Abstract
Background
Clinical and genetic studies suggest circadian clock genes may contribute to biological mechanisms underlying alcohol use disorders (AUD). In particular, the Per2 gene regulates alcohol consumption in mutant animals, and in humans with AUD, the 10870 variant in PER2 has been associated with alcohol consumption. However, with respect to function, the molecular clock remains largely uncharacterized in AUD patients.
Methods
In skin fibroblast cultures from well-characterized human AUD patients (n=19) and controls (n=13), we used a bioluminescent reporter gene (Per2∷luc) to measure circadian rhythms in gene expression at high sampling density for five days. Cells were genotyped for the PER2 10870 variant. The rhythm parameters period and amplitude were then analyzed using a case-control design, and by genetic and clinical characteristics of the AUD subjects.
Results
There were no differences between AUD cases and controls in rhythm parameters. However, period was inversely correlated with illness severity (defined as the number of alcohol dependence criteria met). The PER2 variant 10870 was not associated with differences in rhythm parameters.
Conclusions
Our data suggest that differences in the cellular circadian clock are not pronounced in fibroblasts from AUD cases and controls. However, we found evidence that the circadian clock may be associated with an altered trajectory of AUD, possibly related to illness severity. Future work will be required to determine the mechanistic basis of this association.
Keywords: Circadian Rhythm, Genetics, Alcohol
INTRODUCTION
Socially acceptable alcohol use in humans is restricted to certain times of day, usually the evening hours. However, as alcohol use disorders (AUD) develop, the pattern of drinking widens across the day. Indeed, early morning alcohol drinking (the “eye opener”) is considered a pathological sign of an AUD among clinicians (Ewing, 1984). Although conventions on alcohol drinking may be in part socially constructed, they may also have a basis in biological timing mechanisms. In alcohol dependence, 40–60% of the etiological variance is attributable to genetic factors, likely involving multiple biological systems (Mayfield et al, 2008), possibly including the circadian clock.
Several clinical observations suggest a contributory role for disrupted biological timing and circadian rhythms in AUD. Among subjects at high risk for AUD on the basis of family history, there are disturbances in 24 hr endocrine rhythms (Gianoulakis et al., 2005), and sleep disturbances or daytime fatigue predict subsequent alcohol-related problems (Wong et al., 2010). Subjects with nocturnal chronotype (a preference for evening activity) consume more alcohol than those with morning chronotypes, possibly because they find activity early in the day more stressful, and/or they are better able to synchronize with social cues that support nocturnal drinking behaviors (Adan, 1994; Wittman et al., 2006; 2010). Each of the preceding examples relate to young subjects who did not yet drink heavily, suggesting that genetic abnormalities in the circadian clock may predispose individuals to AUD, independently of alcohol effects on the clock and/or rhythmic behavior. However, among subjects who are already ill and recovering from AUD, sleep problems are common and predict alcohol relapse, even after sustained abstinence, and these too could be related to circadian clock problems (Drummond et al, 1998). These correlates of clock function are suggestive, but can be confounded by non-circadian factors, especially the pharmacological effects of chronic alcohol. Because of this, controlled, mechanistic studies are essential to test the role of the circadian clock in AUD.
In mammals, the circadian clock is a molecular network of ~20 “clock genes” that regulate rhythmic behaviors such as sleep and activity through transcriptional, translational and post-translational mechanisms (Takahashi et al., 2008). As a consequence of the network’s organization, many clock genes (e.g. PER2) are expressed rhythmically. Clock gene associations have been reported for several alcohol related phenotypes. Single nucleotide polymorphisms (SNPs) in the clock genes ARNTL and ARNTL2 are associated with alcohol dependence and alcohol intake in social drinkers, respectively (Kovanen, 2010). The PER2 SNP rs56013859 (commonly called PER2 10870) is associated with high alcohol intake (Spanagel et al., 2005), and an interaction between sleep problems and alcohol intake in adolescent males (Comasco et al., 2010). A PER1 variant is associated with alcohol consumption among adolescents exposed to high stress, and with AUD in adults (Dong et al., 2011). In animal studies, rats’ alcohol consumption follows a circadian rhythm, with intake higher in the dark phase (Garcia-Burgos et al., 2009). Mice selectively bred for high alcohol intake have shorter free-running circadian periods (Hofstetter et al., 2003), whereas rats bred for high alcohol preference have short periods and abnormalities in entrainment to shortening light/dark cycles (Rosenwasser et al., 2005). In rats trained to drink alcohol, pharmacological inhibition of casein kinase Iε/δ, key regulators of the clock, limits the increase in consumption typically observed after a period of abstinence (Perreau-Lenz et al., 2012). In mice, evening alcohol is more intoxicating than morning alcohol, but disruption of the clock gene Per2 eliminates this differential sensitivity, leading to an increase in overall alcohol intake across the 24 hr cycle (Perreau-Lenz et al., 2009). Per1 mutants consume more alcohol than controls following a social stressor (Dong et al., 2011), and Per3 regulatory variants have been associated with alcohol preference and related phenotypes in mice (Wang et al., 2012). Although these studies have largely focused on the sleep/wake aspects of circadian rhythms, clock genes have pleiotropic effects and regulate many additional processes that are not directly related to activity cycles and/or sleep. Clock genes have been implicated in modulating reward through dopamine signaling (McClung et al., 2005), the endorphin response to alcohol (Agapito et al., 2010), and glutamate re-uptake (Beaulé et al., 2009; Spanagel, 2005), suggesting that clock genes could intervene at a number of points to modulate alcohol intake.
