). These dynamic and repeated cell-cell contacts are thought to subserve constitutive functions of synapse regulation and neuronal support (Paolicelli et al. 2011; Schafer et al. 2012b; Vinet et al. 2012). We and others have previously shown that phenotypic features of microglia undergo senescent change in which aged microglia become less ramified and move their processes with decreased dynamism (Sierra et al. 2007; Damani et al. 2011; Tremblay et al. 2012), suggesting a decline in their supportive functions with aging. In addition, aged microglia demonstrate dysregulation in their activation status. Microglia in aged brains show increased signs of activation at baseline (Perry et al. 1993; Sheng et al. 1998) and respond to activating triggers in a manner that is more augmented and prolonged compared to microglia in young brains (Xie et al. 2003; Sierra et al. 2007). In the retina, we have shown that aging microglia, in accumulating increased intracellular lipofuscin, exhibit dysregulated complement activation and increased secretion of inflammatory cytokines (Ma et al. 2013). These findings indicate that microglia are susceptible to a senescent loss of proper regulation in activation in affected tissues.
Molecular mechanisms underlying age-related phenotypic changes in microglia are yet unclear. We investigate this question in the current study by comparing gene expression patterns in microglia isolated ex vivo from mouse retinal tissue obtained from age groups spanning the full range of adult aging. We have focused on microglia located in the retina, a specialized division of the CNS, though findings here may potentially be generalized to microglia elsewhere (de Haas et al. 2008). Analyses of age-related gene expression in the whole retina has been previously performed (Yoshida et al. 2002; Chen et al. 2008a), identifying genes involved inflammatory responses (Chen et al. 2010; Van Kirk et al. 2011) and implicating immunological influences in the overall aging phenotype of the retina (Xu et al. 2009). However, individual contributions of different retinal cell types cannot be discerned in these studies. The current study represents an advance on previous work in its specific analysis of retinal microglia isolated ex vivo, which allows the specific contribution of microglial aging changes to be separately examined.
Our results demonstrate that the overall profile of gene expression in retinal microglia changes as a function of aging and highlight particular molecular pathways regulating microglial constitutive function, microglial activation, and complement regulation. These changes may relate to the development of known senescent microglial phenotypes, including those related with pathological significance. These associations can help direct further investigation into molecular mechanisms underlying age-related immune dysregulation and the search for anti-inflammatory therapeutic approaches to age-related retinal disease.
2. Methods
2.1 Experimental animals of different age groups
Wild type male C57BL/6J mice of four age groups (3 months, 12 months, 18 months, and 24 months) spanning the entire period from adulthood to late senescence were obtained from the NIA Aged Rodent Colonies (National Institutes on Aging, Bethesda, MD, USA). Heterozygous CX3CR1+/gfp transgenic animals, created by breeding CX3CR1gfp/gfp mice (Jung et al. 2000)(The Jackson Laboratory, Bar Harbor, ME) with wild type mice, were used for immunohistochemical studies. All animals were genotyped for the rd8 mutation in the Crb1 gene, a mutation recently found in lines of inbred and transgenic mice (Mattapallil et al. 2012), and were confirmed to lack this mutation. Briefly, detection of the rd8 mutation versus the wild type genotype was performed using two genotyping methods: 1) by PCR using a TaqMan allelic discrimination assay, and 2) by DNA sequencing of the rd8-associated nucleotide deletion using methods and PCR primers as previously described (Mattapallil et al. 2012). All animals were housed in a National Institutes of Health animal facility under a normal 12-hour light-dark cycle. Experiments were conducted according to protocols approved by a local Institutional Animal Care and Use Committee and adhered to the Association for Research in Vision and Ophthalmology (ARVO) Statement for the use of animals in ophthalmic and vision research. Both retinas of each mouse were combined as a single biological replicate; at least four animals from each age-group were used.
2.2 Isolation of retinal microglia
Mice were euthanized and the eyes were immediately enucleated. The globes were immersed in ice-cold Hank’s balanced salt solution (HBSS), the retinas were isolated by dissection and transferred into 0.2% papain solution including glucose (1mg/ml), DNAse1 (Worthington, Lakewood, NJ, 100U/ml), superoxide dismutase (SOD) (Worthington, 5μg/ml) and catalase (Sigma, St Louis, MO, 5μg/ml) in HBSS, and incubated at 8°C for 45 min and then at 28°C for 7 min. The digested tissue was dissociated by trituration and centrifuged at 150G for 5 min at 4°C. The resulting cell pellet was resuspended with neutralization buffer containing glucose (2mg/ml), DNAse1 (100U/ml), SOD (5μg/ml), catalase (5μg/ml), antipain (Roche, Indianopolis, IN, 50μg/ml), d-alpha-tocopheryl acetate (Sigma, 10μg/ml), albumin (40mg/ml), and gentamycin (Sigma, 1μl/ml), and again centrifuged at 150G for 5 min at 4°C. The cellular pellet was resuspended in 100μl of staining buffer (BD Pharmingen, San Diego, CA, Cat#: 554656) containing a FITC-conjugated antibody to CD11b (1:50, eBioscience, San Diego, CA, Cat#11–0112), and incubated for 30 minutes at 4°C to label retinal microglia. The cells were washed twice in 10ml of staining buffer containing 2mM EDTA and suspended with 0.5ml of staining buffer. Labeled retinal microglia were isolated by fluorescence-activated cell sorting (FACS) (BD FACSAria II Flow Cytometer, BD, Franklin Lakes, NJ) at the NEI Flow Cytometry Core Facility (Supplementary Fig 1A). Viability of sorted cells was assessed by labeling with Cy-5-conjugated propidium-iodide; mean percentage of live cells to whole cells was 86.0±4.0%, with no significant differences found between samples from animals of different ages. Approximately 1500–1800 CD11b+ microglial cells were obtained from both retinas of each experimental animal. These were collected into a 1.5ml eppendorf tube containing 200μl of RNAprotect reagent (Qiagen, Valencia, CA, Cat#D-40724) and stored at −80°C for subsequent RNA extraction.
The selectivity of the above CD11b-based method for isolating microglia was verified by additional experiments using CX3CR1+/gfp transgenic mice. Immunohistochemical staining in retinal flat-mounts revealed that all GFP+ retinal microglia in CX3CR1+/gfp animals were also immunopositive for CD11b (Supplementary Fig 1B). Flow cytometric cell sorting of retinal microglia from CX3CR1+/gfp animals using the same CD11b-based method demonstrated that cells isolated on the basis of CD11b were simultaneously positive for GFP (Supplementary Fig 1C), indicating that isolated CD11b+ cells were indeed CX3CR1-expressing microglia.
2.3 Total RNA extraction, microarray hybridization, processing and analysis
Total RNA extraction was performed on FACS-sorted microglia cells with RNeasy Mini kit (Qiagen, Valencia, CA) following the manufacturer’s instruction. The extracted RNA was quantitatively analyzed (Agilent 2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA); the mean RIN from 16 samples was ≥1. Total RNA (10–500 pg) from purified microglia from each biological replicate was used for target generation. cDNA amplification from the extracted RNA was performed using the Ovation One-Direct RNA amplification (NuGen Technologies, San Carlos, CA) according to the manufacturer’s protocol. The amplified cDNA was fragmented and labeled with biotin using the Encore Biotin Module (NuGen Technologies). The labeled target (5 μg) was applied to a GeneChip Mouse Exon 1.0ST Array, hybridized for 21 hours at 45°C, washed and stained on an Affymetrix GeneChip Fluidics Station 450, and then scanned with a GeneChip Scanner 3000 7G (all from Affymetrix, Santa Clara, CA).
Raw data were normalized and analyzed using the GeneSpring GX 11.0.2 software (Silicon Genetics, Redwood City, CA). The Robust Multichip Average (RMA) method (Irizarry et al. 2003) was used for background correction, normalization, and summarization of expression scores for gene expression data. A one-way ANOVA was used to compare gene expression levels between consecutive time-points (3 months to 12 months, 12 months to 18 months, 18 months to 24 months). Gene expression changes that were increased or decreased by a factor of greater than 1.5-fold and had an uncorrected p-value of <0.05 in at least one of the comparisons were identified. From this overall set, subsets of genes that demonstrated a “monotonic-to-age” change in expression across the entire ranges of ages were identified for further analysis. Two subsets of genes were identified: (1) genes that increased monotonically with time (i.e. genes that either increased significantly or remained stable between time-points) and (2) genes that decreased monotonically with time (i.e. genes that either decreased significantly or remained stable between time-points). Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA) was employed to identify affected networks and functional pathways within the gene lists. Microarray data have been deposited into NCBI’s Gene Expression Omnibus (GEO; accession number GSE38739) in compliance with Minimum Information About a Microarray Experiment (MIAME) guidelines.
