Abstract
Pluripotent embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) hold great promise for future use in tissue replacement therapies due to their ability to self-renew indefinitely and to differentiate into all adult cell types. Harnessing this therapeutic potential efficiently requires a much deeper understanding of the molecular processes at work within the pluripotency network. The transcription factors Nanog, Oct4, and Sox2 reside at the core of this network, where they interact and regulate their own expression as well as that of numerous other pluripotency factors. Of these core factors, Nanog is critical for blocking the differentiation of pluripotent cells, and more importantly, for establishing the pluripotent ground state during somatic cell reprogramming. Both mouse and human Nanog are able to form dimers in vivo, allowing them to preferentially interact with certain factors and perform unique functions. Recent studies have identified an evolutionary functional conservation among vertebrate Nanog orthologs from chick, zebrafish, and the axolotl salamander, adding an additional layer of complexity to Nanog function. Here we present a detailed overview of published work focusing on Nanog structure, function, dimerization, and regulation at the genetic and post-translational levels with regard to the establishment and maintenance of pluripotency. The full spectrum of Nanog function in pluripotent stem cells and in cancer is only beginning to be revealed. We therefore use this evidence to advocate for more comprehensive analysis of Nanog in the context of disease, development, and regeneration.
Keywords: ESCs, iPSCs, pluripotency, Nanog, self-renewal, reprogramming
I. Introduction
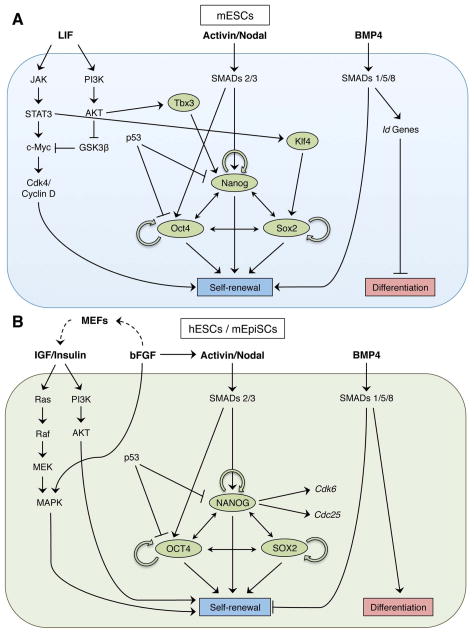
ESCs are derived from the inner cell mass (ICM) of the pre-implantation mammalian embryo and can be maintained indefinitely in culture[1, 2]. Along with their unlimited capacity for self-renewal in vitro, ESCs are also defined by their ability to give rise to all somatic and germ cell lineages of the developing embryo, with the exception of extra-embryonic tissues. Mouse ESCs harvested at embryonic day 3.5 (E3.5) from the naïve epiblast exhibit “ground state” pluripotency and require specific culture conditions for maintenance[3, 4]. The cytokines leukemia inhibitory factor (LIF) and bone morphogenetic protein 4 (BMP4) have been shown to be sufficient for ESC self-renewal in the undifferentiated state in MEF- and serum-free conditions, respectively[5–8]. LIF promotes self-renewal by activating the JAK/STAT3 and PI3K/AKT signaling pathways, and BMP4 up-regulates transcription of inhibitor of differentiation (Id) genes through activation of SMAD proteins 1, 5, and 8 (Fig. 1A). Maintenance of pluripotency in ESCs is governed by the expression of the core transcription factors Nanog, Oct4, and Sox2, as well as a variety of other factors. Nanog, Oct4, and Sox2 have been shown to repress the expression of developmental genes while modulating their own expression levels by binding to each other’s promoter regions[1, 2, 9, 10]. ESCs can give rise to all three germ layers of the developing embryo[3, 4, 11, 12], including the primitive germ cells [5–8, 13, 14]. However, human ESCs exhibit gene expression profiles that are much more akin to mouse epiblast stem cells (mEpiSCs) derived at the post-implantation stage [5, 15, 16]. This “primed” state is a characteristic feature of human ESCs and is also what defines the epiblast at the post-implantation stage[3]. Unlike mouse (m)ESCs, human (h)ESCs (and mEpiSCs) do not require LIF or BMP4 for survival, but instead require basic fibroblast growth factor (bFGF) and insulin or insulin-like growth factor (IGF) signaling[17–19] (Fig. 1B). bFGF activates the mitogen-activated protein kinase (MAPK) as well as the Activin/Nodal signaling pathways, and IGF activates the Ras and PI3K pathways. In mESCs, LIF signaling up-regulates Klf4 and Tbx3 via the JAK/STAT3 and PI3K/AKT pathways, which then go on to activate Sox2 and Nanog, respectively[20]. In hESCs and mEpiSCs, on the other hand, SMADs 2 and 3 propagate Activin/Nodal signaling as well as directly bind and up-regulate NANOG[21]. Taken together, these studies accentuate the elaborate and interconnected relationship between extrinsic survival signals and the transcriptional program in pluripotent stem cells. In this review, we focus on the core transcription factor Nanog and present a broad range of evidence supporting its unique role in regulating pluripotency.
Fig. 1.
Mouse and human ESC survival pathways. (A) Mouse ESCs require LIF and BMP4 for maintenance. (B) Human ESCs and mouse EpiSCs require IGF/insulin and bFGF for maintenance. Human ESC-derived fibroblast-like cells and MEFs are also stimulated by bFGF in culture to secrete IGF (dashed arrows). In both cell types, Nanog, Oct4, and Sox2 form a positive auto-regulatory loop.
II. Genetic and Proteomic Features of Nanog
II.1. Nanog pseudogenes and isoforms
Upon analysis of the NANOG gene in the human genome, eleven pseudogenes were identified aside from the two NANOG alleles (Table 1). Among these, ten are retropseudogenes and one is an expressed tandem duplicate[22]. The ten pseudogenes were named NANOGP2 to NANOGP11, and the duplication pseudogene NANOGP1 (or NANOG2). The same group also uncovered two processed pseudogenes in the mouse genome, which they named NanogPa and NanogPb. Subsequently, Ian Chambers’ group described two novel retrotransposed copies of murine Nanog, named NanogPc and NanogPd[23]. The differences between these two and the previously analyzed pseudogenes reside not only in their chromosomal locations, but also in the fact that NanogPc and NanogPd open reading frames are 98% identical to Nanog and are potentially capable of expressing protein products with roles in ESC maintenance[23]. Zhang et al.[24] demonstrated that the previously identified NANOGP8 pseudogene is actually a retrogene that is expressed in different cancer cell lines, promoting proliferation.
Table 1.