Despite clinical and genetic evidence linking the clock to AUD, there is presently no unequivocal demonstration of functional molecular clock abnormalities in human alcoholics. In a study of 22 subjects undergoing alcohol detoxification, reduced expression of six clock genes was reported in mononuclear blood cells (Huang et al, 2010). However, these measurements were made at insufficient sampling density to assess rhythm patterns. Moreover, as the study used cells from recently drinking individuals, it could not disentangle trait vulnerability factors specific to the clock from generalized effects related to heavy alcohol use. Other attempts at measuring rhythmic gene expression (PER1) in AUD patients were made over a single 24 hr cycle (Dong et al., 2011), limiting the ability to assess rhythm parameters such as period and amplitude. Improving the ability to make these assessments, rhythmic bioluminescent reporter genes have been used to measure gene expression rhythms longitudinally in cells from mice (Welsh et al., 2005) and human donors (Brown et al., 2005; 2008; Pagnani, 2010). In these studies, clock gene expression period correlated with the chronotype of the donor. This suggests that at least some of the genetic mechanisms controlling behavioral rhythms are faithfully reflected in peripheral tissues, and that fibroblasts can serve as a model for some brain disorders, perhaps including those affecting reward and motivation (McCarthy and Welsh, 2012).
To test the role of the circadian clock in AUD, we measured clock gene expression rhythms in fibroblasts from AUD patients and controls over the course of five days. We also examined the impact of the 10870 PER2 variant on rhythms. We found no differences in circadian rhythms between AUD cases and controls, or as a function of PER2 genotype, but among AUD cases there was an inverse correlation between illness severity and fibroblast circadian period length.
METHODS
Human Subjects and AUD Cell Lines
Fibroblast cell lines (N=32) were obtained from subjects who provided a skin biopsy from the upper arm as an elective component of an ongoing parallel design, topiramate vs. placebo trial for alcoholism (cases) or a study of healthy subjects’ response to alcohol ingestion (controls). As part of the study, AUD subjects received a brief counseling intervention to reduce alcohol use. All cell line donors were self-selected, choosing to provide biopsy materials for $80 extra payment. They were not required to donate cells to participate in either of the primary studies. The initial study cohort consisted of cell lines from 19 AUD (N=7 female) and 10 control (no female) donors that were matched for age and race. Three additional cell lines from healthy female subjects were later selected as controls for the female AUD lines. The latter were also matched for donor age/race, anatomical site of biopsy, passage number and storage time, but originated from a different study site and were not assessed for alcohol behaviors beyond the exclusion of AUD. All cell lines used had a passage number <10. Inclusion criteria for AUD subjects were alcohol intake >24 standard drinks/week for men, or >18 drinks/week for women. All control and AUD subjects were evaluated with The Structured Clinical Interview for DSM-IV (SCID) to ascertain psychiatric diagnosis (First et al., 1995). AUD subjects and male controls were also assessed with the 90-day Time Line Follow-back (TLFB) to assess recent alcohol use (Sobell et al., 1992). Most AUD subjects (17/19) met DSM-IV dependence criteria and were classified as being moderately ill. Controls averaged 6 standard drinks per week (range 1–15). Subjects with additional major psychiatric illnesses were excluded, including bipolar disorder, schizophrenia, severe/psychotic major depression, panic, borderline/antisocial personality, eating disorder, drug dependence (other than nicotine) and/or significant risk for suicide/violence. From AUD subjects, detailed clinical history was obtained, including the number of DSM-IV AUD diagnostic criteria met, and the age of AUD onset and cigarette smoking status upon study entry. Illness severity was estimated in two ways. First, narrowly as the number of DSM-IV criteria met for dependence (out of 7), and then as the sum total of DSM-IV criteria met for abuse and dependence (maximum possible score 11). In the latter system, there was no differential weight applied to any of the individual criteria, so abuse and dependence criteria were considered equally important. Family history of AUD and five other psychiatric conditions was obtained with the Family History Assessment Module (Rice et al., 1995), and for each AUD subject and control, a percentage of AUD-affected first-degree family members was calculated.
Per2∷Luc Reporter Gene Construct
The Per2∷luc lentiviral reporter gene was generously provided by Andrew Liu (University of Memphis), and has been described in detail elsewhere (Liu et al., 2007). In brief, the construct consists of the promoter of the clock gene Per2 driving expression of the firefly luciferase reporter. It randomly inserts into the genome of the host cell and reports activation by circadian clock transcriptional activators (i.e., CLOCK/BMAL1) that stimulate E-box elements within the Per2 promoter. Decoy elements limit interference from constitutive (non-rhythmic) transcription factor binding. Single cell imaging studies in fibroblasts suggest infection efficiency ~80–100% (not shown). All experiments used ~1×107 infectious units/plate.
Cell Culture and Luminometry
Cells were initially grown to confluence from frozen stocks in 100 mm plates in standard culture medium [DMEM, fortified with 10% fetal bovine serium (FBS), glutamine, and antibiotics (penicillin/streptomycin/amphotericin)]. After two days, cells were split into identical 35 mm plates and maintained in culture for another 48 hr. Then, infection with the Per2∷luc construct proceeded for 48 hr and cells were again grown to confluence (~1.2×106 cells/plate). Immediately prior to the reporter gene assay, growth medium was replaced with HEPES-buffered, serum-free medium containing 1 mM luciferin (Yamazaki and Takahashi 2005). This procedure was sufficient to synchronize Per2∷luc rhythms without the need for serum shock. For rhythm measurements made in the presence of alcohol, cells were cultured and infected in the absence of alcohol, and then ethanol (0.05% – 1.00%) was added with the recording medium and left on the cells for the duration of the five-day experiment.