2.4 Real-time quantitative reverse transcriptase polymerase chain reaction (rt-PCR) analysis
Quantitative rt-PCR analysis of FACS-purified retinal microglia cells was performed to validate findings from the microarray data. Retinal microglia from four additional independent biological replicates for each of the four aging time-points (3, 12, 18 and 24 months of age) were obtained as described above. RNA was extracted from microglia (RNeasy Mini kit, Qiagen, Valencia, CA), amplified to cDNA (WT-Ovation™ One-Direct System, NuGEN, San Carlos, CA), and purified (MinElute Reaction Cleanup kit, Qiagen) following the manufacturers’ instructions. 2 μl of cDNA was used for real-time PCR using SYBR Green PCR Mastermix on the 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA). β-2 microglobulin(B2M) and ribosomal protein S13 served as internal controls. Primer pairs tested are listed in Supplementary Table 1.
2.5 Immunohistochemistry and image analysis
Retinal and sclerochoroidal flatmounts were prepared from CX3CR1+/gfp mice as described previously (Damani et al. 2011). Sclerochoroidal flatmounts containing a surface monolayer of retinal pigment epithelial (RPE) cells with adhering subretinal microglia were immunostained with the antibodies to the following antigens: C3 (Hycult Biotech, Clone 11H9, Cat#HM1045, The Netherlands, 1:200), complement factor B (CFB) (Santa Cruz Biotechnology, Cat# sc-67141, 1:200), and C3b/iC3b/C3c (mAb 2/11,#HM1078, Hycult, Uden, The Netherlands, 1:200). Secondary antibodies, conjugated to Alexa-633 (Invitrogen, Carlsbad, CA), were added at a 1:200 dilution and incubated for 1–2 hours. Tissue preparations were mounted onto slides and imaged using confocal imaging with a 60x oil objective at a resolution of 1024×1024 pixels; similar imaging settings were employed in the imaging of tissue of different ages. The extents of immunostaining for antibodies to C3, CFB, and iC3b were quantitated using ImageJ software (National Institutes of Health, Bethesda, MD); maximum intensity projections of z-series confocal images stacks were converted to binarized images using a common thresholding function and counts of immunopositive pixels performed. Quantitative analysis of microglial morphological parameters were performed as previously described (Fontainhas et al. 2011). Analysis of microglial soma size was performed by delineating the outline of individual microglia somata using the smooth polygon tool in NIH ImageJ and quantifying the area and perimeter of the circumscribed region. Analysis of microglial dendritic size was performed by circumscribing the area delimited by the termini of dendritic processes of individual microglia and quantifying the area and perimeter of the circumscribed region. Maximum intensity projection image of individual GFP+ microglia was binarized and a topological skeleton derived from the binary image using the “skeletonize” function in ImageJ and total dendritic length was calculated from the skeletonized image.
3. Results
3.1 Profiles of age-associated gene expression in mouse retinal microglial cells
In order to evaluate the differential expression of genes in microglia in the retina, we purified CD11b-positive microglia from the retinas of wild type C57BL/6 mice aged 3, 12, 18, and 24 months (four animals from each age group) using cell sorting with flow cytometry. The number of retinal microglia isolated per experimental animal (using two retinas from each animal) averaged 1805±805 cells (mean±SD). The number of retinal microglia isolated per animal were statistically similar among different age groups (p >0.05 for all comparisons, 1-way ANOVA with Tukey’s multiple comparison test). Age-related differential expression of genes from isolated retinal microglia was analyzed using 16 Affymetrix GeneChip Mouse Exon 1.0ST Arrays. Analysis revealed high expression levels of microglia-associated genes (CD11b, Iba1, P2Y12, and CX3CR1) while genes expressed by macroglia (GFAP), vascular cells (NG2), RPE cells (RPE65), and retinal neurons (calbindin, Brn3a, Brn3b) were expressed at significantly lower background levels (data not shown). Differential expression levels of individual genes between consecutive age groups (i.e. between 3 and 12 months, 12 and 18 months, and 18 and 24 months) were examined to identify microglial genes demonstrating significant changes in mRNA expression across the full range of ages ranging from mature adulthood to late senescence.
We identified a total of 719 genes (out of a total of 16711) that showed differential expression of >1.5-fold change at p<0.05 (uncorrected one-way ANOVA) for at least one of the 3 age-group comparisons (Supplementary Table 2). This method of selection had the goal of identifying genes that are altered in expression level not only at the “end-stage” of late senescence but throughout the entire period of adult aging, from young adulthood through “middle-age” to late senescence. These differentially expressed genes were subjected to a hierarchical cluster analysis to visualize trends in differential expression across individual biological repeats in the 4 age groups (Fig. 1). Individual biological repeats showed a tendency to cluster within their age-group, demonstrating progressive changes in retinal microglial gene expression with age.
Figure 1. Hierarchical clustering heat map for 719 genes demonstrating age-related differential expression (fold-change ≥1.5x and uncorrected p-values <0.05).
Each column represents a biological replicate of a particular age (4 replicates per age group); each column row represents a single gene. Gene expression changes with respect to median changes are represented in direction and magnitude by the legend (bottom): red, up-regulated; blue, down-regulated; gray, unchanged.
Among the 719 genes showing age-dependent expression changes, two subsets of genes were identified that demonstrated either an increasing or a decreasing “monotonic-to-age” pattern of change (i.e. a gene that showed an increasing “monotonic-to-age” pattern would demonstrate a significant increase in expression across one or more consecutive age-groups without significantly decreasing in expression across any consecutive age-group). Of 719 genes, 295 (41%) genes demonstrated a monotonic increase in expression with increasing age, while 284 (39%) genes demonstrated a monotonic decrease in expression with age. The lists of up-regulated and down-regulated genes from each category respectively are shown in Supplementary Tables 3 and 4.
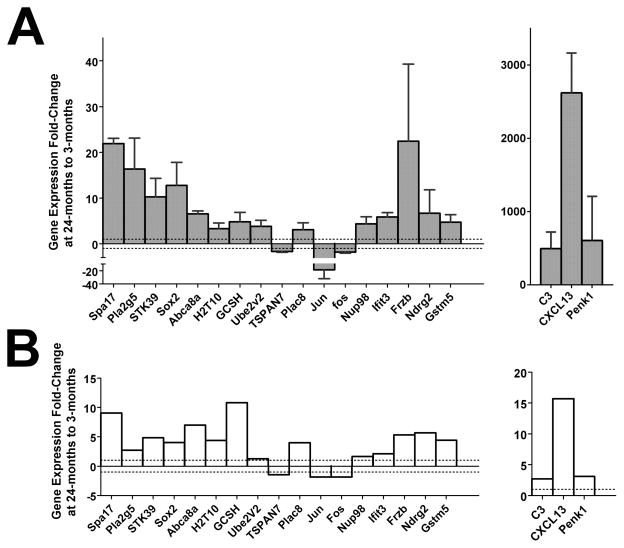
Real-time quantitative rt-PCR (qRT-PCR) was performed for 20 selected genes on cDNA isolated from similarly FACS-sorted retinal microglia. The analysis demonstrated significant age-related changes in gene expression between the 3-month old and 24-month old age groups that were consistent in direction to that observed from the microarray analysis in 19 out of 20 genes (Fig. 2). Also, “housekeeping” genes (RPS13, RPS 9) and functional microglial genes (CD68, Iba1), which were relatively unchanged in expression on microarray analyses, demonstrated a similar relative stability in expression levels on qRT-PCR analyses (Supplementary Fig. 2).
Figure 2. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) validation of selected genes.
Relative gene expression fold change from qRT-PCR from retinal microglia from 24-month old mice (3 -6 biological repeats were used for each experiment, with one sample replicate per biological replicate), normalized relative to those from 3-month old mice (A) is compared to the relative fold changes for the same genes predicted from microarray analysis (B). Error bars indicate ± SEM.
3.2 Biological features of gene expression pattern changes associated with aging in retinal microglia
Gene ontology (GO) enrichment analysis (Sheehan et al. 2008) was used to assign significant age-related gene expression changes to biologically meaningful categories. The GO categories with “Cellular Process” with enrichment scores > 3 and with a representation of at least two differentially-expressed genes were identified and organized according to the broader GO categories in which the subcategories belong (Table 1). In this analysis, genes that demonstrated age-related expression changes involved: 1) biological regulatory mechanisms governing transcription, signal transduction, activity of signaling pathways, phosphorylation, and cell proliferation and growth, 2) lipid biosynthetic process, 3) transport of organic substances and intracellular proteins, and 4) immune system processes, specifically the control of microglial activation.
Table 1.
