SUMMARY
Axon pruning during development is essential for the proper wiring of the mature nervous system, but its regulation remains poorly understood. We have identified an immunoglobulin superfamily (IgSF) transmembrane protein, Plum, that is cell-autonomously required for axon pruning of mushroom body (MB) γ neurons and for ectopic synapse refinement at the developing neuromuscular junction in Drosophila. Plum promotes MB γ neuron axon pruning by regulating the expression of Ecdysone Receptor-B1, a key initiator of axon pruning. Genetic analyses indicate that Plum acts to facilitate signaling of Myoglianin, a glial-derived TGF-β, on MB γ neurons upstream of the type-I TGF-β receptor Baboon. Myoglianin, Baboon, and Ecdysone Receptor-B1 are also required for neuromuscular junction ectopic synapse refinement. Our study highlights both IgSF proteins and TGF-β facilitation as key promoters of developmental axon elimination and demonstrates a mechanistic conservation between MB axon pruning during metamorphosis and the refinement of ectopic larval neuromuscular connections.
INTRODUCTION
Neuronal remodeling is widely used for the maturation and refinement of neural circuits during the development of both vertebrate and invertebrate nervous systems (Luo and O'Leary, 2005). Often, neurons first extend exuberant branches and later remove inappropriate ones through a highly regulated pruning process. Developmental axon pruning can take place by several distinct mechanisms. In distal-to-proximal retraction (Liu et al., 2005; Portera-Cailliau et al., 2005), axonal components are retrieved by the retracting axon. In axosome shedding (Bishop et al., 2004), retracting axons discard axonal debris that are continuously engulfed by nearby cells. In localized degeneration (Watts et al., 2003), spatially-defined segments of axons break into pieces that are later engulfed by surrounding glial cells. Such examples of degenerative developmental axon pruning share molecular and mechanistic similarities with axon degeneration following nerve injury and ‘dying back’ neurodegenerative diseases (Hoopfer et al., 2006; Luo and O'Leary, 2005; Raff et al., 2002). Thus, understanding developmental pruning can provide a deeper and broader insight into axon fragmentation and elimination during development, neurodegenerative disease, and after injury.
The nervous system of Drosophila melanogaster undergoes massive remodeling during metamorphosis between the larval and adult stages (Truman, 1990). During this process, many central and peripheral neurons eliminate specific connections while keeping others intact. Subsequently, they extend new axons and dendrites to form adult specific connections (Kantor and Kolodkin, 2003; Luo and O'Leary, 2005). Drosophila mushroom body (MB) γ neurons have emerged as an excellent model system to study the molecular mechanisms of remodeling, as they undergo highly stereotyped axon and dendrite pruning during metamorphosis (Figure 1A). During the larval stages, γ neurons project bifurcating axons to both the medial and dorsal lobes of the MB. In early pupae, γ neurons completely prune their dendrites, along with the dorsal and medial axonal branches, up to a specific and stereotyped location. Later during development, γ neurons re-extend their axons to an adult-specific medial lobe (Lee et al., 1999; Watts et al., 2003).
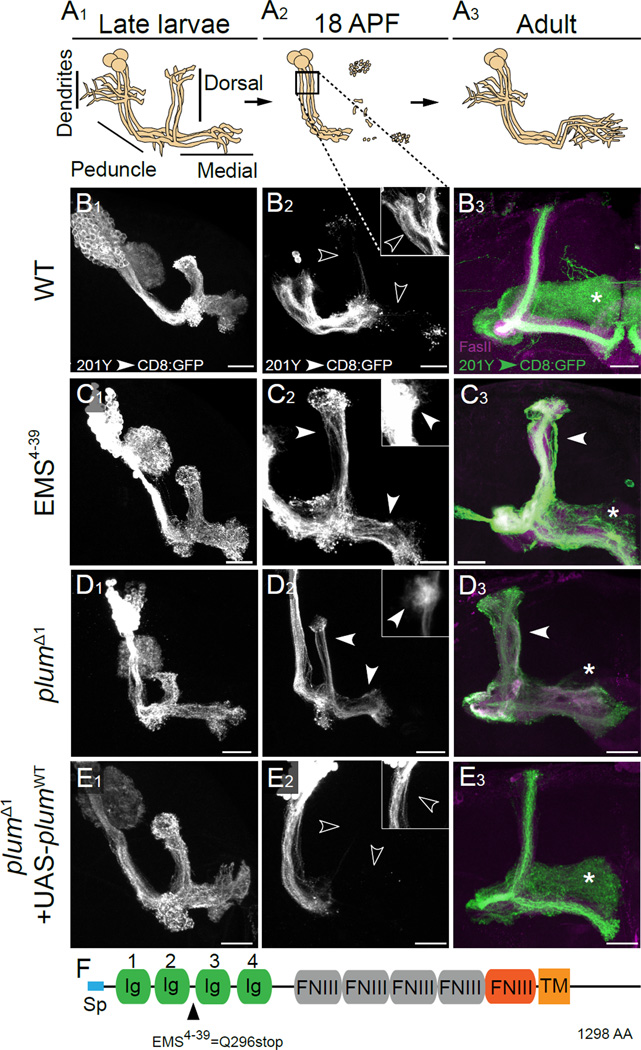
Figure 1. Plum is an IgSF Member Required for Axon Pruning of MB γ Neurons.
(A) Scheme of developmental pruning of MB γ neurons. During embryonic and larval life, each γ neuron extends a single process that branches near the cell body to form dendrites and continues as an axon peduncle that bifurcates to form a dorsal and a medial branch (A1). Both axonal branches, as well as dendrites, are pruned by 18 hrs after puparium formation (APF), whereas the peduncle remains intact (A2). Subsequently, γ neurons extend axons only medially to adult-specific lobes (A3). Square in A2 marks the location of dendrites that are shown in the insets of B2–E2.
(B–E) Confocal Z-projections of (B) WT (n=20), (C) EMS4–39 (n=13), (D) plumΔ1 (n=13) and (E) plumΔ1 additionally expressing a PlumWT transgene (UAS-plumWT) MB neuroblast clones (n=13). MARCM clones are labeled with 201Y–GAL4 driven mCD8∷GFP at (B1–E1) the 3rd instar larval stage, (B2–E2) 18h APF and (B3–E3) in adults. Solid arrowheads indicate unpruned γ axons and dendrites while open arrowheads indicate fragmented γ axons and dendrites. Asterisks indicate the distal tip of the adult γ lobe. White or green, 201Y–GAL4 driven mCD8∷GFP; magenta, anti-FasII. Scale bars, 20 µm. (F) Domain structure of the Plum protein with EMS4–39 depicted (Q296stop). Blue, signal peptide (Sp); Green, Immunoglobulin (Ig) domains; Grey, putative Fibronectin-III (FNIII) domains; Red, FNIII domain; Orange, transmembrane domain (TM).
See also Figure S1.
MB γ neuron pruning is controlled by both intrinsic and extrinsic factors. The cell-autonomous activation of the steroid hormone Ecdysone Receptor B1 (EcR-B1) and its co-receptor Ultraspiracle (Usp) is essential for initiating axon pruning (Lee et al., 2000). EcR-B1 is specifically expressed in γ neurons but not in other MB neurons that do not undergo pruning. EcR-B1 expression in γ neurons is regulated by the TGF-β receptor Baboon (Babo; Zheng et al., 2003), which is activated by the glial-derived TGF-β ligand, Myoglianin (Myo; Awasaki et al., 2011). EcR-B1 expression is also regulated by a post-mitotic function of the cohesin complex (Schuldiner et al., 2008), and by the nuclear receptors Hr39 and Ftz-f1 (Boulanger et al., 2010). While the apoptotic machinery (including the caspase Dronc) is required for dendrite pruning of sensory neurons (Kuo et al., 2006; Williams et al., 2006), it does not appear to be required for dendrite or axon pruning of MB neurons (Watts et al., 2003; our unpublished observations). Following fragmentation, the neuronal debris is engulfed by nearby glia (Awasaki and Ito, 2004; Watts et al., 2004) in a draper (ced-1 homolog)-dependent manner (Awasaki et al., 2006; Hoopfer et al., 2006), and degraded via an endosomal/lysosomal pathway (Watts et al., 2004).
Despite significant progress in the past decade, our understanding of developmental axon pruning remains far from complete. Specifically, very little is known about the nature of cell-cell communication during axon pruning. Through a forward genetic screen, we identified Plum, an immunoglobulin superfamily (IgSF) protein that functions at the cell surface of MB γ neurons and is cell autonomously required for axon pruning. Genetic analyses revealed that Plum promotes pruning by regulating the expression of EcR-B1. Our data suggest that Plum achieves this regulation by facilitating the signal via canonical TGF-β type I/II receptors in response to a glial-derived TGF-β ligand, Myoglianin. Our results also demonstrate molecular conservation in the signaling events that occur in both remodeling of MB neurons during metamorphosis and the refinement of ectopic terminals at the larval neuromuscular junction (NMJ). These underlying similarities indicate Plum as a general regulator of developmental axon elimination.
