Abstract
Previously we reported that Myd88 contributed to tumor progression. To begin to decipher what may be inducing Myd88 dependent signaling we focused on proteins that could function as damage associated molecular pattern molecules (DAMPs) since DAMPs have been reported to be secreted by tumors, and certain DAMPs mediate effects through toll-like receptors. A screen of mammary carcinoma for DAMP expression showed HMGB1 and HSP60 were significantly elevated relative to normal mammary epithelium, and targeting these DAMPs, or receptors for these DAMPs influenced growth of tumor cells. Moreover, analysis using a Myd88 inhibitory peptide suggested that HMGB1 mediated its effects in a Myd88 dependent manner, and inhibiting Myd88 function decreased HMGB1 and HSP60 gene expression. Collectively, these data suggest that HMGB1 and HSP60 contribute to growth of mammary carcinoma cells, HMGB1 accomplishes this, at least in part, through Myd88 dependent signaling, and these DAMPs are expressed in a Myd88 dependent manner.
Keywords: breast cancer, 4T1, HMGB1, HSP60, Myd88
1. Introduction
Previously we investigated the impact of treating tumor cells with toll-like receptor agonists. The data revealed that depending upon the source of agonist, length, and frequency of treatment, the result could be either increased or decreased tumor growth [1]. To decipher the mechanism of action we modulated toll-like receptor (TLR) and myeloid differentiation primary response gene 88 (Myd88) expression in a murine mammary carcinoma. The studies ultimately revealed that even in the absence of an added TLR agonist modulating Myd88 expression alone was sufficient to influence tumor growth and metastasis; tumors with reduced Myd88 levels exhibited decreased growth and metastasis [2]. Because of this we proposed that autocrine signaling in tumor cells may be responsible for the Myd88 dependent effects. Since tumor cells are known to secrete a range of damage associated molecular pattern molecules (DAMPs), and since some DAMPs are capable of mediating effects in a TLR dependent manner [3], we wanted to know which DAMPs were secreted by 4T1 tumor cells and whether modulating expression of these DAMPs would modulate growth of the tumor cells. We focused here on two DAMPs that exhibited elevated expression in the tumor cells relative to normal mammary epithelium; heat shock protein 60 (HSP60) and high mobility group box 1 protein (HMGB1).
HSP60 is normally localized in the mitochondria and is involved in protein folding [4]. However, HSP60 may also increase anti-tumor immunity by stimulating antigen presenting cells in a TLR dependent manner and driving an antigen-specific T cell response [5]. Osterloh et al. [6, 7] similarly reported that HSP60 could facilitate an immune response by enhancing antigen specific IFN-γ release from T cells. Yet, this same DAMP may also have adverse consequences in a tumor setting since HSP60 has been reported to increase survival signals, cellular transformation, and facilitate metastasis [8]. Work by Barazi et al. [9] revealed that HSP60 may facilitate metastasis by its ability to interact with, and activate α 3β 1 integrin.
HMGB1 is normally localized in the nucleus and is involved in DNA folding and transcriptional regulation [10]. Similar to HSP60, HMGB1 can be beneficial to an immune response when released from cells. For example, HMGB1 has been correlated with driving dendritic cell maturation and Th1 responses [11]. Subsequent work by Dumitriu et al. [12] confirmed that HMGB1 could induce dendritic cell maturation and an antigen specific T cell response, and they revealed that HMGB1 mediated maturation of dendritic cells in a RAGE dependent manner. Yet, also like HSP60 there may be adverse consequences of HMGB1 in a tumor setting. For instance, Wild et al. [13] revealed that HMGB1 could enhance regulatory T cell (Treg) functions, results which were confirmed by others [14]. Moreover, HMGB1 has been shown to contribute to proliferation and invasiveness of cancer cells [15–16]. Because of these varying functions, delineating the conditions under which tumor-derived HSP60 and HMGB1 could contribute to anti-tumor immunity versus tumor progression is warranted.
Here we began investigating the role of these DAMPs and Myd88 in tumor-derived chemokine expression and mammary carcinoma cell growth. We report that suppression of HMGB1 and HSP60 using RNA interference significantly influenced growth of the 4T1 tumor cells. Because of their important roles in normal cellular functions we used blocking antibodies to determine whether these DAMPs were mediating effects in an autocrine manner. Indeed, antibodies to HMGB1 or HSP60 also inhibited growth of the tumor cells. Analysis using a Myd88 inhibitory peptide suggested that HMGB1 mediated effects in a Myd88 dependent manner, and inhibition of Myd88 dependent signaling resulted in decreased HMGB1 and HSP60 gene expression. Collectively, these data showed that mammary carcinoma overexpress HMGB1 and HSP60 relative to normal mammary epithelium, their expression influences tumor cell growth, and the effects of HMGB1 are dependent at least in part upon Myd88. Since turning off Myd88 signaling also decreased HMGB1 and HSP60 gene expression, these data further revealed an intricate relationship between HMGB1, HSP60 and Myd88 in the growth of murine mammary carcinoma cells in vitro.
2. Materials and methods
2.1 Cells
4T1, EMT6, 168 and SM1 murine mammary carcinoma cells were maintained in complete RPMI (cRPMI) (RPMI 1640, Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (Lonza), glutamine (2mM, Lonza), penicillin (100U/mL, Lonza), streptomycin (100ug/mL, Lonza), nonessential amino acids (Sigma, St. Lois, MO), 2-mercaptoethanol (5×10−5 M, Sigma), and sodium pyruvate (1mM, Lonza). The cells and experiments were maintained at 37°C, 5% CO2. All cell counts were conducted by hemacytometer and viability was assessed by trypan blue exclusion.
2.2 Quantitative RT-PCR
Gene expression was analyzed by Quantitative RT-PCR (QRT-PCR). First, mRNA was isolated from 1×106 cells using an mRNA isolation kit (Life Technologies, Grand Island, NY). All of the mRNA was precipitated, resuspended in 25ul DEPC treated H2O, and used for cDNA synthesis. RNA from normal mammary epithelium was purchased from Clontech (Mountain View, CA). Complementary DNA was generated using random hexamer primers (0.5ug, Promega, Madison, WI), dithiothreitol (2mM, Promega), dNTP (0.2mM each, Promega) and 200 units of M-MLV reverse transcriptase (Promega) in a 1 hour reaction at 42°C. An aliquot (0.5ul) of cDNA was amplified in a reaction with 1 × iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), and 200nM gene specific primers. Each reaction was run in duplicate or triplicate. The reaction conditions consisted of 40 cycles of a two-step PCR reaction with 94°C for 10 sec., and 68°C for 30 sec., on an iQ5 Real Time PCR Detection System (Bio-RAD). Gene specific primers included gapdh left 5′-cttccgtgttcctacccccaatgt-3′, gapdh right 5′-gcctgcttcaccaccttcttgatgt-3′, hmgb1 left 5′–cctccttcggccttcttcttgttc-3′, hmgb1 right 5′-catagggctgcttgtcatctgctg-3′, s100a8 left 5′-ttcgaggagttccttgcgatgg-3′, s100a8 right 5′-agccctaggccagaagctctgcta-3′, hsp70a1b left 5′-acgccaacggcatcctgaac-3′, hsp70a1b right 5′-ctgcaccatgcgctcgatct-3′, hsp70a1a left 5′-acgccaacggcatcctgaac-3′, hsp70a1a right 5′-tgcaccatgcgctcgatctc-3′, hsp70a4 left 5′-caccagcattctctgcctgtgg-3′, hsp70a4 right 5′-ctgcgctctgcactctcctagtgtt-3′, hsp60 left 5′-ccgaagacgttgacggagaggc-3′, hsp60 right 5′-tgactgccacaacctgaagaccaa-3′, ccl2 left 5′-tcatgcttctgggcctgctgt-3′, ccl2 right 5′-ctcattgggatcatcttgctggtg-3′, mmp9 left 5′-tcgggaaggctctgctgttca-3′, mmp9 right 5′-tccacgcgaatgacgctctg-3′, tlr2 left 5′-cgttcaaggaggtgcggactgt-3′, tlr2 right 5′-cggtgatgcaattcggatgct-3′, tlr4 left 5′-agtgccccgctttcacctctg-3′, tlr4 right 5′-caataaccttccggctcttgtgga-3′, rage left 5′-ttccctcctccttccagccact-3′, rage right 5′-tgacctccttccctcgcctgtt-3′, and E-cadherin left 5′-caaatgcctgctcctgatggtagc-3′, E-cadherin right 5′-tctgactgcctctgcctcctga-3′. The primers were synthesized by Integrated DNA Technologies (Coralville, IA) and analyzed for specificity with the NCBI Blast Program. Standard curves were used to examine efficiency and reproducibility of each reaction, and melt curves were used to validate amplification of single products. The housekeeping gene gapdh was used to establish normalized expression (ΔΔCT).