Gene ontology (GO) enrichment analysis of 719 aging-associated genes in retinal microglia
| Cellular Process | Enrichment Score1 | ||
|---|---|---|---|
| Metabolic process | lipid biosynthetic process | triglyceride metabolic process | 8.21 |
| very long-chain fatty acid biosynthetic process | 7.17 | ||
| platelet activating factor biosynthetic process | 6.01 | ||
| steroid biosynthetic process | 5.13 | ||
| isocitrate metabolic process | 4.46 | ||
| fatty acid metabolic process | 4.34 | ||
| Biological Regulation | positive regulation of transcription, DNA- dependent | positive regulation of transcription from RNA polymerase II promoter | 7.64 |
| negative regulation of DNA binding | 7.34 | ||
| regulation of transcription, DNA-dependent | 5.58 | ||
| regulation of transcription from RNA polymerase II promoter | 3.22 | ||
| regulation of signal transduction | negative regulation of ERK1 and ERK2 cascade | 6.71 | |
| negative regulation of Rho protein signal transduction | 6.34 | ||
| positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway | 4.46 | ||
| positive regulation of I-kappaB kinase/NF-kappaB cascade | 3.7 | ||
| positive regulation of MAPKKK cascade | 3.46 | ||
| Regulation of activity | inactivation of MAPK activity | 6.48 | |
| positive regulation of metalloenzyme activity | 6.01 | ||
| positive regulation of protein serine/threonine kinase activity | 4.78 | ||
| positive regulation of NF-kappaB transcription factor activity | 3.23 | ||
| regulation of phosphorylation | positive regulation of peptidyl-serine phosphorylation | 6.96 | |
| negative regulation of peptidyl-serine phosphorylation | 3.44 | ||
| regulation of homeostatic process | positive regulation of release of sequestered calcium ion into cytosol | 9.97 | |
| negative regulation of cellular process | negative regulation of cell proliferation | 6.06 | |
| negative regulation of cell growth | 5.75 | ||
| regulation of localization | positive regulation of potassium ion transport | 6.72 | |
| regulation of cellular component organization | positive regulation of endocytosis | 5.44 | |
| positive regulation of protein complex assembly | 3.44 | ||
| Transport | organic substance transport | L-glutamate import | 7.71 |
| D-aspartate import | 6.01 | ||
| L-glutamate transport | 5.7 | ||
| L-amino acid transport | 4.26 | ||
| dicarboxylic acid transport | 4.14 | ||
| intracellular protein transport | protein targeting | 4.94 | |
| protein import into nucleus, translocation | 4.26 | ||
| secretion by cell | calcium ion-dependent exocytosis | 5.44 | |
| exocytosis | 3.17 | ||
| Immune system process | microglial cell activation | 5.34 | |
| Locomotion | cell migration | 6.01 | |
| Cellular component organization or biogenesis | histone ubiquitination | 6.72 |
Log transformation of the p-value calculated usinga chi-square test comparing the proportion of genes in the list showing differential expression in a given GO group to the proportion of all genes in the same GO group.
Ingenuity® Pathway Analysis (IPA) which employs a curated genetic database of functional assignments was used to analyze the full set of 719 differentially-expressed genes to discover canonical pathways in microglia influenced by aging. A list of pathways in which age-related microglial genes demonstrate significant representation is shown in Table 2. These pathways included those that have been related to: (1) microglial immune function and regulation (IL-17A, IL1, and IL3 signaling, ceramide signaling, nitric oxide signaling, estrogen receptor signaling, and LPS-stimulated MAPK signaling) (Fig. 3), (2) angiogenesis (TSP1 regulated angiogenesis, and VEGF ligand-receptor interactions) (Fig. 4A, B), and (3) trophic factors (neurotrophin/TRK signaling) (Fig. 4C). These associations indicated that the aging microglial phenotype may involve its ability to regulate immune and inflammatory reactions, influence angiogenesis in its environment, and support neuronal survival and functioning.
Table 2.
Canonical pathway analysis of 719 genes differentially expressed with aging in retinal microglia.
| Canonical Pathways | Age-related genes | Ratio1 | P-value |
|---|---|---|---|
| Pathways related to microglial immune function and regulation | |||
|
| |||
| IL-17A Signaling | FOS, JUN, MAPK10, TNF | 0.1600 | 0.010000 |
| Ceramide Signaling | S1PR4, FOS, JUN, S1PR5, PIK3C2A, TNFRSF1A, PPM1L, PIK3R6, MRAS, PPP2R5E, TNF | 0.1260 | 0.000071 |
| IL-3 Signaling | FOS, PRKCI, J UN, PIK3C2A, CHP, PIK3R6, MRAS, PRKCE | 0.1080 | 0.002951 |
| IL-1 Signaling | GNB1, FOS, JUN, PRKAR2B, MYD88, GNAO1, MRAS, MAPK10, PRKAR2A, GNG3, ADCY7 | 0.1040 | 0.000288 |
| Nitric Oxide Signaling | VEGFA, BDKRB2, PRKAR2B, GUCY1A3, PIK3C2A, KDR, PIK3R6, CAV1, PRKAR2A, SLC7A1 | 0.1000 | 0.000372 |
| Estrogen-Dependent Breast Cancer Signaling | FOS, JUN, PIK3C2A, PIK3R6, MRAS, HSD17B12, HSD17B1 | 0.1000 | 0.004365 |
| LPS-stimulated MAPK Signaling | FOS, PRKCI, JUN, PIK3C2A, PIK3R6, MRAS, MAPK10, PRKCE | 0.0976 | 0.003548 |
|
| |||
| Pathways related to angiogenesis | |||
|
| |||
| Inhibition of Angiogenesis by TSP1 | VEGFA, JUN, GUCY1A3, KDR, MAPK10, CD36 | 0.1540 | 0.000955 |
| VEGF Family Ligand-Receptor Interactions | VEGFA, FOS, PRKCI, PIK3C2A, KDR, PLA2G5, PIK3R6, MRAS, PRKCE | 0.1070 | 0.001175 |
|
| |||
| Pathways related to trophic factors | |||
|
| |||
| Neurotrophin/TRK Signaling | FOS, JUN, Klk1b1, PIK3C2A, SPRY2, PIK3R6, MRAS, SORCS1 | 0.1100 | 0.001698 |
Number of genes in the list of age-related genes in a given canonical pathway divided by the total number of genes mapping to that pathway.
Figure 3. Age-associated gene expression changes in microglial signaling pathways involved with microglial immune function and activation as generated by Ingenuity Pathway analysis.
Signaling pathways involved in the regulation of microglial immune activities were highlighted in the analyses of genes whose expression varied significantly as a function of age. Aging-related genes are indicated either in green (down-regulated) or in red (up-regulated) (for 3-month to 24-month comparisons). The most prominently represented pathways included (A) IL17 signaling, (B) ceramide signaling, (C) IL3 signaling, (D) IL1 signaling, (E) estrogen-dependent signaling, (F) nitric oxide signaling, and (G) lipopolysaccharide (LPS)-stimulated MAPK signaling.
Figure 4. Age-associated gene expression changes in microglial signaling pathways involved with angiogensis and neurotrophic support as generated by Ingenuity Pathway analysis.
(A–B) Two particular signaling pathways involved in angiogenesis were those involving (A) thrombospondin-1 (TSP1), and (B) vascular endothelial growth factor (VEGF). (C) Neurotrophic-TRK receptor signaling pathways were also highlighted by microglial genes demonstrating age-related change. Aging-related genes are indicated either in green (down-regulated) or in red (up-regulated) (for 3-month to 24-month comparisons).
In order to explore the direct interactions between the 719 differentially expressed genes, we employed MetaCore™ from GeneGo, Inc. (Carlsbad, CA) to generate a network construction of interactions (Supplementary Fig. 3). AP1 and Egr1 were identified as “hubs” with the greatest number of interactions between differentially expressed genes.