RESULTS
Plum is an immunoglobulin superfamily protein required for axon pruning
To identify molecules that are required for MB γ neuron pruning, we performed a forward genetic screen using the MARCM technique (Mosaic Analysis with a Repressible Cell Markers; Lee and Luo, 1999). In this screen, mutations were induced by the chemical mutagen EMS and phenotypes were examined in MARCM clones (see Experimental Procedures). To visualize MB γ neurons, we generated neuroblast clones that express a membrane bound GFP (mCD8-GFP) driven by the 201Y–GAL4 driver (Yang et al., 1995), which is expressed in γ neurons during the larval and early pupal stages, and in both γ and a subset of the later born α/β neurons at the adult stage (Schuldiner et al., 2008). We found a mutant, EMS4–39, which caused a severe pruning defect (compare Figure 1C with 1B). In wild type (WT) brains, the dorsal and medial γ-axon branches, as well as dendrites, were completely pruned at 18h after puparium formation (APF; Figure 1A2, open arrowheads in 1B2). In contrast, γ neurons homozygous for EMS4–39 retained these axonal branches as well as their dendrites (see insets for a focus on dendrites, as outlined by the box in Figure 1A2), indicating a failure in pruning (solid arrowheads in Figure 1C2) of both dendrites and axons. Because of the relative technical ease, we have focused our studies below on axon pruning. These unpruned axons persisted into the adult stage as dorsal branches that lie outside the α-lobe (solid arrowhead in Figure 1C3). As a consequence, very few mutant γ neurons innervate the adult γ-lobe (compare asterisks in Figure 1C3 and 1B3). This pruning defect is unlikely to be caused by a secondary effect due to impaired axon growth or guidance defects, as EMS4–39 mutant γ neuron clones appeared normal at the third instar larval stage, prior to the onset of axon pruning (compare Figure 1C1 to 1B1).
Combining SNP and deficiency mapping (see Experimental Procedures), we identified the EMS4–39 mutation as a nonsense mutation (Q296zStop; see Figures 1F and S1) in a previously uncharacterized gene - CG6490. We named the gene plum, because mutant γ neurons do not “prune”. We also generated two small deficiencies - plumΔ1 and plumΔ2 - using cis-FRT recombination (Figure S1; see Experimental Procedures), which confirmed that MB MARCM clones lacking plum exhibited severe pruning defects (compare asterisks in Figure 1D3 to 1B3, results shown for plumΔ1). We used plumΔ1 for most of our subsequent experiments.
plum encodes a transmembrane, immunoglobulin superfamily (IgSF) protein (Figure 1F). Domain analysis of the Plum protein revealed four Immunoglobulin (Ig) domains, one Fibronectin III (FNIII) domain predicted by the SMART algorithm (http://smart.embl-heidelberg.de/), and four additional putative FNIII domains (Figure 1F). Expression of an epitope-tagged, full-length plum transgene (UAS-plumWT:Flag; hereafter termed PlumWT) within plumΔ1 MB MARCM neuroblast clones fully rescued their pruning defect (Figure 1E). Thus, we conclude that Plum is an IgSF protein that plays an essential role during MB γ neuron pruning.
Plum is cell-autonomously required in postmitotic γ neurons for axon pruning
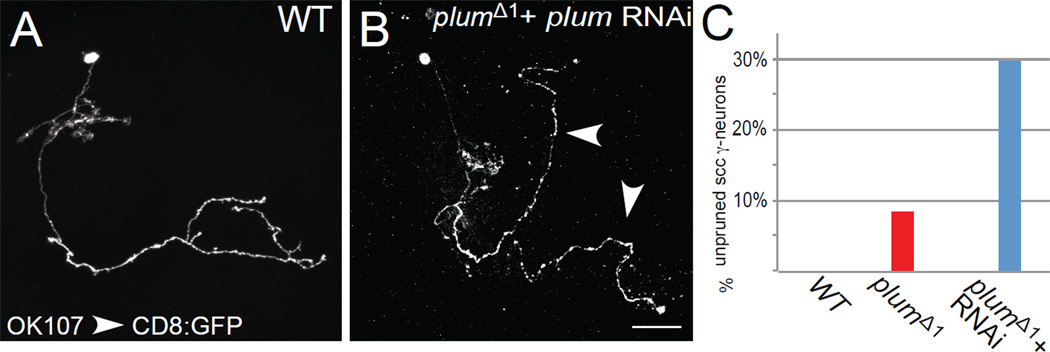
To determine whether Plum is required cell autonomously to regulate pruning, we generated MB γ neuron MARCM single cell clones (SCC) homozygous for plumΔ1 in otherwise heterozygous brains (Figure 2A–B). We found that 8.5% of the plumΔ1 SCC (n=47) (Figure 2C, red bar) retained their larval specific dorsal axon branches into the adult stage, whereas all WT γ neurons pruned these branches (Figure 2A). This low percentage of unpruned SCC might be caused by perdurance of Plum RNA or protein in SCCs (for a more detailed explanation of perdurance, see Experimental Procedures). Indeed, expressing Plum RNAi within mutant single cell clones raised the percentage of unpruned SCCs to 30% (n=115; Figure 2B, quantified in 2C, blue bar). These results indicate that Plum is cell-autonomously required in γ neurons for their axon pruning.
Figure 2. Plum is Cell Autonomously required for Axon Pruning of MB γ Neurons.
Single cell clone analysis: Confocal Z-projections of (A) WT or (B) plumΔ1 additionally expressing plum RNAi (UAS-plumRNAi) transgene. γ single cell clones are labeled by OK 107-GAL4 driving the expression of mCD8:GFP. Arrowheads indicate unpruned γ axon branches that persist into the adult stage. Scale bar, 20 µm. (C) Quantification of pruning defects in single cell clones. Scale bars, 20 µm.
Two additional lines of evidence suggest that Plum functions in postmitotic neurons. First, defects in SCCs resulted from lack of Plum protein only in postmitotic γ neurons. Second, in our rescue experiment (Figure 1E), we used a driver that, in the MB, is only expressed in postmitotic neurons (201Y–GAL4; Schuldiner et al., 2008) to drive the expression of PlumWT in mutant neuroblast clones (Figure 1E). Because GAL4 expression turned on only after the last round of cell division that produces the neuron, these results, along with pruning defects in single cell clones, indicate that Plum functions cell-autonomously in postmitotic γ neurons to promote axon pruning.
The extracellular domain of Plum is required for axon pruning
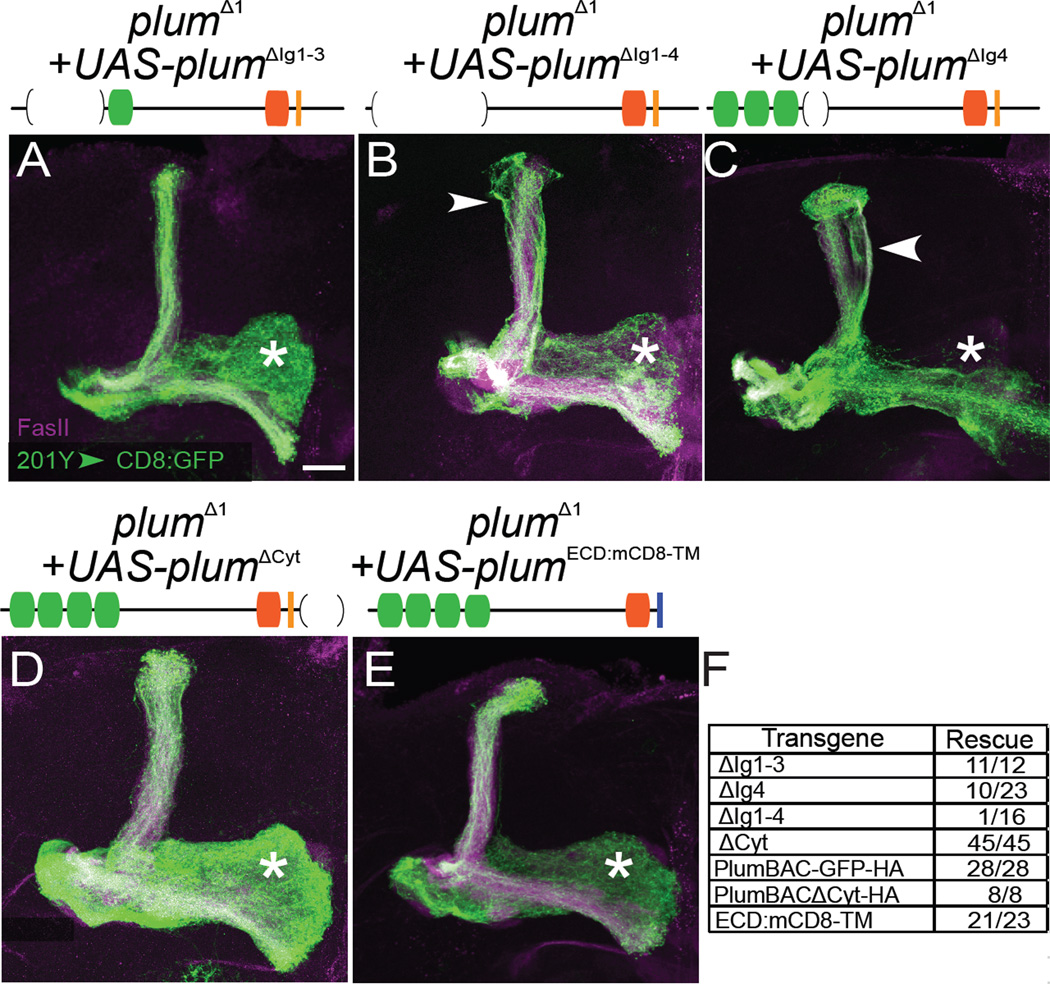
IgSF proteins are implicated in diverse steps of brain development, including neuronal migration, axon pathfinding, target recognition, synapse formation, and in the maintenance and function of adult neuronal networks (Rougon and Hobert, 2003; Vogel et al., 2003). In all cases examined, IgSF proteins mediate cell-cell interactions through their Ig and FNIII domains (Brummendorf and Rathjen, 1996). If Plum functions in cell-cell communication, one or both of these domains should be essential for its function. To assess the role of the Ig domains in the function of Plum during pruning, we performed in vivo structure-function analyses. We generated a series of epitope-tagged UAS-plum transgenes where specific domains were deleted (UAS- plumΔdomain:Flag are abbreviated as PlumΔdomain), and tested their ability to rescue γ neuron pruning defects when expressed within plumΔ1 MARCM neuroblast clones.