Summary of Nanog pseudogenes & isoforms
| Species | Name | Type | References |
|---|---|---|---|
| H. sapiens | NANOGP1 (NANOG2) | Tandem duplicate | [22] |
| H. sapiens | NAN0GP2 | Retropseudogene | [22] |
| H. sapiens | NANOGP3 | Retropseudogene | [22] |
| H. sapiens | NANOGP4 | Retropseudogene | [22] |
| H. sapiens | NANOGP5 | Retropseudogene | [22] |
| H. sapiens | NAN0GP6 | Retropseudogene | [22] |
| H. sapiens | NANOGP7 | Retropseudogene | [22] |
| H, sapiens | NANOGP8 | Retropseudogene | [22, 24] |
| H. sapiens | NANOGP9 | Retropseudogene | [22] |
| H. sapiens | NANOGP10 | Retropseudogene | [22] |
| H. sapiens | NANOGP11 | Retropseudogene | [22] |
| M. fascicuiaris | NanogP | Pseudogene | [22] |
| P. troglodytes | NanogP4 | Pseudogene | [22] |
| M. musculus | NanogPa | Retropseudogene | [22] |
| M. musculus | NanogPb | Retropseudogene | [22] |
| M. musculus | NanogPc | Retropseudogene | [23] |
| M. musculus | NanogPd | Retropseudogene | [23] |
| M. musculus | Nanog a | Isoform | [27] |
| M. musculus | Nanog b | Isoform | [27] |
| M. musculus | Nanog c | Isoform | [27] |
Another way to potentially regulate Nanog function at the post-transcriptional level is through alternative splicing. Previous studies have reported that gene regulation by alternative splicing may affect about half of all genes in mammals[25]. More specifically, computational and experimental analyses have recently revealed that alternative splicing is fundamental for stem cell maintenance, pluripotency, and differentiation[25, 26]. Not surprisingly, a recent study has documented that the Nanog locus, via alternate promoter selection and alternative splicing, encodes two additional previously unknown protein variants, dubbed Nanog b and Nanog c, with reduced functions in mESC maintenance and pluripotency[27] (Table 1). For instance, although Nanog, Nanog b, and Nanog c can dimerize and interact with pluripotency factors such as Oct4 and Sall4, Nanog b cannot execute LIF-independent self-renewal. Both Nanog b and c are also slightly impaired in repressing transcription of primitive endoderm and trophectoderm markers such as Gata6, Gata4, Sox17, and Hand1.
II.2. Post-translational modification of Nanog
Post-translational modification (PTM) of proteins, particularly transcription factors, is a potent way to regulate functions such as transcriptional activity, DNA binding, co-factor association, subcellular localization, and protein stability. In many cellular contexts, important players such as p53 are heavily post-translationally modified by acetylation, phosphorylation, ubiquitination, methylation, and sumoylation, to extensively modulate their functions[28]. In ESCs, however, regulation of self-renewal and pluripotency factors has been broadly investigated at the transcriptional level, but lack of knowledge still exists about how their functions are modulated by PTMs. Nanog in particular has been known for quite some time to be a phosphoprotein in mESCs, since phosphatase treatment caused the disappearance of some slowly migrating forms of Nanog as detected by western blot[29]. Since then, proteomic or site-directed mutagenesis analyses have revealed several Nanog phosphorylation sites in different cellular contexts[30]. Interestingly, only a few reports have investigated Nanog regulation by phosphorylation or by any other PTM in ESCs to date (Table 2). For instance, Moretto-Zita et al.[31] showed that Nanog is phosphorylated at several Ser/Thr-Pro motifs in mESCs. These modified sites are then recognized and bound by the prolyl isomerase Pin1, leading to Nanog protein stabilization by preventing proteasome-mediated degradation. Additionally, they demonstrated that those phosphorylated sites as well as Pin1 activity are important for ESC self-renewal and teratoma formation[31]. In a subsequent report, Ramakrishna and colleagues[32] reported that 3 out of the 4 Ser/Thr-Pro motifs mentioned above reside in a PEST domain that they previously identified, which regulates human NANOG stability by targeting it for proteasomal degradation. A recent report[33] has also shown that Nanog is ubiquitinated in mouse ESCs, which acts to maintain appropriate Nanog levels.
Table 2.
Summary of Nanog post-translational modifications
| Nanog form | Modification | Modified residues | Function | References |
|---|---|---|---|---|
| Mouse | Phosphorylation | Ser52 | Interaction with Pin1, Nanog stabilization | [31] |
| Mouse | Phosphorylation | Ser 56/57 | Interaction with Pin1, Nanog stabilization | [31] |
| Mouse | Phosphorylation | Ser65 | Interaction with Pin1, Nanog stabilization | [31] |
| Mouse | Phosphorylation | Ser 77/78 | Interaction with Pin1, Nanog stabilization | [31] |
| Human | Phosphorylation | Tyr35 | Interaction with FAK, cancer cell motility & invasion | [34] |
| Human | Phosphorylation | Tyr174 | Interaction with FAK, cancer cell motility & invasion | [34] |
| Human | Phosphorylation | Serines (specific residues unknown) | Shuttling to nucleus, breast cancer cell survival & chemoresistance | [35] |
| Human | Ubiquitination | Lys48 | Targeting to 26S proteasome | [32] |
| Human | Ubiquitination | Lys63 | Targeting to 26S proteasome | [32] |
| Mouse | Ubiquitination | Lys112 | Targeting to 26S proteasome | [33] |
| Mouse | Ubiquitination | Lys141 | Targeting to 26S proteasome | [33] |
| Mouse | Ubiquitination | Lys156 | Targeting to 26S proteasome | [33] |
Two additional studies have described a role of Nanog phosphorylation in promoting tumorigenesis. In one, hNANOG was shown to be phosphorylated in vitro and in several cancer cell lines by focal adhesion kinase (FAK)[34]. In another, Bourguignon et al.[35] illustrate that hNANOG, upon phosphorylation by protein kinase Cε (PKCε), translocates to the nucleus and activates miR-21 production to promote tumor progression in breast cancer cells. All things considered, the current knowledge of Nanog regulation by PTMs may merely represent the tip of the iceberg. It would therefore not be surprising if Nanog modifications were as diverse and abundant as those found in p53, owing to the variety of functions that Nanog performs in stem cells and in cancer cells.
