Abstract
Glioblastoma (GBM) is the most aggressive and lethal brain tumor in adults. Its invasive nature currently represents the most challenging hurdle to surgical resection. The mechanism adopted by GBM cells to carry out their invasive strategy is an intricate program that recalls what takes place in embryonic cells during development and in carcinoma cells during metastasis formation, the so-called epithelial-to-mesenchymal transition. GBM cells undergo a series of molecular and conformational changes shifting the tumor toward mesenchymal traits, including extracellular matrix remodeling, cytoskeletal re-patterning, and stem-like trait acquisition. A deeper understanding of the mechanisms driving the whole infiltrative process represents the first step toward successful treatment of this pathology. Here, we review recent findings demonstrating the invasive nature of GBM cancer stem cells, together with novel candidate molecules associated with both cancer stem cell biology and GBM invasion, like doublecortin and microRNAs. These findings may affect the design of effective therapies currently not considered for GBM invasive progression.
Cancer stem cells and neural stem cells: common features with different purposes
Parallels between neurogenesis and the processes contributing to brain tumor formation exist. Neural stem cells (NSCs) are quiescent cells able to self-renew and generate partially committed, highly proliferative progenitors that subsequently undergo complete differentiation into one of the three lineages composing the brain. A recognized hallmark of neural stem/progenitor cells is their ability to migrate, an essential process for recovery after brain injury [1]. The same role exerted by NSCs in the physiological context has been proposed to be played in glioblastoma (GBM) by a rare fraction of self-renewing, multipotent tumor-initiating cells called cancer stem cells (CSCs), responsible for tumor progression, maintenance, and recurrence [2,3]. This sub-population has shown intrinsic resistance to therapy, being able to repopulate the tumor after treatment [4]. Recently, many studies have ascribed to CSCs the infiltrative property of GBM.
Cancer stem cells and invasive cells: two sides of the same coin?
The clinically distinct feature of GBM lies within its infiltrative potential, rendering complete tumor resection nearly impossible. Tumor infiltration is an extremely complex program that requires the steady supply of extra cellular cues, abrogation of cell-cell interactions, and extracellular matrix (ECM) remodeling. Invading GBM cells are particularly resistant to current therapies and are often localized within the neurovascular niche, two features in common with CSCs [5].
Recent experimental data started to suggest that CSCs are responsible for GBM invasiveness. Cells enriched for the putative stem cell marker CD133 display greater migratory and invasive potential in vitro and in vivo when compared with matched CD133-negative tumor cells derived from human primary GBMs, GBM xenografts [4], and brain tumor cell lines [6-9]. We and others reported a marked upregulation of proteins involved in the processes of migration and invasion in GBM CSCs, such as different types of matrix metalloproteinases, or different members of both ADAMs (a disintegrin and metalloproteinases) and ADAMTS (ADAM with thrombospondin motifs) families [4,6-8,10]. Therefore, the highly migrating and invasive ability of GBM CSCs may be due to increased expression of proinvasive genes. Based on the findings that the GBM CSCs are more infiltrative than their diffrentiated descendants, a novel strategy has also been proposed to isolate and enrich CSCs from the whole tumor population by exploiting the tumor cell heterogeneity of invasiveness [11].
Cells at the leading edge of the tumor have been found to be positive for putative stem cell markers such as L1CAM [12], nucleostemin [13], and nestin [14-16], supporting the notion that CSCs are indeed responsible for GBM invasion. A recent paper provided new insights into the role of SOX2 as a novel determinant of the invasive and migration properties of GBM CSCs and glioma cell lines [17]. Recently, many groups have suggested that different CSCs can coexist in the same tumor. Both tumor margin and the corresponding tumor mass contain CSCs that are characterized by different stem cell marker expression, neurosphere formation ability, and in vivo tumorigenic potential [18-20]. Thus, the invasive edge of GBM may function as a new CSC niche [21]. In this scenario, the theory proposed by Brabletz and colleagues [22] about the plasticity of CSCs can also be applied to GBM: CSCs switch from a stationary and proliferative phenotype to a migratory one, and vice versa, ensuring the enlargement of the tumor core and the colonization of the neighboring normal brain tissue.
Epithelial-to-mesenchymal transition in glioblastoma?
During development, embryonic cells undergo a series of trans-differentiation programs collectively assembled under the name of epithelial-to-mesenchymal transition (EMT) [23]. Interestingly, carcinoma cells exploit the same principle to drive invasion and colonization to non-adjacent tissues. EMT is a process that allows a differentiated epithelial cell, entirely settled and patterned to establish stable contacts with neighbor cells, to assume amesenchymal cell phenotype, characterized by loss of cell-cell interactions [24], reduced cellular adhesion [25], active production of ECM proteases [26], increased cytoskeletal dynamics [27], and changes in transcription factor expression [28,29]; all of these events eventually lead to increased migration and invasion ability. Moreover, the acquisition of mesenchymal traits by cancer cells undergoing EMT has been widely reported to be associated with the acquisition of a stem cell program [30]. Thus, the expression of both EMT factors and stem cell markers in selected epithelial tumor cells at the invasive tumor front produces the 'migrating CSCs' [23,31-33].
In GBMs, robust evidence for the existence of the EMT process is still lacking. Recently, Cheng and colleagues [34], analyzing data from The Cancer Genome Atlas (TCGA), demonstrated a strong association in GBM between an epithelial-mesenchymal expression signature, the expression of the putative stem cell marker CD44, and shortened time to recurrence following initial treatment.
The EMT program is regulated mainly by three major groups of transcription factors - the SNAI, TWIST, and Zinc-finger enhancer binding (ZEB) family members - whose upregulation promotes tumor invasiveness and has been associated with poor clinical prognosis in human cancers [28,29,35,36]. The activity of these transcription factors has been reported to be altered in GBMs. SNAI is overexpressed in GBMs [37] and is involved in the regulation of glioma cell proliferation and migration [10,38]. TWIST1 is also upregulated in GBMs, where it promotes cell invasion by the upregulation of genes such as matrix metalloproteinase-2 (MMP-2), hepatocyte growth factor (HGF), fibroblast activation protein (FAP), and SNAI [39]. The ZEB2 expression level is significantly increased in glioma tissues compared with normal brain tissues and is positively correlated with tumor grading. ZEB2 silencing reduces the expression of mesenchymal cell markers such as N-cadherin, Vimentin, and Snail and inhibits glioma cell migration and invasion [40,41]. The overexpression of all of these EMT transcription factors in GBM cell lines may occur following the activation of WNT/β-catenin pathway and results in increased in vitro cell migration and invasion [20,42].