We found that deletion of any or all of the first three Ig domains (Figure 3A, results for single deletions not shown) did not affect the rescue abilities of the transgenes (Figure 3F), indicating that Ig domains 1–3 are not required for Plum’s function during pruning. In contrast, deletion of all four Ig domains (PlumΔIg1–4) nearly abolished the rescue ability of the transgene (Figure 3B, 3F). In addition, deletion of Ig4 domain (PlumΔIg4) partially reduced the rescue ability of that transgene (Figure 3C, 3F). Deletion of these domains resulted in some changes to expression and localization (Figure S2), but these did not correlate with their rescue ability. Our results therefore demonstrate the importance of Plum’s extracellular domain, and specifically the four Ig domains as a whole, in mediating axon pruning.
Figure 3. Structure-function Analysis of Plum.
(A–E) Confocal Z-projections of plumΔ1 MB MARCM neuroblast clones additionally expressing (A) UAS-plumΔIg1–3 (n=12), (B) UAS-plumΔIg1–4 (n=16), (C) UAS-plumΔIg1–4 (n=23), (D) UAS-plumΔCyt (n=45) and (E) UAS-plumECD:CD8TM (n=23). While expression of PlumΔIg1–3 (A), PlumΔCyt (D), and PlumECD:CD8TM (E) rescued the pruning defect, expression of PlumΔIg1–4 (B) or PlumΔIg4 (C) did not. (F) Summary of Plum truncations and their ability to rescue the pruning defects of plumΔ1 MB MARCM neuroblast clones. PlumBAC-GFP:HA and PlumBACΔCyt:HA were expressed under the control of the endogenous promoter (see Figure S4) while expression of other transgenes was driven by 201Y–GAL4. Plum predicted domains are depicted as shown in Figure 1F with brackets representing the deleted domains; the mCD8 transmembrane domain is represented by blue line. Green, 201Y–GAL4 driven mCD8∷GFP; Magenta, FasII; Asterisk marks adult-specific γ-lobe. Scale bar, 20 µm.
See also Figures S2 and S3.
Many IgSF proteins such as Drosophila Fasciclin II, Neuroglian, and DSCAM execute their functions through a trans-homophilic binding mechanism (Agarwala et al., 2000; Islam et al., 2004; Schuster et al., 1996). We explored the formation of Plum trans-homophilic interaction using a Drosophila S2 cell aggregation assay. We found that expression of Plum promoted S2 cell aggregation (Figure S3). However, Plum-mediated trans-homophilic interaction required Ig domains 1–3 (Figure S3), which were not required in vivo to promote pruning (Figure 3). These data suggest that, in vivo, trans-homophilic interaction is not required for Plum’s function in MB γ axon pruning. Therefore, Plum likely interacts with a heterophilic ligand to regulate MB γ neuron axon pruning.
The cytoplasmic domain of Plum is not required for axon pruning
The cell-autonomous requirement for plum in MB γ neurons suggests that it functions genetically as a receptor. We therefore tested the requirement of the cytoplasmic domain for Plum’s function. To our surprise, expressing a Plum transgene lacking its cytoplasmic domain (PlumΔCyt) was sufficient to rescue the pruning defect of plumΔ1 MB γ neurons (Figures 3D, 3F). To confirm that this was not an artifact due to overexpression of the transgene, we generated plumΔ1 homozygous flies expressing plum transgenes with or without its cytoplasmic domain under the control of its own promoter using BAC recombineering (PlumBAC-GFP:HA and PlumBACΔCyt:HA, respectively; Figure S4). We found that both BAC transgenes rescued the mutant phenotype of plumΔ1 neuroblast clones in a similar manner (Figure 3F), confirming that the cytoplasmic domain is not required for pruning.
To exclude the possibility that the PlumΔCyt transgene can elicit a signal to promote pruning by the transmembrane domain or the residual 9 remaining cytoplasmic amino acids, we generated another transgene, encoding for the Plum extracellular domain fused to the mouse CD8 transmembrane domain (UAS-plum-ECD:CD8-TM:Flag; PlumECD:CD8-TM). We found that expression of the PlumECD:CD8-TM transgene also rescued the pruning defect in plumΔ1 MB γ neurons (Figure 3E, 3F). Thus, although Plum functions cell autonomously, neither its cytoplasmic nor its transmembrane domain conveys the pruning signal. These data suggest that Plum transduces signals via the activity of another receptor to direct γ neuron axon pruning.
Despite pan-neural expression, Plum is required only within MB γ neurons for their pruning
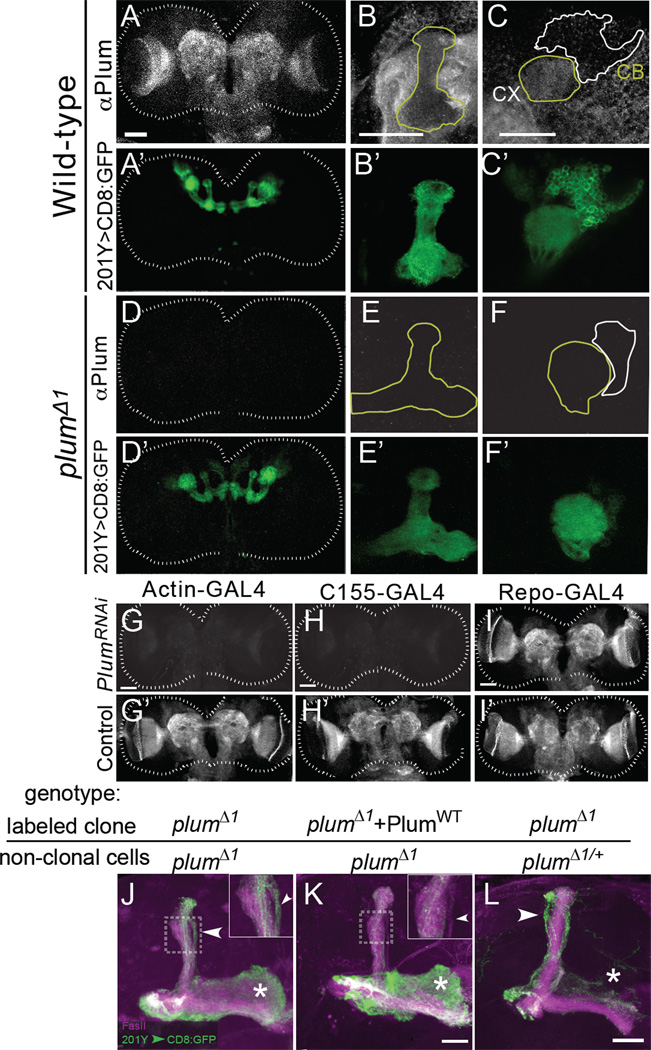
To gain insight into Plum’s function in cellular communication, we generated polyclonal antibodies against the extracellular domain of Plum and examined its endogenous localization in the central brain during development. At 6h APF, a developmental time-point in which MB γ neuron pruning has already begun (Watts et al., 2003), Plum was broadly expressed in the neuropil (Figure 4A–C). The staining was completely eliminated in homozygous plumΔ1 animals (Figure 4D–F), demonstrating the specificity of the antibody. Within the MB, Plum was expressed at a low level and was localized to both axons and dendrites (yellow outlines in Figures 4B, 4C). Plum was detected in very low levels at the cell bodies (white outline in Figure 4C). Interestingly, Plum was expressed at a higher level in the neuropil outside the MB (Figure 4B, outside the yellow outline). This widespread expression was also confirmed by the localization of a GFP-tagged genomic plum transgene (PlumBAC-GFP:HA; Figure S4B). Plum staining in late 3rd instar larval (Figure 4G’, 4H’ and 4I’) and 0h APF pupal brains (data not shown) appeared similar to 6h APF.
Figure 4. Despite Widespread Expression, Plum Is Required Only in MB γ Neurons for Axon Pruning.
(A–F) Confocal Z-projections of (A–C) WT and (D–F) plumΔ1 6h APF brains depicting (A–F) anti-Plum staining or (A–F) MB structure by 201Y–GAL4 driven mCD8∷GFP. (B, E) Close-up view of the MB dorsal lobe (outlined in yellow). (C, F) Close-up of the calyx (CX - outlined in yellow) and cell bodies (CB - outlined in white). Similar results were obtained by examining 15 brains from each genotype.
(G–I) Z-projections of Plum staining in 3rd instar larval brains that express Plum RNAi driven by (G) Actin-GAL4, (H) C155-GAL4 and (I) Repo-GAL4. G’-I’ are controls with the GAL4 transgenes alone. Scale bars in A–I are 50 µm. Similar results were obtained by examining 8~10 brains from each genotype.