II.3. Nanog dimerization enhances ESC self-renewal and pluripotency
Nanog is a homeodomain protein that was discovered in a screen for self-renewal factors that could sustain mESCs in the absence of LIF signaling[9, 36]. Nanog is critical for mammalian development and is required for specification of the ICM in the pre-implantation embryo[37]. Similarly, the successful derivation of ESCs from the mouse blastocyst requires the expression of Nanog[9]. Because of the regulatory cooperation among Nanog, Oct4, and Sox2, it was believed that Nanog interacted with many other key factors in ESCs that govern pluripotency. The notion of ESC maintenance as being a complex and multifaceted process was confirmed by the creation of the first pluripotency protein interaction network in mESCs[38]. This Nanog interactome connects with multiple co-repressors such as the SWI/SNF, NuRD, and Polycomb complexes, and outlines the proteins that physically interact with Nanog, and that are functionally important for ESC maintenance and early development[38].
We[39] and others[40] simultaneously demonstrated that mouse Nanog is able to form functional dimers through its tryptophan-rich (WR) domain. WR domain-mediated dimerization was further verified using an additional mutant form of Nanog containing ten tryptophan to alanine substitutions (10WA) in the WR domain. This mutant form of Nanog was unable to dimerize with the wild-type form, confirming the importance of the WR domain for Nanog dimerization. In this study we also found that the dimeric form of Nanog is essential for mESC self-renewal and pluripotency. We demonstrated by co-immunoprecipitation that factors within the Nanog interactome such as Sall4, Zfp198, Zfp281, Dax1, Nac1, and Oct4 preferentially interacted with wild-type Nanog versus the monomeric Nanog10WA. Whether this differential interaction is due to a sequence-specific or monomer/dimer-specific effect needs to be further distinguished. We showed through colony formation assays and alkaline phosphatase (AP) staining that expression of a tethered Nanog dimer was sufficient to sustain the pluripotent phenotype of mESCs in the absence of LIF. Conversely, expression of a tethered Nanog monomer in the absence of LIF was insufficient to maintain mESCs in culture. Independently, Mullin et al.[40] found that wild-type Nanog only dimerized with Nanog mutants containing intact WR domains. Together these findings have demonstrated that Nanog dimers, but not monomers, are sufficient to support mESC self-renewal in the absence of LIF, and have delineated much of the physical and functional properties of Nanog in ESCs.
Nanog dimers interact with a subset of proteins identified in the Nanog interactome that promote pluripotency[38, 39]. One important Nanog binding partner is the Kruppel-like zinc finger transcription factor Zfp281, which was previously shown by co-immunoprecipitation to preferentially interact with Nanog dimers[39]. Our group recently demonstrated that Zfp281 functions as a transcriptional repressor of key pluripotency genes including Nanog[41] in mESCs. Zfp281 shares many target genes with Oct4, Sox2, and Nanog, and its promoter is also bound by Oct4, Sox2, and Nanog[42, 43]. Targeted deletion of Zfp281 resulted in delayed mESC differentiation as measured by embryoid body formation, likely due to misregulation of Oct4 and Nanog at the transcript and protein levels, compared to wild-type mESCs. Zfp281 was also found to be required for Nanog binding to its own promoter by ChIP-PCR, suggesting that Zfp281 plays a critical role in regulating Nanog expression levels. These findings also suggest that Zfp281 helps to maintain the pluripotent state by fine-tuning Nanog expression in conjunction with other co-repressors (see Section III.3. below) in ESCs.
II.4. Nanog orthologs
The lower vertebrates chick and zebrafish both express Nanog and have been used extensively as developmental model systems. Chick and zebrafish Nanog exhibit low protein sequence similarity to mouse Nanog, due in part to the fact that neither chick Nanog nor zebrafish Nanog contains a WR domain (Fig. 2). Despite this caveat, it was recently shown that zebrafish Nanog is able to dimerize in vitro by a GST pull-down assay and that zebrafish and mouse Nanog can functionally substitute for one another in vivo[44]. Unlike mouse Nanog, zebrafish Nanog requires the N-terminal domain (ND) and the homeodomain (HD) for dimerization. It has not yet been determined, however, whether chick Nanog is able to form dimers or whether chick and mouse Nanog are functionally analogous.
Fig. 2.
Sequence alignment of mouse, human, chick, axolotl salamander, and zebrafish Nanog (top to bottom). All five orthologs contain conserved residues, as indicated by shaded regions (darker = more conserved). All orthologs contain a homeodomain (boxed in red), but only mouse and human Nanog contain WR domains (boxed in green). Alignment created with ClustalW2 and analyzed in Jalview.
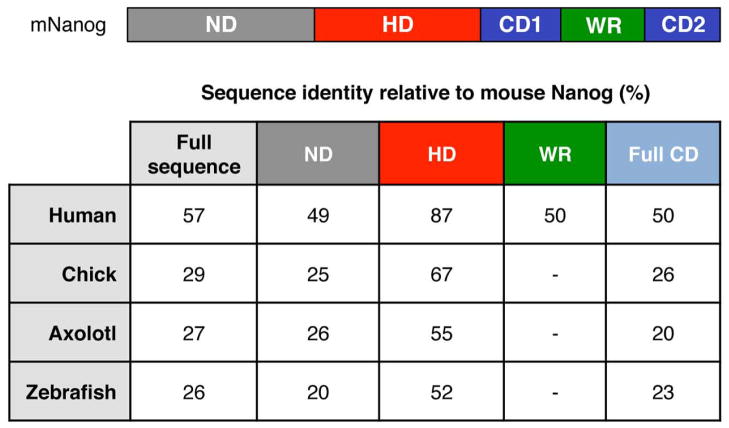
Interestingly, it was recently discovered that chick and zebrafish Nanog can substitute for mouse Nanog during somatic cell reprogramming[45], supporting the idea of a functional conservation among vertebrate Nanog orthologs. Currently, it is believed that mouse Nanog dimers promote mESC self-renewal and maintenance of pluripotency[39, 40], and that Nanog monomers may be sufficient for the establishment of pluripotency[46]. It is not yet known, however, whether the same is true in hESCs, or if inherent differences exist between the inductive capabilities of the monomer and dimer forms in generating iPSCs. Human NANOG contains a WR domain that has 50% sequence identity with the mouse Nanog WR domain (Fig.3). Despite this, it has been demonstrated that human NANOG can dimerize in vivo[47]. The highest percentage identity among mouse, human, chick, zebrafish, and axolotl Nanog sequences lies in the homeodomain, which is also the only structural element in Nanog that has been solved to date by X-ray crystallography[48].
Fig. 3.
Vertebrate Nanog orthologs have conserved domains. Highest sequence identities relative to mouse Nanog reside in the homeodomain. Sequence identity percentages calculated in Jalview. (ND, N-terminal domain; HD, homeodomain; WR, tryptophan-rich domain; CD, C-terminal domain, Full CD = CD1 + WR + CD2).