The SNAI, TWIST, and ZEB transcription factors act by directly repressing the expression of the adhesion molecule E-cadherin, whose functional loss is considered a hallmark of EMT [43]. In parallel, there is an induction of mesenchymal markers (for example, N-cadherin and cadherin-11), reorganization of the cytoskeleton (for example, switch from cytocheratins to vimentin), and production of ECM components and metalloproteases [30]. In general, loss of E-cadherin correlates with high tumor grades and poor prognosis [44], whereas the induction of N-cadherin correlates with increased cell motility [45]. In the brain, E-cadherin expression is rare in both normal and tumoral tissues [46]. Only two recent studies argue that E-cadherin expression inversely correlates with brain tumor grade [40,47]. N-cadherin and cadherin-11 are expressed in different brain regions, including the cortex and the hippocampus, and their expression is upregulated in malignant gliomas [48]. Thus, no switch between cadherins has been described in malignant gliomas, except in some specific cases [49]. However, an alternative cadherin switch, different from the classic E-cadherin-to-N-cadherin switch described in epithelial cells, was recently described in GBMs. Lu and colleagues [50] showed Snail and N-cadherin upregulation in GBM cells, with the concomitant downregulation of T-cadherin. T-cadherin is an atypical member of the cadherin family, downregulated in different tumor types, and associated with a poorer prognosis [51]. T-cadherin expression is absent in invasive GBMs but higher in circumscribed, minimally invasive GBMs [50], and its overexpression in GBM cells inhibits proliferation and migration [52]. Consistent with these findings, Zeb-1 has been demon strated to suppress T-cadherin expression and increase invasion in gallbladder cancer [53], thus reinforcing the role of T-cadherin in the acquisition of EMT phenotype in GBM.
Recently, GBMs have been classified by gene expression signatures in four distinct subtypes (proneural, neural, classic, and mesenchymal) on the basis of a comprehensive analysis of The Cancer Genome Atlas (TCGA) dataset [54], which are not dissimilar from the three subtypes (proneural, proliferative, and mesenchymal) described in the work of Phillips and colleagues [55]. Analysis of these data sets reveals that high expression of genes associated with mesenchyme-derived tissues is common in GBMs and associates with poor overall survival and treatment resistance [54-56]. Consistent with these findings, Joo and colleagues [57] demonstrated that CD133-negative GBM CSCs have a mesenchymal subtype genetic signature compared with CD133-positive GBM CSCs and give rise to tumors characterized by an invasive growth. Similarly, Lottaz and colleagues demonstrated that GBM CSCs positive for the putative stem cell marker CD44 [58] but negative for CD133 possess a mesenchymal gene signature and display enhanced invasive growth both in vitro and in vivo compared with CD44-negative CD133-positive cells, which instead display a proneural gene signature, limited invasiveness, and high proliferation rate [59]. In addition, several human glioma cell lines express genes associated with mesenchyme-derived tissues [60], which are reported to be involved also in migration, invasion, and EMT [61]. Notably, the ectopic coexpression of the two master regulators that define the mesenchymal subtype, CCAAT/enhancer binding protein (C/EBPβ) and signal transducer and activator of transcription 3 (STAT3), reprograms NSCs along the mesenchymal lineage, whereas their silencing in glioma cells abolishes the mesenchymal signature and reduces tumor invasiveness [62]. Similarly, the transcriptional coactivator with PDZ-binding motif (TAZ) drives the mesenchymal differentiation of GBMs: when it is overexpressed in proneural GBM CSCs as well as in NSCs, it induces mesenchymal marker expression and aberrant osteoblastic and chondrocytic differentiation, whereas its silencing in mesenchymal GBM CSCs reduces the expression of mesenchymal markers, invasion, and tumor formation [63]. It is likely that the high expression of mesenchymal genes in a subset of human GBMs can be considered to be reminiscent of the EMT program [54] or that the aberrant activation of EMT factors during gliomagenesis can trigger the mesenchymal shift in GBM [63]. All of these data suggest that GBMs share many features with EMT, but a deeper knowledge of the mechanisms driving this process is indispensable to better tackle the invasive nature of these tumors.
Unconventional molecular determinants of glioblastoma invasion
Cell motility is a complex biological process which is tightly regulated by a multitude of converging signals. Many signaling pathways regulating the stem cell compartment have been reported to be critical for triggering tumor cell migration and invasion. However, their specific involvement in glioma invasion remains poorly understood because of intricate feedback loop mechanisms and missing links between them. Given the large number of detailed reviews describing the role of different signaling pathways in tumor progression (NF-κB, notch, hedgehog-GLI, WNT/β-catenin, RTKs, and AKT are the most relevant), this review focuses on two unconventional molecular determinants of GBM invasion: doublecortin (DCX) and microRNAs.
Why should doublecortin garner cancer scientists' interest?
The identification of single cells responsible for GBM infiltration in normal brain parenchyma is of crucial relevance. Several molecules have been used to identify invading GBM cells (for example, nestin [15,16] and vimentin [64]), but their expression could be altered by the grade of differentiation of the tumors. NSC discovery in the adult brain and the strong relationship between NSCs and CSCs have led to the consideration that these two types of cells may share common markers as well. DCX is one of them.
DCX is a microtubule-associated protein: it promotes microtubule polymerization and stability [65,66], and it is crucial for controlled cell movement, since it promotes nucleus translocation and the maintenance of a bipolar cell shape, preventing branching and nucleokinesis defects [67,68]. In humans, DCX mutations cause type I lissencephaly syndrome and subcortical laminar heterotopia, severe cortical malformations associated with mental retardation and epilepsy [69]. Knockout mouse models display milder symptoms, which are probably due to genetic redundancy with doublecortin-like (DCL) and doublecortin-like kinase (DCLK) and which become more evident when combined with mutations in LIS1, another gene involved in lissencephaly [70].
DCX is a brain-specific gene expressed in differentiating neurons during brain development [69,71], in neuroblasts, in discrete niches along the wall of the lateral ventricles, in the subgranular zone of the hippo-campus, along the rostral migration stream, and in the olfactory bulbs in the adult rodent brain [72-74]. In vitro studies on neurospheres derived from adult rodent SVZ have confirmed the essential function of DCX in promoting cell migration [75].
DCX has also been proposed as a marker for migrating neuroblasts in the adult human brain [76], where it plays a crucial role in pathologic conditions in which the provision of progenitors migrating out of neurogenic areas toward the lesion is needed to replace neurons [77]. In non-injured adult mammalian brain, DCX expression outside the neurogenic areas is very rare [72,78]. Recently, DCX-positive cells have been identified in developing and mature neurons in the cerebellar cortex of guinea pig, cat, and primate [79,80] and, to a minor extent, in cells expressing the astrocytic marker glial fibrillary protein (GFAP) [81,82]; however, these DCX+ GFAP+ cells behave as neural progenitors [82].
Doublecortin: an appealing and promising marker for invasive glioblastoma cells
Microtubule-associated proteins play important roles in cellular functions involving cytoskeletal rearrangement, which is one of the main features of EMT in cancer, thereby allowing cells to detach and move. Interestingly, DCX expression has been found in invasive human brain tumors, with the highest intensity at the invasive front [83], and has been proposed as a specific marker for the identification of infiltrating glioma cells [84]. Furthermore, gene expression profiling of human GBMs has revealed DCX, together with osteonectin (also known as SPARC) and semaphorin3B, as useful genetic markers to predict patient survival: the higher the expression of each of these three genes, the poorer the survival [85]. It is relevant to note that all three of these genes are involved in the regulation of cell migration, thus suggesting a direct link between tumor invasion and patient survival.