3.3 Age-related expression changes in genes regulating complement activation
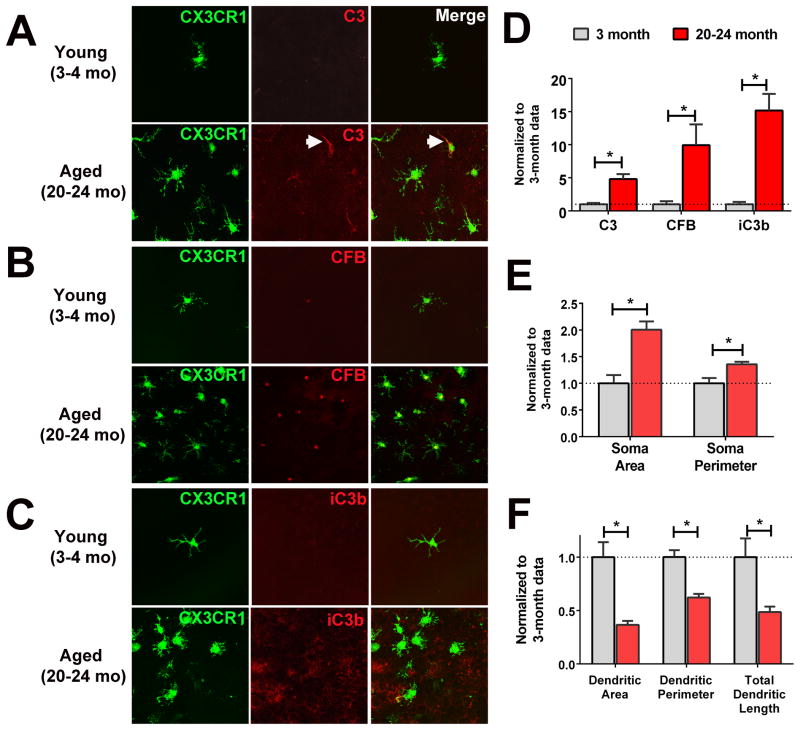
Among the subset of genes which showed monotonic increasing expression with age (295 genes), analysis of direct interactions highlighted those occurring between C3 and CFB. These age-related increases in gene expression as demonstrated on microarray and qPCR analyses (Supplementary Fig. 4) indicate increased expression and secretion of these proteins by aging retinal microglia. As C3 is a complement protein in the alternative pathway and CFB positively regulates complement activation in this pathway, this data indicate that 1) retinal microglia may regulate complement activation locally in the retina and that 2) with aging, the regulatory balance favors complement activation. We performed immunohistochemical analyses on the young adult (3–4 month old) and aged (20–24 month old) retina for C3 and CFB. Within the inner retina, very low levels of C3 immunopositivity were detected in both young and aged animals with no detectable immunopositivity in retinal microglia (data not shown). However, in the subretinal space, while minimal C3 immunopositivity was detectable in the young adult retina, immunopositivity within subretinal microglia was clearly present in cell somata and processes (Fig. 5A). Immunopositivity for CFB was similarly absent within the inner layers of the retina of both ages (data not shown). In the outer retina, CFB staining was weak and sparse in the subretinal microglia of young animals, but present prominently in subretinal microglia of aged animals (Fig. 5B). Immunopositivity for iC3b, formed from complement activation and C3 cleavage, was largely absent in the subretinal space in the young adult retina but prominently deposited on the apical surface of the RPE layer in the aged retina, suggesting an age-related activation of the complement system in the subretinal space resulting from changes in microglial complement regulation (Fig. 5C). The age-dependent changes in the extents of immunopositivity of these complement proteins within the subretinal space were quantitated and shown in Fig. 5D. Subretinal microglia in the aged animals demonstrated larger somata sizes (Fig. 5E) and less ramified morphologies (Fig. 5F).
Figure 5. Age-related changes in the complement gene expression and complement activation in retinal microglia.
Immunohistochemical staining of complement proteins, C3 (A) and CFB (B) in subretinal microglia, and complement activation product iC3b deposition in the subretinal space (C) of young (3–4 month old) and aged (20–24 month old) CX3CR1+/gfp transgenic mice. (A) Immunostaining for C3 (red) in subretinal microglia (green) demonstrated the absence of detectable immunopositivity in young mice. In aged mice, a substantial proportion of subretinal microglia showed immunopositivity, particularly in microglial processes (arrow). (B) Immunostaining for CFB (red) in subretinal microglia (green) demonstrated very weak immunopositivity in young mice but prominent staining of microglial somata in aged mice. (C) Immunopositivity for iC3b (red) was absent from the subretinal space of young mice but found distributed in a punctate manner in that of aged mice (green). (D) Quantification of the extent of immunopositivity of C3, CFB, and iC3b, demonstrating increased levels in the subretinal space in 20–24 month-old mice, relative to 3 month-old mice. (n = 6 representative 60x fields from 4–6 animals in each age group), (E) Morphological analysis of microglial soma sizes, demonstrating enlarged somata in terms of area and perimeter in 20–24 month-old mice, relative to 3 month-old mice. (F) Morphological analysis of microglial dendritic territory sizes, demonstrating decreased ramification in terms of decreased areas, perimeter, and total dendritic lengths of individual microglia in 20–24 month-old mice, relative to 3 month-old mice (n = 9 representative cells from 4–6 animals in each age group). * indicates comparisons for which p <0.05 (unpaired t-test with Welch’s correction).
4. Discussion
4.1 Age-related changes in gene expression relating to constitutive microglial support
In the current study, we carefully isolated microglial cells from the retina of different ages to examine age-related microglial gene expression across the entire span of adult aging. Gene ontology (GO) analysis showed that retinal microglia demonstrate age-related changes in the expression of genes regulating various aspects of mRNA and DNA metabolism, including the regulation of transcription and DNA binding (Table 1). Aging has been previously associated with altered rates of transcript and protein turnover in various cell types (Brewer 2002), including immune cells such as T cells (Cao et al. 2010). A similarly altered mRNA/DNA metabolism appears to be present in aging microglia. We found that genes coding for small GTPases regulatory proteins (Arhgap21, Arhgap22, Arhgef12, Cdc42se2, Farp2, Srgap3) which regulate cellular morphology and cell migration in multiple cell types (Etienne-Manneville & Hall 2002), including microglia (Yan et al. 2012), showed age-related changes. Genes coding for proteins that influence actin and microtubule organization that underlie dynamic process outgrowth and motility were differentially expressed with aging, including formin1 (Chesarone et al. 2010), formin binding protein 1-like (Lee et al. 2010), and Rab13 (Sakane et al. 2010). These changes may underlie age-related decreases in microglial ramification and dynamic behavior (Damani et al. 2011), which may in turn influence the regulation of synapses (Wake et al. 2009; Schafer et al. 2012a) and neuronal activity (Li et al. 2012), as well as neuroprotective functions (Vinet et al. 2012). Canonical pathway analyses highlighted changes in the neurotrophin/TRK signaling pathway (Fig. 2) which has been related to the supportive influence of microglia on neurons and macroglia (Harada et al. 2002). Neurturin, a neutrophic factor required for normal retinal neuronal structure and function (Brantley et al. 2008), was down-regulated with aging. Also, Egr1, a microglia-expressed transcription factor (Langmann et al. 2009) implicated in neuroprotective immune mechanisms (Bakalash et al. 2011; Sharma et al. 2012), was found to decrease monotonically with age. Taken together, altered expressions of genes involved in mRNA/DNA metabolism, cellular motility, and neurotrophin signaling suggest molecular mechanisms related to how microglial support functions decline during aging (Streit & Xue 2009).
4.2 Age-related changes in gene expression relating to altered microglial inflammatory responses
The “inflamm-aging hypothesis” (Franceschi et al. 2000) has proposed that aging changes in microglia drive dysregulated inflammatory responses to stimuli (i.e. priming) (Dilger & Johnson 2008), which result in an increased risk of neurodegeneration in aged animals (Mrak & Griffin 2005). In aged brains, increased microglial activation is evidenced by a greater basal expression of activation markers (Perry et al. 1993; Sheng et al. 1998), elevated basal cytokine profiles (Ye & Johnson 1999; Sierra et al. 2007; Njie et al. 2012), and more severe neurodegeneration following stimulation or injury (Sugama et al. 2003; Godbout et al. 2005; Sandhir et al. 2008; Wasserman et al. 2008). In the aged retina, microglia demonstrate dysfunctional responses to injury and injury signals (e.g. ATP) which are slower to initiate but also more prolonged and less reversible (Damani et al. 2011). These phenomena indicate that the regulation of the onset and resolution of microglial activation are affected by aging. Using gene ontogeny (GO) enrichment analysis, we detected among aging-related genes a significant representation by molecular pathways that regulate microglial activation, including those regulating I-κB kinase and NF-κB transcription factor activity, MAPK activity and ERK1/2 signaling (O’Neill & Kaltschmidt 1997; Koistinaho & Koistinaho 2002; Kim et al. 2004). Canonical pathway analyses highlighted the following inflammatory signaling pathways: (1) signaling involving IL-17, a cytokine demonstrated to increase inflammatory factor production in microglia in vitro (Kawanokuchi et al. 2008) and in animal stroke models in vivo (Lv et al. 2011), (2) ceramide signaling, (3) IL1 signaling, (4) nitric oxide signaling, and (5) lipopolysaccharide-stimulated MAPK signaling. Interestingly, network analysis on age-regulated microglial genes also identified the following genes as “hubs” for multiple direct interactions: (1) Jun and Fos AP-1 proteins, which have been found to be activated in microglia in pro-inflammatory, neurodegenerative contexts (Herdegen & Waetzig 2001), and (2) Egr-1, a regulator of inflammatory gene expression in microglia (Friedle et al. 2011) that is expressed in the retina in injury and disease (Brand et al. 2005; Langmann et al. 2009; Sharma et al. 2012). However, the relationship between these gene expression changes and the eventual activation state of microglia is likely complex and involve altered balances in multiple parallel and interacting regulatory pathways. For example, the expression of MyD88, a gene encoding a key adaptor protein in the activation of NF-κB via TLR receptors, was increased with advancing age. On the other hand, other genes associated with the transcription of pro-inflammatory genes, such as c-Jun and c-Fos of the AP-1 transcription factor complex, were decreased with advancing age. However, the combined consequences of these altered balances also do not appear to result in a strong polarization to a M1- or M2-like state as different individual M1 and M2 markers were found to both increase and decrease with advancing age without a clear polarization towards one state.