Dixon et al.[46] have confirmed that the WR domain of Nanog is required for maintaining pluripotency in mESCs. Sequence alignment with mouse Nanog revealed that axolotl Nanog does not contain a WR domain, but that it still contains a highly conserved homeodomain, a domain important for DNA binding (Figs. 2&3). Axolotl Nanog was also found not to form dimers, as measured by a protein complementation assay (PCA). This group also showed that induced axolotl Nanog dimerization is necessary and sufficient to support mouse ESC self-renewal in the absence of LIF.
III. Regulation of Nanog
III.1. Nanog regulation by transcription factors
Due to the multifaceted functions of Nanog in ESC self-renewal and pluripotency, it does not come as a surprise that Nanog is extensively and promiscuously regulated in ESCs. Indeed, many transcription factors are recruited to the Nanog locus to activate and/or repress Nanog expression (Table 3). Moreover, Nanog expression is primarily monoallelic and fluctuates among mESCs in standard serum/LIF culture conditions, unless cultured in the presence of inhibitors of MAPK and glycogen synthase kinase 3 (GSK3), a condition known as “2i”, with the addition of LIF (2i/LIF)[4, 49]. This suggests that signaling cascades also have important roles in regulating Nanog expression[49, 50]. Soon after Nanog was identified as an important factor for ESC self-renewal and pluripotency, much attention was focused on how the other core pluripotency factors regulate its gene expression. This led to the discovery that the proximal promoter region in the Nanog locus is responsible for most of the positive regulation of Nanog expression in mESCs[51, 52]. Not surprisingly, this region encompasses an Oct-Sox enhancer that is highly conserved among various mammalian species[51], demonstrating that Oct4 and Sox2 are major regulators of Nanog expression in mESCs.
Table 3.
Summary of Nanog regulators
| Factor | Mode of regulation | Effect on Nanog | References |
|---|---|---|---|
| Nanog | Transcriptional & epigenetic | Activator & repressor | [41,62,80,81, 83] |
| Zfp281 | Transcriptional & epigenetic | Repressor | [41,81] |
| Zfp143 | Transcriptional | Activator | [59] |
| Oct4 | Transcriptional & epigenetic | Activator | [51,52,56,59, 62, 73, 77, 80] |
| Sox2 | Transcriptional & epigenetic | Activator | [51,52,62,73, 75] |
| Klf4 | Transcriptional | Activator | [20,60,61,62] |
| Tcf3 | Transcriptional | Activator & repressor | [53, 63, 73] |
| Esrrb | Transcriptional | Activator | [54, 55, 56, 57, 58, 73] |
| Ncoa3 | Transcriptional | Activator | [57, 58] |
| Zic3 | Transcriptional | Activator | [82] |
| Cdx2 | Transcriptional | Repressor | [64] |
| Gcnf | Transcriptional | Repressor | [65] |
| Sp1 | Transcriptional | Activator | [66] |
| Sp3 | Transcriptional | Activator | [66] |
| Timp2 | Transcriptional (Nanog promoter-driven luciferase assay) | Activator | [67] |
| Hig2 | Transcriptional (Nanog promoter-driven luciferase assay) | Activator | [67] |
| Mki67ip | Transcriptional (Nanog promoter-driven luciferase assay) | Activator | [67] |
| Esrrg | Transcriptional (Nanog promoter-driven luciferase assay) | Activator | [67] |
| Dusp7 | Transcriptional (Nanog promoter-driven luciferase assay) | Activator | [67] |
| Spi1 | Transcriptional (Nanog promoter-driven luciferase assay) | Repressor | [67] |
| Prkaca | Transcriptional (Nanog promoter-driven luciferase assay) | Repressor | [67] |
| Jun | Transcriptional (Nanog promoter-driven luciferase assay) | Repressor | [67] |
| Tbx3 | Transcriptional | Activator | [20] |
| Stat3 | Transcriptional | Activator | [68, 73] |
| Brachyury | Transcriptional | Activator | [68] |
| PBAF complex | Transcriptional & epigenetic | Repressor | [73] |
| p53 | Transcriptional | Repressor | [74] |
| Sin3a/HDAC complex | Transcriptional & epigenetic | Activator & repressor | [74, 75] |
| Mof | Epigenetic | Activator | [76] |
| Wdr5 | Transcriptional & epigenetic | Activator | [77] |
| Ezh2 | Epigenetic | Repressor | [78] |
| Satbl | Transcriptional & epigenetic | Repressor | [79] |
| Satb2 | Transcriptional & epigenetic | Activator | [79] |
| NuRD/NODE complexes | Transcriptional & epigenetic | Repressor | [80, 81] |
Fine-tuning the expression of Nanog is also achieved via modulation of the recruitment and activity of additional transcription factors in response to specific cues. In fact, the Wnt signaling-responsive transcriptional regulator Tcf3 binds to an upstream regulatory region in the Nanog locus to down-regulate Nanog levels and to ensure proper differentiation[53]. Recently described as a major downstream target of Nanog in regulating many functions in mESCs and miPSCs[54, 55], Esrrb also directly binds to the Nanog locus and activates its transcription in collaboration with Oct4[56]. To further dissect the mechanism of Esrrb regulation of Nanog transcription, two groups independently demonstrated that the nuclear receptor coactivator 3, Ncoa3, binds directly to Esrrb, is recruited to the Nanog promoter, functions as a coactivator of Esrrb, and couples Esrrb to the basal transcription machinery by binding to RNA polymerase II[57, 58]. While Esrrb requires Oct4 for binding to the Nanog locus, the zinc finger protein Zfp143 stimulates Nanog transcription by modulating Oct4 binding[59]. Klf4 also binds to the distal and proximal promoter regions of Nanog to activate transcription in mESCs[60], and to the proximal promoter in hESCs to up-regulate NANOG levels[61].
Common among the core pluripotency factors is their reciprocal feedback loop regulation[62, 63]. For instance, the two Nanog target genes Sox2 and Oct4 can regulate Nanog expression. However, Nanog shares this feature with another transcription factor, Cdx2, which is not a pluripotency factor, but is instead a lineage specific marker. Indeed, Daley’s group demonstrated that Cdx2 can bind to the Nanog promoter in mESCs and repress its transcription[64]. Another way in which Nanog is down-regulated to trigger differentiation is via the transcriptional repressor Gcnf. Indeed, Gu et al.[65] showed that upon retinoic acid (RA)-induced differentiation, Gcnf binds to its DR0 consensus sequences located 2.5 kb upstream of the transcription start site, and in the 3’ untranslated region in the Nanog locus to directly reduce Nanog expression. To additionally confirm that not only ESC-specific factors, but also ubiquitously expressed transcription factors can regulate Nanog expression in pluripotent cells, Yao’s group[66] demonstrated that Sp1 and Sp3 bind to the Nanog proximal promoter and activate its transcription. To complement studies done on the endogenous Nanog locus, Abujarour et al.[67] utilized a luciferase reporter assay driven by the Nanog promoter coupled with a cDNA library screening. They identified several factors, such as Timp2, Hig2, Mki67ip, Esrrg, and Dusp7 that activated the reporter, and others including Spi1, Prkaca, and Jun that repressed it. However, they did not investigate whether these proteins could bind to the endogenous Nanog locus.