Rhythm measurements by quantitative luminometry were conducted in 35 mm culture plates as previously described (Welsh et al., 2005) using a 32-well luminometer (Actimetrics). Photoemissions were measured from each plate for ~70 seconds, every 10 minutes for ~5 days. Temperature was maintained at 35°C under humidified room air atmospheric conditions. All samples were conducted in duplicate. Samples with poor agreement between duplicates or that appeared to be outliers were repeated in separate trials. Data for each individual are reported as the average of replicates.
DNA preparation and genotyping
Genomic DNA was prepared for each cell line using a Qiagen kit, following the manufacturer’s instructions. A custom Taqman genotyping assay for the PER2 SNP 10870 was purchased using primer and reporter sequence published previously (Kovanen et al., 2010). Genotyping was performed using an ABI 7900 thermocycler.
Analysis of luminometer data
All luminometry data were averaged across replicates and fit to a damped sine curve by the least squares method using commercially available software (Lumicycle Analysis, Actimetrics). Trends were removed by subtracting a 24-hr window moving average. This analysis provides a quantitative estimate of period and amplitude (rhythm parameters). A goodness-of-fit estimate was calculated for each estimate as the proportion of signal variance accounted for by the best fit sine wave. For case-control analyses, each rhythm parameter was analyzed separately using an unpaired, two-tailed t-test with α=0.05. For the case-case studies, a multiple regression analysis was conducted using the rhythm parameters amplitude and period as the independent variables and AUD clinical features as the dependent variables. Multivariate analyses were run with StatistiXL version 1.9, using α=0.05 as the threshold for statistical significance.
RESULTS
Case-control analysis of circadian rhythm parameters
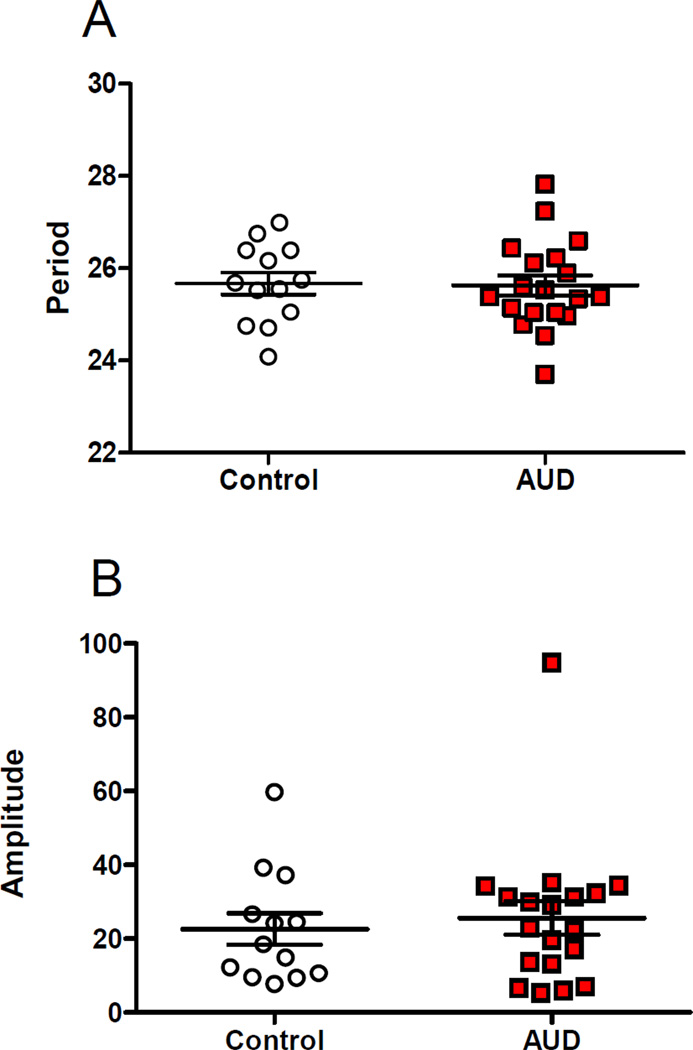
Per2∷luc gene expression rhythms were identified successfully in all cell lines, with a pattern that closely approximates a damped sine wave. Using this function, average goodness of fit was calculated to be 75% and 83% for control and AUD respectively (not statistically different). The period and amplitude were determined for cell lines from AUD cases and controls. Neither rhythm parameter differed significantly by diagnostic group (Table 1, Figure 1A, 1B). In the case of amplitude, there was a single outlier in each group. The results were no different when these individuals were excluded.
Table 1.
Rhythm parameters by clinical status. Data reported as period (hr) and amplitude (counts/sec) ± SEM. In the last row, p-values are indicated for a two tailed t-test between groups for each parameter. None of the parameters was significantly different.
| Period | Amplitude | % Fit | |
|---|---|---|---|
| Control | 25.7 ± 0.2 | 22.6 ± 4.2 | 75.8 ± 4.0 |
| Alcohol | 25.7 ± 0.2 | 25.7 ± 4.6 | 83.0 ± 2.7 |
| p = | 0.88 | 0.64 | 0.13 |
Figure 1.
A) Period and B) amplitude in AUD cases and controls. Rhythm parameters were estimated from Per2∷luc rhythms in fibroblasts obtained from AUD cases (n=19) and controls (n=13). Neither parameter was significantly different between groups.