Recently, we identified RAI (SHC3/SHCC/N-SHC), a member of the family of SHC-like adaptor proteins, as a novel regulator of migration of normal and GBM stem/progenitor cells, affecting DCX protein levels [10]. RAI silencing in cancer stem/progenitor cells isolated from human GBMs reduces cell migration and invasion, both in vitro and in vivo. The rare cells still expressing RAI are localized at the tumor invasion front and express markers for both immature and migratory cells, notably DCX and OLIG2 [86]. We always observed a significant reduction in the number of invading cells positive for DCX in all RAI-interfered GBM xenografts as well as a decreased amount of DCX in both RAI-/- normal stem/progenitor cells and RAI-interfered tumor stem/progenitor cells.
So far, only one group has reported an opposite effect of DCX on glioma cells, with exogenous DCX addition reducing CSC self-renewal [87] and tumor volume [88,89]. Such controversial results could well be related to the usage of GBM cells not expressing DCX. On the contrary, we and others reported endogenous DCX expression in both normal and cancer stem/progenitor cells [10,90].
A deeper knowledge of the molecular mechanisms regulating DCX expression could help in the identification of new targets for therapy. While there is a large amount of literature about DCX regulation in the developing brain, little is known about its regulation in adult normal and cancer brain. DCX activity is regulated by several serine-threonine kinases and phosphatases; upon phosphorylation on Ser297 residue by cyclin-dependent kinase 5, DCX affinity for microtubules decreases, promoting cell migration [91]. c-Jun N-terminal kinase (JNK), which regulates neurite extension, can phosphorylate DCX on Ser332, decreasing its affinity for tubulin and promoting cell migration [92]. Protein kinase A (PKA) and MAP/microtubule affinity-regulating kinase (MARK) regulate DCX binding affinity for microtubules, phosphorylating it on Ser47 [93]. DCX is also spatially regulated: when phosphorylated, it is expressed in the cell soma; when de-phosphorylated (likely by protein phosphatase 2A, or PP2A), it shifts to the tips of the neurites [93]. Another phosphatase acting on DCX is the tumor suppressor phosphatase and tensin homolog (PTEN) [94], which is the major inhibitor of the PI3K/AKT pathway and is frequently deleted in GBMs. A de-regulation of kinases and phosphatases is frequently observed in GBMs, therefore suggesting a possible mechanism enhancing GBM invasion. Another possible mechanism of DCX de regulation in GBM could be linked to its transcriptional regulation. Recent studies have identified an upstream promoter region of DCX containing binding sites for LEF/TCF (lymphoid enhancer factor/T-cell factor) transcription factors [95], which are central effectors of the WNT/β-catenin pathway. This pathway is frequently altered in glioma: nuclear translocation and, therefore, activation of β-catenin have a direct impact on GBM invasion [96]. In both adult normal NSCs and CSCs isolated from human GBMs, DCX is a target of the adaptor protein RAI, which mediates multiple signaling pathways, eventually leading to metalloproteinase upregulation and GBM invasion [10] (Figure 1).
Figure 1.
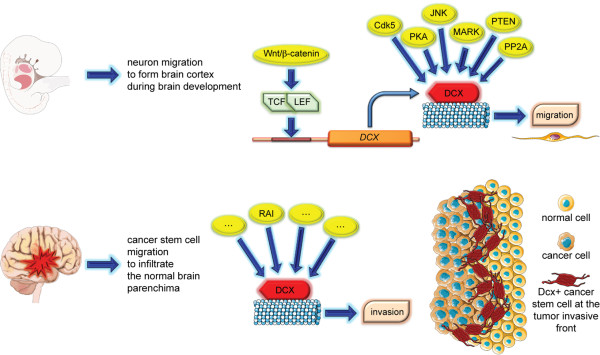
Doublecortin regulators during development and cancer invasion. Several doublecortin regulators have been identified for immature neuron migration during embryonic development (upper panel), whereas in the adult brain little is known, especially in glioblastoma invasion (lower panel), indicating that extensive studies are still required to characterize and address regulators of glioblastoma invasion. RAI is also known as SHC3/SHCC/N-SHC. Figure was produced by using Servier Medical Art. CDK5, cyclin-dependent kinase 5; DCX, doublecortin; JNK, c-Jun N-terminal kinase; LEF, lymphoid enhancer factor; MARK, MAP/microtubule affinity-regulating kinase 3; PKA, protein kinase A; PP2A, protein phosphatase 2; PTEN, phosphatase and tensin homolog; TCF, T-cell factor; WNT, wingless-type MMTV integration site family.
MicroRNAs: a specific role in glioblastoma invasion
MicroRNAs (miRNAs/miRs) are a class of small non-coding RNAs, 21 to 24 nucleotides long, which post-transcriptionally regulate gene expression by either inhibiting mRNA translation or inducing mRNA degradation, binding, via imperfect base pairing, to the 3' untranslated region of mRNAs, allowing the targeting of a high number of mRNAs.
miRNAs play important roles in physiologic conditions as well as in various diseases, including cancer, in which they could be downregulated, acting as tumor suppressors, or upregulated, acting as oncogenes. miRNAs are involved in many aspects of gliomagenesis and GBM CSC biology: proliferation, survival, migration, invasion, and angiogenesis [97]. Moreover, recently, bioinformatics analysis of 82 gliomas indicated the association between some miRNAs and the expression of mesenchymal markers [98].
Table 1 depicts the most intriguing ones found to be directly involved in the GBM invasion process, suggesting interesting putative targets for GBM therapy. Of note, the majority of the literature analyzes the role of miRNAs in GBM infiltration through experiments using glioma cell lines, and very little experimental evidence on the role of miRNAs in migration and invasion of GBM CSCs is available. Given the contribution of CSCs to GBM invasiveness (highlighted in the previous section), we believe that further validation of the properties of specific miRNAs regulating migration and invasion in CSCs is essential.
Table 1.