Finally, to properly regulate Nanog expression, several signaling transduction cascades come into play. For instance, Nanog is activated in response to LIF via two parallel pathways: the JAK/STAT3 pathway via Klf4 and the PI3K/AKT pathway via Tbx3[20]. Interestingly, STAT3 can also directly activate Nanog transcription by binding to an enhancer region upstream of the Nanog promoter, together with Brachyury[68]. Likewise, other signaling cascades, including FGF/MEK[69], GSK3β[70], and TGFβ[71, 72], as well as local changes in chromatin structures by chromatin remodeling complexes such as PBAF[73], are also important for maintaining Nanog levels in undifferentiated cells and for down-regulating Nanog in order to execute differentiation programs.
III.2. Nanog regulation by epigenetic factors
Chromatin modifiers can also modulate Nanog transcription in ESCs (Table 3). For example, the tumor suppressor p53 binds to the Nanog promoter, and upon RA-induced differentiation, recruits the Sin3a/histone deacetylase (HDAC) complex to reduce histone H3 acetylation and to directly repress Nanog expression in mESCs[74]. Interestingly, another group[75] demonstrated that the Sin3a/HDAC complex can activate Nanog when associated with Sox2. The H4 histone acetyltransferase Mof and the H3K4 methyltransferase MLL complex subunit Wdr5 have also been recently implicated in regulating Nanog activity[76, 77]. Unlike other HATs, Mof directly binds to and actively up-regulates transcription of Nanog[76]. Similarly, Wdr5 is recruited to the Nanog promoter in an Oct4-dependent manner to stimulate H3K4 trimethylation as well as to activate Nanog transcription[77]. Another epigenetic regulator, Ezh2, is involved in fine-tuning Nanog expression. In fact, even though the Nanog locus is not bivalent in mESCs, Ezh2 and its catalyzed trimethylation of histone H3 at K27 are both detectable by ChIP assays in the Nanog promoter in mouse ESCs and iPSCs, which inversely correlates with Nanog expression levels[78]. Modulation of higher order chromatin structure is also essential for stemness. In particular, the special AT-rich sequence-binding protein Satb1 binds to and negatively regulates the expression of Nanog. On the other hand, the related factor Satb2 is involved in the positive regulation of Nanog expression[79].
III.3. Nanog auto-regulation
Even though Oct4 and Sox2 protein levels are relatively stable in undifferentiated ESCs, Nanog protein levels fluctuate extensively[50]. Nanog itself can bind to its own promoter and regulate its own transcription either positively, by cooperating with Sox2 and Oct4 for instance, or negatively, by interacting with the transcriptional regulator Zfp281, bound to the NuRD repressor complex[41, 80, 81] (Table 3). Contrary to the common assumption that Nanog up-regulation requires Oct4 and Sox2, Lim et al.[82] found that the transcription factor Zic3 binds to the Nanog promoter in vitro and in vivo, and that it activates Nanog expression even in the absence of Oct4/Sox2 binding regions. Similarly, Navarro et al.[83] recently found that Nanog auto-repression, an endogenous negative feedback loop that prevents over-expression of Nanog, occurs independently of Oct4 and Sox2. This finding confirms our original report of Nanog auto- repression[81] and further emphasizes the dual role of Nanog in transcriptional regulation.
IV. Nanog Function in Stem Cell Pluripotency
The Smith group[84] has coined the term “ground state”, which refers to the pluripotent state of undifferentiated mESCs isolated from the naïve epiblast. Oct4 and Sox2 up-regulate Fgf4 levels, which in turn activates the MAPK pathway and poises ESCs for differentiation[85]. The combination of LIF and BMP4 is sufficient to maintain mESCs in vitro, but these factors are insufficient to block auto-inductive MAPK signaling[4]. In trying to recapitulate the ground state, they hypothesized that the blocking of lineage commitment by LIF and BMP4 was downstream of FGF4-mediated MAPK signaling. To test this, Ying et al.[4] cultured mESCs in 2i/LIF and found that they could be maintained indefinitely in serum- and feeder-free conditions. Nanog is crucial for ICM development, and therefore Nanog−/− embryos are unable to form viable epiblasts[9]. It was later found, however, that conditional deletion of Nanog in cultured mESCs rendered them more prone to differentiation, but that it did not compromise their cellular integrity or pluripotent status[50].
Nanog has been shown to be heterogeneously expressed in mESCs in culture[50, 62, 86]. A recent report[49] indicates that this may be explained by variable allelic expression of Nanog, corresponding to its expression pattern during early embryonic development. Nanog exhibited monoallelic expression from the two-cell blastomere stage to the early blastocyst stage. By the late blastocyst stage, however, Nanog expression transiently became biallelic, coinciding with establishment of the pluripotent ground state in the ICM. A subset of cells also underwent allelic switching of expression, which could explain the heterogeneic expression pattern of Nanog observed in mESCs. Culturing mESCs in 2i/LIF further confirmed that biallelic Nanog expression promotes the transition to ground state pluripotency, as this condition significantly increased the level of biallelic Nanog expression compared to the standard serum plus LIF condition. 2i/LIF treatment also enriched the Nanog locus for trimethylated lysine 4 on histone 3 (H3K4Me3), an active chromatin mark, as well as for RNA polymerase II. Nanog expression is thus controlled by chromatin modifications at each allele, which occurs during pre-implantation embryonic development.
V. Nanog Function in Somatic Cell Reprogramming
The initial report of iPSC generation by Takahashi and Yamanaka did not include Nanog as one of the four canonical reprogramming factors[87]. However, addition of Nanog to the Oct4, Sox2, Klf4, and c-Myc cocktail can enhance reprogramming kinetics in a predominantly cell division rate-independent manner[88]. In addition, initial reprogramming of human fibroblasts by Thomson’s group included NANOG along with OCT4, SOX2, and LIN28[89]. Nanog can also enhance fusion-based reprogramming[90] as well as mouse epiblast stem cell reprogramming[91].
