Abstract
The prevalence of obesity has nearly doubled worldwide over the past three decades, but substantial differences exist between nations. Although these differences are partly due to the degree of westernization, genetic factors also contribute. To date, little is known about whether the same genes contribute to obesity-susceptibility in populations of different ancestry. We review the transferability of obesity-susceptibility loci (identified by genome-wide association studies) using both single nucleotide polymorphism (SNP) and locus-wide comparisons. SNPs in FTO and near MC4R, obesity-susceptibility loci first identified in Europeans, replicate widely across other ancestries. SNP-to-SNP comparisons suggest that more than half of the 36 body mass index-associated loci are shared across European and East Asian ancestry populations, whereas locus-wide analyses suggest that the transferability might be even more extensive. Furthermore, by taking advantage of differences in haplotype structure, populations of different ancestries can help to narrow down loci, thereby pinpointing causal genes for functional follow-up. Larger-scale genetic association studies in ancestrally diverse populations will be needed for in-depth and locus-wide analyses aimed at determining, with greater confidence, the transferability of loci and allowing fine-mapping. Understanding similarities and differences in genetic susceptibility across populations of diverse ancestries might eventually contribute to a more targeted prevention and customized treatment of obesity.
Obesity: an expanding problem globally
Worldwide, the prevalence of adult obesity has almost doubled over the past three decades [1]. The increase in the United States and other high-income countries has drawn attention because of the generally high prevalence of obesity in these countries [2-4], but the obesity epidemic is also spreading to middle- and low-income countries [1]. Globally, the prevalence of obesity increased from 4.8% in 1980 to 9.8% in 2008 for men, and from 7.9% to 13.8% in women, paralleled by an increase in mean body mass index (BMI) of 0.4 to 0.5 kg/m2 per decade (equivalent to 1.2 to 1.4 kg per decade for a person 1.6 to 1.8 m tall) [1]. By 2008, mean BMI had reached 23.8 kg/m2 for men and 24.1 kg/m2 for women [1].
However, substantial differences between nations have been noted; for example, since the 1980s, mean BMI has increased the most in Oceania (1.3 kg/m2 per decade in men, 1.8 kg/m2 per decade in women), whereas no change in BMI was observed in Central Africa (men) and Central and Eastern Europe (women) [1]. By 2008, the mean BMI in many countries from Melanesia, Micronesia and Polynesia was greater than 30 kg/m2, whereas in several countries of sub-Saharan Africa and in East, South and Southeast Asia, it was less than 21.5 kg/m2 [1]. Differences in mean BMI and obesity prevalence across nations are, at least in part, due to the degree that the westernized lifestyle has been adopted [5,6]. In addition, genetic factors contribute to obesity-susceptibility, with heritability estimates ranging between 40 and 70% across populations, without a systematic higher or lower influence for any specific ancestry [7]. Little is known, however, about whether the same genes contribute to obesity-susceptibility across all ancestries or whether there are obesity-susceptibility genes that are unique to specific ancestries.
Large-scale genome-wide association studies (GWAS) have identified at least 58 genetic loci that are robustly associated with obesity-related traits. The majority of loci have been discovered through GWAS in populations of European ancestry, but a growing number of studies are now being performed in populations of non-European ancestry. Here, we review the extent to which obesity-susceptibility loci are shared across populations of different ancestry and summarize the new insights that have been gained by cross-ancestry comparisons.
Why compare obesity-susceptibility across populations of diverse ancestry?
There are at least two reasons for comparing genetic loci across populations of diverse ancestry. First, cross-ancestry comparisons can shed light on the physiology that underlies obesity-susceptibility around the world. Genetic loci that are shared across most ancestries may be indicative of core physiological pathways that are implicated in obesity-susceptibility, irrespective of ancestry. Such pathways might relate to general mechanisms that control the regulation of energy balance and body-fat distribution. Alternatively, loci that are specific to a single ancestry might contribute to differences in obesity-susceptibility between populations. Such loci could explain, for example, why Pima Indians are much more susceptible to gaining weight than other Americans, even though they live in the same westernized environment [8], or why the prevalence of obesity-related comorbidities differ across different races and ethnicities [9].
A second reason for cross-ancestry comparison relates to the fine-mapping of established obesity-susceptibility loci. Genetic loci identified through GWAS often harbor multiple genes, or sometimes no genes at all, which represents a major barrier for the follow-up of discoveries into functional research. By taking advantage of differences in the genetic architecture between ancestries, genetic loci can be narrowed down, eventually to pinpoint the causal gene(s) and/or genetic variant(s) that underlie the observed GWAS association.
We will illustrate both applications using (publicly) available data from large-scale, and thus sufficiently powered, genetic association studies, and we report solely on genetic loci for which the association with the obesity trait reached genome-wide significance (P < 5 × 10-8).
Current status of obesity-susceptibility loci
The search for obesity-susceptibility loci in humans started in the 1990s with candidate gene and genome-wide linkage studies [10,11]. Despite the large number of studies conducted over the past 30 years, these two approaches have identified only a few genetic loci that are robustly associated with common obesity- or adiposity-related traits [10,11]. The limitation of these approaches can often be ascribed to the small-scale of most of the studies and consequently insufficient power to identify the expected small genetic effects, and to the limited knowledge of human genetic variation and linkage disequilibrium (LD), particularly in the early years.
The advent of the genome-wide association approach in 2005, however, has substantially accelerated the pace of gene discovery for many common diseases and traits [12,13], including obesity-related traits [11]. We focus our review on the GWAS-identified loci because of the robustness of the associations and the availability of GWAS across populations of diverse ancestry.
Obesity-susceptibility loci identified in populations of white-European ancestry
GWAS of obesity-related traits have been performed predominantly in populations of European ancestry, and have, to date, identified 35 loci for BMI, three loci for body-fat percentage, five loci for waist circumference, 14 loci for BMI-adjusted waist-to-hip ratio (WHRadjBMI), three loci for abdominal subcutaneous tissue (SAT) and visceral adipose tissue (VAT), six loci for extreme and early-onset obesity and nine loci for common childhood obesity. After accounting for the overlap of loci across traits, GWAS in populations of European ancestry have identified 54 obesity-susceptibility loci (Additional file 1; Figure 1).
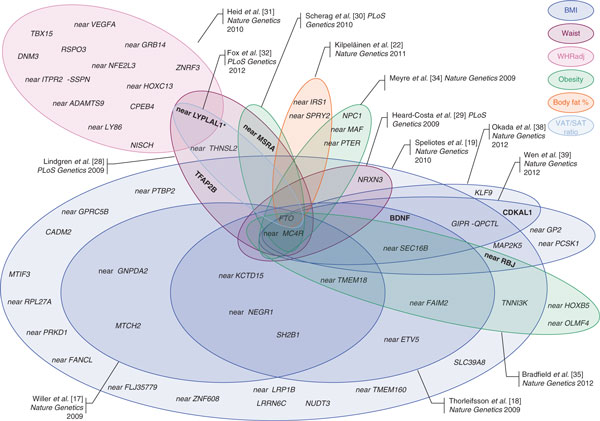
Figure 1.
Obesity-susceptibility loci discovered through genome-wide association studies (GWAS) for body mass index (blue), three waves of GWAS for waist circumference and waist-to-hip ratio (pink) and two waves of GWAS for extreme and early onset of obesity (green). Each Venn diagram represents the loci from one paper, except for papers that discovered only one locus, that is, the fat mass and obesity associated gene FTO [14,15,33] and the near-MC4R loci [16,36], for which no Venn diagram was drawn. An additional three BMI-associated loci (TOMM40-APOE-APOC1, SREBF2, and NTRK2) were identified using the gene-centric ITMAT-Broad-Candidate Gene Association Resource (IBC) array [21]; these are not depicted. Figure modified and updated from Loos [11].
Body mass index
To date, 32 BMI-associated loci have been identified over the course of four consecutive waves of GWAS meta-analyses; each subsequent wave being larger and more fruitful than the preceding. The first obesity-susceptibility locus, in the first intron of FTO, was identified by two relatively small GWAS (NGWAS ~5,000) in 2007 [14,15]. One year later, a GWAS meta-analysis for BMI of nearly 17,000 individuals confirmed the FTO locus and identified a second locus near MC4R [16]. In 2009, the GIANT (Genetic Investigation of ANthropometric Traits) consortium was established to further increase the sample size, combining 15 GWAS for BMI (NGWAS ~32,000) [17]. At the same time, the deCODE Genetics research group performed a GWAS for BMI of a similar size [18]. Both studies confirmed the FTO and near-MC4R loci, and identified an additional ten loci [17,18]. In 2010, the GIANT Consortium expanded further by combining 46 GWAS for BMI, including around 124,000 individuals in total, which confirmed the 12 previously established BMI loci and identified an additional 20 loci [19].