(J, K) Confocal Z-projections of a MARCM-labeled MB neuroblast clone from a brain in which all cells are plumΔ1 mutant in the absence (J) or presence (K) of an additional PlumWT rescue transgene expressed within the clone. 7/8 clones in (J) and 1/14 in (K) exhibited unpruned projections. Insets show a close-up view of the dorsal lobe. Arrowheads point to the unpruned dorsal lobe of γ axons, which fall outside the FasII staining (magenta, marker of α/β axons). Asterisks indicate adult-specific γ axons in the medial lobe.
(L) Confocal Z-projections of a plumΔ1 MARCM neuroblast clone in an otherwise plumΔ1/+ brain (n=13). Compared with (J), there are more unpruned γ axons in the dorsal lobe (arrowhead) and fewer adult-specific γ axons (asterisk), indicating a much more severe pruning defect in Panel L. Green is mCD8∷GFP; Magenta is FasII staining.
Scale bars in J–L are 20 µm. Figure S5A shows the result of a blind rank order to compare the phenotypes in 3J and 3L.
See also Figures S4 and S5.
To determine whether the non-MB Plum staining originates from neurons or glia, we knocked down Plum’s expression specifically in both or either one of these cells by using GAL4 drivers that are ubiquitous (Actin5c–GAL4), pan-neuronal (C155-GAL4), or pan-glial (Repo-GAL4) to drive the expression of a UAS-plumRNAi transgene. We found that ubiquitous or pan- neuronal RNAi knockdown of Plum eliminated the antibody staining (Figure 4G, 4H) while pan-glial knockdown did not (Figure 4I). Consistently, using a GAL4-independent MB fluorescence reporter (Figure S4C–L), we found that ubiquitous and pan-neuronal, but not glial, knock-down of Plum caused strong pruning defects (n=20 for each genotype; data not shown). Therefore, Plum is expressed predominantly in neurons in the developing brain.
Since Plum is highly expressed in the neuropil adjacent to MB axons, we tested whether it has an additional, non-cell-autonomous, role in γ axon pruning. We first examined this possibility by determining whether Plum expression within γ neurons is sufficient to promote pruning. We took advantage of the fact that plumΔ1 flies are homozygous viable, and generated MARCM neuroblast clones expressing PlumWT in an otherwise plumΔ1 mutant brain. Because MB neurons are born from four identical neuroblasts (Ito et al., 1997), expression of the PlumWT transgene within a MARCM clone would result in one clone of positively labeled plumΔ1 neurons expressing PlumWT, while the remaining ¾ of MB neurons (as well as the rest of the brain), would be unlabeled, homozygous mutants. We found that expressing PlumWT in a neuroblast clone was sufficient to rescue the pruning defect in plumΔ1 mutant brains (Figure 4K, compared to Figure 4J), demonstrating that Plum is only required within γ neurons to promote axon pruning.
Interestingly, labeled plumΔ1 MB neuroblast clones within homozygous mutant brains (where all cells were plumΔ1, but only a neuroblast clone was labeled; Figure 4J) displayed markedly weaker pruning defects compared to plumΔ1 clones within otherwise plum heterozygous brains (Figure 4L). In plumΔ1 homozygous mutant brains, MB neuroblast clones displayed both unpruned dorsal larval axons as well as normal adult axons (Figure 4J, asterisk), indicating that a significant proportion of γ neurons pruned normally. By contrast, in otherwise plum heterozygous brains, plumΔ1 γ neuron MARCM clones did not contain any adult medial axons (Figures 4L and 1D3, asterisk), suggesting that all γ neurons within the clone failed to prune. Indeed, a blind rank order test based on pruning severity clearly separated the two genetic conditions (Figure S5A). Because the labeled axons in both cases were of the same genotype, their phenotypic differences suggest that Plum expression outside the clone negatively regulates MB γ axon pruning. A likely interpretation is that Plum outside the MB competes for a ligand used by Plum within the MB (see Discussion).
Plum promotes axon pruning by regulating EcR-B1 expression
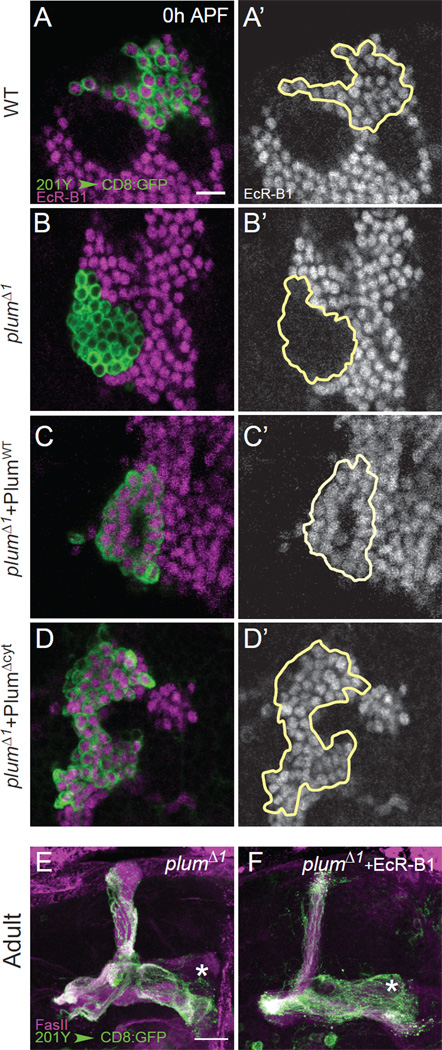
To investigate the mechanism by which Plum regulates axon pruning, we tested its relationship with other molecular pathways required for MB γ neuron axon pruning. The steroid hormone receptor EcR-B1 is a major initiator of axon pruning (Lee et al., 2000), whose expression is regulated by multiple mechanisms (see Introduction). Remarkably, we found that Plum was also required for EcR-B1 expression, as EcR-B1 expression was absent in plumΔ1 MARCM γ neuron clones (compare Figure 5B with 5A). Transgenic expression of PlumWT (Figure 5C) or PlumΔCyt (Figure 5D) within plumΔ1 mutant MARCM clones restored EcR-B1 expression. Interestingly, in plumΔ1 homozygous mutant brains, where only a subset of MB γ neurons failed to prune (Figure 4J), we also observed a corresponding partial loss of EcR-B1 expression in MB γ neurons (Figure S6). Finally, overexpression of EcR-B1 within plumΔ1 MARCM clones largely rescued their pruning defect (compare Figure 5F with 5E; see Figure S5B for a blind rank order test). These results indicate that a major function of Plum in axon pruning lies in its regulation of EcR-B1 expression.
Figure 5. Plum Controls Axon Pruning by Regulating EcR-B1 Expression.
(A–D) Representative single confocal sections of the cell body regions of 0 hr APF MB MARCM neuroblast clones in (A) WT (n=6), (B) plumΔ1 (n=10) and (C) plumΔ1 additionally expressing PlumWT (n=6) or (D) PlumΔCyt (n=5). Left panels show both MARCM clones labeled by 201Y–GAL4 driven mCD8∷GFP (green) and EcR-B1 expression (magenta). Right panels show EcR-B1 expression (gray), clones are outlined. Scale bar: 10 µm. n is number of clones examined.
(E–F) Confocal Z-projections of (E) plumΔ1 (n=14) and (F) plumΔ1 additionally expressing UAS-EcR-B1 (n=15). Green, 201Y–GAL4 driven mCD8∷GFP; magenta, FasII; asterisks mark the adult-specific γ lobe; scale bars are 20 µm. Figure S5B shows the result of a blind rank order to compare the phenotypes in 4E and 4F.
See also Figures S5 and S6.
Plum acts upstream of the TGF-β receptor Baboon
The only cell surface proteins known to be involved in MB pruning are the TGF-β receptors, Babo (type I receptor), and Wishful thinking (Wit) and Punt (type II receptors). Together, they function to up-regulate the expression of EcR-B1 prior to axon pruning (Zheng et al., 2003). Given the similarity between the functions of Plum and Babo, and our finding that Plum relies on another receptor for its signaling, we hypothesized that the two may function together to regulate MB pruning.