We found a significant representation of genes involved in lipid biosynthesis among aging-related genes (Table 1), including those implicated in immune cell regulation. Ch25H, the gene for the enzyme that catalyzes the formation of 25-hydroxycholesterol (25Ch) from cholesterol, was upregulated with aging. In macrophages, Ch25 was found to regulate survival (Joseph et al. 2004; Diczfalusy et al. 2009), cytokine expression (Joseph et al. 2003), and signaling to the adaptive immune system (Bauman et al. 2009). Pla2g5, which codes for a secreted phospholipase A2 that regulates the synthesis of inflammatory mediators (arachidonic acid, prostaglandins, leukotrienes, and platelet-activating factor) (Murakami et al. 1997), the mobilization of macrophages (Ruiperez et al. 2009)and leucocytes (Lapointe et al. 2010), and microglial inflammatory responses (Yang et al. 2009) was also increased in expression with aging. Genes involved in the production of neurosteroids which control the magnitude and duration of microglial activation (Sierra et al. 2008; Gottfried-Blackmore et al. 2010; Saijo et al. 2011) were also altered in expression. A down-regulated gene, Hsd17b1, codes for 11β-hydroxysteroid dehydrogenase type 1, which activates the glucocorticoid cortisone (Seckl & Walker 2001) and induces the production of steroid hormone 5-androsten-3β,17β-diol (ADIOL) (Moeller & Adamski 2009), a key factor in limiting microglial activation (Gottfried-Blackmore et al. 2010; Saijo et al. 2011). As a result, age-related changes in these lipid biosynthetic pathways may underlie altered activation responses in aged microglia.
Other studies have previously examined the effect of aging on the expression of inflammatory genes on the level of the entire retina. One study found that inflammatory chemokines (e.g. CCL2, CCL12) and cytokines (e.g. TNF-α) (Chen et al. 2010) were up-regulated with aging, while another failed to find similar changes (Chen et al. 2008a). While we found similar aging trends between microglial-specific and whole retina transcripts for some genes (e.g. Egr1, C3), we discovered opposite trends in other genes (e.g. TNF-α). CXCL13, a chemokine associated with B-cell signaling (Lalor & Segal 2010) and a biomarker of neuroinflammatory conditions (Alvarez et al. 2013), was markedly up-regulated with aging in microglia-specific transcripts, a trend not detected in previous whole-retina analyses. These disparities may arise from different molecular aging changes in microglia versus other retinal cell types, a difference that is likely to be apparent given the relative scarcity of microglia among all retinal cells. How these specific changes result in altered interactions between senescent microglia and other retinal cells will be a focus for our future functional studies.
4.3 Age-related changes in gene expression relating to disease susceptibility
We related the results of our current analyses to what is currently known about the physiology and genetic susceptibility to age-related retinal disease. Age-related macular degeneration (AMD), the largest cause of blindness in the Western hemisphere (Resnikoff et al. 2004), demonstrates a strong age-related prevalence (Friedman et al. 2004a) and a suggestive relationship to microglial senescence (Ma et al. 2013). In this study, we discovered several age-related microglial genes that have also been previously associated with AMD pathobiology. Canonical pathways involved in VEGF ligand-receptor signaling and TSP-1 signaling (Table 2, Fig. 4A, B), which have been implicated in general angiogenesis (de Fraipont et al. 2001; Chung & Ferrara 2011)and in ocular angiogenesis (Witmer et al. 2003; Hiscott et al. 2006), were highlighted, underscoring a connection between aging microglial changes and neovascular AMD disease. We also found that mRNA expression of VEGF and VEGFR2 were significantly increased during retinal microglial aging, possibly reflecting an increased ability of aged retinal microglia to promote angiogenesis (Fantin et al. 2010; Fischer et al. 2011), contributing in some part to the increased risk of neovascular AMD with age.
We also examined how the results of the current analysis relate to known genetic susceptibilities to AMD, in particular to genes in the complement pathway (Swaroop et al. 2009). Given that elevated levels of complement and complement activation products colocalize with AMD lesions (Mullins et al. 2000; Hageman et al. 2005; Lommatzsch et al. 2008), risk polymorphisms in complement genes likely result in increased complement activation, driving aspects of disease progression (Scholl et al. 2008; Montes et al. 2009; Hecker et al. 2010). However, how complement and complement activated becomes dysregulated in the aged retina is unclear. We and others have previously demonstrated that retina microglia are able to synthesize complement and complement regulatory proteins, and may indeed regulate complement activation locally (Collier et al. 2011; Luo et al. 2011; Rutar et al. 2011; Ma et al. 2013). We had also shown that accumulation of an age-related ocular lipofuscin constituent A2E alters complement protein expression to favor increased complement activation (Ma et al. 2013). In the current analysis, we discovered that two central complement genes, C3 and CFB, exhibited a monotonic-to-age increase in retinal microglia. We confirmed these findings in immunohistochemical studies where increased labeling of both C3 and CFB were observed in the outer retina of aged mice that colocalized with subretinal microglia. The deposition of activated C3 breakdown product, iC3b, was increased in the subretinal space in the areas of microglial accumulation.
Previous microarray studies examining age-related changes in ocular tissues such as in the whole retina (Chen et al. 2008b) and in the RPE-choroid complex (Chen et al. 2008a), have also described in increases in the expression of C3 and CFB, as well as other complement proteins. However the cellular source for these increases was undemonstrated. Our data here indicate that aging retinal microglia indeed constitute a cellular source of increased complement proteins, and can drive increased complement activation in the outer retina in ways that contribute causally to AMD progression. Corroboratively, early stage up-regulation of complement has also been identified in the retinas of a mouse model of glaucoma, another age-related retinal disease (Howell et al. 2011). Given the presence of early microglial activation in this mouse model (Bosco et al. 2011), and that interventions that decrease microglial activation can alleviate glaucomatous change (Bosco et al. 2008; Bosco et al. 2012), it will be interesting in future studies to confirm whether microglia indeed constitute the cellular locus for complement gene dysregulation in glaucoma. If true, complement dysregulation in aging retinal microglia may constitute a common cellular mechanism driving multiple age-related neurodegenerative diseases in the retina.
In conclusion, the findings from the current analysis indicate a number of molecular mechanisms that underlie functional and pathogenic changes in microglial aging including changes in constitutive supportive microglial functions and altered regulation of microglial activation. Compellingly, the results also highlight complement genes that have previously been identified as conferring risk for the age-related retinal diseases of AMD and glaucoma. Future work will focus on validating these potential molecular mechanisms in regulating features of the senescent phenotype in microglia, with the goal of understanding and therapeutically addressing the various forms of age-related neurodegeneration in the retina.
Supplementary Material
Primers used in real time reverse-transcriptase polymerase chain reactions (rt-PCR).
List of 719 genes showing differential expression with aging at at least one of the following consecutive age-groups comparisons: 3- to 12- months, 12- to 18- months, and 18- to 24- months.
List of differentially regulated genes demonstrating a monotonic-to-age increase in expression
List of differentially regulated genes demonstrating a monotonic-to-age decrease in expression
(A) Retina of wild type mice of different ages (3 month old and 24 month old shown here) were subject to a dissociation protocol and the resulting cells were immunolabeled using a FITC-conjugated CD11b antibody. Live cells were sorted according to fluorescence in the FITC channel and isolated a separate population of CD11bhi cells. (B) Sorted CD11bhi cells are likely to be also CX3CR1+ microglia as immunostaining of CD11b in CX3CR1+/GFP mouse retina showed a co-localization of GFP expression and CD11b immunopositivity in all microglia. (C) Retinal tissue from a CX3CR1+/GFP mouse was subjected to the same dissociation protocol as used in (A) and resulting cells immunolabeled with a PE-Cy5-conjugated CD11b antibody. Live cells were sorted according to fluorescence in the PE-Cy5 channel and the GFP channel. A single population of CD11bhiGFP+ cells was identified (red box) indicating that CD11bhicells comprise of GFP+ microglia.
Gene expression fold change from qRT-PCR analysis of retinal microglia from 12-, 18-, and 24-month old mice (n = 3 for each experiment) were assessed relative the housekeeping gene β-2 microglobulin (B2M). Results were normalized to gene expression levels in 3-month old mice. These RT-PCR results (A) demonstrate a similar stability in these genes across aging as found in parallel microarray analyses (B). Error bars indicate ± SEM.
Network analysis of 719 genes from retinal microglia showing age-related gene expression. Direction interactions between genes analyzed using MetaCore and depicted by connecting arrows. The most prominent “hubs” in this network, indicating genes that interact most extensively with other genes within this set of age-related genes, were AP-1 and Egr-1.