Besides GWAS, the IBC (ITMAT-Broad-Candidate Gene Association Resource (CARe)) gene-centric array, comprising nearly 50,000 single nucleotide polymorphisms (SNPs) across approximately 2,100 metabolic and cardiovascular-related loci, has been used to identify obesity-susceptibility loci [20]. A meta-analysis of the IBC array of nearly 93,000 individuals of European descent identified three additional BMI loci (TOMM40/APOE/APOC1, SREBF2, and NTRK2) and confirmed seven of the previous GWAS-identified BMI loci [21].
Taken together, a total of 35 BMI-associated loci have been reported for populations of European ancestry.
Body-fat percentage
While BMI is generally a good indicator of overall adiposity, it does not distinguish fat from lean mass. Hence, a GWAS meta-analysis for body-fat percentage, as a more accurate estimate of body composition, involving data from 15 studies (NGWAS ~36,500) was performed [22]. Besides the locus in FTO, loci near IRS1 and near SPRY2 were identified. Interestingly, the fat-percentage-increasing allele of the near-IRS1 locus is associated with a lower risk of type 2 diabetes [23] and cardiovascular disease [24], as well as with a favorable lipid profile [25]. This association may be mediated through the fact that the fat-percentage-increasing allele is associated with increased SAT, but not with the metabolically more harmful VAT [22]. These effects are significantly more pronounced in men than in women [22]. The near-SPRY2 locus that associates with body-fat percentage is 50 kb upstream of the gene and seems independent (LD r2 <0.01) of the near-SPRY2 locus found to be associated with type 2 diabetes in East Asians [26] and Europeans [27], which locates approximately 200 kb downstream of SPRY2.
Waist circumference
On the basis of the hypothesis that distinct physiological pathways govern overall adiposity and abdominal adiposity, GWAS have been performed using proxy traits for body-fat distribution. The first two GWAS meta-analyses, by the GIANT consortium [28] and the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) consortium [29], focused on waist circumference. All five identified loci were later confirmed by GWAS for BMI and for early-onset extreme obesity [19,30], suggesting that these loci are associated with overall adiposity, rather than specifically with body-fat distribution.
WHR adjusted for BMI
When the GIANT consortium expanded in 2010, WHR (instead of waist circumference) was chosen as the parameter used to assess fat distribution [31]. In addition, to account for possible influences of overall adiposity, WHR was further adjusted for BMI. This meta-analysis combined data from 32 GWAS (NGWAS ~77,000), and identified 14 WHRadjBMI loci [31]. Seven of the 14 loci have a significantly more pronounced effect in women than in men [31].
Abdominal subcutaneous and visceral adipose tissue
While WHR is a non-invasive and inexpensive estimate of abdominal obesity, it does not allow discrimination between SAT and VAT. This distinction is important as a propensity to store fat viscerally rather than subcutaneously (as assessed by VAT/SAT ratio) is metabolically more harmful. A GWAS meta-analysis of data on VAT and SAT, quantified using computed tomography (CT), from more than 10,000 individuals identified three loci [32]. The FTO locus was associated with SAT; the LYPLAL1 locus, previously identified in association with WHRadjBMI, was associated with VAT/SAT ratio; and a novel locus near THNSL2 was associated with VAT in women but not men [32].
Extreme and early-onset obesity
Individuals with early-onset and extreme obesity might be enriched for genetic variants that predispose to common obesity in the general population. Thus, GWAS comparing extremely obese individuals with lean controls might have greater statistical power than typical GWAS of quantitative obesity traits in the general population. Such GWAS have, to date, identified three additional obesity-susceptibility loci, while confirming the FTO, near-MC4R, and near MSRA loci [30,33,34].
Common childhood obesity
A meta-analysis of 14 GWAS of childhood obesity, defined as a BMI ≥95th percentile for a given age (<18 years of age), identified two loci that had not been identified before in studies of adults and confirmed seven previously identified (adult) BMI loci [35].
Obesity-susceptibility loci identified in populations of Indian Asian ancestry
In 2008, a GWAS for waist circumference in 2,684 Indian Asians identified the locus near-MC4R [36] at the same time that this locus was identified for BMI in Europeans [16]. No other or larger GWAS in populations of Indian Asian ancestry has been reported to date.
Obesity-susceptibility loci identified in populations of East Asian ancestry
The first GWAS in individuals of East Asian ancestry was performed in 2009. Genotypes of more than 8,800 Koreans were analyzed for association with eight metabolic and cardiovascular traits, including BMI and WHR [37]. This study confirmed the FTO locus but no new loci were identified for BMI [37]. For WHR, a SNP (rs2074356) in HECTD4 reached genome-wide significance [37]. While common in East Asians - minor allele frequency (MAF) 11% in CHB (Chinese Hans of Beijing), 20% in JPT (Japanese of Tokyo), 15% in Koreans - this SNP is not polymorphic in individuals of European or African descent. It is in moderate LD (LD r2CHB/JPT = 0.58) with the Glu504Lys coding variant in ALDH2, which encodes an enzyme involved in alcohol metabolism.
In 2012, two GWAS meta-analyses for BMI in East Asians were reported, each involving around 27,000 individuals (Additional file 1; Figure 1) [38,39]. Associations of 11 loci reached genome-wide significance, and four of these loci (KLF9, CDKAL1, near GP2 and near PCSK1) had not previously been reported as obesity-susceptibility loci in European ancestry populations [38,39]. The previously identified non-synonymous SNPs in a candidate gene study of PCSK1 in Europeans [40] seem independent of the locus reported in the GWAS of East Asian ancestry individuals (LD r2 < 0.10 in both the CHB/JPT and CEU (Utah residents with northern and western European ancestry) populations). We describe the overlap between European and East Asian ancestry BMI loci in greater detail below.
Taken together, GWAS in East Asians have identified one locus associated with WHR and four loci associated with BMI that had not been identified before in populations of European ancestry.
Obesity-susceptibility loci identified in populations of other ancestry
Two relatively small (NGWAS ~1,800) GWAS for BMI in individuals of African ancestry [41,42] and a GWAS of 1,792 Filipino women [43] confirmed some of the loci previously identified in GWAS of European ancestry populations, but none were able to identify any new loci. A low-density GWAS of 413 Pima Indians identified a locus in A2BP1 that is associated with body-fat percentage, which was not replicated in Old Order Amish or Europeans [44].
Large-scale GWAS meta-analyses will be needed to establish more convincingly whether the identified obesity-susceptibility loci are specific to these ancestries or whether they are shared with other ancestries.
Transferability of obesity-susceptibility loci across populations of different ancestry
Since the first GWAS discoveries, numerous studies have examined whether the identified obesity-susceptibility loci are transferable across populations of other ancestry. As more than 90% of the current obesity-susceptibility loci were first identified in populations of European ancestry, almost all studies reported to date aimed to replicate these European-identified loci in non-European ancestry populations. The majority of these replication studies limited their analyses to the GWAS-identified (index) SNP only. Few examined the wider locus surrounding the index SNP to account for potential differences in genetic architecture between the different ancestries. A major challenge that many studies face is the often limited statistical power to replicate associations as sample sizes tend to be relatively small. After all, the majority of obesity-susceptibility loci have been identified in GWAS meta-analyses of 30,000 to 124,000 individuals. Hence, large-scale studies will be required to confirm or refute with confidence whether a SNP or locus associates with obesity-related traits in other-ancestry populations.
We have focused our review of the transferability of obesity-susceptibility loci on observations from larger studies (N > 5,000 or the largest study for a given ancestry, typically N > 1,500) that examined the BMI-associated loci, for which there are convincing data from an already large body of literature. We have also taken advantage of the available data from the recent, and currently largest, GWAS of BMI in populations of East Asian [38,39] and European ancestry [19] to illustrate how the SNP-to-SNP versus locus-wide comparison can lead to different conclusions.
Transferability assessed by SNP-to-SNP comparisons
Studies that assess the transferability of the obesity-susceptibility loci by SNP-to-SNP analyses only test whether the GWAS-identified SNP shows association with obesity-related traits in their population. SNP-to-SNP analyses are cost-efficient, as only one SNP per locus needs to be genotyped. If, however, replication is tested in an ancestry that has a different genetic architecture (for example, haplotype structure differences or allelic heterogeneity) from the ancestry in which the discovery was made, then the interpretation of transferability is limited. For example, when making a discovery, we assume that the GWAS-identified (index) SNP is in strong LD with the causal variant (that was not genotyped) in the discovery population. If replication of this index SNP is tested in a second population with a different ancestry, it is possible that the LD between the index SNP and the causal SNP is weaker in this second ancestry population than that in the discovery population, resulting in a weaker or no association with the obesity trait. Yet, other SNPs in the same locus might show strong LD with the causal SNP in the second population, and if tested, they would show association with obesity-related traits. If they are not tested for association, as is the case in SNP-to-SNP comparisons, one may wrongly conclude that the locus does not transfer across ancestries. Thus, SNP-to-SNP comparison allows confirmation of the transferability of a locus when the index SNP shows association in other ancestries. If, however, the index SNP shows no association in other ancestries, transferability is not refuted. This is an important limitation to be kept in mind when interpreting the replication studies in the literature.