Biological functions of the different microRNAs associated with glioblastoma invasion
| microRNA | Biological function | Reference |
|---|---|---|
| miR-7 | Downregulated in GBM. Acts as a tumor suppressor in GBM, preventing GBM CSC invasion, proliferation, and survival by repressing EGFR and its downstream pathway AKT on one side and FAK and the production of MMP-2 and MMP-9 on the other side. | [108,109] |
| miR-10b | Acts as oncogene: it is overexpressed in glioma samples. Induces glioma cell invasion in vitro by decreasing HOXD10 protein levels, which in turn control MMP-14 and uPAR expression, resulting in their upregulation. | [101] |
| miR-21 | Acts as oncogene: it is overexpressed in GBM, where it represses the MMP inhibitors TIMP3 and RECK, thus resulting in metalloproteinase MMP-2 upregulation, promoting glioma invasion and angiogenesis. High levels of this miR are associated with poor patient survival. | [102,103] |
| miR-23b | Acts as a tumor suppressor and is downregulated at the invasive edge in GBM. Pyk2, whose expression is higher in high-grade gliomas, is one of its direct targets. | [110] |
| miR-34a | Acts as a tumor suppressor and is downregulated in human glioma tumors as compared with normal brain. Its overexpression exerts a suppressive effect in GBM CSCs by inhibiting proliferation and migration while inducing differentiation (Guessous et al. [111]). | [111] |
| miR-101 | Acts as a tumor suppressor and is downregulated in GBM. Its target EZH2, a histone metyl transferase belonging to the Polycomb Group of proteins, is therefore overexpressed, promoting cell migration, neo-angiogenesis, and cell growth. | [112] |
| miR-124a | Acts as a tumor suppressor and is downregulated in GBM, in association with a shorter patient survival. In vitro, it negatively regulates cell migration and invasion through the negative regulation of its direct targets IQGAP1, laminin 1, and integrin β1. In vivo, it inhibits glioma invasion by targeting SNAI2. It inhibits glioma stem cell traits through SNAI2 and promotes neuronal differentiation, antagonizing the transcriptional repressor RE1 silencing transcription factor (REST). |
[113,114] |
| miR-137 | It is downregulated in GBM and acts as a tumor suppressor. In vitro, it negatively regulates glioma cell migration, invasion, and proliferation, reducing MMP-9 levels and directly targeting Cox-2 expression. | [116] |
| miR-145 | Not clear yet. Suggested as pro-invasive oncogene by Koo et al. [124] but as a tumor suppressor inhibiting glioma cell invasion by Lee et al. [123]. | [123,124] |
| miR-146b | Acts as a tumor suppressor. Represses both EGFR and MMP expression, reducing glioma cell migration and invasion. | [117,118] |
| miR-195 | Acts as a tumor suppressor. It is downregulated in GBM, where it promotes proliferation and invasion through deregulation of its targets E2F3 and cyclin D3, respectively. | [119] |
| miR- 221/222 | Act as oncogenes: their overexpression in glioma cells causes downregulation of connexin 43, a major component of gap junctions, inducing cell proliferation and invasion. Their overexpression is induced by de-regulated NF-κB and c-Jun transcription factors, resulting in the repression of the cell cycle inhibitor p27kip1 and in the activation of the Akt pathway leading to, among several targets, MMP activation and thus promoting glioma cell proliferation and invasion. |
[104-107] |
| miR-302/367 | Act as a tumor suppressor: when expressed, they suppress stemness gene signature, CSC self-renewal, infiltration, and tumorigenic capacity through a drastic inhibition of the CXCR4 pathway (Fareh et al. [120]). | [120] |
| miR-410 | Acts as tumor suppressor in GBM, decreasing cell proliferation and invasion by targeting MET and consequently AKT signaling and MMP-9 levels. | [122] |
AKT, serine/threonine-specific protein kinase; CSC, cancer stem cell; EGFR, epidermal growth factor receptor; FAK, focal adhesion kinase; GBM, glioblastoma; HOXD10, homeobox D10; miRNA/miR, microRNA; MMP, matrix metalloproteinase; NF-κB, nuclear factor-kappa-B; RECK, Reversion-inducing cysteine-rich protein with Kazal motifs precursor; TIMP, tissue inhibitors of metalloproteinase; uPAR, urokinase-type plasminogen activator receptor.
MicroRNAs acting as oncogenes in glioblastoma
TWIST1, a transcription factor involved in EMT, has been shown to bind the upstream portion of the miR-10 hairpin region, modulating its expression [99]. MiR-10b, as well as TWIST [100], is overexpressed in glioma samples, with the highest peaks in GBM as compared with low-grade astrocytomas, and induces glioma cell invasion in vitro by decreasing HOXD10 (homeobox D10) protein levels, which in turn control MMP-14 and uPAR (urokinase receptor) expression, resulting in their upregulation [101]. Other metalloproteinases, such as MMP-2, are upregulated in cancer as a consequence of the repression of MMP inhibitors, such as TIMP3 (metalloproteinase inhibitor 3) and RECK, which are themselves direct targets of miR-21, found to be overexpressed in GBM, thus promoting glioma invasion and angiogenesis [102]. Moreover, analysis of miRNAs in gliomas belonging to different grades associated high miR-21 with poor patient survival [103].
Overexpression of the miR-221/222 cluster in malignant glioma cells results in the reduced expression of the tumor suppressor connexin 43 (Cx43), leading to the reduction of gap junction intercellular communication and, as a result, to an increase in proliferation and invasion of glioma cells [104]. In a highly intricate regulatory system, other important transcription factors, involved in cancer development when de-regulated, have been linked to miRNAs, such as NF-κB and c-Jun. These factors can induce glioma cell proliferation and invasion by promoting overexpression of miR-221/222 [105], resulting in the repression of the cell cycle inhibitor p27kip1 and in the activation of the AKT pathway leading to, among several targets, MMP activation [106,107].
MicroRNAs acting as tumor suppressors in glioblastoma
Mir-7 blocks cancer invasion, proliferation, and survival. It has been found strongly downregulated in GBM samples compared with normal brain, and in vitro experiments showed that it inhibits GBM CSC proliferation and invasion by signaling on different targets: it directly inhibits the EGFR expression and therefore the downstream AKT pathway [108] and directly inhibits focal adhesion kinase (FAK), which has a role in the production of metalloproteinases such as MMP-2 and MMP-9 [109]. MiR-23b is downregulated at the invasive edge of human GBM samples as opposed to the tumor core and suppresses glioma cell migration and invasion in in vitro and ex vivo experiments. It directly targets Pyk2 (proline-rich tyro sine kinase 2), the expression of which correlates positively with tumor grade and is known to promote GBM cell migration and invasion [110]. MiR-34a is downregulated in human glioma tumors as compared with normal brain. When overexpressed in GBM CSCs, it exerts a suppressive effect by inhibiting proliferation and migration while inducing differentiation [111].
MiR-101, when downregulated in GBM, causes not only cell migration and invasion but also neo-angiogenesis and cell growth [112]. MiR-124a expression was found to be downregulated in a wide range of GBM samples, in association with shorter patient survival. In vitro studies showed that it negatively regulates cell migration and invasion, through the negative regulation of its direct targets, IQ motif-containing GTPase-activating protein 1 (IQGAP1), laminin 1 (LAMC1), and integrin β1 (ITGB1) [113]. Recently, miR-124 was shown to inhibit glioma cell invasion and stem cell traits through SNAI2, which is often upregulated in glioma and enriched in the stem cell population [114]. The fact that miR-124a also promotes neuronal differentiation, antagonizing the transcriptional repressor RE1 silencing transcription factor (REST) [115], is even more promising for the therapeutic use of miR-124a, which could block GBM invasion and induce tumor cell differentiation at the same time. MiR-137 expression inversely correlates with glioma grade. In vitro experiments on glioma cell lines showed that it inhibits migration and invasion, decreasing MMP-9 levels, and proliferation, directly targeting Cox-2, which is more expressed in high-grade tumors and correlates with poorer prognosis [116]. MiR-146b represses both EGFR and MMP expression, reducing glioma cell migration and invasion [117,118]. MiR-195 has been found to be strongly downregulated in human GBM cells, causing upregulation of its targets E2F3 and cyclin D3, thus promoting glioma cell proliferation and invasion, respectively [119].