Relative gene expression fold change from qRT-PCR (top) and (bottom) microarray analyses from retinal microglia from 12-, 18-, and 24-month old mice, normalized relative to those from 3-month old mice were performed for (A) C3 (n = 6 biological repeats, each with one sample replicate) and for (B) CFB (n = 3 biological repeats, each with 3 sample replicates).
Acknowledgments
This study was supported by the NEI Intramural by the National Eye Institute Intramural Research Program and a grant from the American Health Assistance Foundation (AHAF). Acknowledgement is made to the donors of ADR, a program of the American Health Assistance Foundation, for support of this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The data in this manuscript is published or submitted elsewhere, and has been approved by all contributing authors. Procedures involving animals here have been previously approved by institutional review boards.
Footnotes
Disclosure statement
All authors indicate that they do not have conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Wenxin Ma, Email: mawenxin@nei.nih.gov.
Radu Cojocaru, Email: cojocarur@nei.nih.gov.
Norimoto Gotoh, Email: eyegotoh@kuhp.kyoto-u.ac.jp.
Linn Gieser, Email: gieserl@nei.nih.gov.
Rafael Villasmil, Email: villasmilr@nei.nih.gov.
Tiziana Cogliati, Email: cogliatitp@nei.nih.gov.
Anand Swaroop, Email: swaroopa@nei.nih.gov.
Wai T. Wong, Email: wongw@nei.nih.gov.
References
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Alvarez E, Piccio L, Mikesell RJ, Klawiter EC, Parks BJ, Naismith RT, Cross AH. CXCL13 is a biomarker of inflammation in multiple sclerosis, neuromyelitis optica, and other neurological conditions. Multiple sclerosis. 2013 doi: 10.1177/1352458512473362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalash S, Pham M, Koronyo Y, Salumbides BC, Kramerov A, Seidenberg H, Berel D, Black KL, Koronyo-Hamaoui M. Egr1 expression is induced following glatiramer acetate immunotherapy in rodent models of glaucoma and Alzheimer’s disease. Invest Ophthalmol Vis Sci. 2011;52:9033–9046. doi: 10.1167/iovs.11-7498. [DOI] [PubMed] [Google Scholar]
- Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci U S A. 2009;106:16764–16769. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco A, Crish SD, Steele MR, Romero CO, Inman DM, Horner PJ, Calkins DJ, Vetter ML. Early reduction of microglia activation by irradiation in a model of chronic glaucoma. PLoS One. 2012;7:e43602. doi: 10.1371/journal.pone.0043602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco A, Inman DM, Steele MR, Wu G, Soto I, Marsh-Armstrong N, Hubbard WC, Calkins DJ, Horner PJ, Vetter ML. Reduced retina microglial activation and improved optic nerve integrity with minocycline treatment in the DBA/2J mouse model of glaucoma. Invest Ophthalmol Vis Sci. 2008;49:1437–1446. doi: 10.1167/iovs.07-1337. [DOI] [PubMed] [Google Scholar]
- Bosco A, Steele MR, Vetter ML. Early microglia activation in a mouse model of chronic glaucoma. J Comp Neurol. 2011;519:599–620. doi: 10.1002/cne.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand C, Burkhardt E, Schaeffel F, Choi JW, Feldkaemper MP. Regulation of Egr-1, VIP, and Shh mRNA and Egr-1 protein in the mouse retina by light and image quality. Mol Vis. 2005;11:309–320. [PubMed] [Google Scholar]
- Brantley MA, Jr, Jain S, Barr EE, Johnson EM, Jr, Milbrandt J. Neurturin-mediated ret activation is required for retinal function. J Neurosci. 2008;28:4123–4135. doi: 10.1523/JNEUROSCI.0249-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G. Messenger RNA decay during aging and development. Ageing Res Rev. 2002;1:607–625. doi: 10.1016/s1568-1637(02)00023-5. [DOI] [PubMed] [Google Scholar]
- Buschini E, Piras A, Nuzzi R, Vercelli A. Age related macular degeneration and drusen: neuroinflammation in the retina. Prog Neurobiol. 2011;95:14–25. doi: 10.1016/j.pneurobio.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Cao JN, Gollapudi S, Sharman EH, Jia Z, Gupta S. Age-related alterations of gene expression patterns in human CD8+ T cells. Aging Cell. 2010;9:19–31. doi: 10.1111/j.1474-9726.2009.00534.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu B, Lukas TJ, Neufeld AH. The aged retinal pigment epithelium/choroid: a potential substratum for the pathogenesis of age-related macular degeneration. PLoS One. 2008a;3:e2339. doi: 10.1371/journal.pone.0002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Muckersie E, Forrester JV, Xu H. Immune activation in retinal aging: a gene expression study. Invest Ophthalmol Vis Sci. 2010;51:5888–5896. doi: 10.1167/iovs.09-5103. [DOI] [PubMed] [Google Scholar]
- Chen M, Muckersie E, Robertson M, Forrester JV, Xu H. Up-regulation of complement factor B in retinal pigment epithelial cells is accompanied by complement activation in the aged retina. Exp Eye Res. 2008b;87:543–550. doi: 10.1016/j.exer.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chung AS, Ferrara N. Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol. 2011;27:563–584. doi: 10.1146/annurev-cellbio-092910-154002. [DOI] [PubMed] [Google Scholar]
- Collier RJ, Wang Y, Smith SS, Martin E, Ornberg R, Rhoades K, Romano C. Complement deposition and microglial activation in the outer retina in light-induced retinopathy: inhibition by a 5-HT1A agonist. Invest Ophthalmol Vis Sci. 2011;52:8108–8116. doi: 10.1167/iovs.10-6418. [DOI] [PubMed] [Google Scholar]
- Combadiere C, Feumi C, Raoul W, Keller N, Rodero M, Pezard A, Lavalette S, Houssier M, Jonet L, Picard E, Debre P, Sirinyan M, Deterre P, Ferroukhi T, Cohen SY, Chauvaud D, Jeanny JC, Chemtob S, Behar-Cohen F, Sennlaub F. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117:2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon N, O’Colmain B, Klaver CC, Klein R, Munoz B, Friedman DS, Kempen J, Taylor HR, Mitchell P. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122:477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- Congdon NG, Friedman DS, Lietman T. Important causes of visual impairment in the world today. JAMA. 2003;290:2057–2060. doi: 10.1001/jama.290.15.2057. [DOI] [PubMed] [Google Scholar]
- Damani MR, Zhao L, Fontainhas AM, Amaral J, Fariss RN, Wong WT. Age-related alterations in the dynamic behavior of microglia. Aging Cell. 2011;10:263–276. doi: 10.1111/j.1474-9726.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- de Fraipont F, Nicholson AC, Feige JJ, Van Meir EG. Thrombospondins and tumor angiogenesis. Trends Mol Med. 2001;7:401–407. doi: 10.1016/s1471-4914(01)02102-5. [DOI] [PubMed] [Google Scholar]
- de Haas AH, Boddeke HW, Biber K. Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia. 2008;56:888–894. doi: 10.1002/glia.20663. [DOI] [PubMed] [Google Scholar]
- Diczfalusy U, Olofsson KE, Carlsson AM, Gong M, Golenbock DT, Rooyackers O, Flaring U, Bjorkbacka H. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J Lipid Res. 2009;50:2258–2264. doi: 10.1194/jlr.M900107-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilger RN, Johnson RW. Aging, microglial cell priming, and the discordant central inflammatory response to signals from the peripheral immune system. J Leukoc Biol. 2008;84:932–939. doi: 10.1189/jlb.0208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW, Ruhrberg C. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116:829–840. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F, Martin G, Agostini HT. Activation of retinal microglia rather than microglial cell density correlates with retinal neovascularization in the mouse model of oxygen-induced retinopathy. J Neuroinflammation. 2011;8:120. doi: 10.1186/1742-2094-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontainhas AM, Wang M, Liang KJ, Chen S, Mettu P, Damani M, Fariss RN, Li W, Wong WT. Microglial morphology and dynamic behavior is regulated by ionotropic glutamatergic and GABAergic neurotransmission. PLoS One. 2011;6:e15973. doi: 10.1371/journal.pone.0015973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Friedle SA, Brautigam VM, Nikodemova M, Wright ML, Watters JJ. The P2X7-Egr pathway regulates nucleotide-dependent inflammatory gene expression in microglia. Glia. 2011;59:1–13. doi: 10.1002/glia.21071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, de Jong PT, Nemesure B, Mitchell P, Kempen J. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004a;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- Friedman DS, Wolfs RC, O’Colmain BJ, Klein BE, Taylor HR, West S, Leske MC, Mitchell P, Congdon N, Kempen J. Prevalence of open-angle glaucoma among adults in the United States. Arch Ophthalmol. 2004b;122:532–538. doi: 10.1001/archopht.122.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout JP, Chen J, Abraham J, Richwine AF, Berg BM, Kelley KW, Johnson RW. Exaggerated neuroinflammation and sickness behavior in aged mice following activation of the peripheral innate immune system. FASEB J. 2005;19:1329–1331. doi: 10.1096/fj.05-3776fje. [DOI] [PubMed] [Google Scholar]