Upon discovery that mESCs could be maintained in the absence of extrinsic factors, the Smith group set out to determine if the 2i condition could enhance iPSC generation. They found that the initial products of somatic cell reprogramming existed in a “pre-iPSC” state, resting on the threshold of pluripotency[17]. Pre-iPSCs exhibit qualities quite different from ESCs. For example, they incompletely express pluripotency markers, retain silencing of an X chromosome in female cells, are unresponsive to LIF, and are unable to contribute to chimeras. Remarkably, Silva et al.[17] found that serum- and feeder-free medium supplemented with 2i/LIF was able to drive pre-iPSCs towards ground state pluripotency to become bona fide iPSCs. Shortly afterward, this group[91] determined that Nanog is not required for the early stages of iPSC generation, but that it is required for the final transition from the pre-iPSC state to the fully induced ground state (Fig. 4). Using Nanog−/− neural stem cells and three (Oct4, Klf4, and c-Myc) of the four Yamanaka factors, they observed that 2i/LIF medium was insufficient for establishing ground state iPSCs. Contrastingly, upon addition of a floxed Nanog transgene as a reprogramming factor in the same conditions, pre-iPSCs were then able to fully transition to the ground state. Cre recombinase-mediated excision of the Nanog transgene had no effect on these iPSCs once pluripotency was established, as they were then able to contribute to chimeras. These results are also consistent with previous findings in Nanogflox/flox ESCs, wherein Nanog excision did not affect pluripotency[50]. Interestingly, it was recently shown that Esrrb, a direct downstream target of Nanog, can drive pre-iPSCs to the pluripotent ground state[54], further supporting the critical role of Nanog in establishing pluripotency. We also recently demonstrated that Nanog co-localizes with the methylcytosine hydroxylases Tet1 and Tet2 to a subset of pluripotency genes in mESCs, and that Nanog synergizes with these key epigenetic regulators during somatic cell reprogramming[92]. How Nanog precisely orchestrates the genetic and epigenetic events during the pre-iPSC to iPSC transition is only beginning to be defined.
Fig. 4.
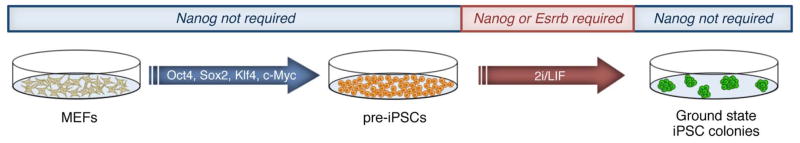
Nanog or its direct target Esrrb is required in the final stages of somatic cell reprogramming. Mouse embryonic fibroblasts (MEFs) transduced with the Yamanaka factors yield pre-iPSCs. Nanog is required in the pre-iPSC to ground state iPSC transition, as shown in red. Once the pluripotent ground state is established, Nanog is no longer required.
VI. Conclusions
Nanog dimers have been shown to be critical for maintenance of mESCs, however the specific functions of Nanog monomers and dimers in ESCs and during somatic cell reprogramming is not yet clear. Though it has been shown that human NANOG can form dimers in vivo, it is not yet known whether the dimeric form is sufficient for self-renewal of hESCs. Zfp281 preferentially interacts with Nanog dimers in mESCs, and is required for Nanog auto-repression. Interestingly, vertebrate Nanog orthologs can bind to and activate transcription of mouse Nanog target genes[45]. This and the fact that these orthologs can replace mouse Nanog in reprogramming Nanog−/− pre-iPSCs demonstrates a functional conservation among Nanog orthologs. Additional functional studies of Nanog orthologs in ESCs and iPSCs could create a novel platform for interrogating Nanog function. The lack of functional data regarding Nanog post-translational modifications emphasizes the importance of future studies designed to assess the implications of these modifications in regulating self-renewal and pluripotency. A number of Nanog phosphorylation sites have already been identified, yet it is unknown how phosphorylation or other modifications at these sites plays a role in Nanog protein-protein interactions or transcriptional activity.
iPSCs exhibit striking similarities to ESCs, that is, the capacity for unlimited self-renewal and multi-lineage differentiation. Although Nanog is crucial for the establishment of ground state pluripotency, it appears that it is not required for maintaining this state once it is established. Because of the positive and negative transcriptional activity that Nanog exerts, it is likely that Nanog cooperates with epigenetic activators or repressors to enhance the establishment of pluripotency. Further investigation into the interactions between Nanog and epigenetic regulators aside from Tet1 and Tet2 may point to additional synergistic effects on somatic cell reprogramming upon co-expression of Nanog and these regulators. Future work aimed at delineating the behavior of Nanog in ESCs and iPSCs will provide much needed mechanistic insights into the establishment and maintenance of pluripotency, and importantly, will enhance our understanding of the highly dynamic process of somatic cell reprogramming.