The cluster miR-302/367 is induced during serum-mediated stemness suppression in GBM CSCs; when expressed, it suppresses stemness gene signature, CSC self-renewal, infiltration, and tumorigenic capacity through adrastic inhibition of the CXCR4 pathway [120]. The blockage of CXCR4 signaling leads to a drastic repression of the sonic hedgehog (SHH)-GLI-NANOG network, which has been demonstrated to regulate self-renewal and expression of the embryonic stem cell-like signature [121].
MiR-410 was recently characterized as a direct regulator of MET (the receptor for hepatocyte growth factor receptor, or HGF), downregulating MET-induced AKT signal transduction, with a negative effect on cell proliferation and invasion, decreasing P-AKT and MMP-9 levels [122].
Contrasting pieces of evidence have recently been reported for miR-145. MiR-145 has been characterized as a possible tumor suppressor gene in GBM, negatively acting on the expression of several metastasis-related genes, such as PLAUR (plasminogen activator urokinase receptor), SPOCK3, ADAM22, SLC7A5 (solute carrier family 7 member 5), and FASCN1, and inhibiting glioma cell invasion in vitro [123]. On the other hand, miR-143 and -145 (which are expressed by the same genetic locus) have been suggested as possible oncogenes and, in particular, have been shown to exert pro-invasive function in in vitro experiments [124].
Conclusions and perspectives
Highly diffuse and infiltrative growth is an important effector of GBM pathogenesis and represents the main limitation in the feasibility of current treatments. In recent years, the molecular players and the signaling cascades triggering the motility and invasion of glioma cells have been widely investigated in order to increase our understanding of the mechanism of glioma spreading and eventually improve the survival of patients with glioma. Following the classification of gliomas in subtypes based upon molecular signatures [54,55], a similar approach based upon microRNA expression has been proposed. Gene expression profile analysis of gliomas revealed a capacity to effectively distinguish GBMs from low-grade gliomas, which overexpressed different classes of microRNAs [125], but also to establish a signature able to predict the survival of patients with GBM on the basis of the expression of 10 microRNAs [126]. In this scenario, the analysis of both GBM molecular signatures and miRNA expression signatures may contribute to cancer diagnosis, prognosis, patient stratification, and a more personalized treatment.
This review lends further weight to these findings, highlighting the role of GBM CSCs as novel determinants of the infiltrative behavior of GBMs. Of further interest, specific effectors implicated in the process of migration and invasion (that is, EMT-associated factors, DCX, and microRNAs) are known to be associated with CSC biology. Given the resistance of CSCs to traditional therapies, approaches designed to target CSCs and their effectors will represent exciting therapeutic targets effective in suppressing the CSC-mediated GBM invasion.
Recently, GBMs have been shown to contain micro-vesicles carrying angiogenic proteins and inhibitors of MMPs, of EGFRvIII mRNA, and of miRNA-21, responsible for GBM invasion [102,127]. These microvesicles have been used as biomarkers since they carry brain tumor-specific antigens and cytokines and are present in the serum of patients with high-grade glioma [128]. Furthermore, these microvesicles could be exploited as possible vehicles for innovative anti-miR therapy of GBM since short-hairpin RNAs have been efficiently delivered to the brain by systemic injection [129]. The possibility to limit the GBM infiltrative component is likely to have a large impact on therapies designed to block tumor progression.
Abbreviations
CSC: cancer stem cell; DCX: doublecortin; ECM: extracellular matrix; EMT: epithelial-to-mesenchymal transition; GBM: glioblastoma; miRNA/miR: microRNA; MMP: matrix metalloproteinase; NSC: neural stem cell.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All authors contributed equally to the manuscript. BO prepared the figure and the table. All authors read and approved the final manuscript.
Contributor Information
Barbara Ortensi, Email: barbara.ortensi@ieo.eu.
Matteo Setti, Email: matteo.setti@ieo.eu.
Daniela Osti, Email: daniela.osti@ieo.eu.
Giuliana Pelicci, Email: giuliana.pelicci@ieo.eu.
Acknowledgements
The work was supported by the Association for International Cancer Research (08-0562 to GP).
References
- Imitola J, Raddassi K, Park KI, Mueller FJ, Nieto M, Teng YD, Frenkel D, Li J, Sidman RL, Walsh CA, Snyder EY, Khoury SJ. Directed migration of neural stem cells to sites of CNS injury by the stromal cell-derived factor 1alpha/CXC chemokine receptor 4 pathway. Proc Natl Acad Sci USA. 2004;101:18117–18122. doi: 10.1073/pnas.0408258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Annabi B, Rojas-Sutterlin S, Laflamme C, Lachambre MP, Rolland Y, Sartelet H, Beliveau R. Tumor environment dictates medulloblastoma cancer stem cell expression and invasive phenotype. Mol Cancer Res. 2008;6:907–916. doi: 10.1158/1541-7786.MCR-07-2184. [DOI] [PubMed] [Google Scholar]
- Inoue A, Takahashi H, Harada H, Kohno S, Ohue S, Kobayashi K, Yano H, Tanaka J, Ohnishi T. Cancer stem-like cells of glioblastoma characteristically express MMP-13 and display highly invasive activity. Int J Oncol. 2010;37:1121–1131. doi: 10.3892/ijo_00000764. [DOI] [PubMed] [Google Scholar]
- Annabi B, Lachambre MP, Plouffe K, Sartelet H, Beliveau R. Modulation of invasive properties of CD133+ glioblastoma stem cells: a role for MT1-MMP in bioactive lysophospholipid signaling. Mol Carcinog. 2009;48:910–919. doi: 10.1002/mc.20541. [DOI] [PubMed] [Google Scholar]
- Yu SP, Yang XJ, Zhang B, Ming HL, Chen C, Ren BC, Liu ZF, Liu B. Enhanced invasion in vitro and the distribution patterns in vivo of CD133+ glioma stem cells. Chin Med J (Engl) 2011;124:2599–2604. [PubMed] [Google Scholar]
- Ortensi B, Osti D, Pellegatta S, Pisati F, Brescia P, Fornasari L, Levi D, Gaetani P, Colombo P, Ferri A, Nicolis S, Finocchiaro G, Pelicci G. Rai is a new regulator of neural progenitor migration and glioblastoma invasion. Stem Cells. 2012;30:817–832. doi: 10.1002/stem.1056. [DOI] [PubMed] [Google Scholar]
- Yu SC, Bian XW. Enrichment of cancer stem cells based on heterogeneity of invasiveness. Stem Cell Rev. 2009;5:66–71. doi: 10.1007/s12015-008-9047-8. [DOI] [PubMed] [Google Scholar]