- Gottfried-Blackmore A, Sierra A, McEwen BS, Ge R, Bulloch K. Microglia express functional 11 beta-hydroxysteroid dehydrogenase type 1. Glia. 2010;58:1257–1266. doi: 10.1002/glia.21007. [DOI] [PubMed] [Google Scholar]
- Gupta N, Brown KE, Milam AH. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp Eye Res. 2003;76:463–471. doi: 10.1016/s0014-4835(02)00332-9. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Harada C, Kohsaka S, Wada E, Yoshida K, Ohno S, Mamada H, Tanaka K, Parada LF, Wada K. Microglia-Muller glia cell interactions control neurotrophic factor production during light-induced retinal degeneration. J Neurosci. 2002;22:9228–9236. doi: 10.1523/JNEUROSCI.22-21-09228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker LA, Edwards AO, Ryu E, Tosakulwong N, Baratz KH, Brown WL, Charbel Issa P, Scholl HP, Pollok-Kopp B, Schmid-Kubista KE, Bailey KR, Oppermann M. Genetic control of the alternative pathway of complement in humans and age-related macular degeneration. Hum Mol Genet. 2010;19:209–215. doi: 10.1093/hmg/ddp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdegen T, Waetzig V. AP-1 proteins in the adult brain: facts and fiction about effectors of neuroprotection and neurodegeneration. Oncogene. 2001;20:2424–2437. doi: 10.1038/sj.onc.1204387. [DOI] [PubMed] [Google Scholar]
- Hiscott P, Paraoan L, Choudhary A, Ordonez JL, Al-Khaier A, Armstrong DJ. Thrombospondin 1, thrombospondin 2 and the eye. Prog Retin Eye Res. 2006;25:1–18. doi: 10.1016/j.preteyeres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Howell GR, Macalinao DG, Sousa GL, Walden M, Soto I, Kneeland SC, Barbay JM, King BL, Marchant JK, Hibbs M, Stevens B, Barres BA, Clark AF, Libby RT, John SW. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest. 2011;121:1429–1444. doi: 10.1172/JCI44646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O’Connell RM, Cheng G, Saez E, Miller JF, Tontonoz P. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanokuchi J, Shimizu K, Nitta A, Yamada K, Mizuno T, Takeuchi H, Suzumura A. Production and functions of IL-17 in microglia. J Neuroimmunol. 2008;194:54–61. doi: 10.1016/j.jneuroim.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Kim SH, Smith CJ, Van Eldik LJ. Importance of MAPK pathways for microglial pro-inflammatory cytokine IL-1 beta production. Neurobiol Aging. 2004;25:431–439. doi: 10.1016/S0197-4580(03)00126-X. [DOI] [PubMed] [Google Scholar]
- Koistinaho M, Koistinaho J. Role of p38 and p44/42 mitogen-activated protein kinases in microglia. Glia. 2002;40:175–183. doi: 10.1002/glia.10151. [DOI] [PubMed] [Google Scholar]
- Krady JK, Basu A, Allen CM, Xu Y, LaNoue KF, Gardner TW, Levison SW. Minocycline reduces proinflammatory cytokine expression, microglial activation, and caspase-3 activation in a rodent model of diabetic retinopathy. Diabetes. 2005;54:1559–1565. doi: 10.2337/diabetes.54.5.1559. [DOI] [PubMed] [Google Scholar]
- Lalor SJ, Segal BM. Lymphoid chemokines in the CNS. J Neuroimmunol. 2010;224:56–61. doi: 10.1016/j.jneuroim.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmann T, Ebert S, Walczak Y, Weigelt K, Ehrengruber MU, Stiewe T, Weber BH. Induction of early growth response-1 mediates microglia activation in vitro but is dispensable in vivo. Neuromolecular Med. 2009;11:87–96. doi: 10.1007/s12017-009-8061-6. [DOI] [PubMed] [Google Scholar]
- Lapointe S, Brkovic A, Cloutier I, Tanguay JF, Arm JP, Sirois MG. Group V secreted phospholipase A2 contributes to LPS-induced leukocyte recruitment. J Cell Physiol. 2010;224:127–134. doi: 10.1002/jcp.22106. [DOI] [PubMed] [Google Scholar]
- Lee JE, Liang KJ, Fariss RN, Wong WT. Ex vivo dynamic imaging of retinal microglia using time-lapse confocal microscopy. Invest Ophthalmol Vis Sci. 2008;49:4169–4176. doi: 10.1167/iovs.08-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. 2010;329:1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Du XF, Liu CS, Wen ZL, Du JL. Reciprocal Regulation between Resting Microglial Dynamics and Neuronal Activity In Vivo. Dev Cell. 2012;23:1189–1202. doi: 10.1016/j.devcel.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Lommatzsch A, Hermans P, Muller KD, Bornfeld N, Bird AC, Pauleikhoff D. Are low inflammatory reactions involved in exudative age-related macular degeneration? Morphological and immunhistochemical analysis of AMD associated with basal deposits. Graefes Arch Clin Exp Ophthalmol. 2008;246:803–810. doi: 10.1007/s00417-007-0749-4. [DOI] [PubMed] [Google Scholar]
- Luo C, Chen M, Xu H. Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Mol Vis. 2011;17:1588–1597. [PMC free article] [PubMed] [Google Scholar]
- Lv M, Liu Y, Zhang J, Sun L, Liu Z, Zhang S, Wang B, Su D, Su Z. Roles of inflammation response in microglia cell through Toll-like receptors 2/interleukin-23/interleukin-17 pathway in cerebral ischemia/reperfusion injury. Neuroscience. 2011;176:162–172. doi: 10.1016/j.neuroscience.2010.11.066. [DOI] [PubMed] [Google Scholar]
- Ma W, Coon S, Zhao L, Fariss RN, Wong WT. A2E accumulation influences retinal microglial activation and complement regulation. Neurobiol Aging. 2013;34:943–960. doi: 10.1016/j.neurobiolaging.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Bruck W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- Moeller G, Adamski J. Integrated view on 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2009;301:7–19. doi: 10.1016/j.mce.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Montes T, Tortajada A, Morgan BP, Rodriguez de Cordoba S, Harris CL. Functional basis of protection against age-related macular degeneration conferred by a common polymorphism in complement factor B. Proc Natl Acad Sci U S A. 2009;106:4366–4371. doi: 10.1073/pnas.0812584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak RE, Griffin WS. Glia and their cytokines in progression of neurodegeneration. Neurobiol Aging. 2005;26:349–354. doi: 10.1016/j.neurobiolaging.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- Murakami M, Nakatani Y, Atsumi G, Inoue K, Kudo I. Regulatory functions of phospholipase A2. Crit Rev Immunol. 1997;17:225–283. doi: 10.1615/critrevimmunol.v17.i3-4.10. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Njie EG, Boelen E, Stassen FR, Steinbusch HW, Borchelt DR, Streit WJ. Ex vivo cultures of microglia from young and aged rodent brain reveal age-related changes in microglial function. Neurobiol Aging. 2012;33:195 e191–112. doi: 10.1016/j.neurobiolaging.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Kaltschmidt C. NF-kappa B: a crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20:252–258. doi: 10.1016/s0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Perry VH, Matyszak MK, Fearn S. Altered antigen expression of microglia in the aged rodent CNS. Glia. 1993;7:60–67. doi: 10.1002/glia.440070111. [DOI] [PubMed] [Google Scholar]
- Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- Ruiperez V, Astudillo AM, Balboa MA, Balsinde J. Coordinate regulation of TLR-mediated arachidonic acid mobilization in macrophages by group IVA and group V phospholipase A2s. J Immunol. 2009;182:3877–3883. doi: 10.4049/jimmunol.0804003. [DOI] [PubMed] [Google Scholar]
- Rutar M, Natoli R, Kozulin P, Valter K, Gatenby P, Provis JM. Analysis of complement expression in light-induced retinal degeneration: synthesis and deposition of C3 by microglia/macrophages is associated with focal photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2011;52:5347–5358. doi: 10.1167/iovs.10-7119. [DOI] [PubMed] [Google Scholar]