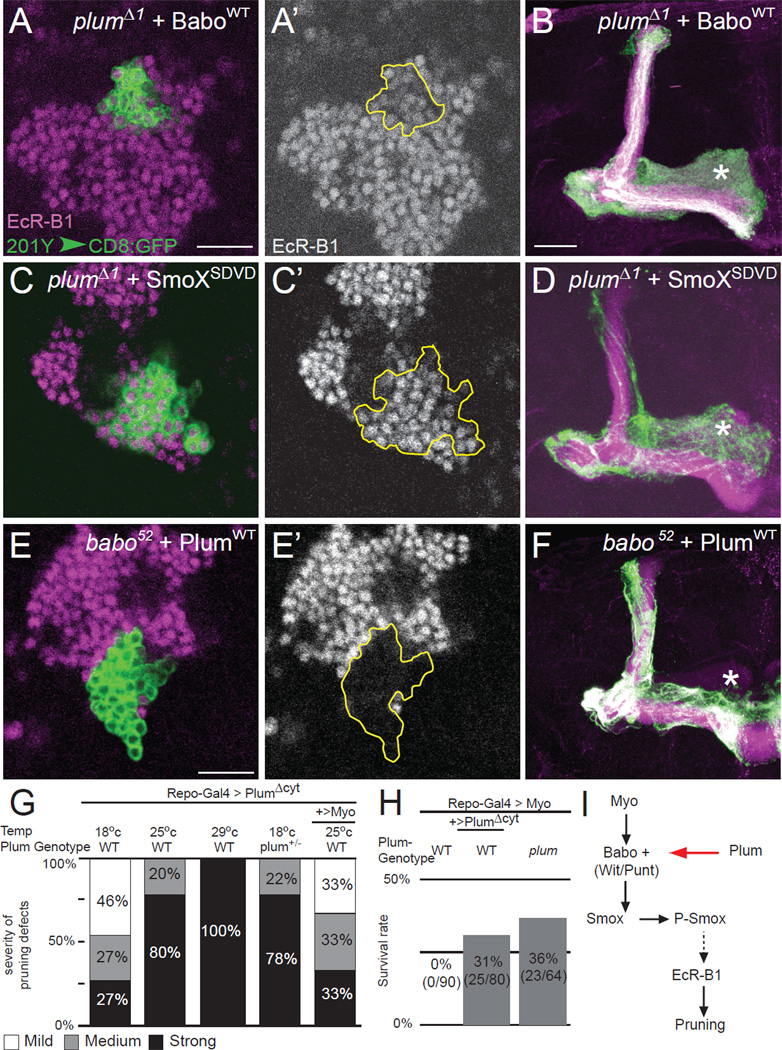
To determine the relationship between Plum and Babo, we overexpressed UAS transgenes corresponding to each gene in a mutant MARCM clone of the other. We found that overexpression of either BaboWT or constitutively active Babo (UAS-baboQ302D; Brummel et al., 1999) in plumΔ1 mutant MARCM clones rescued both EcR-B1 expression (Figure 6A, A’; data not shown, respectively) and the pruning defects (Figure 6B; Figure S5C). These results indicate that upregulation of TGF-β signaling can compensate for the loss of Plum. Similarly, we also found that overexpression of a phosphomimetic active form (SmoXSDVD) of dSmad2 (also known as SmoX; Gesualdi and Haerry, 2007), the factor downstream of Babo signaling, also partially rescued EcR-B1 expression (Figure 6C, C’) and the pruning defects (Figure 6D; Figure S5C) of plumΔ1 mutant MARCM clones. The converse is not true: overexpression of PlumWT within babo MARCM γ neuron mutant clones did not rescue the loss of EcR-B1 expression (Figure 6E, E’’) or the pruning defects (Figure 6F). These results suggest that Plum acts upstream of Babo to regulate EcR-B1 expression and axon pruning (Figure 6I).
Figure 6. Plum Facilitates TGF-β Signaling.
(A–F) Single confocal sections (A, C, E) of 0 hr APF MB cell bodies or (B, D, F) confocal Z-projections of adult MB neurons of the following genotypes: (A, B) plumΔ1 MB MARCM neuroblast clones additionally expressing BaboWT (n=8) or (C, D) SmoXSDVD (n=11) or (E, F) babo52 neuroblast clones additionally expressing PlumWT (n=26). Green, 201Y–GAL4 driven mCD8∷GFP; magenta/gray in A, C and E, EcR-B1; magenta in B, D and F, FasII; asterisks mark the adult-specific γ lobe; scale bars are 20 µm. Figure S5C shows the result of a blind rank order test to compare the phenotypes in 5B, 5D, and 5F.
(G) Quantification of the pruning defects seen in animals of different genotypes and rearing conditions. PlumΔCyt was overexpressed in glia in WT, plumΔ1/+ or coexpressed together with Myo in WT animals. Sample sizes (n) scored for each condition are (from left to right): 22, 18, 20, 32, 25. See Experimental Procedures for definitions of the different severity levels.
(H) Survival rates of WT or plumΔ1 animals overexpressing Myo alone or together with PlumΔCyt in glia (See Experimental Procedures).
(I) Scheme of Plum integration into the TGF-β signaling pathway based on genetic data.
See also Figure S5.
Plum interacts genetically with the TGF-β ligand Myoglianin
Since Babo receives its signal from the glial-derived Myo (Awasaki et al., 2011), we tested whether Plum also interacts with Myo. Indeed, we found strong genetic interactions between Plum and Myo in three separate experiments. First, ectopic, pan-glial expression of PlumΔCyt (using Repo-GAL4) in WT animals resulted in an axon-pruning defect (Figure 6G). This effect was dose-dependent, as higher PlumΔCyt expression (caused by higher GAL4 activity when animals were raised at higher temperatures) resulted in stronger pruning defects (Figure 6G, compare 1st, 2nd and 3rd columns). At the same time, reducing the endogenous plum gene dose also resulted in a stronger pruning defect (Figure 6G, compare 4th to 1st column). Interestingly, co-overexpression of Myo and PlumΔCyt in glia partially suppressed this pruning defect (Figure 6G, compare 5th to 2nd column). The simplest interpretation for these results is that PlumΔCyt overexpression in glia causes a pruning defect because of Myo sequestration. Reduction of endogenous Plum exacerbated, whereas glial co-expression of Myo alleviated, this sequestration effect, and thus enhanced or suppressed the pruning defect, respectively.
Second, previous findings indicated that ubiquitous expression of Myo causes lethality (Awasaki et al., 2011). We found that pan-glial overexpression of Myo recapitulated this lethality (Figure 6H, first column); most animals died as larvae. However, co-expression of PlumΔCyt together with Myo in glia resulted in 31% of the animals surviving to adulthood (Figure 6H, second column). This suggests that glial overexpression of PlumΔCyt mitigates lethality by sequestering excessive Myo, which by itself is toxic.
Third, lethality caused by glial overexpression of Myo was also suppressed in homozygous plum mutants (Figure 6H, 3rd column), indicating that endogenous Plum is required to transduce the Myo-derived signals that cause lethality. Since MB γ axon pruning is not essential for viability, this third experiment indicates that Plum interacts with Myo in a more general context, beyond MB axon pruning.
Plum regulates ectopic motoneuron projections at the NMJ through EcR-B1
At the mammalian neuromuscular junction (NMJ), the mature pattern of connectivity is achieved via a general process of synaptic refinement that eliminates weak connections and prevents improper ones (Nguyen and Lichtman, 1996). At the fly larval NMJ, developmental synaptic refinement also occurs to produce normal neuromuscular connectivity. While some of the mechanistic bases for refinement by synapse retraction are known (Eaton et al., 2002; Massaro et al., 2009; Pielage et al., 2005), other forms involving the prevention of off-target, ectopic contacts (Carrillo et al., 2010; Jarecki and Keshishian, 1995; Kopczynski et al., 1996) are not as well understood.
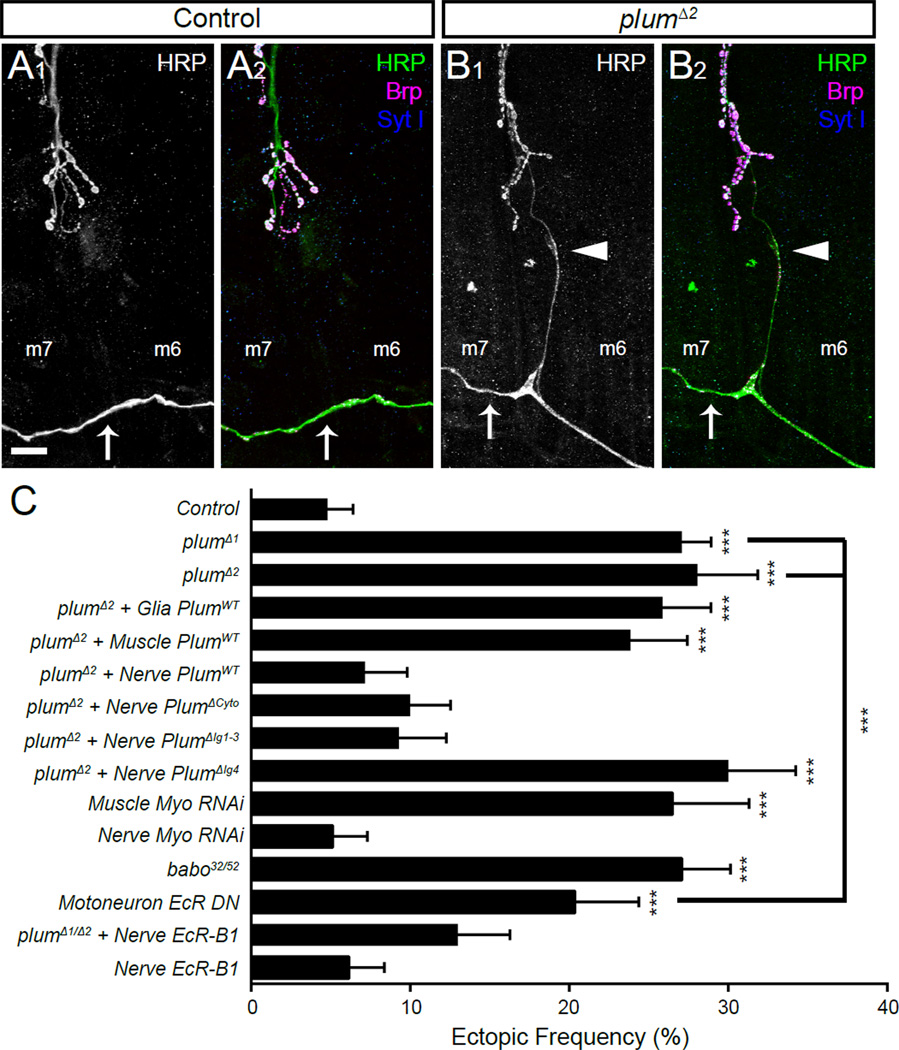
To examine whether Plum is required for normal neuromuscular connectivity, we stained the NMJs of WT and plum mutant 3rd instar larvae with antibodies to HRP (Figure 7A–B) and quantified the number of ectopically placed connections onto muscles 6 and 7. We found that in plumΔ1 or plumΔ2 larvae, the frequency of improper, ectopic connections was increased nearly 6-fold (27.1% and 28.1%, respectively) over WT (4.8%) larvae (Figure 7C). While the majority of these ectopic connections were made by the transverse nerve, we also observed improper connections from other motoneurons that normally innervate muscles 13, 15 and 28 (data not shown). The increased frequency of ectopic connections was not a secondary effect of altered connectivity at normal (i.e., not ectopic) NMJs, as plum mutant NMJs displayed normal targeting and bouton number, as compared to WT larvae (Figure S7F–H).