- Cheng L, Wu Q, Guryanova OA, Huang Z, Huang Q, Rich JN, Bao S. Elevated invasive potential of glioblastoma stem cells. Biochem Biophys Res Commun. 2011;406:643–648. doi: 10.1016/j.bbrc.2011.02.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamase A, Muraguchi T, Naka K, Tanaka S, Kinoshita M, Hoshii T, Ohmura M, Shugo H, Ooshio T, Nakada M, Sawamoto K, Onodera M, Matsumoto K, Oshima M, Asano M, Saya H, Okano H, Suda T, Hamada J, Hirao A. Identification of tumor-initiating cells in a highly aggressive brain tumor using promoter activity of nucleostemin. Proc Natl Acad Sci USA. 2009;106:17163–17168. doi: 10.1073/pnas.0905016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Kitai R, Horita R, Sato K, Yoshida K, Arishima H, Higashino Y, Hashimoto N, Takeuchi H, Kubota T, Kikuta K. Nestin expression in astrocytic tumors delineates tumor infiltration. Brain Tumor Pathol. 2010;27:17–21. doi: 10.1007/s10014-009-0261-0. [DOI] [PubMed] [Google Scholar]
- Ishiwata T, Teduka K, Yamamoto T, Kawahara K, Matsuda Y, Naito Z. Neuroepithelial stem cell marker nestin regulates the migration, invasion and growth of human gliomas. Oncol Rep. 2011;26:91–99. doi: 10.3892/or.2011.1267. [DOI] [PubMed] [Google Scholar]
- Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, Urquiza L, Jauregi P, Lopez de Munain A, Sampron N, Aramburu A, Tejada-Solis S, Vicente C, Odero MD, Bandres E, Garcia-Foncillas J, Idoate MA, Lang FF, Fueyo J, Gomez-Manzano C. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo SG, Combi R, Cajola L, Patrizi A, Redaelli S, Bentivegna A, Baronchelli S, Maira G, Pollo B, Mangiola A, DiMeco F, Dalpra L, Vescovi AL. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Dietz S, Madhu B, Griffiths J, Price SJ, Collins VP, Watts C. Fluorescence-guided surgical sampling of glioblastoma identifies phenotypically distinct tumour-initiating cell populations in the tumour mass and margin. Br J Cancer. 2012;107:462–468. doi: 10.1038/bjc.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Jeon HY, Joo KM, Kim JK, Jin J, Kim SH, Kang BG, Beck S, Lee SJ, Kim JK, Park AK, Park WY, Choi YJ, Nam DH, Kim H. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 2011;71:3066–3075. doi: 10.1158/0008-5472.CAN-10-1495. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 2011;8:482–485. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Przybylo JA, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition: tumor progression at Snail's pace. Int J Biochem Cell Biol. 2007;39:1082–1088. doi: 10.1016/j.biocel.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Levayer R, Lecuit T. Breaking down EMT. Nat Cell Biol. 2008;10:757–759. doi: 10.1038/ncb0708-757. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell States in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WY, Kandel JJ, Yamashiro DJ, Canoll P, Anastassiou D. A multi-cancer mesenchymal transition gene expression signature is associated with prolonged time to recurrence in glioblastoma. PLoS One. 2012;7:e34705. doi: 10.1371/journal.pone.0034705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71:245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Menon LG, Black PM, Carroll RS, Johnson MD. SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer. 2010;10:301. doi: 10.1186/1471-2407-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SP, Kim JH, Han ME, Sim HE, Kim KS, Yoon S, Baek SY, Kim BS, Oh SO. SNAI1 is involved in the proliferation and migration of glioblastoma cells. Cell Mol Neurobiol. 2011;31:489–496. doi: 10.1007/s10571-010-9643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L, Maxwell JP, Glackin CA, Wakimoto H, Gonzalez-Herrero I, Sanchez-Garcia I, Silber JR, Horner PJ, Rostomily RC. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Hu M, Wang J, Xu Y, Chen X, Su L. Identification of the role of Smad interacting protein 1 (SIP1) in glioma. J Neurooncol. 2010;97:225–232. doi: 10.1007/s11060-009-0015-1. [DOI] [PubMed] [Google Scholar]
- Qi S, Song Y, Peng Y, Wang H, Long H, Yu X, Li Z, Fang L, Wu A, Luo W, Zhen Y, Zhou Y, Chen Y, Mai C, Liu Z, Fang W. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS One. 2012;7:e38842. doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert UD, Maciaczyk D, Doostkam S, Orr BA, Simons B, Bogiel T, Reithmeier T, Prinz M, Schubert J, Niedermann G, Brabletz T, Eberhart CG, Nikkhah G, Maciaczyk J. Activation of canonical WNT/beta-catenin signaling enhances in vitro motility of glioblastoma cells by activation of ZEB1 and other activators of epithelial-to-mesenchymal transition. Cancer Lett. 2012;325:42–53. doi: 10.1016/j.canlet.2012.05.024. [DOI] [PubMed] [Google Scholar]
- Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.E08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama K, Shapiro I, Guttman M, Hazan RB. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2:301–314. doi: 10.1016/S1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- Utsuki S, Sato Y, Oka H, Tsuchiya B, Suzuki S, Fujii K. Relationship between the expression of E-, N-cadherins and beta-catenin and tumor grade in astrocytomas. J Neurooncol. 2002;57:187–192. doi: 10.1023/A:1015720220602. [DOI] [PubMed] [Google Scholar]
- Motta FJ, Valera ET, Lucio-Eterovic AK, Queiroz RG, Neder L, Scrideli CA, Machado HR, Carlotti-Junior CG, Marie SK, Tone LG. Differential expression of E-cadherin gene in human neuroepithelial tumors. Genet Mol Res. 2008;7:295–304. doi: 10.4238/vol7-2gmr424. [DOI] [PubMed] [Google Scholar]
- Asano K, Duntsch CD, Zhou Q, Weimar JD, Bordelon D, Robertson JH, Pourmotabbed T. Correlation of N-cadherin expression in high grade gliomas with tissue invasion. J Neurooncol. 2004;70:3–15. doi: 10.1023/b:neon.0000040811.14908.f2. [DOI] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Rodriguez F, Giannini C, Scheithauer B, Necela BM, Sarkaria JN, Anastasiadis PZ. Misregulated E-cadherin expression associated with an aggressive brain tumor phenotype. PLoS One. 2010;5:e13665. doi: 10.1371/journal.pone.0013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, Isachenko N, Fouse SD, Phillips JJ, Cheresh DA, Park M, Bergers G. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer. 2010;49:775–790. doi: 10.1002/gcc.20787. [DOI] [PubMed] [Google Scholar]
- Huang ZY, Wu Y, Hedrick N, Gutmann DH. T-cadherin-mediated cell growth regulation involves G2 phase arrest and requires p21(CIP1/WAF1) expression. Mol Cell Biol. 2003;23:566–578. doi: 10.1128/MCB.23.2.566-578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Takeuchi T, Nagayama T, Ohtsuki Y, Furihata M. Zeb1-mediated T-cadherin repression increases the invasive potential of gallbladder cancer. FEBS Lett. 2009;583:430–436. doi: 10.1016/j.febslet.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Colman H, Aldape K. Molecular predictor s in glioblastoma: toward personalized therapy. Arch Neurol. 2008;65:877–883. doi: 10.1001/archneur.65.7.877. [DOI] [PubMed] [Google Scholar]
- Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, Jin J, Hong SC, Park WY, Lee DS, Kim H, Nam DH. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, Cuartas I, Raventos C, Martinez-Ricarte F, Poca MA, Garcia-Dorado D, Lahn MM, Yingling JM, Rodon J, Sahuquillo J, Baselga J, Seoane J. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J, Spang R, Storch A, Beier CP. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- Tso CL, Shintaku P, Chen J, Liu Q, Liu J, Chen Z, Yoshimoto K, Mischel PS, Cloughesy TF, Liau LM, Nelson SF. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4:607–619. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, Lee Y, Dia EQ, Yoshimoto K, Mischel PS, Liau LM, Cloughesy TF, Nelson SF. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, Turski A, Azodi Y, Yang Y, Doucette T, Colman H, Sulman EP, Lang FF, Rao G, Copray S, Vaillant BD, Aldape KD. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan HJ, Lee CC, Shih YL, Hueng DY, Ma HI, Lai JH, Wei HW, Lee HM. Osteopontin regulates human glioma cell invasiveness and tumor growth in mice. Neuro Oncol. 2010;12:58–70. doi: 10.1093/neuonc/nop013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Perderiset M, Francis F, Chelly J, Houdusse A, Milligan RA. Mechanism of microtubule stabilization by doublecortin. Mol Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Moores CA, Perderiset M, Kappeler C, Kain S, Drummond D, Perkins SJ, Chelly J, Cross R, Houdusse A, Francis F. Distinct roles of doublecortin modulating the microtubule cytoskeleton. Embo J. 2006;25:4448–4457. doi: 10.1038/sj.emboj.7601335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi H, Higginbotham H, Poon T, Tanaka T, Brinkman BC, Gleeson JG. Doublecortin maintains bipolar shape and nuclear translocation during migration in the adult forebrain. Nat Neurosci. 2006;9:779–786. doi: 10.1038/nn1704. [DOI] [PubMed] [Google Scholar]
- Kappeler C, Saillour Y, Baudoin JP, Tuy FP, Alvarez C, Houbron C, Gaspar P, Hamard G, Chelly J, Metin C, Francis F. Branching and nucleokinesis defects in migrating interneurons derived from doublecortin knockout mice. Hum Mol Genet. 2006;15:1387–1400. doi: 10.1093/hmg/ddl062. [DOI] [PubMed] [Google Scholar]
- des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, Catala M, Kahn A, Beldjord C, Chelly J. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/S0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Pramparo T, Youn YH, Yingling J, Hirotsune S, Wynshaw-Boris A. Novel embryonic neuronal migration and proliferation defects in Dcx mutant mice are exacerbated by Lis1 reduction. J Neurosci. 2010;30:3002–3012. doi: 10.1523/JNEUROSCI.4851-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis F, Koulakoff A, Boucher D, Chafey P, Schaar B, Vinet MC, Friocourt G, McDonnell N, Reiner O, Kahn A, McConnell SK, Berwald-Netter Y, Denoulet P, Chelly J. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/S0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- Brown JP, Couillard-Despres S, Cooper-Kuhn CM, Winkler J, Aigner L, Kuhn HG. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 2003;467:1–10. doi: 10.1002/cne.10874. [DOI] [PubMed] [Google Scholar]
- Yang HK, Sundholm-Peters NL, Goings GE, Walker AS, Hyland K, Szele FG. Distribution of doublecortin expressing cells near the lateral ventricles in the adult mouse brain. J Neurosci Res. 2004;76:282–295. doi: 10.1002/jnr.20071. [DOI] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, Aigner L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur J Neurosci. 2005;21:1–14. doi: 10.1111/j.1460-9568.2004.03813.x. [DOI] [PubMed] [Google Scholar]
- Ocbina PJ, Dizon ML, Shin L, Szele FG. Doublecortin is necessary for the migration of adult subventricular zone cells from neurospheres. Mol Cell Neurosci. 2006;33:126–135. doi: 10.1016/j.mcn.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu F, Liu YY, Zhao CH, You Y, Wang L, Zhang J, Wei B, Ma T, Zhang Q, Zhang Y, Chen R, Song H, Yang Z. Identification and characterization of neuroblasts in the subventricular zone and rostral migratory stream of the adult human brain. Cell Res. 2011;21:1534–1550. doi: 10.1038/cr.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YW, Curtis MA, Gibbons HM, Mee EW, Bergin PS, Teoh HH, Connor B, Dragunow M, Faull RL. Doublecortin expression in the normal and epileptic adult human brain. Eur J Neurosci. 2008;28:2254–2265. doi: 10.1111/j.1460-9568.2008.06518.x. [DOI] [PubMed] [Google Scholar]
- Omori Y, Suzuki M, Ozaki K, Harada Y, Nakamura Y, Takahashi E, Fujiwara T. Expression and chromosomal localization of KIAA0369, a putative kinase structurally related to Doublecortin. J Hum Genet. 1998;43:169–177. doi: 10.1007/s100380050063. [DOI] [PubMed] [Google Scholar]
- Xiong K, Luo DW, Patrylo PR, Luo XG, Struble RG, Clough RW, Yan XX. Doublecortin-expressing cells are present in layer II across the adult guinea pig cerebral cortex: partial colocalization with mature interneuron markers. Exp Neurol. 2008;211:271–282. doi: 10.1016/j.expneurol.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Xiong K, Chu Y, Luo DW, Luo XG, Yuan XY, Struble RG, Clough RW, Spencer DD, Williamson A, Kordower JH, Patrylo PR, Yan XX. Doublecortin expression in adult cat and primate cerebral cortex relates to immature neurons that develop into GABAergic subgroups. Exp Neurol. 2009;216:342–356. doi: 10.1016/j.expneurol.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwer RW, Sluiter AA, Balesar RA, Baayen JC, Noske DP, Dirven CM, Wouda J, van Dam AM, Lucassen PJ, Swaab DF. Mature astrocytes in the adult human neocortex express the early neuronal marker doublecortin. Brain. 2007;130(Pt 12):3321–3335. doi: 10.1093/brain/awm264. [DOI] [PubMed] [Google Scholar]
- Bloch J, Kaeser M, Sadeghi Y, Rouiller EM, Redmond DE Jr, Brunet JF. Doublecortin-positive cells in the adult primate cerebral cortex and possible role in brain plasticity and development. J Comp Neurol. 2011;519:775–789. doi: 10.1002/cne.22547. [DOI] [PubMed] [Google Scholar]
- Daou MC, Smith TW, Litofsky NS, Hsieh CC, Ross AH. Doublecortin is preferentially expressed in invasive human brain tumors. Acta Neuropathol. 2005;110:472–480. doi: 10.1007/s00401-005-1070-0. [DOI] [PubMed] [Google Scholar]
- Masui K, Mawatari SY, Suzuki SO, Iwaki T. Evaluation of sensitivity and specificity of doublecortin immunostatining for the detection of infiltrating glioma cells. Brain Tumor Pathol. 2008;25:1–7. doi: 10.1007/s10014-007-0225-1. [DOI] [PubMed] [Google Scholar]
- Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, Dobra A, Dressman HK, Bigner DD, Nevins JR, West M. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M, Santra S, Buller B, Santra K, Nallani A, Chopp M. Effect of doublecortin on self-renewal and differentiation in brain tumor stem cells. Cancer Sci. 2011;102:1350–1357. doi: 10.1111/j.1349-7006.2011.01952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M, Zhang X, Santra S, Jiang F, Chopp M. Ectopic doublecortin gene expression suppresses the malignant phenotype in glioblastoma cells. Cancer Res. 2006;66:11726–11735. doi: 10.1158/0008-5472.CAN-06-1978. [DOI] [PubMed] [Google Scholar]