2.3 siRNA treatment
For siRNA treatment tumor cells were cultured in cRPMI (without antibiotics) in 24-well culture dishes (Corning, Corning, NY) at 1×104 cells/well. After 24 hours, the media was replaced. For siRNA delivery 20, 40 or 80pmole of siRNA was mixed with Optimem (Life Technologies) for a final volume of 50ul in a sterile microcentrifuge tube and incubated for 5 min. at room temperature. In a separate microcentrifuge tube 12ul oligofectamine (Invitrogen) was mixed with 3ul Optimem, and incubated at room temperature for 5 min. The contents of the tubes were mixed, incubated at room temperature for 20 min., and then added to the cells which were harvested 48 hours later for analysis. To determine optimal gene inhibition conditions 80pmole of siRNA was used while the amount of oligofectamine was varied (3, 6 or 12ul of oligofectamine was mixed with 12, 9 or 3ul Optimem) and the cells were treated for 24, 48 or 72 hours. The scrambled control and Myd88 specific siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
2.4 Antibody treatment
For antibody treatment the tumor cells were cultured in cRPMI in 24-well culture dishes (Corning) at 1×104 cells/well. To neutralize HMGB1 and HSP60 2.5ug of antibody specific for these proteins or an isotype control was added, and the cells were cultured for 24 to 72 hours before analysis. For the 48 and 72 hour time points 2.5ug of antibody was added daily. Similar conditions were used to neutralize TLR2, TLR4 and RAGE. The anti-RAGE antibody was purchased from Millipore (Temecula, CA). All other antibodies, and the isotype controls were purchased from Santa Cruz Biotechnology.
2.5 ELISA
ELISAs were used to quantitate secretion of proinflammatory mediators. For this purpose, supernatants were harvested from cells treated with DAMP specific antibodies, centrifuged at 350×g to remove particulate materials, and stored at −20°C. For analysis, samples were assayed for CCL2 and pro-MMP9 using Quantikine Sandwich ELISAs (R&D Systems, Minneapolis, MN). HMGB1 and HSP60 secretion were analyzed from supernatants harvested from 1×106 tumor cells cultured for 24 hours in a single well of a 24 well plate. For analysis, samples were assayed for HMGB1 and HSP60 using specific ELISAs (Novateinbio, Cambridge, MA).
2.6 Cell cycle analysis
To determine whether neutralizing the DAMPs impacted progression of cells through the cell cycle propidium iodide staining was used. For this purpose 1×105 cells were cultured in T75 tissue culture flasks (Corning) with 10ml cRPMI and 2.5ug antibody/ml. Another dose of antibody was added at 24 hours, and then the cells were harvested at 48 hours for cell cycle analysis. Following a wash with 10ml cold phosphate buffered saline (PBS), the cells were resuspended in 200ul cold PBS, and then slowly added to 4ml cold 70% ethanol while vortexing. Following a 90 min. incubation on ice, the cells were centrifuged at 450×g, resuspended in 500ul PI/RNase solution (BD Biosciences, San Jose, CA), and sent to Hershey Medical Center (Hershey, PA) for analysis.
2.7 Apoptosis analysis
To determine whether neutralizing DAMPs induced apoptosis annexin V staining was used. For this purpose tumor cells were cultured in cRPMI in 24-well culture dishes (Corning) at 1×104 cells/well on poly-L-lysine coated coverslips (BD Biosciences) with 2.5ug antibody/well. Another dose of antibody was added at 24 hours, and then cells were analyzed at 48 hours. Two hours before staining the positive controls were treated with staurosporine (2uM, Fisher Scientific, Pittsburg, PA). To stain for apoptosis the media was removed and then 500ul apoptosis binding buffer and 5ul annexin V Alexa Fluor 488 (Life Technologies) were added to each well. Following a 30 min. incubation at 4°C in the dark, the cells were washed with Hanks Balanced Salt Solution (HBSS, Lonza) twice and analyzed using confocal microscopy (Confocal Microscope C1, Nikon Instruments, Melville, NY).
2.8 Peptide treatment
A Myd88 specific inhibitory peptide was used in combination with HMGB1 and HSP60 neutralizing antibodies to determine whether the DAMPs mediated effects in a Myd88 dependent manner. For this purpose tumor cells were cultured in cRPMI in 24-well culture dishes at 1×104 cells/well. To neutralize HMGB1 and HSP60 2.5ug of antibodies specific for these proteins or an isotype control was added at time 0 and 24 hours, and the cells were cultured for 48 hours before analysis. In addition to the antibodies, 100uM of a Myd88 specific inhibitory peptide or 100uM control peptide lacking the Myd88 binding domain (Imgenex, San Diego, CA) was added. The cells were harvested following 48 hours of treatment and evaluated for cell number and viability.
2.9 Analysis of genes modulated by inhibiting Myd88-dependent signaling
To determine how targeting the Myd88 signaling cascade influenced DAMP expression 4T1 were treated with 100uM of the Myd88 inhibitory or control peptide for 48 hours, and alterations in gene expression were assessed using the RT2 Profiler PCR Array (Qiagen, Valencia, CA). RNA isolation, cDNA synthesis, and PCR arrays were carried out according to manufacturer’s suggestions. Briefly, following 48 hours of treatment, the cells were harvested and total RNA was isolated and treated with RNase-free DNase using the RNAeasy kit (Qiagen). Complementary DNA was synthesized using 8ul of total RNA and subsequently amplified by quantitative PCR on an iQ5 Real Time PCR Detection System (Bio-RAD). The housekeeping gene gapdh was used to establish normalized expression (ΔΔCT).