The FTO locus was first identified in European ancestry populations in which each additional minor allele increases BMI by 0.39 kg/m2 (equivalent to 1,230 g for a person 1.7m tall) and increases the risk of obesity by 1.2-fold [19]. Of all currently identified loci, the FTO locus has the largest effect on obesity-susceptibility and its BMI-increasing allele is common in European ancestry populations: approximately 18% carry two BMI-increasing alleles and another 49% carry one BMI-increasing allele. Even though the effect of this locus is the largest known and the allele frequency is common, the FTO locus explains only 0.31% of the inter-individual variation in BMI (Table 1) [19]. Because FTO was the first identified obesity-susceptibility locus, many replication efforts in non-European ancestry populations have been conducted. Small sample sizes may have led to inconsistent results in early studies, but in the past few years, there has been growing and convincing support for the transferability of the FTO locus across almost all ancestries that have been studied. More specifically, SNPs that cluster in the FTO locus have been found to associate with increased obesity-susceptibility in the Chinese [39,45-51], the Japanese [38,52-55], Koreans [37,56], Filipinos [43], the Vietnamese [57], Malays [46,49], Indian Asians [45,49,58-63], Pima Indians [64], Hispanics [65-69] and, although somewhat less consistently, also in Africans and African Americans [69-75].
Table 1.
Characteristics of the associations between GWAS-identified SNPs and BMI in populations of European and East Asian Ancestry
| Marker info | Results in European ancestry populations (derived from Speliotes et al. [19]) | Results in East Asian ancestry populations (derived from Wen et al. [39]) | Ancestry of discovery population | Reference(s) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | Effect allele | Other allele | Effect allele frequency | Effect size | Standard error | P-value | Explained variance (%) | Effect allele frequency | Effect size | Standard error | P-value | Explained variance (%) | ||
| FTO | rs17817449 | g | t | 40% | 0.080 | 0.004 | 1.4E-97 | 0.31% | 17% | 0.085 | 0.008 | 4.6E-27 | 0.20% | European | [14,15] |
| near MC4R | rs6567160 | c | t | 24% | 0.060 | 0.004 | 2.4E-41 | 0.13% | 21% | 0.056 | 0.007 | 2.8E-15 | 0.11% | European and Indian Asian | [16.36] |
| near TMEM18 | rs11127485 | t | c | 83% | 0.060 | 0.005 | 1.3E-33 | 0.10% | 91% | 0.051 | 0.010 | 1.2E-06 | 0.04% | [17,18] | |
| near SEC16B | rs574367 | t | g | 20% | 0.049 | 0.005 | 1.7E-25 | 0.08% | 20% | 0.066 | 0.007 | 9.5E-20 | 0.14% | European | [18] |
| BDNF | rs6265 | c | t | 81% | 0.043 | 0.005 | 2.1E-19 | 0.06% | 44% | 0.045 | 0.006 | 3.6E-13 | 0.10% | European | [18] |
| near GNPDA2 | rs10938397 | g | a | 43% | 0.041 | 0.004 | 2.2E-26 | 0.08% | 29% | 0.037 | 0.007 | 9.7E-08 | 0.06% | European | [17] |
| near ETV5 | rs7647305 | c | t | 79% | 0.039 | 0.005 | 1.2E-16 | 0.05% | 95% | 0.026 | 0.015 | 0.09 | 0.01% | European | [18] |
| near NEGR1 | rs2568958 | a | g | 61% | 0.033 | 0.004 | 8.7E-18 | 0.05% | 92% | 0.023 | 0.011 | 0.04 | 0.01% | European | [17,18] |
| near FAIM2 | rs7138803 | a | g | 38% | 0.032 | 0.004 | 4.2E-17 | 0.05% | 31% | 0.020 | 0.006 | 1.7E-03 | 0.02% | European | [18] |
| SH2B1 | rs4788102 | a | g | 40% | 0.029 | 0.004 | 1.0E-13 | 0.04% | 13% | 0.024 | 0.009 | 6.5E-03 | 0.01% | European | [17,18] |
| MTCH2 | rs3817334 | t | c | 41% | 0.024 | 0.004 | 1.9E-10 | 0.03% | 31% | 0.024 | 0.006 | 1.8E-04 | 0.02% | European | [17] |
| near KCTD15 | rs29941 | g | a | 67% | 0.021 | 0.004 | 1.1E-07 | 0.02% | 23% | 0.007 | 0.007 | 0.30 | 0.00% | European | [17,18] |
| near PRKD1 | rs11847697 | t | c | 4% | 0.053 | 0.010 | 2.8E-07 | 0.02% | 0% | European | [29] | ||||
| SLC39A8 | rs13107325 | t | c | 7% | 0.046 | 0.008 | 2.6E-08 | 0.03% | 0% | European | [29] | ||||
| TFAP2B | rs4715210 | t | c | 17% | 0.043 | 0.005 | 3.9E-18 | 0.05% | 21% | 0.038 | 0.007 | 1.6E-07 | 0.05% | European | [29] |
| near GPRC5B | rs12444979 | c | t | 86% | 0.042 | 0.006 | 2.5E-13 | 0.04% | 0% | European | [29] | ||||
| GIPR-QPTCL | rs2287019 | c | t | 80% | 0.032 | 0.005 | 1.4E-09 | 0.03% | 79% | 0.016 | 0.008 | 0.05 | 0.01% | European | [29] |
| MAP2K5 | rs2241423 | g | a | 78% | 0.032 | 0.004 | 1.0E-12 | 0.03% | 38% | 0.031 | 0.006 | 2.3E-09 | 0.05% | European | [29] |
| near ADCY3/RBJ | rs713586 | c | t | 47% | 0.030 | 0.004 | 3.1E-15 | 0.05% | 46% | 0.029 | 0.007 | 1.4E-13 | 0.04% | European | [29] |
| near FLJ35779 (HMGCR) | rs2112347 | t | g | 63% | 0.029 | 0.005 | 1.0E-13 | 0.04% | 44% | 0.018 | 0.006 | 3.9E-03 | 0.02% | European | [29] |
| near TMEM160 | rs3810291 | a | g | 67% | 0.028 | 0.005 | 5.1E-11 | 0.04% | 24% | 0.035 | 0.008 | 2.7E-05 | 0.04% | European | [29] |
| near LRP1B | rs2890652 | c | t | 18% | 0.028 | 0.005 | 3.6E-08 | 0.02% | 0% | European | [29] | ||||
| MTIF3 | rs4771122 | g | a | 24% | 0.027 | 0.005 | 2.0E-09 | 0.03% | 22% | 0.005 | 0.009 | 0.62 | 0.00% | European | [29] |
| CADM2 | rs13078807 | g | a | 20% | 0.026 | 0.005 | 3.9E-08 | 0.02% | 0% | European | [29] | ||||
| near FANCL | rs887912 | t | c | 29% | 0.024 | 0.004 | 6.3E-09 | 0.02% | 0% | European | [29] | ||||
| LRRN6C | rs10968576 | g | a | 32% | 0.024 | 0.004 | 2.5E-09 | 0.03% | 19% | 0.009 | 0.008 | 0.28 | 0.00% | European | [29] |
| NRXN3 | rs10150332 | c | t | 22% | 0.024 | 0.005 | 1.5E-07 | 0.02% | 0% | European | [29] | ||||
| near PTBP2 | rs1555543 | c | a | 59% | 0.024 | 0.005 | 5.2E-10 | 0.03% | 87% | 0.005 | 0.009 | 0.57 | 0.00% | European | [29] |
| TNNI3K | rs1514175 | a | g | 43% | 0.023 | 0.005 | 1.0E-09 | 0.03% | 79% | 0.029 | 0.007 | 6.4E-05 | 0.03% | European | [29] |
| NUDT3 | rs206936 | g | a | 20% | 0.020 | 0.005 | 3.3E-05 | 0.01% | 56% | 0.008 | 0.006 | 0.22 | 0.00% | European | [29] |
| near ZNF608 | rs4836133 | a | c | 48% | 0.018 | 0.005 | 6.4E-06 | 0.02% | 0% | European | [29] | ||||
| near RPL27A | rs4929949 | c | t | 52% | 0.018 | 0.005 | 1.6E-06 | 0.02% | 43% | 0.018 | 0.007 | 0.01 | 0.02% | European | [29] |
| GIPR/QPTCL | rs11671664 | g | a | 89% | 0.029 | 0.009 | 1.2E-03 | 0.02% | 50% | 0.044 | 0.006 | 5.9E-14 | 0.10% | East Asian | [38,39] |
| near KLF9 | rs11142387 | c | a | 53% | 0.003 | 0.005 | 0.45 | 0.00% | 46% | 0.040 | 0.007 | 1.3E-09 | 0.08% | East Asian | [38] |
| near GP2 | rs12597579 | c | t | 95% | 0.028 | 0.011 | 9.0E-03 | 0.01% | 80% | 0.036 | 0.006 | 1.0E-08 | 0.04% | East Asian | [39] |
| CDKAL1 | rs9356744 | t | c | 68% | 0.007 | 0.005 | 0.19 | 0.00% | 58% | 0.034 | 0.005 | 2.0E-11 | 0.06% | East Asian | [38,39] |
| near PCSK1 | rs261967 | c | a | 43% | 0.015 | 0.005 | 1.6E-03 | 0.01% | 41% | 0.031 | 0.005 | 5.1E-09 | 0.05% | East Asian | [39] |
The table includes 32 SNPs identified in European Ancestry populations [14-19,36] and four SNPs identified in East Asian ancestry populations [38,39], sorted by year of discovery and subsequently by effect size in the population in which discovery was made. Data were obtained and adapted from Speliotes et al. [19] and from Wen et al. [39].