- Saijo K, Collier JG, Li AC, Katzenellenbogen JA, Glass CK. An ADIOL-ERbeta-CtBP transrepression pathway negatively regulates microglia-mediated inflammation. Cell. 2011;145:584–595. doi: 10.1016/j.cell.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane A, Honda K, Sasaki T. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol. 2010;30:1077–1087. doi: 10.1128/MCB.01067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhir R, Onyszchuk G, Berman NE. Exacerbated glial response in the aged mouse hippocampus following controlled cortical impact injury. Exp Neurol. 2008;213:372–380. doi: 10.1016/j.expneurol.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012a;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Stevens B. The “quad-partite” synapse: Microglia-synapse interactions in the developing and mature CNS. Glia. 2012b doi: 10.1002/glia.22389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl HP, Charbel Issa P, Walier M, Janzer S, Pollok-Kopp B, Borncke F, Fritsche LG, Chong NV, Fimmers R, Wienker T, Holz FG, Weber BH, Oppermann M. Systemic complement activation in age-related macular degeneration. PLoS One. 2008;3:e2593. doi: 10.1371/journal.pone.0002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckl JR, Walker BR. Minireview: 11beta-hydroxysteroid dehydrogenase type 1- a tissue-specific amplifier of glucocorticoid action. Endocrinology. 2001;142:1371–1376. doi: 10.1210/endo.142.4.8114. [DOI] [PubMed] [Google Scholar]
- Sharma YV, Cojocaru RI, Ritter LM, Khattree N, Brooks M, Scott A, Swaroop A, Goldberg AF. Protective gene expression changes elicited by an inherited defect in photoreceptor structure. PLoS One. 2012;7:e31371. doi: 10.1371/journal.pone.0031371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan B, Quigley A, Gaudin B, Dobson S. A relation based measure of semantic similarity for Gene Ontology annotations. BMC Bioinformatics. 2008;9:468. doi: 10.1186/1471-2105-9-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng JG, Mrak RE, Griffin WS. Enlarged and phagocytic, but not primed, interleukin-1 alpha-immunoreactive microglia increase with age in normal human brain. Acta Neuropathol. 1998;95:229–234. doi: 10.1007/s004010050792. [DOI] [PubMed] [Google Scholar]
- Sierra A, Gottfried-Blackmore A, Milner TA, McEwen BS, Bulloch K. Steroid hormone receptor expression and function in microglia. Glia. 2008;56:659–674. doi: 10.1002/glia.20644. [DOI] [PubMed] [Google Scholar]
- Sierra A, Gottfried-Blackmore AC, McEwen BS, Bulloch K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia. 2007;55:412–424. doi: 10.1002/glia.20468. [DOI] [PubMed] [Google Scholar]
- Streit WJ, Xue QS. Life and death of microglia. J Neuroimmune Pharmacol. 2009;4:371–379. doi: 10.1007/s11481-009-9163-5. [DOI] [PubMed] [Google Scholar]
- Sugama S, Yang L, Cho BP, DeGiorgio LA, Lorenzl S, Albers DS, Beal MF, Volpe BT, Joh TH. Age-related microglial activation in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced dopaminergic neurodegeneration in C57BL/6 mice. Brain Res. 2003;964:288–294. doi: 10.1016/s0006-8993(02)04085-4. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Kern TS. Inflammation in diabetic retinopathy. Prog Retin Eye Res. 2011;30:343–358. doi: 10.1016/j.preteyeres.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay ME, Zettel ML, Ison JR, Allen PD, Majewska AK. Effects of aging and sensory loss on glial cells in mouse visual and auditory cortices. Glia. 2012;60:541–558. doi: 10.1002/glia.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kirk CA, VanGuilder HD, Young M, Farley JA, Sonntag WE, Freeman WM. Age-related alterations in retinal neurovascular and inflammatory transcripts. Mol Vis. 2011;17:1261–1274. [PMC free article] [PubMed] [Google Scholar]
- Vinet J, van Weering HR, Heinrich A, Kalin RE, Wegner A, Brouwer N, Heppner FL, van Rooijen N, Boddeke HW, Biber K. Neuroprotective function for ramified microglia in hippocampal excitotoxicity. J Neuroinflammation. 2012;9:27. doi: 10.1186/1742-2094-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bernhardi R, Tichauer JE, Eugenin J. Aging-dependent changes of microglial cells and their relevance for neurodegenerative disorders. J Neurochem. 2010;112:1099–1114. doi: 10.1111/j.1471-4159.2009.06537.x. [DOI] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman JK, Yang H, Schlichter LC. Glial responses, neuron death and lesion resolution after intracerebral hemorrhage in young vs. aged rats. Eur J Neurosci. 2008;28:1316–1328. doi: 10.1111/j.1460-9568.2008.06442.x. [DOI] [PubMed] [Google Scholar]
- Wax MB, Tezel G. Immunoregulation of retinal ganglion cell fate in glaucoma. Exp Eye Res. 2009;88:825–830. doi: 10.1016/j.exer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Witmer AN, Vrensen GF, Van Noorden CJ, Schlingemann RO. Vascular endothelial growth factors and angiogenesis in eye disease. Prog Retin Eye Res. 2003;22:1–29. doi: 10.1016/s1350-9462(02)00043-5. [DOI] [PubMed] [Google Scholar]
- Xie Z, Morgan TE, Rozovsky I, Finch CE. Aging and glial responses to lipopolysaccharide in vitro: greater induction of IL-1 and IL-6, but smaller induction of neurotoxicity. Exp Neurol. 2003;182:135–141. doi: 10.1016/s0014-4886(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Xu H, Chen M, Forrester JV. Para-inflammation in the aging retina. Prog Retin Eye Res. 2009;28:348–368. doi: 10.1016/j.preteyeres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhou X, Guo JJ, Mao L, Wang YJ, Sun J, Sun LX, Zhang LY, Zhou XF, Liao H. Nogo-66 inhibits adhesion and migration of microglia via GTPase Rho pathway in vitro. J Neurochem. 2012;120:721–731. doi: 10.1111/j.1471-4159.2011.07619.x. [DOI] [PubMed] [Google Scholar]
- Yang CS, Yuk JM, Shin DM, Kang J, Lee SJ, Jo EK. Secretory phospholipase A2 plays an essential role in microglial inflammatory responses to Mycobacterium tuberculosis. Glia. 2009;57:1091–1103. doi: 10.1002/glia.20832. [DOI] [PubMed] [Google Scholar]
- Ye SM, Johnson RW. Increased interleukin-6 expression by microglia from brain of aged mice. J Neuroimmunol. 1999;93:139–148. doi: 10.1016/s0165-5728(98)00217-3. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Yashar BM, Hiriyanna S, Swaroop A. Microarray analysis of gene expression in the aging human retina. Invest Ophthalmol Vis Sci. 2002;43:2554–2560. [PubMed] [Google Scholar]
- Yuan L, Neufeld AH. Activated microglia in the human glaucomatous optic nerve head. J Neurosci Res. 2001;64:523–532. doi: 10.1002/jnr.1104. [DOI] [PubMed] [Google Scholar]
- Zeng HY, Green WR, Tso MO. Microglial activation in human diabetic retinopathy. Arch Ophthalmol. 2008;126:227–232. doi: 10.1001/archophthalmol.2007.65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in real time reverse-transcriptase polymerase chain reactions (rt-PCR).
List of 719 genes showing differential expression with aging at at least one of the following consecutive age-groups comparisons: 3- to 12- months, 12- to 18- months, and 18- to 24- months.
List of differentially regulated genes demonstrating a monotonic-to-age increase in expression
List of differentially regulated genes demonstrating a monotonic-to-age decrease in expression
(A) Retina of wild type mice of different ages (3 month old and 24 month old shown here) were subject to a dissociation protocol and the resulting cells were immunolabeled using a FITC-conjugated CD11b antibody. Live cells were sorted according to fluorescence in the FITC channel and isolated a separate population of CD11bhi cells. (B) Sorted CD11bhi cells are likely to be also CX3CR1+ microglia as immunostaining of CD11b in CX3CR1+/GFP mouse retina showed a co-localization of GFP expression and CD11b immunopositivity in all microglia. (C) Retinal tissue from a CX3CR1+/GFP mouse was subjected to the same dissociation protocol as used in (A) and resulting cells immunolabeled with a PE-Cy5-conjugated CD11b antibody. Live cells were sorted according to fluorescence in the PE-Cy5 channel and the GFP channel. A single population of CD11bhiGFP+ cells was identified (red box) indicating that CD11bhicells comprise of GFP+ microglia.
Gene expression fold change from qRT-PCR analysis of retinal microglia from 12-, 18-, and 24-month old mice (n = 3 for each experiment) were assessed relative the housekeeping gene β-2 microglobulin (B2M). Results were normalized to gene expression levels in 3-month old mice. These RT-PCR results (A) demonstrate a similar stability in these genes across aging as found in parallel microarray analyses (B). Error bars indicate ± SEM.
Network analysis of 719 genes from retinal microglia showing age-related gene expression. Direction interactions between genes analyzed using MetaCore and depicted by connecting arrows. The most prominent “hubs” in this network, indicating genes that interact most extensively with other genes within this set of age-related genes, were AP-1 and Egr-1.
Relative gene expression fold change from qRT-PCR (top) and (bottom) microarray analyses from retinal microglia from 12-, 18-, and 24-month old mice, normalized relative to those from 3-month old mice were performed for (A) C3 (n = 6 biological repeats, each with one sample replicate) and for (B) CFB (n = 3 biological repeats, each with 3 sample replicates).