Figure 7. Neuronal plum and TGF-β Signaling is Required for Refinement of Connections at the Third Instar Neuromuscular Junction.
(A) Representative confocal image of the lower portion of the neuromuscular junction (NMJ) between muscles 6 and 7 in a control third instar larva stained with antibodies to HRP (A1 and green in A2), Brp (magenta) and Synaptotagmin I (blue). Under normal conditions, the transverse nerve (arrow) does not make any connections to the muscle, which is otherwise normally innervated.
(B) Representative confocal image in a plumΔ2 third instar larvae stained as in (A). Here, the transverse nerve (arrow) has made an inappropriate ectopic connection onto muscle 6 (arrowhead); this connection possesses release machinery and synaptic vesicles.
(C) Quantification (mean ± SEM) of the frequency of larval hemisegments with at least one ectopic neuromuscular projection expressed as a percentage of the total hemisegments examined in third instar larvae with various genotypes listed on the left. In all cases, n ≥ 5 larvae and 60 hemisegments scored.
See also Figure S7.
Plum acts in neurons to regulate normal connectivity, as restoring neuronal (but not muscle or glia) expression of Plum within a plumΔ2 background was sufficient to restore the number of hemisegments with ectopic connections to WT levels (Figure 7C). Moreover, neuronal knockdown of Plum using a UAS-plum-RNAi transgene phenocopied the increase in ectopic connections seen in plumΔ1 flies, whereas muscle knockdown did not (Figure S7). Therefore, Plum acts in motoneurons via a heterophilic ligand to ensure normal connectivity.
We also conducted a structure-function analysis of Plum at the NMJ, similar to the MB (Figure 4). As neuronal Plum is required for normal NMJ connectivity, we drove expression of the PlumΔCyt, PlumΔIg4 and PlumΔIg1–3 transgenes within the nervous system of plumΔ2 larvae using the pan-neural elav-GAL4 driver. Similar to the MB, we observed a rescue of the mutant phenotype with the PlumΔCyt and PlumΔIg1–3 transgenes but not with the PlumΔIg4 transgene (Figure 7C). Thus, at the NMJ as in the MB, the C-terminus of Plum is dispensable for its function while the extracellular domain is essential.
Both the type I TGF-β receptor Babo and the ecdysone receptor EcR-B1 function at the NMJ in regulating synaptic growth (Ellis et al., 2010) and in the dismantling of synapses during metamorphosis (Liu et al., 2010), respectively. However, ensuring normal neuromuscular connectivity by preventing ectopic connections may be a qualitatively different process. Given the similarities of Plum action in MB γ axon pruning and in ensuring normal motoneuron connectivity in larvae, we tested whether Myo, Babo, and EcR-B1 play a role in the latter process. We found that RNAi knockdown of Myoglianin in muscle, but not in neurons, resulted in a significant increase of ectopic projection frequency, to the same extent as in the plum mutant (Figure 7C). babo32/52 mutants also exhibited quantitatively similar phenotypes, as did larvae expressing a dominant-negative EcR transgene (Cherbas et al., 2003) driven by OK6-GAL4 in motoneurons (Figure 7C). Thus, both the TGF-β and ecdysone signaling pathways regulate larval neuromuscular connectivity.
To determine whether these effects were related to Plum, we examined ectopic projections in plumΔ1/Δ2 larvae overexpressing EcR-B1 in all neurons. This manipulation significantly rescued the ectopic projection phenotype of plum mutants but did not itself have an ectopic phenotype (Figure 7C). In summary, as in the MB, Plum plays a role in developmental synaptic remodeling at the NMJ via the EcR-B1, likely in response to muscle-derived TGF-β (Myo) signaling through the Baboon receptor. In all, we conclude that Plum is not only required for MB neuronal remodeling, but is also essential for normal neuromuscular connectivity, likely via the refinement of off-target ectopic connections of peripheral motoneuron terminals.
DISCUSSION
Using a forward genetic screen, we identified Plum, an IgSF transmembrane protein, as a key player in promoting the elimination of axonal and dendritic processes in mushroom body (MB) neurons. Plum acts cell-autonomously in postmitotic neurons to promote axon pruning by regulating the expression of steroid hormone ecdysone receptor EcR-B1. Genetic and cell biological analyses indicate that Plum regulates EcR-B1 expression by facilitating TGF-β signaling, acting upstream of the type I receptor Baboon (Figure 6I). Remarkably, we found that the same pathway also functions in synapse refinement at the larval NMJ, highlighting its broad usage and conservation.
Possible models by which Plum facilitates TGF-β signaling in MB axon pruning
Cell-cell interactions play an important role during neuronal remodeling to specify the temporal, and possibly spatial, extent of pruning (Bagri et al., 2003; Nikolaev et al., 2009; Zheng et al., 2003). Previous studies have identified the TGF-β type I receptor Babo and either of the type II receptors Wit or Punt as required for the initiation of MB γ axon pruning by regulating the expression of steroid hormone receptor EcR-B1 (Zheng et al., 2003). The strong genetic interactions between Plum, Babo, and Myo raised several possible models for Plum to participate in TGF-β signaling.
Plum could affect Babo cell surface expression by, for example, functioning as a chaperone. This is consistent with our genetic epistasis experiments in which we could rescue the plum mutant phenotype by overexpressing Baboon in MB neurons. However, this model does not fit well with our finding that ectopic expression of PlumΔCyt in glia affects pruning in MB neurons (Figure 6G). Moreover, glial or panneural ectopic overexpression of a secreted version of Plum’s entire extracellular domain (PlumECD) similarly caused axon pruning defects in neighboring MB neurons (data not shown). These data argue against upregulation of Babo cell surface expression as the primary mechanism of Plum action.
Plum could also act in a signaling pathway parallel to the canonical type I and II TGF-β receptors, Babo and Wit/Punt. In this model, the sum of signaling from both pathways could determine the expression levels of EcR-B1 and thus, the pruning status. However, this model is not consistent with experiments in which pruning was inhibited by ectopically overexpressing PlumΔCyt or PlumECD in glial cells, unless the assumed Plum-dependent/Baboon-independent pathway is also activated by the TGF-β ligand Myo.
In another possible model, Plum functions by stabilizing the TGF-β receptor complex and/or facilitating TGF-β ligand binding, analogous to the roles played by accessory TGF-β receptors that have been described in mammals. Previous work in mammalian systems demonstrated that TGF-β type I/II receptor classic signaling is subject to modulation by accessory receptors, sometimes called type III receptors (Massague, 1998). The TGF-β type III receptors Betaglycan and Endoglin are the most well studied accessory receptors. Betaglycan is a membrane-anchored proteoglycan that facilitates binding of TGF-β2 to TβRII (Gatza et al., 2010). The role of TGF-β accessory receptors is not yet well understood and they have been implicated in both facilitating, as well as inhibiting, TGF-β signaling (Shi and Massague, 2003). A recent study has found that the Betaglycan extracellular ZP-C region adopts an immunoglobulin-like fold, despite sharing no sequence homology with Ig proteins, and possessing different disulfide linkages (Lin et al., 2011). The model by which Plum functions by stabilizing the TGF-β receptor complex and/or facilitating TGF-β ligand binding satisfactorily accounts for all the genetic interaction data, including the suppression of the plum phenotype by Baboon overexpression, as well as the pruning defect caused by glial misexpression of PlumΔCyt or PlumECD. To test this hypothesis, we examined whether Plum physically interacts with the conventional type I/II receptors (Babo or Wit, respectively) or the TGF-β ligand (Myo). However, we have not been able to detect physical interactions of Plum with Myo, Babo, Wit, or their combinations under physiological conditions. Thus, while our genetic results suggest that Plum may act in an analogous fashion as the TGF-β accessory receptor described in previous mammalian studies, we could not support this model with conclusive biochemical data. Future studies are required to elucidate the exact mechanisms by which Plum relates to the canonical TGF-β receptors Babo, Wit/Punt, and the TGF-β ligand Myo.
Regardless of detailed mechanisms, our study has established a close connection between an IgSF protein and the TGF-β signaling pathway. Plum is a distant homologue of Protogenin and Nope, members of the DCC family that have been implicated in developmental processes but their precise role is far from being understood (Salbaum and Kappen, 2000; Wong et al., 2010). Because of the broad roles of the TGF-β pathway in development and disease, it will be of great interest to determine whether other IgSF proteins act by facilitating TGF-β signaling.
Non-MB Plum may regulate the availability of the TGF-β ligand Myoglianin
The availability and accessibility of TGF-β superfamily ligands fall under intricate regulation during animal development, with gastrulation being a hallmark example (De Robertis, 2009). Our results suggest that Plum might function within this network of regulation, in the context of MB axon pruning.