2.10 Western blot analysis
To determine how targeting the Myd88 signaling cascade influenced DAMP protein expression 4T1 were treated with 100uM of the Myd88 inhibitory or control peptide for 48 hours, and alterations in protein expression were assessed by western blot. For this purpose an equivalent number of cells were washed 3 times with ice-cold PBS, resuspended in 150uL of buffer A (10mM Hepes (Sigma), 10mM KCl (Sigma), 0.1mM EDTA (Sigma), 0.1mM EGTA (Sigma)), supplemented with the protease inhibitors aprotinin, leupeptin, chymostatin, and pefabloc (Roche Molecular Biochemicals, Indianapolis, IN) and placed on ice. Following a 15 min. incubation, 10uL of 10% Nonidet P-40 (Sigma) was added. The samples were vortexed for 10 sec., centrifuged 12,000×g at 4°C for 1 min, and supernatants containing the cytoplasmic proteins were collected. The pellets were washed one time with buffer A and after adding 50ul buffer C (25% glycerol, 20mM Hepes, 0.4M NaCl, 1.0mM EDTA, 1.0mM EGTA), supplemented with the protease inhibitors aprotinin, leupeptin, chymostatin, and pefabloc the samples were sonicated for 30 sec. on ice. Following a 10 min. centrifugation at 12,000×g at 4°C the nuclear proteins were collected. NuPAGE LDS sample buffer (Invitrogen) was added to the proteins and the samples were stored at −20°C. SDS PAGE gels (12.5%, Invitrogen) were loaded with 15uL of proteins, electrophoresed, and transferred to PVDF membranes (Invitrogen). The membranes were blocked in PBS with 5% powdered milk and 0.05% Tween 20 (Sigma) for two hours. Primary antibodies (10ug) specific for actin, HMGB1, or HSP60 (Santa Cruz Biotechnology, Santa Cruz, CA) were added, and the blots were incubated at room temperature for 2 hours and then overnight at 4°C. After washing 2 times with blocking buffer the following day, a horse-radish-peroxidase-conjugated secondary antibody (Santa Cruz) was added and the blots were incubated for 1 hour at room temperature. Following 4 washes, proteins were visualized by enhanced chemiluminesence on an Alpha Innotech Gel Documentation System (Alpha Innotech Corp., San Leandro, CA).
3. Results
3.1 Screening DAMP expression by tumor cells
The first step in determining whether tumor-derived DAMPs were mediating autocrine signaling in a Myd88 dependent manner was to determine which DAMPs were expressed by the tumor cells. For this purpose we screened four different murine mammary carcinoma lines for DAMP expression using QRT-PCR and compared this to DAMP expression in normal mammary epithelium. Of the seven different DAMPs that were investigated only S100A9 was not expressed at appreciative levels (data not shown). Six other DAMPs were expressed in all four of the tumor cell lines and normal mammary epithelium (Fig. 1). Of these DAMPs only HMGB1 and HSP60 exhibited a relative expression of ≥ 2 in all four of the cancer cell lines relative to normal mammary epithelium. Since E-cadherin expression is often decreased in breast cancer [17] we included analysis of this gene in order to validate that its expression would be greater in normal mammary epithelium than the tumors, which was indeed the case (Fig. 1). Collectively, these data suggested that HMGB1 and HSP60 warranted further study.
Fig. 1.
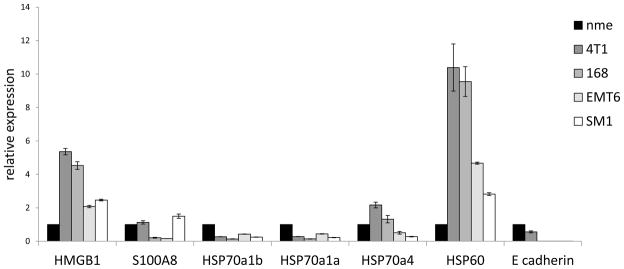
DAMP mRNA expression among murine mammary carcinoma. Messenger RNA was isolated from tumor cells, converted to cDNA, and used to screen for DAMP expression in four different mammary carcinomas by QRT-PCR using gapdh as the reference gene. Data were normalized to DAMP expression in normal mammary epithelium (nme). All data represent the average and standard error of three separate experiments.
3.2 siRNA targeting both HMGB1 and HSP60 inhibits tumor cell growth and CCL2 and MMP-9 expression
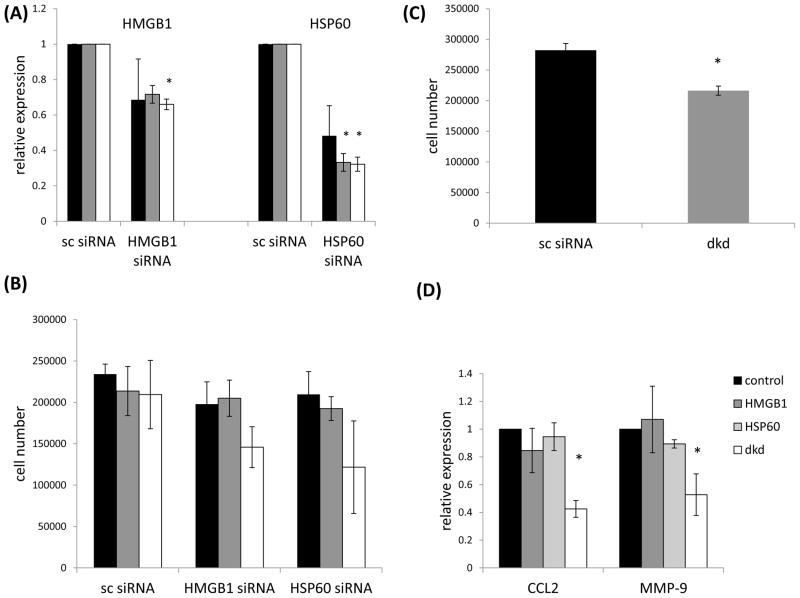
To determine whether HMGB1 and HSP60 influenced growth of the tumor cells we used RNA interference to target these DAMPs. Initial dose titration analysis showed that 20, 40 and 80 pmole of siRNA could down regulate HMGB1 and HSP60 mRNA levels relative to control siRNA, and the highest dose of siRNA led to a decrease in cell number (Figs. 2A, 2B). While 209,416 +/− 41,280 cells were recovered following treatment with 80pmole of control siRNA, 145,833 +/− 24,710, and 121,666 +/− 55,941 cells were recovered from the HMGB1 and HSP60 siRNA treated cells respectively (Fig. 2B). Due to the high standard error these decreases were not statistically significant. However, we did find a significant decrease in cell number when both HMGB1 and HSP60 were targeted together. After targeting both HMGB1 and HSP60 216,250 +/− 7,447 cells were recovered compared to 282,187 +/− 11,091 cells following control siRNA treatment (Fig. 2C). Since we previously showed that targeting Myd88 led to a significant decrease in CCL2 and CCL5 expression [2] we wanted to know whether inhibiting HMGB1 and HSP60 would have a similar effect. While downregulating the DAMPs had no impact on CCL5 expression (data not shown), CCL2 was significantly down regulated following treatment of the tumor cells with both HMGB1 and HSP60 siRNA (double knockdown (dkd), Fig. 2D). We also observed a significant reduction in MMP-9 gene expression (Fig. 2D).
Fig. 2.