Acknowledgments
The work is funded by a grant from the NIH (1R01-GM095942-01A1), a grant from NY state Dept. of Health (NYSTEM#C026420), and a seed fund from the Black Family Stem Cell Institute to J.W.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. NATURE. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. PROC NATL ACAD SCI US A. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols J, Smith A. Naive and Primed Pluripotent States. STEM CELL. 2009;4(6):487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Ying Q-L, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. NATURE. 2008;453(7194):519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. NATURE. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 6.Smith AG, Heath JK, Donaldson DD, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. NATURE. 1988;336(6200):688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 7.Williams RL, Hilton DJ, Pease S, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. NATURE. 1988;336(6200):684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 8.Ying Q-L, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. CELL. 2003;115(3):281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 9.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. CELL. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 10.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. CELL. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson J, Itskovitz-Eldor J, Shapiro S. Embryonic stem cell lines derived from human blastocysts. SCIENCE. 1998 doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 12.Bradley A, Evans M, Kaufman MH, et al. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. NATURE. 1984;309(5965):255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 13.Tilgner K, Atkinson SP, Golebiewska A, et al. Isolation of primordial germ cells from differentiating human embryonic stem cells. STEM CELLS. 2008;26(12):3075–3085. doi: 10.1634/stemcells.2008-0289. [DOI] [PubMed] [Google Scholar]
- 14.Geijsen N, Horoschak M, Kim K, et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells : Abstract : Nature. NATURE. 2003 doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 15.Brons IGM, Smithers LE, Trotter MWB, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. NATURE. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 16.Najm FJ, Chenoweth JG, Anderson PD, et al. Isolation of Epiblast Stem Cells from Preimplantation Mouse Embryos. STEM CELL. 2011;8(3):318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva J, Barrandon O, Nichols J, et al. Promotion of Reprogramming to Ground State Pluripotency by Signal Inhibition. Goodell MA, ed. PLOS BIOL. 2008;6(10):e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallier L. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. JOURNAL OF CELL SCIENCE. 2005;118(19):4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 19.Bendall SC, Stewart MH, Menendez P, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. NATURE. 2007;448(7157):1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 20.Niwa H, Ogawa K, Shimosato D, et al. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. NATURE. 2009;460(7251):118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 21.Vallier L, Mendjan S, Brown S, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. DEVELOPMENT. 2009;136(8):1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Booth HAF, Holland PWH. Eleven daughters of NANOG3. GENOMICS. 2004;84(2):229–238. doi: 10.1016/j.ygeno.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Robertson M, Stenhouse F, Colby D, et al. Nanog retrotransposed genes with functionally conserved open reading frames. MAMM GENOME. 2006;17(7):732–743. doi: 10.1007/s00335-005-0131-y. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Wang X, Li M, et al. NANOGP8 is a retrogene expressed in cancers. FEBS J. 2006;273(8):1723–1730. doi: 10.1111/j.1742-4658.2006.05186.x. [DOI] [PubMed] [Google Scholar]
- 25.Pritsker M, Doniger TT, Kramer LC, et al. Diversification of stem cell molecular repertoire by alternative splicing. PROC NATL ACAD SCI USA. 2005;102(40):14290–14295. doi: 10.1073/pnas.0502132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomonis N, Schlieve CR, Pereira L, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. PROC NATL ACAD SCI USA. 2010;107(23):10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, Jena S, Levasseur DN. Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. JOURNAL OF BIOLOGICAL CHEMISTRY. 2011;286(49):42690–42703. doi: 10.1074/jbc.M111.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu B, Zhu W-G. Surf the post-translational modification network of p53 regulation. INT J BIOL SCI. 2012;8(5):672–684. doi: 10.7150/ijbs.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yates A, Chambers I. The homeodomain protein Nanog and pluripotency in mouse embryonic stem cells. BIOCHEM SOC TRANS. 2005;33(Pt 6):1518–1521. doi: 10.1042/BST0331518. [DOI] [PubMed] [Google Scholar]
- 30.Hornbeck PV, Kornhauser JM, Tkachev S, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. NUCLEIC ACIDS RESEARCH. 2012;40(Database issue):D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moretto-Zita M, Jin H, Shen Z, et al. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. PROC NATL ACAD SCI USA. 2010;107(30):13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramakrishna S, Suresh B, Lim K-H, et al. PEST Motif Sequence Regulating Human NANOG for Proteasomal Degradation. STEM CELLS AND DEVELOPMENT. 2011;20(9):1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- 33.Buckley SM, Aranda-Orgilles B, Strikoudis A, et al. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. CELL STEM CELL. 2012 doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho B, Olson G, Figel S, et al. Nanog increases focal adhesion kinase (FAK) promoter activity and expression and directly binds to FAK protein to be phosphorylated. JOURNAL OF BIOLOGICAL …. 2012 doi: 10.1074/jbc.M111.322883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourguignon LYW, Spevak CC, Wong G, et al. Hyaluronan-CD44 Interaction with Protein Kinase C Promotes Oncogenic Signaling by the Stem Cell Marker Nanog and the Production of MicroRNA-21, Leading to Down-regulation of the Tumor Suppressor Protein PDCD4, Anti-apoptosis, and Chemotherapy Resistance in Breast Tumor Cells. JOURNAL OF BIOLOGICAL CHEMISTRY. 2009;284(39):26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. CELL. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 37.Rossant J, Tam PPL. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. DEVELOPMENT. 2009;136(5):701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. NATURE. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Levasseur DN, Orkin SH. Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. PROC NATL ACAD SCI USA. 2008;105(17):6326–6331. doi: 10.1073/pnas.0802288105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullin NP, Yates A, Rowe AJ, et al. The pluripotency rheostat Nanog functions as a dimer. BIOCHEM J. 2008;411(2):227–231. doi: 10.1042/BJ20080134. [DOI] [PubMed] [Google Scholar]
- 41.Fidalgo M, Shekar PC, Ang Y-S, et al. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. STEM CELLS. 2011;29(11):1705–1716. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Chu J, Shen X, et al. An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. CELL. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z-X, Teh CH-L, Chan CM-Y, et al. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. STEM CELLS. 2008;26(11):2791–2799. doi: 10.1634/stemcells.2008-0443. [DOI] [PubMed] [Google Scholar]
- 44.Schuff M, Siegel D, Philipp M, et al. Characterization of Danio rerio Nanog and Functional Comparison to Xenopus Vents. STEM CELLS AND DEVELOPMENT. 2012;21(8):111003133356005. doi: 10.1089/scd.2011.0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theunissen TW, Costa Y, Radzisheuskaya A, et al. Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. DEVELOPMENT. 2011;138(22):4853–4865. doi: 10.1242/dev.068775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon JE, Allegrucci C, Redwood C, et al. Axolotl Nanog activity in mouse embryonic stem cells demonstrates that ground state pluripotency is conserved from urodele amphibians to mammals. DEVELOPMENT. 2010;137(18):2973–2980. doi: 10.1242/dev.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang DF, Tsai SC, Wang XC, et al. Molecular Characterization of the Human NANOG Protein. STEM CELLS. 2009;27(4):812–821. doi: 10.1634/stemcells.2008-0657. [DOI] [PubMed] [Google Scholar]