The locus near-MC4R was the second GWAS-identified obesity-susceptibility locus, discovered in European [16] and Indian Asian [36] ancestry populations. This BMI-increasing allele has a substantially smaller effect on BMI (0.23 kg/m2/allele, equivalent to 665 g for a person 1.70 m tall) and obesity risk (odds ratio 1.11/allele) and is much less frequent (5% of Europeans carry two and 36% carry one BMI-increasing allele) when compared to the FTO locus (Table 1) [19]. The evidence for transferability of the near-MC4R locus, although not yet as strong as that for the FTO locus, is growing for a number of different ancestries, including the Chinese [39,51,76-79], the Japanese [38,80], Koreans [37,81], Filipinos [43], Indian Asians [60,82,83], Hispanics [84], and Africans and African Americans [18,74,85,86].
SNPs identified by the more recent GWAS meta-analyses, which included more than 30,000 individuals at the discovery stage, show much less consistency in SNP-to-SNP comparisons between ancestries. This is most probably due to too small sample sizes and thus a lack of statistical power to confirm associations, but it could also be due to differences in the genetic architecture of the different ancestry populations.
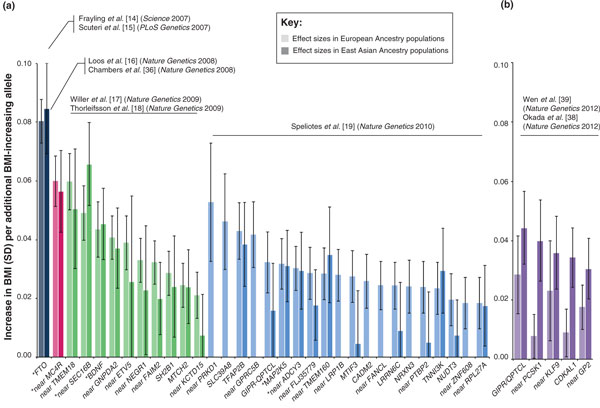
The recent two GWAS meta-analyses for BMI in East Asians reported association results that allowed comprehensive SNP-to-SNP comparisons [38,39]. More specifically, they provided association results from their GWAS-meta-analyses (N > 64,000) for all 32 previously GWAS-identified SNPs. In addition, they provided results from European ancestry GWAS meta-analyses for the newly identified loci in their GWAS (Table 1 and Figures 2 and 3). Of the 32 BMI-associated SNPs identified first in European ancestry populations, eight SNPs (in or near PRKD1, SLC39A8, GPRC5B, LRP1B, FANCL, NRXN3, CADM2 and ZNF608) are not polymorphic in East Asians (Table 1 and Figures 2a and 3b). Thus, on the basis of the SNP-to-SNP comparison, they are considered non-transferable. The associations for the remaining 24 SNPs with BMI in East Asians are all directionally consistent with those observed in European-ancestry populations (Figures 2a and 3a). For 6 of these 24 European ancestry-identified loci (in or near FTO, MC4R, BDNF, SEC16B, MAP2K5 and ADCY3/RBJ/POMC), associations reached genome-wide significance in East Asians and are therefore considered to be transferable across European and East Asian ancestry (Table 1 and Figures 1 and 2a). Even though the transferability of the remaining 18 loci identified in European populations seems less clear-cut, the effect sizes of 12 of these loci (in or near TMEM18, GNPDA2, ETV5, NEGR1, FAIM2, SH2B1, MTCH2, TFAP2B, FLJ35779, TMEM160, TNNI3K and RPL27A) on BMI in East Asians are comparable to those observed for European-ancestry populations (Table 1 and Figure 2a), with most reaching nominal significance and some approaching genome-wide significance (Table 1). The effects of the six other loci (in or near KCTD15, GIPR/QPTCL, MTIF3, LRRN6C, PTBP2 and NUDT3) are much less pronounced (P-values >0.20) in East Asian than in European ancestry populations (Table 1 and Figure 2a and 3b). Thus, of the 32 European-ancestry identified BMI loci, SNP-to-SNP comparisons suggest that 18 SNPs (56%) show comparable effects on BMI in European and East Asian ancestry populations, eight SNPs (25%) are non-polymorphic in East Asians, and for six SNPs (19%) there is as yet no evidence of transferability to East Asian ancestry populations.
Figure 2.
Effect sizes for (a) the 32 BMI-associated SNPs identified in European ancestry populations and (b) the five BMI-associated SNPs identified in East Asian ancestry populations. The lighter shaded bars represent the effect sizes (and 95% confidence interval (CI)) in European ancestry populations [19] (in both (a) and (b)), the darker shaded bars represent the effect sizes (and 95% CI) in East Asian ancestry populations [38,39]. Data were obtained and adapted from Wen et al. [39] and are presented in Table 1. Asterisks indicate SNPs that reached genome-wide significance in GWAS meta-analyses of East Asian ancestry populations [39]. SD, standard deviation.
Figure 3.
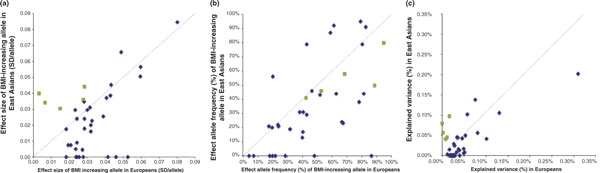
SNP-to-SNP comparison of (a) effects on BMI, (b) frequency of BMI-increasing alleles, and (c) explained BMI variance in East Asian ancestry populations (y-axis) and European ancestry populations (x-axis) of the 32 BMI-associated loci identified in European ancestry populations (blue diamonds) and five BMI-loci additionally identified in East Asian ancestry populations (green squares). Data were obtained and adapted from Wen et al. [39] and are presented in Table 1. SD, standard deviation.
As some of the BMI loci were identified by a GWAS meta-analysis of more than 123,000 European ancestry individuals, a larger sample size of East Asian ancestry individuals might be needed to provide firmer evidence of the transferability of the European ancestry BMI loci. Furthermore, as noted above, the SNP-to-SNP comparison has major limitations as it does not account for differences in genetic architecture between the two ancestries. This limitation is illustrated by the GIPR/QPTCL locus, which was first identified in European ancestry populations [19]. At first sight, the GIPR/QPTCL locus does not seem to be transferable to East Asian populations as the effect of the SNP (rs2287019) representing this locus in European ancestry populations is much less pronounced in East Asians [38,39] (Table 1 and Figure 2a). However, the GWAS in East Asian ancestry populations identified another SNP (rs11671664) in the same GIPR/QPTCL locus, the effect of which is less pronounced in European ancestry populations [19] (Table 1 and Figure 2b). The LD between the two SNPs is very low in East Asian ancestries (r2CHB/JPT = 0.08) and modest in European ancestries (r2CEU = 0.43), which suggests that the GIPR/QPTCL locus is an obesity-susceptibility locus in both European and East Asian ancestry populations, represented by ancestry-specific SNPs that each may be in LD with the same causal variant. A more in-depth analysis that takes advantage of differences in genetic architecture between ancestries will be required to pinpoint the causal variant(s).
Besides the GIPR/QPTCL locus, the recent GWAS meta-analyses in East Asian ancestry populations identified four BMI-associated loci that had not been identified before in large-scale GWAS of European ancestry populations [38,39] (Table 1 and Figures 1 and 2b). SNPs representing each of the four loci in East Asian ancestry populations show directionally consistent association in European ancestry populations (Figures 2b and 3a). For the near KLF9 locus, however, the association with BMI in European ancestry populations is not significant (P-value = 0.45; Table 1 and Figure 2b). For the CDKAL1 locus, the rs9356744 SNP identified by Wen et al. [39] suggests no evidence of transferability to European ancestry populations (P-value = 0.18) (Table 2 and Figure 2b). However, the rs2206734 SNP identified by Okada et al. [38] would suggest transferability of this locus (P-value = 0.005). While the LD between these two CDKAL1 SNPs is high in East Asian ancestry populations (r2CHB/JPT = 0.93), it is much more modest in European ancestry populations (r2CEU = 0.36), emphasizing the need for locus-wide comparisons (see also next section and Figure 4c,d). Even though the effect sizes of the two other loci (near GP2 and PCSK1) are relatively small, the associations reach nominal significance (Table 1 and Figure 2b). Taken together, the SNP-to-SNP comparisons suggest that two of the four loci identified in East Asian ancestry populations show some evidence of transferability to European ancestry populations, albeit this evidence is not as convincing as for the transferability of European-identified loci to East Asian ancestries.