Analysis of circadian rhythms in AUD subjects by clinical features
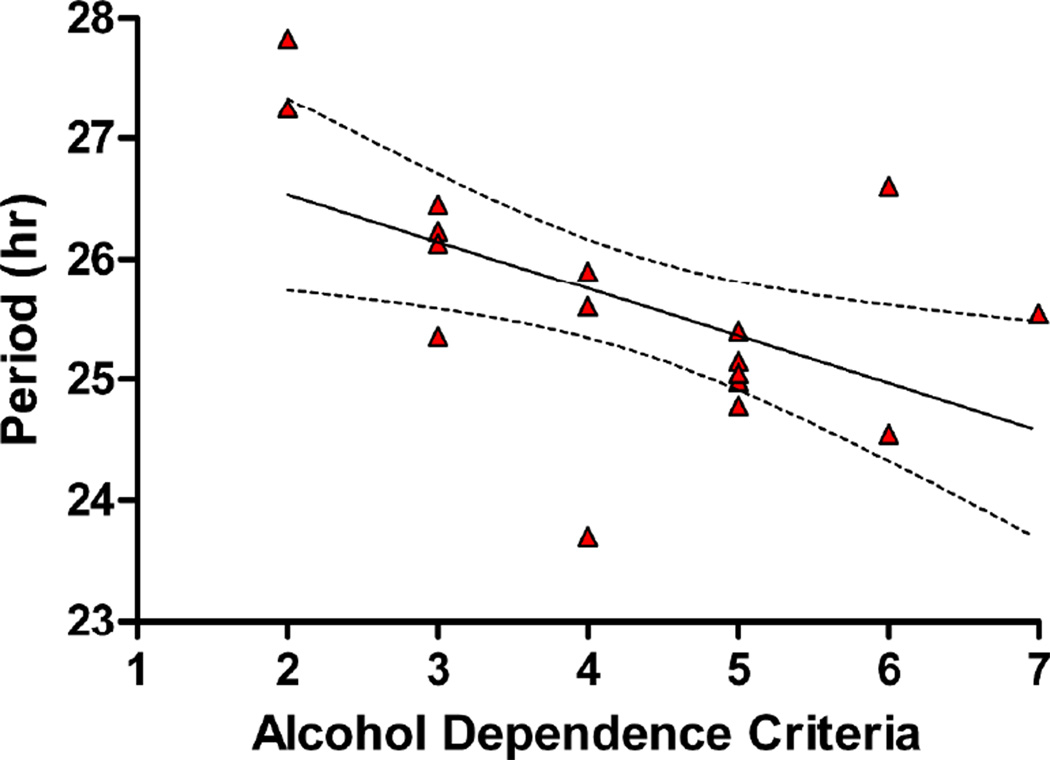
Rhythm parameters were analyzed in AUD cases to identify correlations between circadian rhythm parameters and age of onset, illness severity, and family history. When narrowly defined (i.e., using only alcohol dependence criteria), illness severity was inversely correlated to period length (r = −0.56, p = 0.01, Figure 2). This suggests shorter periods are associated with greater illness severity (Figure 3). When broadly defined (using alcohol abuse and dependence criteria), illness severity remained negatively correlated to period, but the relationship was less robust (r =−0.44, p = 0.06). A multiple regression analysis of narrowly defined illness severity using period and amplitude was significant, explaining 24% of the variance (adjusted r2 = 24%, F2,16= 3.82, p = 0.04). The model was no longer significant when the broad definition was employed (not shown). In contrast to illness severity, neither age of AUD onset, smoking status, nor family history (scored either categorically as positive/negative or continuously as 0–100% of family members affected) were correlated with any of the rhythm parameters.
Figure 2.
Relationship between circadian rhythm period and illness severity. Data for all AUD subjects are shown (triangles). Solid line indicates best fit ± 95% confidence intervals (dotted line).
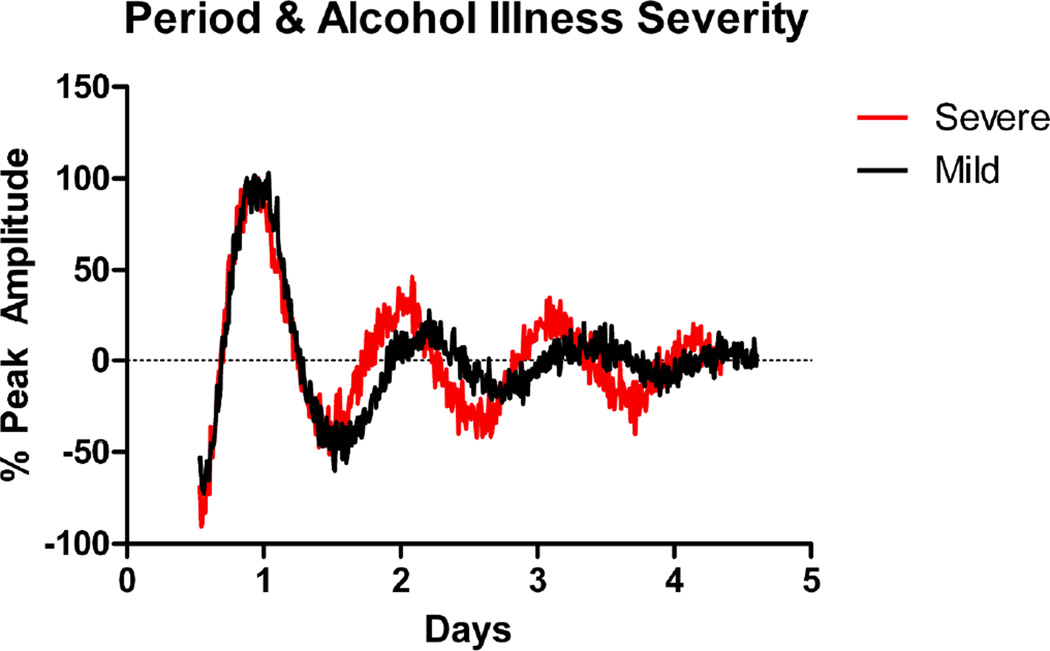
Figure 3.