- Santra M, Zheng X, Roberts C, Santra S, Lu M, Panda S, Jiang F, Chopp M. Single doublecortin gene therapy significantly reduces glioma tumor volume. J Neurosci Res. 2010;88:304–314. doi: 10.1002/jnr.22207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Wang X, Jin K, Zhu J, Wang Y, Xiong S, Mao Y, Zhou L. B7-H4 is preferentially expressed in non-dividing brain tumor cells and in a subset of brain tumor stem-like cells. J Neurooncol. 2008;89:121–129. doi: 10.1007/s11060-008-9601-x. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Tseng HC, Kulkarni AB, Tsai LH, Gleeson JG. Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron. 2004;41:215–227. doi: 10.1016/S0896-6273(03)00852-3. [DOI] [PubMed] [Google Scholar]
- Jin J, Suzuki H, Hirai S, Mikoshiba K, Ohshima T. JNK phosphorylates Ser332 of doublecortin and regulates its function in neurite extension and neuronal migration. Dev Neurobiol. 2010;70:929–942. doi: 10.1002/dneu.20833. [DOI] [PubMed] [Google Scholar]
- Schaar BT, Kinoshita K, McConnell SK. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 2004;41:203–213. doi: 10.1016/S0896-6273(03)00843-2. [DOI] [PubMed] [Google Scholar]
- Li L, He F, Litofsky NS, Recht LD, Ross AH. Profiling of genes expressed by PTEN haploinsufficient neural precursor cells. Mol Cell Neurosci. 2003;24:1051–1061. doi: 10.1016/j.mcn.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Piens M, Muller M, Bodson M, Baudouin G, Plumier JC. A short upstream promoter region mediates transcriptional regulation of the mouse doublecortin gene in differentiating neurons. BMC Neurosci. 2010;11:64. doi: 10.1186/1471-2202-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wang J, Fang B, Fang X, Lu Z. alpha-Catenin inhibits glioma cell migration, invasion, and proliferation by suppression of beta-catenin transactivation. J Neurooncol. 2011;103:445–451. doi: 10.1007/s11060-010-0413-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci. 2012;17:700–712. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Yoshimoto K, Guan Y, Hata N, Mizoguchi M, Sagata N, Murata H, Kuga D, Amano T, Nakamizo A, Sasaki T. Associations between microRNA expression and mesenchymal marker gene expression in glioblastoma. Neuro Oncol. 2012;14:1153–1162. doi: 10.1093/neuonc/nos145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Elias MC, Tozer KR, Silber JR, Mikheeva S, Deng M, Morrison RS, Manning TC, Silbergeld DL, Glackin CA, Reh TA, Rostomily RC. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005;7:824–837. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, Wang X, You Y, Yang Z, Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Zhang C, Zhang A, Wang K, Jia Z, Wang G, Han L, Kang C, Pu P. miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol Rep. 2012;27:1504–1510. doi: 10.3892/or.2012.1652. [DOI] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Farace MG, Ciafre SA. NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 2011;39:3892–3902. doi: 10.1093/nar/gkr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Agami R. Diverse w ays to control p27Kip1 function: miRNAs come into play. Cell Cycle. 2007;6:2742–2749. doi: 10.4161/cc.6.22.4900. [DOI] [PubMed] [Google Scholar]
- Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–920. doi: 10.3892/ijo_00000570. [DOI] [PubMed] [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- Wu DG, Wang YY, Fan LG, Luo H, Han B, Sun LH, Wang XF, Zhang JX, Cao L, Wang XR, You YP, Liu N. MicroRNA-7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chin Med J (Engl) 2011;124:2616–2621. [PubMed] [Google Scholar]
- Loftus JC, Ross JT, Paquette KM, Paulino VM, Nasser S, Yang Z, Kloss J, Kim S, Berens ME, Tran NL. miRNA expression profiling in migrating glioblastoma cells: regulation of cell migration and invasion by miR-23b via targeting of Pyk2. PLoS One. 2012;7:e39818. doi: 10.1371/journal.pone.0039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, Purow B, Abounader R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, Noske DP, Tannous BA, Wurdinger T. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A, Thomson D, Giles K, Maleki S, Mreich E, Wheeler H, Leedman P, Biggs M, Cook R, Little N, Robinson B, McDonald K. miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur J Cancer. 2011;47:953–963. doi: 10.1016/j.ejca.2010.11.026. [DOI] [PubMed] [Google Scholar]
- Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, Leung GK, Lu G, Chan DT, Bian XW, Kung HF, Poon WS, Lin MC. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang X, Wang H, Li Y, Yan W, Han L, Zhang K, Zhang J, Wang Y, Feng Y, Pu P, Jiang T, Kang C, Jiang C. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer. 2012;48:3104–3111. doi: 10.1016/j.ejca.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, Ge R, Jiang S, Li G, Chen Y, He ML, Kung HF, Lai L, Lin MC. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- Katakowski M, Zheng X, Jiang F, Rogers T, Szalad A, Chopp M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest. 2010;28:1024–1030. doi: 10.3109/07357907.2010.512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ, Yao Q, Zhou H, Qu LH. MicroRNA-195 plays a tumor-suppressor role in human glioblastoma cells by targeting signaling pathways involved in cellular proliferation and invasion. Neuro Oncol. 2012;14:278–287. doi: 10.1093/neuonc/nor216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, Paquis P, Preynat-Seauve O, Krause KH, Chneiweiss H, Virolle T. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. Embo J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang J, Feng Y, Li R, Sun X, Du W, Piao X, Wang H, Yang D, Sun Y, Li X, Jiang T, Kang C, Li Y, Jiang C. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012;44:1711–1717. doi: 10.1016/j.biocel.2012.06.027. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Kim SJ, Seo HH, Shin SP, Kim D, Park CS, Kim KT, Kim YH, Jeong JS, Kim IH. Over-expression of miR-145 enhances the effectiveness of HSVtk gene therapy for malignant glioma. Cancer Lett. 2012;320:72–80. doi: 10.1016/j.canlet.2012.01.029. [DOI] [PubMed] [Google Scholar]
- Koo S, Martin GS, Schulz KJ, Ronck M, Toussaint LG. Serial selection for invasiveness increases expression of miR-143/miR-145 in glioblastoma cell lines. BMC Cancer. 2012;12:143. doi: 10.1186/1471-2407-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lages E, Guttin A, El Atifi M, Ramus C, Ipas H, Dupre I, Rolland D, Salon C, Godfraind C, Defraipont F, Dhobb M, Pelletier L, Wion D, Gay E, Berger F, Issartel JP. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One. 2011;6:e20600. doi: 10.1371/journal.pone.0020600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. Proteomic and immunologic analyses of brain tumor exosomes. Faseb J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]