Inhibiting HMGB1 and HSP60 expression using RNA interference decreases tumor cell number. (A) 4T1 were treated with (20 (■), 40 (■) or 80 (□) pmole) siRNA specific for HMGB1 or HSP60 and screened for expression of these genes by QRT-PCR using gapdh as the reference gene. Data were normalized to DAMP expression in cells treated with scrambled siRNA (sc siRNA). (B) Forty eight hours after siRNA treatment the cells were harvested and counted. (C) Cells treated with both HMGB1 and HSP60 siRNA (double knockdown (dkd)) exhibited a small yet significant reduction in number. (D) Cells treated with siRNA specific for HMGB1, HSP60 or both (dkd) were screened for CCL2 and MMP9 gene expression by QRT-PCR using gapdh as the reference gene. Where indicated (*) p < 0.05 using Student’s t-Test relative to control. All data represent the average and standard error of three separate experiments.
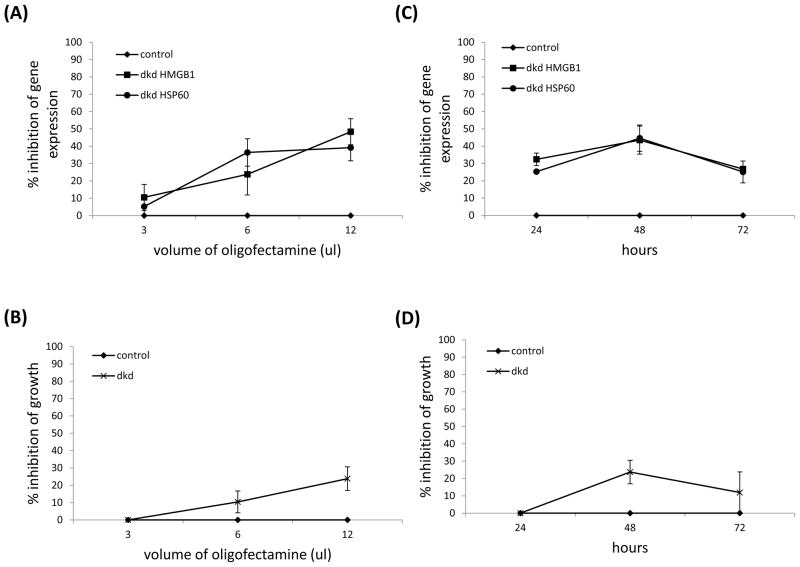
In order to optimize inhibition of gene expression we next varied the dose of the lipid transfection reagent (oligfectamine) and the length of siRNA treatment. The results showed that 12ul of oligofectamine and 48 hours of treatment with both siRNA led to the greatest reduction in gene expression and cell number (Fig. 3). These data revealed a clear relationship between DAMP expression and cell growth; lower levels of HMGB1 and HSP60 mRNA resulted in lower cell numbers. Collectively, these data revealed that targeting mRNA encoding HMGB1 and HSP60 could impact tumor cell number, and CCL2 and MMP-9 expression, but only in a significant manner when both DAMPs were targeted.
Fig. 3.
Inhibiting both HMGB1 and HSP60 expression using RNA interference correlates with decreased cell number. 4T1 were treated with 80pmole of siRNA using different amounts of the lipid transfection reagent oligofectamine and screened for success of gene knockdown using QRT-PCR with gapdh as the reference gene (A), or percent inhibition of growth (B) after 48 hours of treatment. 4T1 were treated with 80pmole of siRNA using 12ul oligofectamine and screened for success of gene knockdown using QRT-PCR with gapdh as the reference gene (C), or percent inhibition of growth (D) after 24, 48 or 72 hours of treatment. All data were normalized to DAMP expression in cells treated with scrambled siRNA (control), and represent the average and standard error of three separate experiments.
3.3 Neutralizing HMGB1 and HSP60 influences tumor cell growth, but not CCL2 or MMP9 secretion
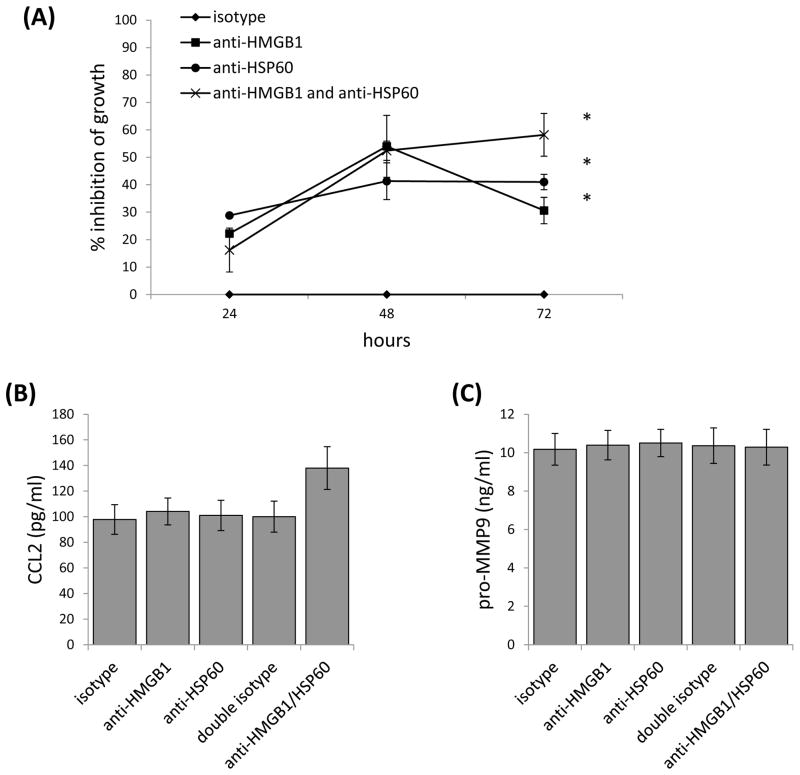
Since HMGB1 and HSP60 have important normal cellular functions we wanted to know whether we could similarly influence growth and chemokine expression using HMGB1 and HSP60 neutralizing antibodies. An initial dose titration analysis using 2.5, 5.0, 10.0 or 20.0ug/ml of antibody showed that 2.5ug of the antibody had the greatest effect on cell number. Unlike the RNA interference approach, targeting individual DAMPs with neutralizing antibodies led to a significant inhibition of tumor cell growth compared to controls (Fig. 4A). Treatment with antibody to HMGB1for 48 hours resulted in 54% inhibition of growth, and treatment with antibody to HSP60 resulted in 41% inhibition of growth (Fig. 4A). Maximum inhibition of growth when both antibodies were added was seen at 72 hours of treatment with 58% inhibition of growth which was significantly greater than that achieved with antibody to HMGB1 alone (30% inhibition), but not HSP60 alone (41% inhibition) at this time point (Fig. 4A). Another difference we found using neutralizing antibodies versus RNA interference was that neither CCL2 nor MMP-9 expression were significantly impacted by neutralizing these DAMPs (Figs. 4B, 4C). Collectively, these data confirmed that HMGB1 and HSP60 could influence growth of the tumor cells in an autocrine manner, but not CCL2 or MMP-9 secretion.
Fig. 4.
Neutralizing HMGB1 and HSP60 using antibodies impacts cell number but not cytokine expression. (A) 4T1 were treated with antibodies to HMGB1, HSP60 or both and then analyzed for inhibition of growth after 24 to 72 hours. Data were normalized to 4T1 treated with an isotype control. Supernatants harvested from the 48 hour time points were screened for CCL2 (B) and pro-MMP9 (C) levels by ELISA. Where indicated (*) p < 0.05 using Student’s t-Test relative to the isotype control. All data represent the average and standard error of three to six separate experiments.