Representative data for Per2∷luc rhythms obtained from two subjects with differing AUD severity. The red line indicates a severely affected individual (meets five DSM-IV criteria for alcohol dependence). The black line indicates a less severely affected individual (meets two DSM-IV criteria for alcohol dependence). For each curve, amplitude is normalized to the first day’s peak value to facilitate direct comparison.
Effects of ethanol on circadian rhythms
Rhythm parameters that differ by AUD severity could be related to cumulative alcohol exposure or a more severe genetic pre-disposition to the illness. To examine the possibility that alcohol may affect the clock directly, we measured Per2∷luc rhythms in the presence or absence of an alcohol concentration that might be expected to occur in human alcoholics. We found no consistent effect on rhythm parameters or cell viability in the range that would be expected for blood levels following pharmacological alcohol exposure (0.05–0.6%). At ethanol concentrations exceeding the pharmacological range (1%), alcohol caused a modest increase in rhythm amplitude, but no change in period (not shown).
PER2 genotype and rhythm parameters
The rare G allele at the PER2 10870 has been associated previously with alcohol intake (Comasco et al., 2010; Spanagel et al., 2005), but remains poorly characterized functionally. AUD and control subjects in our study were genotyped at this locus to determine whether circadian period or amplitude were quantitative traits associated with PER2 genotype. Among control subjects, only a single individual carried the rare allele. In contrast, among alcoholics 6 of 19 individuals were carriers (Table 2). Despite the apparent discrepancy in allele frequencies (8% vs. 32%, Odds ratio = 5.07), this difference was not statistically significant, presumably due to the small sample size (χ2 = 2.27, p = 0.13). Nonetheless, we re-analyzed Per2∷luc expression by genotype, comparing carriers of the rare allele (AG/GG genotypes) to those homozygous for the common allele (AA genotype). No rhythm parameter differed significantly by PER2 10870 genotype (Table 3). This was true whether or not the control subject with the PER2 variant was included in the analysis, and when the single subject with GG genotype was excluded (not shown).
Table 2.
PER2 10870 Genotypes for AUD cases and controls. Number of subjects with each PER2 10870 genotype is shown. DNA was not available for two control cell lines. There was no significant difference in allele frequency between AUD cases and controls.
| Diagnosis | GG | AG | AA |
|---|---|---|---|
| Control | 11 | 1 | 0 |
| AUD | 13 | 5 | 1 |
Table 3.
Rhythm parameters by PER2 10870 Genotype. Rhythm parameters were re-analyzed by genotype. Data reported as period (hr) and amplitude (counts/sec) ± SEM. In the last row, p-values are indicated for a two tailed t-test between groups for each parameter. None of the parameters was significantly different.
| PER2 10870 Genotype |
Period | Amplitude | % Fit |
|---|---|---|---|
| AA | 25.5 ± 0.2 | 26.4 ± 4.0 | 79.7 ± 2.8 |
| AG/GG | 25.7 ± 0.4 | 21.8 ± 5.0 | 82.2 ± 4.3 |
| p = | 0.58 | 0.56 | 0.66 |
PER2 genotype and clinical features
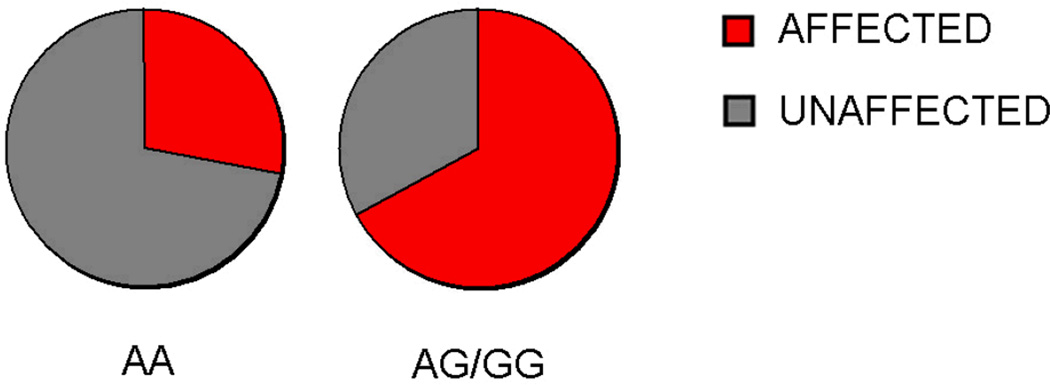
We examined the clinical features of our sample to determine whether PER2 10870 is associated with any additional AUD sub-phenotypes. PER2 genotype was not associated with age of onset, illness severity, gender, or family history. However, among AUD subjects with a positive family history of AUD, carriers of the PER2 SNP had a significantly greater proportion of affected family members than non-carriers (28% affected in AA genotype vs. 67% AG/GG genotype respectively, t = 3.322, df = 10, p = 0.008, Figure 4). Among controls with a positive family history (n=4), 25% of first-degree relatives were affected by AUD. All of these control subjects were homozygous common for the PER2 variant.
Figure 4.
PER2 10870 genotype and AUD family prevalence. The proportion of family members affected by AUD was assessed for each proband with a positive family history. On average, probands with the common AA variant (N=9) had 28% (range: 9–67%) of family members affected while AG/GG subjects (N=3) had 64% of family members affected (range: 50–75%).