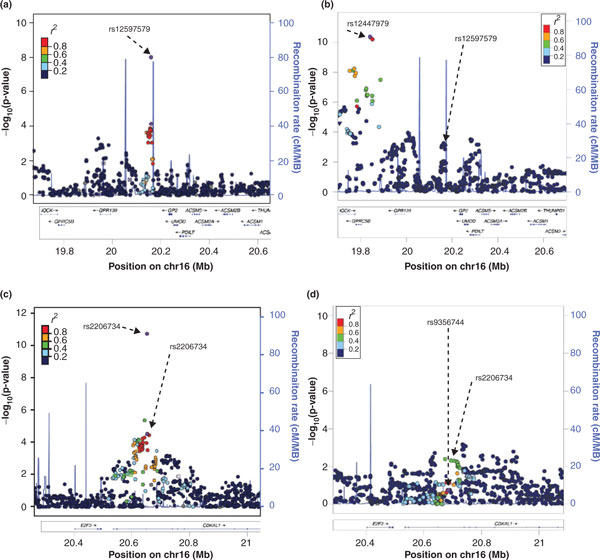
Figure 4.
Regional plots of (a,b) the GP2/GPRC5B and (c,d) the CDKAL1 loci in East Asian ancestry populations [39]and European ancestry populations, respectively [19]. SNPs are plotted by position on the chromosome against association with BMI (-log10 P-value). Recombination rates (from HapMap) are plotted in blue to reflect the local LD structure. The SNPs surrounding the most significant SNP (in purple) are color coded to reflect their LD with this SNP (r2 values from the HapMap CHB/JPT and CEU data, respectively).
In an overall SNP-to-SNP comparison of the 36 BMI-associated loci (Figure 3), we see that SNPs that were first identified in European ancestry populations have generally a larger effect and greater explained variance in European ancestry populations than in East Asian ancestry populations and, vice versa, that SNPs first identified in East Asian ancestry populations have a larger effect and explained variance in East Asian than in European ancestry populations. This is not a surprise and is consistent with the fact that SNPs with larger effects and greater explained variance are statistically more easily discovered (Figure 3a,c). The frequency of the BMI-increasing allele of European-identified SNPs tends to be higher in European than in East Asian ancestries. Interestingly, the frequency of the BMI-increasing allele of East Asian-identified SNPs is also higher in European than in East Asian ancestry populations (Figure 3b).
Taken together, the SNP-to-SNP comparison suggests that around 50% of the BMI-associated loci identified in European ancestry populations transfer to East Asian ancestry populations and vice versa. However, the non-transferability of the remaining 50% of BMI-associated loci cannot be substantiated because of the relatively small sample size and thus the insufficient statistical power of the East Asian GWAS meta-analyses to replicate some of the recently identified loci in Europeans, and also because of the limitations of the SNP-to-SNP comparison.
Transferability assessed by locus-wide comparisons
In locus-wide comparisons, not only is the GWAS-identified (index) SNP examined for transferability to another ancestry but also the SNPs surrounding the index SNP are tested. Locus-wide comparisons are more comprehensive and account for differences in the genetic architecture between ancestries. The data needed for such comparisons are not, however, always available. For example, we showed earlier that of the 32 GWAS-identified BMI-associated SNPs found in European ancestry populations, 8 SNPs were non-polymorphic and another 6 showed no convincing evidence of association in East Asian ancestry populations. To firmly exclude the transferability of these 14 loci, not only the index SNP but also all other SNPs in the region must be examined, but these data are not publicly available at present.
Using the publicly available data from the latest GWAS meta-analysis for BMI from the GIANT consortium [19], we illustrate how the BMI-associated loci identified in East Asian ancestry populations, of which the SNP-to-SNP transferability to European ancestry populations was doubtful, do show evidence of transferability when the whole locus is examined. The first example focuses on the GP2 locus, of which the rs12597579 SNP is associated with BMI at genome-wide significant levels in East Asians, whereas the association in European ancestry populations, though directionally consistent with that in East Asians, is not as convincing (P = 9 × 10-3; Table 1). When examining the wider locus, however, SNPs at less than 325 kb downstream of GP2-rs12597579 show genome-wide significant association in European ancestry individuals, of which the most significant SNP (rs12444979 near GPRC5B) is not polymorphic in East Asians (Table 1 and Figure 4a,b). Thus, although the SNP-to-SNP comparisons discussed earlier suggested possible transferability of GP2-rs1257579 to Europeans and no evidence of transferability of GPRC5B-rs12444979 to East Asians (Table 1 and Figure 2), a locus-wide comparison depicts a more comprehensive picture, suggesting that GP2 and GPRC5B might be part of the same locus, which possibly represents one causal gene that is common to European and East Asian ancestry populations. Interestingly, in European ancestry populations, GPRC5B-rs12444979 is in perfect LD with a copy number variant [19].
Our second example focuses on rs9356744 in CDKAL1, which showed genome-wide significant association with BMI in East Asian ancestry populations [38,39] but not in European ancestry populations (P-value = 0.19) [19] (Table 1 and Figures 2 and 4). When examining the locus surrounding rs9356744, however, there is evidence that other SNPs that are not in LD with rs9356744 are also associated with BMI in European ancestry populations (P-values <10-3) [19] (Figure 4d), supporting the transferability of the CDKAL1 locus.
More rigorous analyses of each of the extended loci will be needed to establish transferability across ancestries, but these examples of locus-wide examinations provide preliminary insights to suggest that the transferability of obesity-susceptibility loci across ancestries might be more extensive than is suggested by SNP-to-SNP comparisons.
Fine mapping of obesity-susceptibility loci using data from populations of different ancestry
Differences in the genetic architecture and LD structure of different ancestries can help to narrow down the locus in order to pinpoint the causal gene or variants, at least for loci that are transferable across populations of different ancestry. As the LD between SNPs in African ancestry populations is generally less extensive than that in European and East Asian ancestry populations, African populations are particularly suited for fine-mapping efforts. To date, the FTO locus has been the focus of most fine-mapping efforts [70,75,87], mainly because, of all the obesity-susceptibility loci, FTO has SNPs that show the strongest associations with obesity-related traits, and the FTO locus is one of the most generally transferable obesity-susceptibility loci. The most recent and largest fine-mapping project for the FTO locus has been reported by the PAGE (Population Architecture using Genomics and Epidemiology) study group [75]. The FTO locus is represented by a cluster of SNPs in the first intron of FTO at chr16q12.2. In the PAGE study, the genotypes of 3,756 SNPs across a 646-kb region in the first intron were interrogated for association with BMI in more than 20,000 African Americans [75]. The most significant association with BMI in African Americans was observed for rs56137030. This SNP represents a cluster (r2CEU >0.50) of 103 SNPs in Europeans, but in African Americans - due to weaker correlations between SNPs - this cluster includes only 29 SNPs, spanning 44.5 kb [75]. Six of the 29 SNPs locate within intronic regulatory elements, two of which are predicted to have allele-specific binding affinities for different transcription factors [75].
This example illustrates the great value of locus-wide comparisons using populations of different ancestries. The weaker the LD structure and the larger the sample size of the other-ancestry population, the greater the power to narrow-down a locus. It should be noted that this approach requires the disease-locus to be transferable across the ancestries.
Conclusions and future steps
Using the data currently available, we have shown that of all GWAS-identified loci, at least the FTO and near-MC4R loci seem to affect obesity-susceptibility in a wide range of ancestries. While much less information is available for other obesity-susceptibility loci, SNP-to-SNP comparisons of associations between European and East Asian ancestry populations suggest that at least half of the BMI-associated loci are shared across these two ancestries. Locus-wide comparisons suggest that the transferability might be even more extensive, but more rigorous analyses with larger samples will be required to establish these preliminary observations more firmly. The transferability of these loci is not unexpected: we might speculate that the general physiological pathways underlying obesity-susceptibility are shared across populations of different ancestries, whereas ancestry-specific susceptibilities are more likely to be caused by low-frequency variants that have not yet been identified. With the new genotyping arrays that focus on low-frequency and rare variants, studying populations of diverse ancestry will become even more important as such variants are more likely to be ancestry-specific.