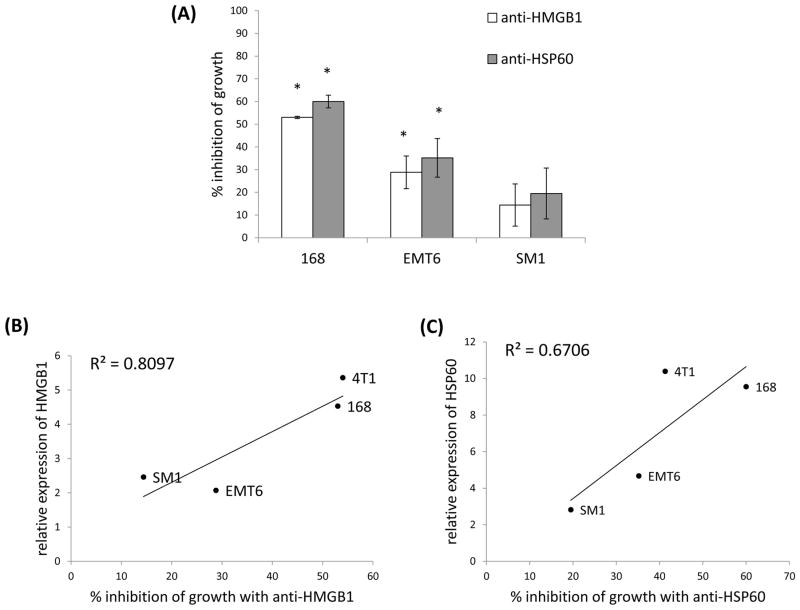
To determine whether these findings were limited to the 4T1 tumor cell line we screened 3 additional mammary carcinomas for effects of neutralizing HMGB1 and HSP60. Similar to 4T1, when antibodies to HMGB1 or HSP60 were added to 168 or EMT6 tumor cell cultures there was significant inhibition of growth (Fig. 5A). Although the tumor cell line SM1 also exhibited reduced growth in the presence of these antibodies, the inhibition was not statistically significant (Fig. 5A). Because these tumor cell lines expressed different levels of HMGB1 and HSP60 (Fig. 1), and exhibited different levels of inhibition of growth in the presence of the neutralizing antibodies, we looked at whether there was a correlation between the two. Significantly, the two correlated fairly well together (Figs. 5B, 5C). In essence, the more HMGB1 or HSP60 the tumor cell line made the more susceptible the tumor cell line was to inhibition of growth when treated with DAMP specific neutralizing antibodies. These data support the contention that HMGB1 and HSP60 could function as autocrine growth factors for murine mammary carcinoma.
Fig. 5.
The ability to inhibit tumor cell number achieved with neutralizing HMGB1 and HSP60 antibodies correlated with the amount of HMGB1 or HSP60 expressed by the tumors. (A) Three additional murine mammary carcinomas were treated with antibodies to HMGB1 or HSP60 and then analyzed for inhibition of growth after 48 hours. Data were normalized to cells treated with an isotype control. Where indicated (*) p < 0.05 using Student’s t-Test relative to the isotype control. Relative expression of HMGB1 (B) or HSP60 (C) in four different mammary carcinoma lines were correlated with the amount of inhibition of growth that could be achieved with DAMP specific neutralizing antibodies. All data represent the average and standard error of three separate experiments.
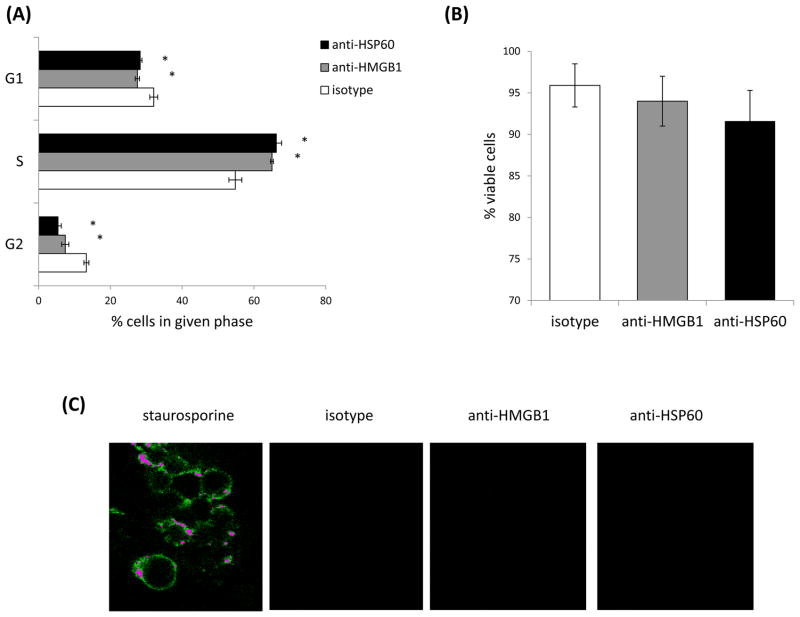
3.4 Inhibition of HMGB1 and HSP60 leads to an alteration in the cell cycle, not cell death
The inhibition of growth evident upon treatment with neutralizing antibodies specific for HMGB1 or HSP60 may be attributed to an increase in cell death and/or an alteration in the cell cycle. To determine whether there was an alteration in cell cycle we examined whether treatment with antibodies to HMGB1 or HSP60 modulated the percentage of cells in different stages of the cell cycle. Interestingly, there was a small, yet significant increase in the percentage of cells in the S phase of the cell cycle, and a corresponding decrease in the percentage of cells in the G1 and G2 phases of the cell cycle upon treatment with antibodies specific for HMGB1 or HSP60 (Fig. 6A). To determine whether neutralizing the DAMPs influenced cell death we examined viability and annexin V staining (as a marker for cells undergoing apoptosis) following treatment with the antibodies. While there was a small drop in cell viability following treatment with the antibodies, the decrease was not statistically significant (Fig. 6B), and none of the cells stained with annexin V (Fig. 6C). These data suggested that neutralizing HMGB1 or HSP60 could influence progression of the cells through the cell cycle, but not apoptotic cell death.
Fig. 6.
Neutralizing HMGB1 or HSP60 alters the cell cycle, not cell death. 4T1 were treated with antibodies to HMGB1 or HSP60 for 48 hours and then analyzed for cell cycle by PI/RNase staining (A), viability by trypan blue exclusion (B), or apoptosis by annexin V staining (C). All data represent the average and standard error of three separate experiments. Where indicated (*) p < 0.05 using Student’s t-Test relative to the isotype control.