Axon pruning of MB γ neurons can be strongly influenced by nearby neurons, which also express Plum. The requirement for Plum within MB γ neurons during pruning is drastically reduced when neighboring neurons also lack Plum, compared to the situation where all other cells are heterozygous for Plum (Figure 4J, L). Our Plum-Myo genetic interaction data (Figure 6G, H) suggest that Plum sequesters Myo, which can satisfactorily explains the above phenomenon. In a heterozygous background, Plum outside the γ-neuron clones sequesters Myo such that not enough ligand is available within the plum homozygous mutant clone to enable pruning. In a plum homozygous mutant animal, Myo is not sequestered. Therefore, higher levels of Myo reach the MB and are sufficient to partly activate pruning. This sequestration model is further supported by our finding that overexpression of PlumΔCyt or PlumECD (data not shown) by glia inhibits pruning in a dose-dependent manner and that concurrent glial overexpression of Myo suppresses the defect.
Plum as a general regulator of axon remodeling during development
Our identification of Plum as a cell-surface regulator of axon pruning has enriched our understanding of the mechanism by which extracellular signals trigger axon pruning. In addition to a role in MB γ neuron axon pruning, we also show that plum is involved in ensuring normal larval neuromuscular connectivity prior to metamorphosis. During motoneuron outgrowth in the embryo, both proper and off-target neuromuscular connections are formed. Off-target ectopic projections initially form as filopodial contacts (Jarecki and Keshishian, 1995), but are quickly removed. Delayed innervation (Kopczynski et al., 1996), impaired electrical activity (Jarecki and Keshishian, 1995) or chemorepulsion (Carrillo et al., 2010) cause a higher frequency of ectopic projections. Such connections are indistinguishable at the filopodial stage from normal growth cones (Halpern et al., 1991; Johansen et al., 1989; Murray et al., 1998) but persist and stabilize into ectopic connections. Several lines of evidence suggest that they are the result of reduced motoneuron pruning rather than improper sprouting (Carrillo et al., 2010). When normal innervation is delayed, the frequency of ectopic projections is increased (Kopczynski et al., 1996). However, after the muscle is finally innervated by its normal motoneuron, these connections are withdrawn. Similarly, increased ectopic connections due to reduced electrical activity are readily withdrawn following the restoration of activity during a critical period (Jarecki and Keshishian, 1995). Moreover, the requirement for chemorepulsion in preventing ectopic connections (Carrillo et al., 2010) suggests an active process of exclusion, inconsistent with improper sprouting. We found that in plum mutant, the increased ectopic connectivity is not accompanied by changes in bouton or branch number at normal NMJs (Figure S7). Taken together, these data suggest that normal connectivity at the NMJ and the prevention of off-target ectopic contacts arises through a process of synaptic refinement involving some form of motoneuron pruning, though not precisely analogous to the MB.
Our identification of plum suggests that the mechanism of larval neuromuscular refinement shares common molecular mechanisms with developmental MB axon pruning. Genetic studies suggest that, as in the MB, Plum functions to facilitate a TGF-β signal from the muscle upstream of EcR-B1 activity. Structure-function analysis identified the same domain requirement of Plum in promoting the refinement of ectopic motor axons as MB γ neuron pruning. Cell type-specific rescue experiments are consistent with neuronal Plum interacting via a heterophilic partner. This heterophilic partner may indeed be Myoglianin, as muscle knockdown of Myo phenocopies the plum mutant. Such a mechanism is consistent with the involvement of other muscle-derived ligands in preventing ectopic connection formation (Winberg et al., 1998). At the NMJ, Plum regulates one aspect of synaptic refinement, conveying a signal that can cooperate with others to maintain normal connectivity. Our findings mechanistically connect two disparate processes of developmental axon remodeling, and highlight the general involvement of the Plum-Babo-Myo-EcR pathway in neuronal process refinement during development.
EXPERIMENTAL PROCEDURES
MARCM-based forward genetic screen
We used ethyl methane sulfonate (EMS; 25 mM in sucrose solution) to mutagenize male flies carrying FRT2A and FRT82B, which are sites for the FLP-mediated recombination on the left and right arms of the third chromosome, respectively. After establishing individual mutant stocks and confirming the lethality of mutations located on FRT-containing third chromosomes, we crossed these mutants to a “MARCM-ready fly stock” (for the third chromosome right arm screen: y, w, hsFlp122, UAS-mCD8-GFP; 201Y–GAL4, UAS-mCD8-GFP / CyO; FRT82B, tubP-Gal80 / TM6, Tb). Cross progeny were heat-shocked for 40–60 minutes at 37°C at 20–28 hr after egg laying. We then dissected out the adult fly brains of the appropriate genotype as previously described (Wu and Luo, 2006) and analyzed MB γ neurons by visualizing expression of mCD8-GFP in whole mount live or fixed brains using a compound fluorescent microscope.
An intrinsic feature of mosaic analysis with MARCM is that once a clone is generated, no new functional mRNA or protein is made in mutant cells. Nevertheless, preexisting mRNAs and proteins inherited from heterozygous parental cells can persist and function normally for a certain period of time, resulting in perdurance. Therefore, the amount of protein perdurance depends on the number of divisions the parental cell undergoes before the postmitotic cell is born. Because single cell clones are generated from a single cell division of the ganglion mother cell (in which the mitotic recombination occurs) the mRNA and proteins are diluted only by a factor of two. In contrast, neuroblast clones include neurons that are at least two cell divisions from the neuroblast in which the recombination occurred. Thus, protein perdurance could strongly affect single cell clones.
Generation and imaging MARCM clones
MB MARCM neuroblast or single cell clones were generated by heat shocking newly hatched larvae and examined as described previously (Lee et al., 1999). Brains were mounted in Slowfade (Invitrogen) while larvae (for NMJ analysis) were mounted in Vectashield (Vector Laboratories) and imaged on Zeiss LSM510 or LSM710 confocal microscopes.
Genetic mapping of plum
Genetic mapping for the causal gene in EMS4–39 was performed by first using SNP-based recombination mapping (Berger et al., 2001). This mapping narrowed down the suspected region to a cytological location between 97A10 and 97C5 and at the same time eliminated the lethal mutation(s) on the third chromosome, revealing that EMS4–39 was actually not homozygous lethal. At this point, due to a lack of more informative SNPs within the suspected region, we continued the mapping by generating molecularly defined chromosomal deletions and testing the phenotypes of EMS4–39/deletions compound heterozygous flies. We used FLP-mediated trans- recombination to generate deletions as previously described (Parks et al., 2004). The two informative deletions, as well as the flies used to generate them, are depicted in Figure S1.
Cloning plum
At the time we cloned plum, the annotated CG6490 sequence was incomplete and did not contain a signal peptide. We therefore performed 5’ RACE (rapid amplification of cDNA ends) using the Invitrogen GeneRacer™ kit. We obtained the full length RNA of plum and uncovered two additional upstream exons, encoding a protein that contained a signal peptide. The plum full-length sequence is available in Genbank, under accession number JF268497.
Neuromuscular ectopic projection analysis
Wandering third instar larvae were dissected and processed as described (Mosca and Schwarz, 2010). During imaging, the HRP channel was artificially enhanced to reveal low-level background staining of the underlying muscles. Two kinds of ectopic projections were scored: improper connections from the transverse nerve, which runs along the segment border, onto muscles 6 or 7 or improper connections onto muscle 6 from the other motoneurons that normally avoid this muscle. Phenotypes were calculated as the number of body wall hemisegments containing one or more ectopic connections. Statistical significance was calculated using an ANOVA with a Dunnett post-hoc comparison to the control sample.
Immunofluorescence antibody conditions
Rat monoclonal anti-mouse CD8 α subunit, 1:100 (Caltag, Burlingame, CA); rabbit polyclonal anti-HA (ab9110), 1:2000 (Abcam, Cambridge, MA); mouse monoclonal anti-HA (12CA5), 1:100 (Abcam, Cambridge, MA); mouse monoclonal anti-Flag M2 antibody (F1804), 1:100 (Sigma); mouse α-Brp, 1:250 (Wagh et al., 2006); rabbit α-Synaptotagmin I, 1:4000 (Mackler et al., 2002); mouse monoclonal anti-FasII (1D4), 1:50 (Developmental Studies Hybridoma Bank). Alexa 488, Alexa546 or Alexa633 conjugated secondary antibodies were used at 1:300 (Invitrogen). FITC-conjugated antibodies to horseradish peroxidase (HRP) were used at 1:100 to visualize nerves (Jackson ImmunoResearch). For staining of plumBAC-GFP:HA, samples were first incubated with mouse anti-HA (12CA5), then H2O2 treated for 10 min, incubated with Biotin conjugated Goat anti Mouse (Jackson ImmunoResearch), signal amplified with ABC amplification kit (Vector Labs, Inc.), and incubated with Cy5-tyramide (1:400) for 5 min.
Characterization of pruning defect severity and fly survival
The severity of γ MARCM clone pruning defects in the adult brain is categorized based on whether the clone innervates the adult γ lobe and whether it contains dorsal unpruned axonal branches. The adult γ lobe is defined by moderate FasII immunoreactivity, usually labeled with an asterisk in figures. Dorsal unpruned axonal branches are those that are significantly outside the α lobe, which is defined by strong FasII immunoreactivity.
For Figure 6H, γ neurons were visualized by FasII antibody staining. The different categories of pruning defect severity are defined as such: strong = no γ neurons innervate the adult γ lobe; medium = innervation of the adult γ lobe is seen, but the majority of γ neurons are unpruned; mild = the adult γ lobe is substantially innervated, unpruned γ neurons are sparse. For figure 6I, the survival rate was calculated as the number of flies surviving to adulthood divided by the genetically expected number of flies of a given genotype.