At present, the publicly available data on the genetic susceptibility to obesity in non-European ancestry populations is still limited. More and larger-scale collaborations, involving more ancestrally diverse populations with genome-wide genotype data of common and low-frequency variants, will be needed to establish the transferability of obesity-susceptibility loci to other ancestries. Furthermore, cross-ancestry comparison has the potential to fine-map loci that harbor multiple genes and eventually to pinpoint one or a few genes for functional follow-up analyses. Fine-mapping of loci is a critical step in the translation of GWAS discoveries into functional research, and more large-scale efforts such as those for the FTO locus by the PAGE study will be important. Eventually, studying genetic susceptibility across populations of different ancestry will contribute to a better understanding of the mechanisms underlying obesity at a global level. Furthermore, understanding similarities and differences in genetic susceptibility across populations of diverse ancestries might eventually contribute to a more targeted prevention and customized treatment of obesity.
Abbreviations
BMI: body mass index; CHB: Chinese Hans of Beijing; CEU: Utah residents with northern and western European ancestry; GIANT Consortium: Genetic Investigation of ANthropometric Traits Consortium; GWAS: genome-wide association study; JPT: Japanese of Tokyo; LD: linkage disequilibrium; SAT: subcutaneous adipose tissue; SNP: single nucleotide polymorphism; VAT: visceral adipose tissue; WHR: waist-to-hip ratio.
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
Table S1. Overview of single nucleotide polymorphisms (SNPs) associated with obesity-related traits (P < 5 × 10-8) identified by genome-wide association studies (GWAS) and GWAS-meta-analyses, sorted by chromosome and position.
Contributor Information
Yingchang Lu, Email: kevin.lu@mssm.edu.
Ruth JF Loos, Email: ruth.loos@mssm.edu.
References
- Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, Singh GM, Gutierrez HR, Lu Y, Bahalim AN, Farzadfar F, Riley LM, Ezzati M. Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Body Mass Index) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999-2004. JAMA. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- International Association for the Study of Obesity. http://www.iaso.org/resources/obesity-data-portal/resources/trends/
- Ng SW, Popkin BM. Time use and physical activity: a shift away from movement across the globe. Obes Rev. 2012;13:659–680. doi: 10.1111/j.1467-789X.2011.00982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popkin BM, Adair LS, Ng SW. Global nutrition transition and the pandemic of obesity in developing countries. Nutr Rev. 2012;70:3–21. doi: 10.1111/j.1753-4887.2011.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, Ong KK. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Pettitt DJ, Saad MF, Charles MA, Nelson RG, Howard BV, Bogardus C, Bennett PH. Obesity in the Pima Indians: its magnitude and relationship with diabetes. Am J Clin Nutr. 1991;53:1543S–1551S. doi: 10.1093/ajcn/53.6.1543S. [DOI] [PubMed] [Google Scholar]
- Cossrow N, Falkner B. Race/ethnic issues in obesity and obesity-related comorbidities. J Clin Endocrinol Metab. 2004;89:2590–2594. doi: 10.1210/jc.2004-0339. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Pérusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Loos RJF. In: Adipose Tissue Biology. Symonds ME, editor. New York: Springer; 2012. Genetics determinants of adiposity. pp. 317–378. [Google Scholar]
- Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A Catalog of Published Genome-wide Association Studies. http://www.genome.gov/gwastudies/
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLos Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SI. Prostate, Lung, Colorectal and Ovarian (PLCO)Cancer Screening Trial; Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS. et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segrè AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpeläinen TO. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT. et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lanktree MB, Taylor KC, Hakonsarson H, Lange LA, Keating BJ. IBC 50K SNP array BMI Consortium. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum Mol Genet. 2013;22:184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpeläinen TO, Zillikens MC, Stančákova A, Finucane FM, Ried JS, Langenberg C, Zhang W, Beckmann JS, Luan J, Vandenput L, Styrkarsdottir U, Zhou Y, Smith AV, Zhao JH, Amin N, Vedantam S, Shin SY, Haritunians T, Fu M, Feitosa MF, Kumari M, Halldorsson BV, Tikkanen E, Mangino M, Hayward C, Song C, Arnold AM, Aulchenko YS, Oostra BA, Campbell H. et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rung J, Cauchi S, Albrechtsen A, Shen L, Rocheleau G, Cavalcanti-Proença C, Bacot F, Balkau B, Belisle A, Borch-Johnsen K, Charpentier G, Dina C, Durand E, Elliott P, Hadjadj S, Järvelin MR, Laitinen J, Lauritzen T, Marre M, Mazur A, Meyre D, Montpetit A, Pisinger C, Posner B, Poulsen P, Pouta A, Prentki M, Ribel-Madsen R, Ruokonen A, Sandbaek A. et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P. et al. Genomewide association analysis of coronary artery disease. New Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu XO, Long J, Cai Q, Qi L, Xiang YB, Cho YS, Tai ES, Li X, Lin X, Chow WH, Go MJ, Seielstad M, Bao W, Li H, Cornelis MC, Yu K, Wen W, Shi J, Han BG, Sim XL, Liu L, Qi Q, Kim HL, Ng DP, Lee JY, Kim YJ, Li C, Gao YT, Zheng W, Hu FB. Identification of new genetic risk variants for Type 2 diabetes. PLoS Genet. 2010;6:e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, Speliotes EK, Thorleifsson G, Willer CJ, Herrera BM, Jackson AU, Lim N, Scheet P, Soranzo N, Amin N, Aulchenko YS, Chambers JC, Drong A, Luan J, Lyon HN, Rivadeneira F, Sanna S, Timpson NJ, Zillikens MC, Zhao JH, Almgren P, Bandinelli S, Bennett AJ, Bergman RN, Bonnycastle LL. et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard-Costa NL, Zillikens MC, Monda KL, Johansson A, Harris TB, Fu M, Haritunians T, Feitosa MF, Aspelund T, Eiriksdottir G, Garcia M, Launer LJ, Smith AV, Mitchell BD, McArdle PF, Shuldiner AR, Bielinski SJ, Boerwinkle E, Brancati F, Demerath EW, Pankow JS, Arnold AM, Chen YD, Glazer NL, McKnight B, Psaty BM, Rotter JI, Amin N, Campbell H, Gyllensten U. et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, Müller TD, Grallert H, Wichmann HE, Balkau B, Heude B, Jarvelin MR, Hartikainen AL, Levy-Marchal C, Weill J, Delplanque J, Körner A, Kiess W, Kovacs P, Rayner NW, Prokopenko I, McCarthy MI, Schäfer H, Jarick I, Boeing H, Fisher E, Reinehr T, Heinrich J, Rzehak P, Berdel D. et al. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in french and german study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Mägi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpeläinen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL. et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K. GIANT Consortium; MAGIC Consortium; GLGC Consortium; Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, Muller TD, Grallert H, Illig T, Wichmann HE, Rief W, Schäfer H, Hebebrand J. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyre D, Delplanque J, Chèvre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proença C, Gaget S, Körner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, Hypponen E, Holst C, Valcarcel B, Thiering E, Salem RM, Schumacher FR, Cousminer DL, Sleiman PM, Zhao J, Berkowitz RI, Vimaleswaran KS, Jarick I, Pennell CE, Evans DM, St Pourcain B, Berry DJ, Mook-Kanamori DO, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, van der Valk RJ, de Jongste JC, Postma DS. et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, Balding D, Scott J, Kooner JS. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK. et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, Hosono N, Maeda S, Wen W, Dorajoo R, Go MJ, Zheng W, Kato N, Wu JY, Lu Q. GIANT consortium; Tsunoda T, Yamamoto K, Nakamura Y, Kamatani N, Tanaka T. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet. 2012;44:302–306. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L, Chen CH, Delahanty RJ, Okada Y, Tabara Y, Gu D, Zhu D, Haiman CA, Mo Z, Gao YT, Saw SM, Go MJ, Takeuchi F, Chang LC, Kokubo Y, Liang J, Hao M, Le Marchand L, Zhang Y, Hu Y, Wong TY, Long J, Han BG, Kubo M, Yamamoto K. et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44:307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E, Guerardel A, Boutin P, Jouret B, Heude B, Balkau B, Tichet J, Marre M, Potoczna N, Horber F, Le Stunff C, Czernichow S, Sandbaek A, Lauritzen T, Borch-Johnsen K, Andersen G, Kiess W, Körner A, Kovacs P, Jacobson P, Carlsson LM, Walley AJ, Jørgensen T, Hansen T, Pedersen O. et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Chiang CW, Palmer CD, Tayo BO, Lettre G, Butler JL, Hackett R, Adeyemo AA, Guiducci C, Berzins I, Nguyen TT, Feng T, Luke A, Shriner D, Ardlie K, Rotimi C, Wilks R, Forrester T, McKenzie CA, Lyon HN, Cooper RS, Zhu X, Hirschhorn JN. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MC, Hester JM, Wing MR, Li J, Xu J, Hicks PJ, Roh BH, Lu L, Divers J, Langefeld CD, Freedman BI, Palmer ND, Bowden DW. Genome-wide association of BMI in African Americans. Obesity (Silver Spring) 2012;20:622–627. doi: 10.1038/oby.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau-Chonka DC, Marvelle AF, Lange EM, Lee NR, Adair LS, Lange LA, Mohlke KL. Genome-wide association study of anthropometric traits and evidence of interactions with age and study year in Filipino women. Obesity (Silver Spring) 2011;19:1019–1027. doi: 10.1038/oby.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Hanson RL, Traurig MT, Muller YL, Kaur BP, Perez JM, Meyre D, Fu M, Körner A, Franks PW, Kiess W, Kobes S, Knowler WC, Kovacs P, Froguel P, Shuldiner AR, Bogardus C, Baier LJ. Evaluation of A2BP1 as an obesity gene. Diabetes. 2010;59:2837–2845. doi: 10.2337/db09-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kilpeläinen TO, Liu C, Zhu J, Liu Y, Hu C, Yang Z, Zhang W, Bao W, Cha S, Wu Y, Yang T, Sekine A, Choi BY, Yajnik CS, Zhou D, Takeuchi F, Yamamoto K, Chan JC, Mani KR, Been LF, Imamura M, Nakashima E, Lee N, Fujisawa T, Karasawa S, Wen W, Joglekar CV, Lu W, Chang Y. et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55:981–995. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JT, Dorajoo R, Seielstad M, Sim XL, Ong RT, Chia KS, Wong TY, Saw SM, Chew SK, Aung T, Tai ES. FTO variants are associated with obesity in the Chinese and Malay populations in Singapore. Diabetes. 2008;57:2851–2857. doi: 10.2337/db08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, Chen Z, Yu L, Yang Y, Feng G, Li J, Zhang J, Liu S, Zhang Z, He L, Xu H. Meta-analysis added power to identify variants in FTO associated with type 2 diabetes and obesity in the Asian population. Obesity. 2010;18:1619–1624. doi: 10.1038/oby.2009.469. [DOI] [PubMed] [Google Scholar]
- Ng MCY, Tam CHT, So WY, Ho JSK, Chan AW, Lee HM, Wang Y, Lam VKL, Chan JCN, Ma RCW. Implication of genetic variants near NEGR1, SEC16B, TMEM18, ETV5/DGKG, GNPDA2, LIN7C/BDNF, MTCH2, BCDIN3D/FAIM2, SH2B1, FTO, MC4R, and KCTD15 with obesity and type 2 diabetes in 7705 Chinese. J Clin Endocrinol Metab. 2010;95:2418–2425. doi: 10.1210/jc.2009-2077. [DOI] [PubMed] [Google Scholar]
- Dorajoo R, Blakemore AI, Sim X, Ong RT, Ng DP, Seielstad M, Wong TY, Saw SM, Froguel P, Liu J, Tai ES. Replication of 13 obesity loci among Singaporean Chinese, Malay and Asian-Indian populations. Int J Obes (Lond) 2012;36:159–163. doi: 10.1038/ijo.2011.86. [DOI] [PubMed] [Google Scholar]
- Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, Li HY, Kuo SS, Lee KC, Chuang LM. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57:2245–2252. doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CYY, Tso AWK, Cheung BMY, Xu A, Ong KL, Fong CHY, Wat NMS, Janus ED, Sham PC, Lam KSL. Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a Chinese population. J Clin Endocrinol Metab. 2010;95:1395–1403. doi: 10.1210/jc.2009-1465. [DOI] [PubMed] [Google Scholar]
- Hotta K, Nakata Y, Matsuo T, Kamohara S, Kotani K, Komatsu R, Itoh N, Mineo I, Wada J, Masuzaki H, Yoneda M, Nakajima A, Miyazaki S, Tokunaga K, Kawamoto M, Funahashi T, Hamaguchi K, Yamada K, Hanafusa T, Oikawa S, Yoshimatsu H, Nakao K, Sakata T, Matsuzawa Y, Tanaka K, Kamatani N, Nakamura Y. Variations in the FTO gene are associated with severe obesity in the Japanese. J Hum Genet. 2008;53:546–553. doi: 10.1007/s10038-008-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, Kawamori R, Nakamura Y, Maeda S. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–795. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
- Tabara Y, Osawa H, Guo H, Kawamoto R, Onuma H, Shimizu I, Takara Y, Nishida W, Yamamoto M, Makino H, Kohara K, Miki T. Prognostic significance of FTO genotype in the development of obesity in Japanese: the J-SHIPP study. Int J Obes. 2009;33:1243–1248. doi: 10.1038/ijo.2009.161. [DOI] [PubMed] [Google Scholar]
- Karasawa S, Daimon M, Sasaki S, Toriyama S, Oizumi T, Susa S, Kameda W, Wada K, Muramatsu M, Fukao A, Kubota I, Kawata S, Kayama T, Kato T. Association of the common fat mass and obesity associated (FTO) gene polymorphism with obesity in a Japanese population. Endocr J. 2010;57:293–301. doi: 10.1507/endocrj.K09E-305. [DOI] [PubMed] [Google Scholar]
- Cha SW, Choi SM, Kim KS, Park BL, Kim JR, Kim JY, Shin HD. Replication of genetic effects of FTO polymorphisms on BMI in a Korean population. Obesity (SilverSpring) 2008;16:2187–2189. doi: 10.1038/oby.2008.314. [DOI] [PubMed] [Google Scholar]
- Binh TQ, Phuong PT, Nhung BT, Thoang DD, Lien HT, Thanh DV. Association of the common FTO-rs9939609 polymorphism with type 2 diabetes, independent of obesity-related traits in a Vietnamese population. Gene. 2013;513:31–35. doi: 10.1016/j.gene.2012.10.082. [DOI] [PubMed] [Google Scholar]
- Yajnik CS, Janipalli CS, Bhaskar S, Kulkarni SR, Freathy RM, Prakash S, Mani KR, Weedon MN, Kale SD, Deshpande J, Krishnaveni GV, Veena SR, Fall CH, McCarthy MI, Frayling TM, Hattersley AT, Chandak GR. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia. 2009;52:247–252. doi: 10.1007/s00125-008-1186-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramya K, Radha V, Ghosh S, Majumder PP, Mohan V. Genetic variations in the FTO gene are associated with type 2 diabetes and obesity in south Indians (CURES-79). Diabetes Technol Ther. 2011;13:33–42. doi: 10.1089/dia.2010.0071. [DOI] [PubMed] [Google Scholar]
- Taylor AE, Sandeep MN, Janipalli CS, Giambartolomei C, Evans DM, Kranthi Kumar MV, Vinay DG, Smitha P, Gupta V, Aruna M, Kinra S, Sullivan RM, Bowen L, Timpson NJ, Davey Smith G, Dudbridge F, Prabhakaran D, Ben-Shlomo Y, Reddy KS, Ebrahim S, Chandak GR. Associations of FTO and MC4R variants with obesity traits in Indians and the role of rural/urban environment as a possible effect modifier. J Obes. 2011;2011:307542. doi: 10.1155/2011/307542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan G, Tabassum R, Mahajan A, Dwivedi OP, Mahendran Y, Kaur I, Nigam S, Dubey H, Varma B, Madhu SV, Mathur SK, Ghosh S, Tandon N, Bharadwaj D. Common variants of FTO and the risk of obesity and type 2 diabetes in Indians. J Hum Genet. 2011;56:720–726. doi: 10.1038/jhg.2011.87. [DOI] [PubMed] [Google Scholar]
- Vasan SK, Fall T, Neville MJ, Antonisamy B, Fall CH, Geethanjali FS, Gu HF, Raghupathy P, Samuel P, Thomas N, Brismar K, Ingelsson E, Karpe F. Associations of variants in FTO and near MC4R with obesity traits in South Asian Indians. Obesity (Silver Spring) 2012;20:2268–2277. doi: 10.1038/oby.2012.64. [DOI] [PubMed] [Google Scholar]
- Sanghera D, Ortega L, Han S, Singh J, Ralhan S, Wander G, Mehra N, Mulvihill J, Ferrell R, Nath S, Kamboh M. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Hanson RL, Ortiz D, Wiedrich C, Kobes S, Knowler WC, Bogardus C, Baier LJ. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761 and CDKN2B with type 2 diabetes and related quantitative traits in Pima indians. Diabetes. 2008. p. db08-0877. [DOI] [PMC free article] [PubMed]