3.5 Targeting DAMP receptors also influences growth of the tumor cells
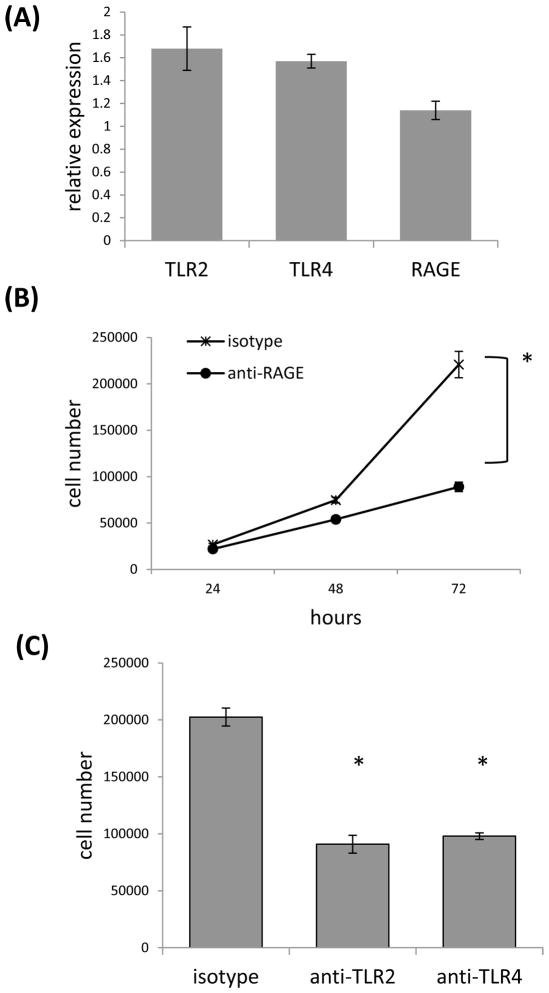
If HMGB1 and HSP60 were influencing tumor cell growth in an autocrine manner then blocking receptors for these DAMPs should also interfere with cell growth. For this reason we screened expression of TLR2, TLR4 and receptor for advanced glycation end product (RAGE), which have been reported to function as receptors for HMGB1 and HSP60 [12, 18, 19]. Analysis of 4T1 cells by QRT-PCR showed expression of all three receptors (Fig. 7A), and treatment of tumor cells with RAGE, TLR2 or TLR4 specific neutralizing antibodies significantly affected growth of the tumor cells. Maximum inhibition of growth following treatment with anti-RAGE antibody was achieved at 72 hours with 89,027 +/− 5,058 cells recovered compared to 220,833 +/− 14,177 cells recovered after treatment with the isotype control (Fig. 7B). Similarly, antibodies to TLR2 and TLR4 significantly impacted growth of the tumor cells relative to treatment with the isotype control. After 72 hours of treatment with the isotype control there were 202,500 +/− 7,877 cells, whereas 90,833 +/− 7,813 and 97,916 +/− 2,902 cells were recovered following treatment with TLR2 and TLR4 specific antibodies respectively (Fig. 7C). These data help support the contention that HMGB1 and HSP60 can modulate growth of the tumor cells in an autocrine manner.
Fig. 7.
Neutralizing antibodies specific for TLR2, TLR4 or RAGE significantly inhibits cell number. (A) QRT-PCR was used to assess TLR2, TLR4 and RAGE gene expression using gapdh as the reference gene. (B) Anti-RAGE specific antibody was added to 4T1 cultures and cells were counted at 24 to 72 hours. (C) Anti-TLR-2 or TLR-4 specific antibody was added to 4T1 cultures and cells were counted at 72 hours. Where indicated (*) p < 0.05 using Student’s t-Test relative to the control.
3.6 Relationship between HMGB1, HSP60 and Myd88
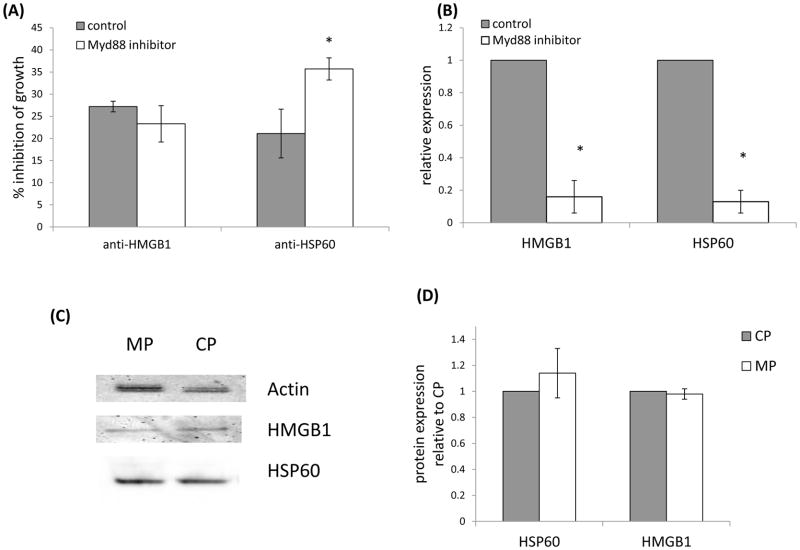
Finally, since we initiated this study to investigate proteins that may be mediating affects in a Myd88 dependent manner, and since we previously we reported that treatment with a Myd88 inhibitory peptide resulted in decreased growth of the tumor cells [2], we asked whether targeting HMGB1 or HSP60 could enhance the inhibitory effect of targeting Myd88 alone. If the DAMPs mediated affects in a Myd88 independent manner then targeting both the DAMP and Myd88 should result in enhanced inhibition of growth. The data revealed that the inhibition of growth evident by neutralizing HMGB1 was not enhanced when Myd88 was also targeted, whereas the inhibition of growth evident upon neutralizing HSP60 was enhanced when Myd88 was also targeted (Fig. 8A). These data suggest that HMGB1 mediates effects in a Myd88 dependent manner, whereas HSP60 mediates effects in a Myd88 independent manner. Finally, we found that turning off the Myd88 signaling cascade using the Myd88 inhibitory peptide also decreased HMGB1 and HSP60 gene expression. When tumor cells were treated with the Myd88 inhibitory peptide for 48 hours there was an 84% inhibition of HMGB1 gene expression, and 87% inhibition of HSP60 gene expression (Fig. 8B). However, despite the alteration in gene expression we did not find a decrease in protein expression (Figures 8C, 8D) perhaps related to the slow turnover rate of these proteins [20, 21]. Regardless, these data suggest that Myd88 contributes to expression of mRNA encoding HMGB1 and HSP60, and the decreased growth evident upon blocking HMGB1 is at least in part mediated through the Myd88 signaling pathway.
Fig. 8.
Relationship between HMGB1, HSP60 and Myd88. (A) A Myd88 inhibitory peptide (Myd88 inhibitor) or control peptide (control) was added to 4T1 cultures and the percent inhibition of growth was determined after 48 hours in the presence or absence of antibody to HMGB1 or HSP60. (B) The Myd88 inhibitory peptide (Myd88 inhibitor) or control peptide (control) were added to 4T1 cultures and HMGB1 and HSP60 gene expression were assessed after 48 hours. The data represent the average and standard error of three to six separate experiments. Where indicated (*) p < 0.05 using Student’s t-Test relative to the control. (C) Cytoplasmic and nuclear lysates were examined for HSP60 and HMGB1 protein levels respectively following treatment with the Myd88 inhibitory peptide (MP) or control peptide (CP). (D) The average and standard error of the densitometry data from three separate experiments are shown.
Discussion
Initially we began this study to determine whether DAMPs were responsible for the effects we previously attributed to Myd88; particularly decreased growth and chemokine expression [2]. We found that HMGB1 and HSP60 were overexpressed by murine mammary carcinomas and that siRNA specific for these DAMPs, as well as neutralizing antibodies specific for them, or their receptors were capable of decreasing tumor cell growth, but not chemokine expression.