Supplementary Material
Highlights.
An IgSF transmembrane protein, Plum, is cell-autonomously required for axon pruning
Plum promotes mushroom body axon pruning by regulating ecdysone receptor expression
Plum regulates ecdysone receptor expression by facilitating TGF-β signaling
Plum regulates ectopic terminal projections of developing larval motoneurons
Acknowledgements
We thank L. Attisano, T. Lee, M. B. O’Connor, N. Reist, the Vienna Drosophila RNAi Center, and the Bloomington stock center for reagents; the FasII (1D4) and Brp (nc82) monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa; R. Watts and E. Hoopfer for many hours of joint screening; N. Goriatcheva, D. Luginbuhl, N. Issman and O. Fuchs for technical assistance; O. Golani for help with MATLAB programming; M. Hortsch and M. Serpe for discussions; K. Shen, A. Yaron, M. Schuldiner, and members of the Luo and Schuldiner labs for discussions and critical readings of the manuscript. This work was supported by NIH grant R37-NS041044 to L.L and by ISF grants 1864/08 and 683/11 (Bio-med Legacy program), funds from the Irving B. Harrison foundation, the Estate of Florence and Charles Cuevas and the Adelis foundation to O. S. L.L. and K.C.G. are investigators of the Howard Hughes Medical Institute. O. S. is the incumbent of the Rothstein Career Development Chair of Genetic Diseases. TJM was supported by Epilepsy, Neonatology and Developmental Biology Training Grants (NIH 5T32 NS 007280 and HD007249). Xiaomeng M. Yu was a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG1993-08).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agarwala KL, Nakamura S, Tsutsumi Y, Yamakawa K. Down syndrome cell adhesion molecule DSCAM mediates homophilic intercellular adhesion. Brain Res Mol Brain Res. 2000;79:118–126. doi: 10.1016/s0169-328x(00)00108-x. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Huang Y, O’Connor MB, Lee T. Glia instruct developmental neuronal remodeling through TGF-beta signaling. Nat Neurosci. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Bagri A, Cheng HJ, Yaron A, Pleasure SJ, Tessier-Lavigne M. Stereotyped pruning of long hippocampal axon branches triggered by retraction inducers of the semaphorin family. Cell. 2003;113:285–299. doi: 10.1016/s0092-8674(03)00267-8. [DOI] [PubMed] [Google Scholar]
- Berger J, Suzuki T, Senti KA, Stubbs J, Schaffner G, Dickson BJ. Genetic mapping with SNP markers in Drosophila. Nat Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–661. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Boulanger A, Clouet-Redt C, Farge M, Flandre A, Guignard T, Fernando C, Juge F, Dura JM. ftz-f1 and Hr39 opposing roles on EcR expression during Drosophila mushroom body neuron remodeling. Nat Neurosci. 2010 doi: 10.1038/nn.2700. [DOI] [PubMed] [Google Scholar]
- Brummel T, Abdollah S, Haerry TE, Shimell MJ, Merriam J, Raftery L, Wrana JL, O’Connor MB. The Drosophila activin receptor baboon signals through dSmad2 and controls cell proliferation but not patterning during larval development. Genes Dev. 1999;13:98–111. doi: 10.1101/gad.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummendorf T, Rathjen FG. Structure/function relationships of axon-associated adhesion receptors of the immunoglobulin superfamily. Curr Opin Neurobiol. 1996;6:584–593. doi: 10.1016/s0959-4388(96)80089-4. [DOI] [PubMed] [Google Scholar]
- Carrillo RA, Olsen DP, Yoon KS, Keshishian H. Presynaptic activity and CaMKII modulate retrograde semaphorin signaling and synaptic refinement. Neuron. 2010;68:32–44. doi: 10.1016/j.neuron.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Spemann's organizer and the self-regulation of embryonic fields. Mech Dev. 2009;126:925–941. doi: 10.1016/j.mod.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BA, Fetter RD, Davis GW. Dynactin is necessary for synapse stabilization. Neuron. 2002;34:729–741. doi: 10.1016/s0896-6273(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Ellis JE, Parker L, Cho J, Arora K. Activin signaling functions upstream of Gbb to regulate synaptic growth at the Drosophila neuromuscular junction. Dev Biol. 2010;342:121–133. doi: 10.1016/j.ydbio.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal. 2010;22:1163–1174. doi: 10.1016/j.cellsig.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesualdi SC, Haerry TE. Distinct signaling of Drosophila Activin/TGF-beta family members. Fly (Austin) 2007;1:212–221. doi: 10.4161/fly.5116. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Chiba A, Johansen J, Keshishian H. Growth cone behavior underlying the development of stereotypic synaptic connections in Drosophila embryos. J Neurosci. 1991;11:3227–3238. doi: 10.1523/JNEUROSCI.11-10-03227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DD, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Islam R, Kristiansen LV, Romani S, Garcia-Alonso L, Hortsch M. Activation of EGF receptor kinase by L1-mediated homophilic cell interactions. Mol Biol Cell. 2004;15:2003–2012. doi: 10.1091/mbc.E03-05-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Keshishian H. Role of neural activity during synaptogenesis in Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15:8177–8190. doi: 10.1523/JNEUROSCI.15-12-08177.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Halpern ME, Keshishian H. Axonal guidance and the development of muscle fiber-specific innervation in Drosophila embryos. J Neurosci. 1989;9:4318–4332. doi: 10.1523/JNEUROSCI.09-12-04318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor DB, Kolodkin AL. Curbing the excesses of youth: molecular insights into axonal pruning. Neuron. 2003;38:849–852. doi: 10.1016/s0896-6273(03)00364-7. [DOI] [PubMed] [Google Scholar]
- Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Liu XB, Low LK, Jones EG, Cheng HJ. Stereotyped axon pruning via plexin signaling is associated with synaptic complex elimination in the hippocampus. J Neurosci. 2005;25:9124–9134. doi: 10.1523/JNEUROSCI.2648-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen Y, Wang D, Wang S, Zhang YQ. Distinct presynaptic and postsynaptic dismantling processes of Drosophila neuromuscular junctions during metamorphosis. J Neurosci. 2010;30:11624–11634. doi: 10.1523/JNEUROSCI.0410-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, O'Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- Mackler JM, Drummond JA, Loewen CA, Robinson IM, Reist NE. The C(2)B Ca(2+)-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature. 2002;418:340–344. doi: 10.1038/nature00846. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massaro CM, Pielage J, Davis GW. Molecular mechanisms that enhance synapse stability despite persistent disruption of the spectrin/ankyrin/microtubule cytoskeleton. J Cell Biol. 2009;187:101–117. doi: 10.1083/jcb.200903166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrosinski J, Krajewska WM. [TGF beta signalling accessory receptors] Postepy Biochem. 2008;54:264–273. [PubMed] [Google Scholar]
- Mosca TJ, Schwarz TL. The nuclear import of Frizzled2-C by Importins-beta11 and alpha2 promotes postsynaptic development. Nature neuroscience. 2010;13:935–943. doi: 10.1038/nn.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MJ, Merritt DJ, Brand AH, Whitington PM. In vivo dynamics of axon pathfinding in the Drosophilia CNS: a time-lapse study of an identified motorneuron. Journal of neurobiology. 1998;37:607–621. [PubMed] [Google Scholar]
- Nguyen QT, Lichtman JW. Mechanism of synapse disassembly at the developing neuromuscular junction. Current opinion in neurobiology. 1996;6:104–112. doi: 10.1016/s0959-4388(96)80015-8. [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Pielage J, Fetter RD, Davis GW. Presynaptic spectrin is essential for synapse stabilization. Curr Biol. 2005;15:918–928. doi: 10.1016/j.cub.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Weimer RM, De Paola V, Caroni P, Svoboda K. Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 2005;3:e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Whitmore AV, Finn JT. Axonal self-destruction and neurodegeneration. Science. 2002;296:868–871. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- Rougon G, Hobert O. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu Rev Neurosci. 2003;26:207–238. doi: 10.1146/annurev.neuro.26.041002.131014. [DOI] [PubMed] [Google Scholar]
- Salbaum JM, Kappen C. Cloning and expression of nope, a new mouse gene of the immunoglobulin superfamily related to guidance receptors. Genomics. 2000;64:15–23. doi: 10.1006/geno.2000.6114. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Goumans MJ, Pardali E. Endoglin in angiogenesis and vascular diseases. Angiogenesis. 2008;11:79–89. doi: 10.1007/s10456-008-9101-9. [DOI] [PubMed] [Google Scholar]
- Truman JW. Metamorphosis of the central nervous system of Drosophila. J Neurobiol. 1990;21:1072–1084. doi: 10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- Vogel C, Teichmann SA, Chothia C. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development. 2003;130:6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- Winberg ML, Mitchell KJ, Goodman CS. Genetic analysis of the mechanisms controlling target selection: complementary and combinatorial functions of netrins, semaphorins, and IgCAMs. Cell. 1998;93:581–591. doi: 10.1016/s0092-8674(00)81187-3. [DOI] [PubMed] [Google Scholar]
- Wong YH, Lu AC, Wang YC, Cheng HC, Chang C, Chen PH, Yu JY, Fann MJ. Protogenin defines a transition stage during embryonic neurogenesis and prevents precocious neuronal differentiation. J Neurosci. 2010;30:4428–4439. doi: 10.1523/JNEUROSCI.0473-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nature protocols. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O'Connor MB, Lee CH, Lee T. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112:303–315. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.