- Villalobos-Comparán M, Teresa Flores-Dorantes M, Teresa Villarreal-Molina M, Rodríguez-Cruz M, García-Ulloa AC, Robles L, Huertas-Vázquez A, Saucedo-Villarreal N, López-Alarcón M, Sánchez-Muñoz F, Domínguez-López A, Gutiérrez-Aguilar R, Menjivar M, Coral-Vázquez R, Hernández-Stengele G, Vital-Reyes VS, Acuña-Alonzo V, Romero-Hidalgo S, Ruiz-Gómez DG, Riaño-Barros D, Herrera MF, Gómez-Pérez FJ, Froguel P, García-García E, Teresa Tusié-Luna M, Aguilar-Salinas CA, Canizales-Quinteros S. The FTO gene is associated with adulthood obesity in the Mexican population. Obesity (Silver Spring) 2008;16:2296–2301. doi: 10.1038/oby.2008.367. [DOI] [PubMed] [Google Scholar]
- Dong C, Beecham A, Slifer S, Wang L, McClendon MS, Blanton SH, Rundek T, Sacco RL. Genome-wide linkage and peak-wide association study of obesity-related quantitative traits in Caribbean Hispanics. Hum Genet. 2011;129:209–219. doi: 10.1007/s00439-010-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing MR, Ziegler J, Langefeld CD, Ng MC, Haffner SM, Norris JM, Goodarzi MO, Bowden DW. Analysis of FTO gene variants with measures of obesity and glucose homeostasis in the IRAS family study. Hum Genet. 2009;125:615–626. doi: 10.1007/s00439-009-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, You NC, Hsu YH, Howard BV, Langer RD, Manson JE, Nathan L, Niu T, Tinker L, Liu S. FTO polymorphisms are associated with obesity but not diabetes risk in postmenopausal women. Obesity (Silver Spring) 2008;16:2472–2480. doi: 10.1038/oby.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing MR, Ziegler JM, Langefeld CD, Roh BH, Palmer ND, Mayer-Davis EJ, Rewers MJ, Haffner SM, Wagenknecht LE, Bowden DW. Analysis of FTO gene variants with obesity and glucose homeostasis measures in the multiethnic Insulin Resistance Atherosclerosis Study cohort. Int J Obes (Lond) 2011;35:1173–1182. doi: 10.1038/ijo.2010.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Li M, Bradfield JP, Kim CE, Annaiah K, Santa E, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Shaner JL, Smith RM, Imielinski M, Eckert AW, Chiavacci RM, Berkowitz RI, Hakonarson H. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS ONE. 2008;3:e1746. doi: 10.1371/journal.pone.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Chen G, Zhou J, Shriner D, Doumatey A, Huang H, Rotimi C. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes. 2010;59:1549–1554. doi: 10.2337/db09-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhu H, Lagou V, Gutin B, Stallmann-Jorgensen I, Treiber F, Dong Y, Snieder H. FTO variant rs9939609 is associated with body mass index and waist circumference, but not with energy intake or physical activity in European- and African-American youth. BMC Med Genet. 2010;11:57. doi: 10.1186/1471-2350-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollepalli S, Dolan LM, Deka R, Martin LJ. Association of FTO gene variants with adiposity in African-American adolescents. Obesity. 2010;18:1959–1963. doi: 10.1038/oby.2010.82. [DOI] [PubMed] [Google Scholar]
- Cauchi S, Ezzidi I, El Achhab Y, Mtiraoui N, Chaieb L, Salah D, Nejjari C, Labrune Y, Yengo L, Beury D, Vaxillaire M, Mahjoub T, Chikri M, Froguel P. European genetic variants associated with type 2 diabetes in North African Arabs. Diabetes Metab. 2012;38:316–323. doi: 10.1016/j.diabet.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Peters U, North KE, Sethupathy P, Buyske S, Haessler J, Jiao S, Fesinmeyer MD, Jackson RD, Kuller LH, Rajkovic A, Lim U, Cheng I, Schumacher F, Wilkens L, Li R, Monda K, Ehret G, Nguyen KD, Cooper R, Lewis CE, Leppert M, Irvin MR, Gu CC, Houston D, Buzkova P, Ritchie M, Matise TC, Le Marchand L, Hindorff LA, Crawford DC. et al. A systematic mapping approach of 16q12.2/FTO and BMI in more than 20,000 African Americans narrows in on the underlying functional variation: results from the Population Architecture using Genomics and Epidemiology (PAGE) study. PLoS Genet. 2013;9:e1003171. doi: 10.1371/journal.pgen.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Long J, Gao YT, Lu W, Cai Q, Wen W, Zheng Y, Yu K, Xiang YB, Hu FB, Zheng W, Shu XO. Evaluation of genetic susceptibility loci for obesity in Chinese women. Am J Epidemiol. 2010;172:244–254. doi: 10.1093/aje/kwq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sun Y, Sun J. Combined effects of FTO rs9939609 and MC4R rs17782313 on obesity and BMI in Chinese Han populations. Endocrine. 2011;39:69–74. doi: 10.1007/s12020-010-9413-6. [DOI] [PubMed] [Google Scholar]
- Xi B, Cheng H, Shen Y, Chandak GR, Zhao X, Hou D, Wu L, Wang X, Mi J. Study of 11 BMI-associated loci Identified in GWAS for associations with central obesity in the Chinese children. PLoS ONE. 2013;8:e56472. doi: 10.1371/journal.pone.0056472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi B, Chandak GR, Shen Y, Wang Q, Zhou D. Association between common polymorphism near the MC4R gene and obesity risk: a systematic review and meta-analysis. PLoS ONE. 2012;7:e45731. doi: 10.1371/journal.pone.0045731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Nakamura M, Nakamura T, Matsuo T, Nakata Y, Kamohara S, Miyatake N, Kotani K, Komatsu R, Itoh N, Mineo I, Wada J, Masuzaki H, Yoneda M, Nakajima A, Funahashi T, Miyazaki S, Tokunaga K, Kawamoto M, Ueno T, Hamaguchi K, Tanaka K, Yamada K, Hanafusa T, Oikawa S, Yoshimatsu H, Nakao K, Sakata T, Matsuzawa Y, Kamatani N, Nakamura Y. Association between obesity and polymorphisms in SEC16B, TMEM18, GNPDA2, BDNF, FAIM2 and MC4R in a Japanese population. J Hum Genet. 2009;54:727–731. doi: 10.1038/jhg.2009.106. [DOI] [PubMed] [Google Scholar]
- Sull JW, Lee M, Jee SH. Replication of genetic effects of MC4R polymorphisms on body mass index in a Korean population. Endocrine. 2013. in press . [DOI] [PubMed]
- Been LF, Nath SK, Ralhan SK, Wander GS, Mehra NK, Singh J, Mulvihill JJ, Sanghera DK. Replication of association between a common variant near melanocortin-4 receptor gene and obesity-related traits in Asian Sikhs. Obesity. 2009;18:425–429. doi: 10.1038/oby.2009.254. [DOI] [PubMed] [Google Scholar]
- Dwivedi OP, Tabassum R, Chauhan G, Kaur I, Ghosh S, Marwaha RK, Tandon N, Bharadwaj D. Strong influence of variants near MC4R on adiposity in children and adults: a cross-sectional study in Indian population. J Hum Genet. 2013;58:27–32. doi: 10.1038/jhg.2012.129. [DOI] [PubMed] [Google Scholar]
- Mejía-Benítez A, Klünder-Klünder M, Yengo L, Meyre D, Aradillas C, Cruz E, Pérez-Luque E, Malacara JM, Garay ME, Peralta-Romero J, Flores-Huerta S, García-Mena J, Froguel P, Cruz M, Bonnefond A. Analysis of the contribution of FTO, NPC1, ENPP1, NEGR1, GNPDA2 and MC4R genes to obesity in Mexican children. BMC Med Genet. 2013;14:21. doi: 10.1186/1471-2350-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Chiang CW, Palmer CD, Tayo BO, Lettre G, Butler JL, Hackett R, Adeyemo AA, Guiducci C, Berzins I, Nguyen TT, Feng T, Luke A, Shriner D, Ardlie K, Rotimi C, Wilks R, Forrester T, McKenzie CA, Lyon HN, Cooper RS, Zhu X, Hirschhorn JN. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester JM, Wing MR, Li J, Palmer ND, Xu J, Hicks PJ, Roh BH, Norris JM, Wagenknecht LE, Langefeld CD, Freedman BI, Bowden DW, Ng MC. Implication of European-derived adiposity loci in African Americans. Int J Obes (Lond) 2012;36:465–473. doi: 10.1038/ijo.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanein MT, Lyon HN, Nguyen TT, Akylbekova EL, Waters K, Lettre G, Tayo B, Forrester T, Sarpong DF, Stram DO, Butler JL, Wilks R, Liu J, Le Marchand L, Kolonel LN, Zhu X, Henderson B, Cooper R, McKenzie C, Taylor HA Jr, Haiman CA, Hirschhorn JN. Fine mapping of the association with obesity at the FTO locus in African-derived populations. Hum Mol Genet. 2010;19:2907–2916. doi: 10.1093/hmg/ddq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Overview of single nucleotide polymorphisms (SNPs) associated with obesity-related traits (P < 5 × 10-8) identified by genome-wide association studies (GWAS) and GWAS-meta-analyses, sorted by chromosome and position.