DISCUSSION
Longitudinal examination of Per2∷luc gene expression in fibroblasts allowed us to estimate circadian rhythm parameters of the molecular clock in AUD patients. Our findings provide an important bridge between previous human genetic studies that provided evidence of association between clock genes and AUD, but did not assess function; and animal studies that showed important functional differences after alcohol exposure in clock gene mutants, but cannot readily be generalized to human subjects with more subtle genetic abnormalities. We found overlapping distributions of rhythm amplitude and period between AUD cases and controls, with ranges similar to those reported previously in non-clinical populations (Brown et al, 2005). Although we did not find gene expression rhythm differences between AUD cases and controls, we did find that among subjects with AUD, illness severity correlates inversely with rhythm period. The latter indicates that AUD subjects with shorter circadian periods are more severely alcohol dependent than those with longer circadian periods. Our small sample size is under-powered for subgroup analyses, and key underlying mechanisms remain to be elucidated. Nonetheless, our findings suggest that while defects in the circadian clock may not directly influence susceptibility to AUD, they may influence the course and severity of the illness. That is, clock gene variants may be insufficient to cause AUD per se, but may be important co-factors in illness progression, perhaps by mediating the stress response. For example, at baseline Per1 mutant mice do not prefer alcohol over wild types (Zghoul et al., 2007), but after social defeat stress, they preferentially increase intake. Similarly, among adolescent humans, a PER1 association with drinking was revealed only after considering the gene×environment interaction with stress (Dong et al., 2011).
Alcohol, period, and chronotype
When maintained in a 24-hr light/dark cycle, human subjects with short circadian periods tend towards circadian phase advances and morning chronotype, whereas those with long periods tend towards circadian phase delays and evening chronotype (Brown et al., 2008). For this reason, our result of short period association with greater AUD severity is at odds with two human studies suggesting that evening chronotype is a risk factor for high alcohol intake (Adan, 1994; Wittman et al., 2010). On the other hand, among AUD patients dually diagnosed with bipolar disorder, an association with morning chronotype has been reported that supports our finding (Hatonen et al, 2008). Our findings also agree with two animal studies that examined wheel-running activity in rodents bred for high alcohol preference. Under constant dark conditions, high alcohol-preferring mice showed shorter circadian periods in running activity than their low alcohol-preferring counterparts (Hofstetter et al., 2003). Short periods were also reported in alcohol preferring rats, but with variability attributed to differences in animal strain and light conditions (Rosenwasser et al., 2005). In both cases, the animals were alcohol naïve, implicating shared genetic factors regulating both alcohol preference and circadian rhythms, but not alcohol exposure history.
At least two factors need to be addressed before resolving the discrepancy between our studies and past chronotype assessments. First, many of the chronotype associations with alcohol intake were reported in adolescents, whereas our AUD subjects were adults ranging from 38–61 years old (mean 50.7 yr), with a mean illness duration of 12 yr. Chronotype, while genetically influenced, varies considerably by age, with the greatest eveningness reported among adolescents and young adults, and progressively lower eveningness as age increases into the eight decade (Roenneberg et al., 2004). In adolescents, this apparent switch occurs during an important time in brain maturation, and could be affected by alcohol use (De Bellis et al., 2000), potentially altering developmental chronotype trajectories. In addition, chronotype may be affected by environmental or social factors that mask circadian function. Therefore, chronotypes in adult alcoholics may differ in unpredictable ways from those of abstinent controls and/or alcohol consuming adolescents.
The second point to consider is that cell-specific differences may limit the applicability of our conclusions to the brain clock. We did not find direct effects of alcohol on the fibroblast clock, suggesting that the observed differences are due to intrinsic properties of the cells, and not alcohol exposure. However, we exposed the cells to alcohol for < 5 days, and did not test neurons, the cell type most relevant to AUD. In rats, neonatal alcohol exposure was reported to alter amplitude and phase of Cry1 and Per2 in the brain and liver (Farnell et al., 2008), suggesting that under some circumstances, alcohol does affect the clock directly. It remains possible that among adult humans either intoxicated by or withdrawing from the substance, alcohol could engage with neuron-specific signaling systems upstream from the clock (e.g., GABA, glutamate), causing changes in rhythm parameters that would not be detectable in fibroblasts.
Genetic variation at the PER2 locus and alcohol
The PER2 10870 has been associated with alcohol intake > 300 g/day in adult alcoholics, and sleep problems associated with alcohol in adolescent boys (Comasco et al., 2010; Spanagel et al., 2005). However, the association of PER2 10870 with alcohol intake has not been replicated universally, and has failed to associate with a related, but distinct phenotype (alcohol dependence) in previous studies (Comasco et al., 2010; Kovanen et al., 2010, Sjoholm et al., 2010). We were unable to detect any difference in rhythm parameters associated with PER2 10870 when carriers of the rare G allele were compared to subjects homozygous for the common A allele. However, due to sample size, we were limited to studying heterozygous carriers of the rare variant. It is possible that GG homozygotes are more severely affected and thus, analysis of such individuals is warranted. In contrast, we did find preliminary evidence that PER2 10870 may be associated with the penetrance of AUD in families with a genetic history of the illness; but due to the small sample size, this finding should be interpreted cautiously. Our case-control association analysis of PER2 10870 neither refutes nor supports the previous associations of PER2 10870 with alcohol intake since we used a different phenotype than those from the earlier positive reports (AUD instead of alcohol intake). However, the trend towards association of the rare G allele with AUD is opposite to the direction expected based on claims that the G allele is protective against high alcohol intake (Comasco et al., 2010; Spanagel et al., 2005).