Interestingly, targeting both HMGB1 and HSP60 with siRNA resulted in a significant decrease in CCL2 and MMP-9 expression which was not evident when the cells were treated with neutralizing antibodies. These data may be attributed to “off-target” effects of the siRNA, or a stress response brought about in the cells due to a decrease in two important proteins; HMGB1 being necessary for DNA folding and transcriptional regulation [10], and HSP60 being necessary for protein folding [4]. Since altered CCL2/MMP-9 expression was not seen with the control scrambled siRNA, and a BLAST search failed to identify additional sequences which the HMGB1 or HSP60 siRNA would bind, the latter seems a more reasonable explanation. The results may also be due in part to different ways in which the DAMPs could function. For instance, Chun et al. [22] showed that cytosolic HSP60 could initiate a NFκ B dependent signaling pathway. In particular, HSP60 interacted with IKKα/β and initiated a signaling cascade culminating in cell survival [22]. Thus, one may predict different outcomes using RNA interference versus neutralizing antibodies; the siRNA approach may diminish intracellular as well as extracellular initiated signaling, whereas neutralizing antibodies may have more of an effect on signaling initiated at the plasma membrane.
In order to validate release of HMGB1 and HSP60 that could potentially bind receptors on the cell surface we attempted to quantitate the level of these DAMPs in tumor cell supernatants. Although we detected HMGB1 and HSP60 in supernatants from the tumor cells, the levels were too close to the sensitivities of the ELISAs to obtain reliable levels (3.1ng/ml for the HMGB1 ELISA, and 1.5ng/ml for the HSP60 ELISA). Regardless, because we found different outcomes with regard to CCL2 expression following siRNA versus antibody treatment, and because we were mostly interested in the potential autocrine effects of the DAMPs, for the remainder of the study we used neutralizing antibodies rather than siRNA.
Besides CCL2, CCL5 and MMP9 we also investigated whether IL-10 expression was modulated by neutralizing HMGB1 since HMGB1 has been associated with IL-10 expression in dendritic cells and regulatory T cells [14, 23]. However, we found no detectable IL-10 levels expressed by the 4T1 tumor cells by QRT-PCR (data not shown). Similarly, IL-12, which has been reported to be induced in an HMGB1 dependent manner [11, 12], was below detection levels (data not shown). While 4T1 express appreciable levels of TGF-β, VEGF and G-CSF, their expression was not modulated by neutralizing HMGB1 or HSP60 (data not shown).
With respect to the decrease in cell number evident upon neutralizing HMGB1 or HSP60, our data suggest that it is unlikely a consequence of apoptosis or necrosis since the cells showed high viability and no annexinV staining. We did however find a small, yet significant alteration in the percentage of cells in different stages of the cell cycle. Blocking HMGB1 or HSP60 increased the percentage of tumor cells in the S phase of the cell cycle. The correlating decrease in the percentage of cells in the G1 phase of the cell cycle agree with the findings of Jiao et el. [24] who showed that increasing HMGB1 in a human breast cancer cell line (MCF7) resulted in more cells in the G1 phase of the cell cycle. Yet, interestingly, they reported the opposite findings with respect to the relationship between HMGB1 and cell number; cells transfected with HMGB1 exhibited decreased growth. With respect to the relationship between HMGB1 levels and cell number Song et al. [15] reported results similar to those presented here. They showed that decreasing HMGB1 using siRNA resulted in decreased cell proliferation. Yet, Song et al. [15] also reported that decreasing HMGB1 was accompanied by an increase in the number of cells in the G0/G1 phases of the cell cycle, and a decrease in the number of cells in the S and G2/M phases; opposite results from those here with respect to the cell cycle. The differences found in these three studies are difficult to reconcile, but may be because of differences in the methods used to increase or decrease HMGB1, or even the different cell lines used. Some of the differing effects with HMGB1 may also be attributed to differences in post translational processing which can influence HMGB1 function [25]. Despite the fact that further studies are necessary to determine the extent to which these findings are relevant to other types of cancer cells, the decrease in cell number we observed upon blocking receptors that could function for these DAMPs helps to substantiate the possibility that HMGB1 and HSP60 have a role in the autocrine growth of murine mammary carcinoma, at least in vitro.
Interestingly, Liu et al. [14] showed that targeting HMGB1 in 4T1 using siRNA did not affect tumor growth in vivo, although it did influence IL-10 and regulatory T cells. Thus, in addition to complications that arise from trying to compare results obtained with different cell lines in vitro, the impact of HMGB1 and HSP60 released from tumors in vivo is likely more complicated, and may change as a tumor progresses. For instance, early on the release of HMGB1 and HSP60 may contribute to cell proliferation and invasiveness, and therefore contribute to tumor progression and metastasis [8, 15, 16]. At the same time, release of these DAMPs may foster DC maturation and a Th1 response [5–7, 11, 12] to the tumor. As time progresses the response may give way to regulatory T cells and myeloid derived suppressor cells (MDSC) [13, 14] and an ensuing tumor-induced immune suppression. This possibility would make sense in light of a great deal of literature that shows the accumulation of these suppressor cells as tumors progress, despite evidence of tumor-specific immune responses early on [26, 27].
Another finding worthy of note from this study was that it appeared that 50–60% inhibition of growth was about the maximum achievable when targeting DAMPs or their receptors since combining antibody treatments did not yield a greater inhibition of growth than treatment with single antibodies alone (data not shown). One possible explanation for this is that the cells switch to an alternative autocrine signaling mechanism. The Myd88 independent signaling pathway is a possibility, and we have found that there is a >10 fold increase in TICAM1 expression upon inhibition of Myd88 function (data not shown). Thus, in the absence of Myd88 signaling cells may compensate with an upregulation of proteins involved with the Myd88 independent signaling pathway. However, more studies are necessary to determine whether this is indeed the case.
In conclusion, it is clear that targeting HMGB1 and HSP60 does not recapitulate all of the findings of targeting Myd88 that we previously reported [2], and results from this study do not help to make sense of the different effects these DAMPs may have in a tumor setting. Although, the findings that Myd88 drives HMGB1 and HSP60 gene expression, and murine mammary carcinoma overexpress HMGB1 and HSP60 relative to normal mammary epithelium are significant in their own right. Thus, given these data, in combination with the evidence that Myd88 contributes to tumor progression and the cell cycle [28–30], it is important to consider these DAMPs and Myd88 together as studies move forward investigating the role of DAMPs and PAMPs, and how they may be relevant to patients with breast cancer.
Highlights.