- 48.Jauch R, Ng CKL, Saikatendu KS, et al. Crystal Structure and DNA Binding of the Homeodomain of the Stem Cell Transcription Factor Nanog. JOURNAL OF MOLECULAR BIOLOGY. 2008;376(3):758–770. doi: 10.1016/j.jmb.2007.11.091. [DOI] [PubMed] [Google Scholar]
- 49.Miyanari Y, Torres-Padilla M-E. Control of ground-state pluripotency by allelic regulation of Nanog. NATURE. 2012 doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- 50.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. NATURE. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 51.Rodda DJ. Transcriptional Regulation of Nanog by OCT4 and SOX2. JOURNAL OF BIOLOGICAL CHEMISTRY. 2005;280(26):24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 52.Kuroda T, Tada M, Kubota H, et al. Octamer and Sox Elements Are Required for Transcriptional cis Regulation of Nanog Gene Expression. … AND CELLULAR BIOLOGY. 2005 doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira L, Yi F, Merrill BJ. Repression of Nanog Gene Transcription by Tcf3 Limits Embryonic Stem Cell Self-Renewal. MOLECULAR AND CELLULAR BIOLOGY. 2006 doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Festuccia N, Osorno R, Halbritter F, et al. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. CELL STEM CELL. 2012;11(4):477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martello G, Sugimoto T, Diamanti E, et al. Esrrb Is a Pivotal Target of the Gsk3/Tcf3 Axis Regulating Embryonic Stem Cell Self-Renewal. CELL STEM CELL. 2012;11(4):491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Berg DLC, Zhang W, Yates A, et al. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. MOLECULAR AND CELLULAR BIOLOGY. 2008;28(19):5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Percharde M, Lavial F, Ng J-H, et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. GENES & DEVELOPMENT. 2012;26(20):2286–2298. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Z, Yang M, Liu H, et al. Role of nuclear receptor coactivator 3 (ncoa3) in pluripotency maintenance. JOURNAL OF BIOLOGICAL CHEMISTRY. 2012;287(45):38295–38304. doi: 10.1074/jbc.M112.373092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, Fang F, Liou Y-C, et al. Zfp143 regulates Nanog through modulation of Oct4 binding. STEM CELLS. 2008;26(11):2759–2767. doi: 10.1634/stemcells.2008-0398. [DOI] [PubMed] [Google Scholar]
- 60.Zhang P, Andrianakos R, Yang Y, et al. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. JOURNAL OF BIOLOGICAL CHEMISTRY. 2010;285(12):9180–9189. doi: 10.1074/jbc.M109.077958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan KK-K, Zhang J, Chia N-Y, et al. KLF4 and PBX1 directly regulate NANOG expression in human embryonic stem cells. STEM CELLS. 2009;27(9):2114–2125. doi: 10.1002/stem.143. [DOI] [PubMed] [Google Scholar]
- 62.MacArthur Ben D, Sevilla A, Lenz M, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. NAT CELL BIOL. 2012;14(11):1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cole MF, Johnstone SE, Newman JJ, et al. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. GENES & DEVELOPMENT. 2008;22(6):746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L, Yabuuchi A, Eminli S, et al. Cross-regulation of the Nanog and Cdx2 promoters. NATURE PUBLISHING GROUP. 2009;19(9):1052–1061. doi: 10.1038/cr.2009.79. [DOI] [PubMed] [Google Scholar]
- 65.Gu P, LeMenuet D, Chung AC-K, et al. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. MOLECULAR AND CELLULAR BIOLOGY. 2005;25(19):8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 66.Wu DY, Yao Z. Functional analysis of two Sp1/Sp3 binding sites in murine Nanog gene promoter. CELL RES. 2006;16(3):319–322. doi: 10.1038/sj.cr.7310040. [DOI] [PubMed] [Google Scholar]
- 67.Abujarour R, Efe J, Ding S. Genome-wide gain-of-function screen identifies novel regulators of pluripotency. STEM CELLS. 2010;28(9):1487–1497. doi: 10.1002/stem.472. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki A, Raya A, Kawakami Y, et al. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. PROC NATL ACAD SCI USA. 2006;103(27):10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Santostefano KE, Hamazaki T, Pardo CE, et al. Fibroblast Growth Factor Receptor 2 Homodimerization Rapidly Reduces Transcription of the Pluripotency Gene Nanog without Dissociation of Activating Transcription Factors. JOURNAL OF BIOLOGICAL CHEMISTRY. 2012;287(36):30507–30517. doi: 10.1074/jbc.M112.388181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo Y, Lim CL, Nichols J, et al. Cell signalling regulates dynamics of Nanog distribution in embryonic stem cell populations. J R SOC INTERFACE. 2012 doi: 10.1098/rsif.2012.0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu R-H, Sampsell-Barron TL, Gu F, et al. NANOG Is a Direct Target of TGFß/Activin-Mediated SMAD Signaling in Human ESCs. CELL STEM CELL. 2008;3(2):196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galvin-Burgess KE, Travis ED, Pierson KE, et al. TGF-beta-Superfamily Signaling Regulates Embryonic Stem Cell Heterogeneity: Self-Renewal as a Dynamic and Regulated Equilibrium. STEM CELLS. 2012 doi: 10.1002/stem.1252. N/A–N/A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaniel C, Ang Y-S, Ratnakumar K, et al. Smarcc1/Baf155 Couples Self-Renewal Gene Repression with Changes in Chromatin Structure in Mouse Embryonic Stem Cells. STEM CELLS. 2009 doi: 10.1002/stem.223. N/A–N/A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. NAT CELL BIOL. 2005;7(2):165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 75.Baltus GA, Kowalski MP, Tutter AV, et al. A positive regulatory role for the mSin3A-HDAC complex in pluripotency through Nanog and Sox2. J BIOL CHEM. 2009;284(11):6998–7006. doi: 10.1074/jbc.M807670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X, Li L, Pandey R, et al. The Histone Acetyltransferase MOF Is a Key Regulator of the Embryonic Stem Cell Core Transcriptional Network. CELL STEM CELL. 2012;11(2):163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ang Y-S, Tsai S-Y, Lee D-F, et al. Wdr5 Mediates Self-Renewal and Reprogramming via the Embryonic Stem Cell Core Transcriptional Network. CELL. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Villasante A, Piazzolla D, Li H, et al. Epigenetic regulation of Nanog expression by Ezh2 in pluripotent stem cells. CELL CYCLE. 2011;10(9):1488–1498. doi: 10.4161/cc.10.9.15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savarese F, Dávila A, Nechanitzky R, et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. GENES & DEVELOPMENT. 2009;23(22):2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. NAT CELL BIOL. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 81.Fidalgo M, Faiola F, Pereira C-F, et al. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. PROC NATL ACAD SCI USA. 2012 doi: 10.1073/pnas.1208533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim LS, Hong FH, Kunarso G, et al. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. STEM CELLS. 2010;28(11):1961–1969. doi: 10.1002/stem.527. [DOI] [PubMed] [Google Scholar]
- 83.Navarro P, Festuccia N, Colby D, et al. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. THE EMBO JOURNAL. 2012:1–16. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Silva J, Smith A. Capturing Pluripotency. CELL. 2008;132(4):532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Niwa H. How is pluripotency determined and maintained? DEVELOPMENT. 2007;134(4):635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 86.Singh AM, Hamazaki T, Hankowski KE, et al. A heterogeneous expression pattern for Nanog in embryonic stem cells. STEM CELLS. 2007;25(10):2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 87.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. CELL. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 88.Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. NATURE. 2009;462(7273):595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. SCIENCE. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 90.Silva J, Chambers I, Pollard S, et al. Nanog promotes transfer of pluripotency after cell fusion. NATURE. 2006;441(7096):997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 91.Silva J, Nichols J, Theunissen TW, et al. Nanog is the gateway to the pluripotent ground state. CELL. 2009;138(4):722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Costa Y, Ding J, Theunissen TW, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. NATURE. 2013 doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]