Peripheral clocks and alcohol
Clock genes are expressed almost ubiquitously throughout the mammalian body, and may be important for AUD in additional organs besides the brain. We have primarily taken the perspective that peripheral cell lines model aspects of brain function relevant to the illness. We are supported in this assumption by studies that have measured rhythms in dissociated single cells in various clock gene mutants, suggesting that there are few genetic differences in the intracellular clock between neurons and peripheral cells (Liu et al., 2007). However, it remains likely that there are differences between neurons and fibroblasts in electrochemical inputs to the clock, and in the formation of higher order, multi-cellular networks. With this is mind, our work could also model aspects of the liver clock. The liver is the principal site of alcohol metabolism and also is a site of robust circadian regulation. Liver genes involved in ethanol metabolism such as alcohol dehydrogenases (ADH) and aldehyde dehydrogenases (ALDH) may modulate AUD susceptibility and are under the control of the circadian clock (Frank et al., 2012; Hughes et al., 2009; Mayfield et al., 2008). For this reason, we remain cautious about generalizing the results from fibroblasts to a specific organ system, and acknowledge AUD severity could be modulated by clock dysfunction not only in the brain, but also in other organs.
Conclusions
These results partially support a role for the circadian clock in AUD, and suggest that further work aimed at understanding clock-based mechanisms involved in AUD progression may be fruitful. Human clinical studies in AUD rarely characterize circadian rhythm phenotypes in detail. Our work suggests that detailed chronotype assessments, actigraphic recordings or measurements of endocrine rhythms in AUD may be useful directions for future research. In conjunction with longitudinal clinical assessment and additional molecular imaging, this work could allow for more detailed examinations of the circadian clock and its relationship to AUD illness trajectory.
ACKNOWLEDGEMENTS
MJM is supported by a Veterans Affairs Career Development Award (1IK2BX001275). JMC receives support from NIH (R21AA017584). HRK receives support from NIH (R01AA021164). DKW receives support from a Veterans Affairs Merit Award (1I01BX001146), NIMH (R01 MH082945), and a NARSAD Young Investigator Award. The funders had no role in the analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- Adan A. Chronotype and personality factors in the daily consumption of alcohol and psychostimulants. Addiction. 1994;89:455–462. doi: 10.1111/j.1360-0443.1994.tb00926.x. [DOI] [PubMed] [Google Scholar]
- Agapito M, Mian N, Boyadjieva NI, Sarkar DK. Period 2 gene deletion abolishes beta-endorphin neuronal response to ethanol. Alcohol Clin Exp Res. 2010;34:1613–1618. doi: 10.1111/j.1530-0277.2010.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaule C, Swanstrom A, Leone MJ, Herzog ED. Circadian modulation of gene expression, but not glutamate uptake, in mouse and rat cortical astrocytes. PLoS.One. 2009;4:e7476. doi: 10.1371/journal.pone.0007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Fleury-Olela F, Nagoshi E, Hauser C, Juge C, Meier CA, Chicheportiche R, Dayer JM, Albrecht U, Schibler U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS.Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Kunz D, Dumas A, Westermark PO, Vanselow K, Tilmann-Wahnschaffe A, Herzel H, Kramer A. Molecular insights into human daily behavior. Proc Natl Acad Sci U S A. 2008;105:1602–1607. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comasco E, Nordquist N, Gokturk C, Aslund C, Hallman J, Oreland L, Nilsson KW. The clock gene PER2 and sleep problems: association with alcohol consumption among Swedish adolescents. Ups J Med Sci. 2010;115:41–48. doi: 10.3109/03009731003597127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bellis MD, Clark DB, Beers SR, Soloff PH, Boring AM, Hall J, Kersh A, Keshavan MS. Hippocampal volume in adolescent-onset alcohol use disorders. Am J Psychiatry. 2000;157:737–744. doi: 10.1176/appi.ajp.157.5.737. [DOI] [PubMed] [Google Scholar]
- Dong L, Bilbao A, Laucht M, Henriksson R, Yakovleva T, Ridinger M, Desrivieres S, Clarke TK, Lourdusamy A, Smolka MN, Cichon S, Blomeyer D, Treutlein J, Perreau-Lenz S, Witt S, Leonardi-Essmann F, Wodarz N, Zill P, Soyka M, Albrecht U, Rietschel M, Lathrop M, Bakalkin G, Spanagel R, Schumann G. Effects of the circadian rhythm gene period 1 (per1) on psychosocial stress-induced alcohol drinking. Am J Psychiatry. 2011;168:1090–1098. doi: 10.1176/appi.ajp.2011.10111579. [DOI] [PubMed] [Google Scholar]
- Drummond SP, Gillin JC, Smith TL, DeModena A. The sleep of abstinent pure primary alcoholic patients: natural course and relationship to relapse. Alcohol Clin Exp Res. 1998;22:1796–1802. [PubMed] [Google Scholar]
- Ewing JA. Detecting alcoholism. The CAGE questionnaire. JAMA. 1984;252:1905–1907. doi: 10.1001/jama.252.14.1905. [DOI] [PubMed] [Google Scholar]