This is the first report HMGB1 and HSP60 are expressed in a Myd88 dependent manner
The manuscript shows mammary carcinoma overexpress HMGB1 and HSP60
The manuscript shows HMGB1 and HSP60 expression influences tumor cell growth
Results contribute to a better understanding of DAMPs and PAMPs in a tumor setting
Acknowledgments
This work was supported by the National Institutes of Health 2R15CA137858-02 to R.A.K. The Department of Biology and Lafayette College Excel Scholar program also supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palha De Sousa C, Blum CM, Sgroe EP, Crespo AM, Kurt RA. Murine mammary carcinoma cells and CD11c+ dendritic cells elicit distinct responses to lipopolysaccharide and exhibit differential expression of genes required for TLR4 signaling. Cell Immunol. 2010;266:67–75. doi: 10.1016/j.cellimm.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egunsola AT, Zawislak CL, Akuffo AA, Chalmers SA, Ewer JC, Vail CM, Lombardo JC, Perez DN, Kurt RA. Growth, metastasis, and expression of CCL2 and CCL5 by murine mammary carcinomas are dependent upon Myd88. Cell Immunol. 2012;272:220–229. doi: 10.1016/j.cellimm.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srikrishna G, Freeze HH. Endogenous damage-associated molecular pattern molecules at the crossroads of inflammation and cancer. Neoplasia. 2009;11:615–628. doi: 10.1593/neo.09284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer MP. Gymnastics of molecular chaperones. Molecular Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 5.More SH, Breloer M, von Bonin A. Eukaryotic heat shock proteins as molecular links in innate and adaptive immune responses: HSP60-mediated activation of cytotoxic T cells. Int Immunol. 2001;13:1121–1127. doi: 10.1093/intimm/13.9.1121. [DOI] [PubMed] [Google Scholar]
- 6.Osterloh A, Meier-Stiegen F, Veit A, Fleischer B, von Bonin A, Breloer M. Lipopolysaccharide-free heat shock protein 60 activates T cells. J Biol Chem. 2004;279:47906–47911. doi: 10.1074/jbc.M408440200. [DOI] [PubMed] [Google Scholar]
- 7.Osterloh A, Kalinke U, Weiss S, Fleischer B, Breloer M. Synergistic and differential modulation of immune responses by HSP60 and lipopolysaccharide. J Biol Chem. 2007;282:4669–4680. doi: 10.1074/jbc.M608666200. [DOI] [PubMed] [Google Scholar]
- 8.Tsai Y, Yang M, Huang C, Chang S, Chen P, Liu C, Teng S, Wu K. Interaction between HSP60 and β-catenin promotes metastasis. Carcinogenesis. 2009;30:1049–1057. doi: 10.1093/carcin/bgp087. [DOI] [PubMed] [Google Scholar]
- 9.Barazi HO, Zhou L, Templeton NS, Krutzsch HC, Roberts DD. Identification of heat shock protein 60 as a molecular mediator of α 3β 1 integrin activation. Cancer Res. 2002;62:1541–1548. [PubMed] [Google Scholar]
- 10.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Messmer D, Yang H, Telusma G, Knoll F, Li J, Messmer B, Tracey KJ, Chiorazzi N. High mobility group box protein 1: an endogenous signal for dendritic cell maturation and Th1 polarization. J Immunol. 2004;173:307–313. doi: 10.4049/jimmunol.173.1.307. [DOI] [PubMed] [Google Scholar]
- 12.Dumitriu IE, Baruah P, Valentinis B, Voll RE, Herrmann M, Nawroth PP, Arnold B, Bianchi ME, Manfredi AA, Rovere-Querini P. Release of high mobility group box 1 by dendritic cells controls T cell activation via the receptor for advanced glycation end products. J Immunol. 2005;174:7506–7515. doi: 10.4049/jimmunol.174.12.7506. [DOI] [PubMed] [Google Scholar]
- 13.Wild CA, Bergmann C, Fritz G, Schuler P, Hoffmann TK, Lotfi R, Westendorf A, Brandau S, Lang S. HMGB1 conveys immunosuppressive characteristics on regulatory and conventional T cells. Int Immunol. 2012;24:485–494. doi: 10.1093/intimm/dxs051. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Falo LD, You Z. Knockdown of HMGB1 in tumor cells attenuates their ability to induce regulatory T cells and uncovers naturally acquired CD8 T cell-dependent antitumor immunity. J Immunol. 2011;187:118–125. doi: 10.4049/jimmunol.1003378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song B, Song WG, Li ZJ, Xu ZF, Wang XW, Wang CX, Liu J. Effect of HMGB1 silencing on cell proliferation, invasion and apoptosis of MGC-803 gastric cancer cells. Cell Biochem Funct. 2012;30:11–17. doi: 10.1002/cbf.1811. [DOI] [PubMed] [Google Scholar]
- 16.Yan W, Chan Y, Liang X, Cardinal JS, Huang H, Thome SH, Monga SP, Geller DA, Lotze MT, Tsung A. High mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology. 2012;55:1863–1875. doi: 10.1002/hep.25572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews JL, Kim AC, Hens JR. The role and function of cadherins in the mammary gland. Breast Cancer Res. 2012;14:203. doi: 10.1186/bcr3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Si R, Feng Y, Chen HH, Zou L, Wang E, Zhang M, Warren HS, Sosnovik DE, Chao W. Myocardial ischemia activates an injurious innate immune signaling via cardiac heat shock protein 60 and toll-like receptor 4. J Biol Chem. 2011;286:30308–31319. doi: 10.1074/jbc.M111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JS, Gamboni-Robertson F, He Q, Svetkauskaite D, Kim J, Strassheim D, Sohn J, Yamada S, Maruyama I, Banerjee A, Ishizaka A, Abraham E. High mobility group box 1 protein interacts with multiple toll-like receptors. Am J Physiol Cell Physiol. 2006;290:917–924. doi: 10.1152/ajpcell.00401.2005. [DOI] [PubMed] [Google Scholar]
- 20.Rechsteiner M, Kuehl L. Microinjection of nonhistone chromosomal protein HMG1 into bovine fibroblasts and HeLa cells. Cell. 1979;16:901–908. doi: 10.1016/0092-8674(79)90105-3. [DOI] [PubMed] [Google Scholar]
- 21.Lanks KW. Metabolite regulation of heat shock protein levels. Proc Natl Acad Sci. 1983;80:5325–5329. doi: 10.1073/pnas.80.17.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun JN, Choi B, Lee KW, Lee DJ, Kang DH, Lee JY, Song IS, Kim HI, Lee S, Kim HS, Lee NK, Lee SY, Lee K, Kim J, Kang SW. Cytosolic Hsp60 is involved in the NF-kB-dependent survival of cancer cells via IKK regulation. PLoS One. 2010;5:1–15. doi: 10.1371/journal.pone.0009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Yao Y, Yan Y, Dong N, Sheng Z. High mobility group box 1 protein suppresses T cell-mediated immunity via CD11clow CD45RBhigh dendritic cell differentiation. Cytokine. 2011;54:205–211. doi: 10.1016/j.cyto.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Jiao Y, Wang H, Fan S. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol Sin. 2007;28:1957–1967. doi: 10.1111/j.1745-7254.2007.00669.x. [DOI] [PubMed] [Google Scholar]
- 25.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe MA, Oda JM, Amarante MK, Cesar Voltarelli J. Regulatory T cells and breast cancer: implications for immunopathogenesis. Cancer Met Rev. 2010;29:569–79. doi: 10.1007/s10555-010-9247-y. [DOI] [PubMed] [Google Scholar]
- 27.Montero AJ, Diaz-Montero CM, Kyriakopoulos CE, Bronte V, Mandruzzato S. Myeloid-derived suppressor cells in cancer patients: a clinical perspective. J Immunother. 2012;35:107–115. doi: 10.1097/CJI.0b013e318242169f. [DOI] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through adaptor protein Myd88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 29.Naugler WE, Sakurai T, Kim S, Maeda S, Kim KH, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in Myd88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 30.Coste I, Le Corf K, Kfoury A, Hmitou I, Druillennec S, Hainaut P, Eychene A, Lebecque S, Renno T. Dual function of Myd88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest. 2010;120:3663–3667. doi: 10.1172/JCI42771. [DOI] [PMC free article] [PubMed] [Google Scholar]