- Farnell YZ, Allen GC, Nahm SS, Neuendorff N, West JR, Chen WJ, Earnest DJ. Neonatal alcohol exposure differentially alters clock gene oscillations within the suprachiasmatic nucleus, cerebellum, and liver of adult rats. Alcohol Clin Exp Res. 2008;32:544–552. doi: 10.1111/j.1530-0277.2007.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV Axis I Disorders - Patient Edition (SCID - I/P, Version 2.0) New York: Biometrics Research Department, New York State Psychiatric Institute; 1995. [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mossner R, Gaebel W, Dahmen N, Scherbaum N, Schmal C, Steffens M, Lucae S, Ising M, Muller-Myhsok B, Nothen MM, Mann K, Kiefer F, Rietschel M. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict.Biol. 2012;17:171–180. doi: 10.1111/j.1369-1600.2011.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Burgos D, Gonzalez F, Manrique T, Gallo M. Patterns of ethanol intake in preadolescent, adolescent, and adult Wistar rats under acquisition, maintenance, and relapse-like conditions. Alcohol Clin Exp Res. 2009;33:722–728. doi: 10.1111/j.1530-0277.2008.00889.x. [DOI] [PubMed] [Google Scholar]
- Gianoulakis C, Dai X, Thavundayil J, Brown T. Levels and circadian rhythmicity of plasma ACTH, cortisol, and beta-endorphin as a function of family history of alcoholism. Psychopharmacology (Berl) 2005;181:437–444. doi: 10.1007/s00213-005-0129-x. [DOI] [PubMed] [Google Scholar]
- Hofstetter JR, Grahame NJ, Mayeda AR. Circadian activity rhythms in high-alcohol-preferring and low-alcohol-preferring mice. Alcohol. 2003;30:81–85. doi: 10.1016/s0741-8329(03)00095-8. [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanen L, Saarikoski ST, Haukka J, Pirkola S, Aromaa A, Lonnqvist J, Partonen T. Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol Alcohol. 2010;45:303–311. doi: 10.1093/alcalc/agq035. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, III, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MJ, Welsh DK. Cellular circadian clocks in mood disorders. J Biol Rhythms. 2012;27:339–352. doi: 10.1177/0748730412456367. [DOI] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, Nestler EJ. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci U S A. 2005;102:9377–9381. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Semenova EA, Moriggi E, Revell VL, Hack LM, Lockley SW, Arendt J, Skene DJ, Meier F, Izakovic J, Wirz-Justice A, Cajochen C, Sergeeva OJ, Cheresiz SV, Danilenko KV, Eckert A, Brown SA. The physiological period length of the human circadian clock in vivo is directly proportional to period in human fibroblasts. PLoS One. 2010;5:e13376. doi: 10.1371/journal.pone.0013376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau-Lenz S, Zghoul T, de Fonseca FR, Spanagel R, Bilbao A. Circadian regulation of central ethanol sensitivity by the mPer2 gene. Addict Biol. 2009;14:253–259. doi: 10.1111/j.1369-1600.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- Rice JP, Reich T, Bucholz KK, Neuman RJ, Fishman R, Rochberg N, Hesselbrock VM, Nurnberger JI, Jr, Schuckit MA, Begleiter H. Comparison of direct interview and family history diagnoses of alcohol dependence. Alcohol Clin Exp Res. 1995;19:1018–1023. doi: 10.1111/j.1530-0277.1995.tb00983.x. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Kuehnle T, Pramstaller PP, Ricken J, Havel M, Guth A, Merrow M. A marker for the end of adolescence. Curr Biol. 2004;14:R1038–R1039. doi: 10.1016/j.cub.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Rosenwasser AM, Fecteau ME, Logan RW, Reed JD, Cotter SJ, Seggio JA. Circadian activity rhythms in selectively bred ethanol-preferring and nonpreferring rats. Alcohol. 2005;36:69–81. doi: 10.1016/j.alcohol.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Sjoholm LK, Kovanen L, Saarikoski ST, Schalling M, Lavebratt C, Partonen T. CLOCK is suggested to associate with comorbid alcohol use and depressive disorders. J. Circadian. Rhythms. 2010;8:1. doi: 10.1186/1740-3391-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell LC, Sobell MB. Measuring alcohol consumption: Psychosocial and Biochemical Methods. Humana Press: R. Litten and J. Allen; 1992. Timeline follow-back: A technique for assessing self-reported alcohol consumption; pp. 41–42. [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mozhui K, Li Z, Mulligan MK, Ingels JF, Zhou X, Hori RT, Chen H, Cook MN, Williams RW, Lu L. A promoter polymorphism in the Per3 gene is associated with alcohol and stress response. Transl Psychiatry. 2012;2:e73. doi: 10.1038/tp.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- Wittmann M, Dinich J, Merrow M, Roenneberg T. Social jetlag: misalignment of biological and social time. Chronobiol Int. 2006;23:497–509. doi: 10.1080/07420520500545979. [DOI] [PubMed] [Google Scholar]
- Wittmann M, Paulus M, Roenneberg T. Decreased psychological well-being in late 'chronotypes' is mediated by smoking and alcohol consumption. Subst Use Misuse. 2010;45:15–30. doi: 10.3109/10826080903498952. [DOI] [PubMed] [Google Scholar]
- Wong MM, Brower KJ, Nigg JT, Zucker RA. Childhood sleep problems, response inhibition, and alcohol and drug outcomes in adolescence and young adulthood. Alcohol Clin Exp Res. 2010;34:1033–1044. doi: 10.1111/j.1530-0277.2010.01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 2005;393:288–301. doi: 10.1016/S0076-6879(05)93012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghoul T, Abarca C, Sanchis-Segura C, Albrecht U, Schumann G, Spanagel R. Ethanol self-administration and reinstatement of ethanol-seeking behavior in Per1(Brdm1) mutant mice. Psychopharmacology (Berl) 2007;190:13–19. doi: 10.1007/s00213-006-0592-z. [DOI] [PubMed] [Google Scholar]