Abstract
The Norway spruce genome provides key insights into the evolution of plant genomes, leading to testable new hypotheses about conifer, gymnosperm, and vascular plant evolution.
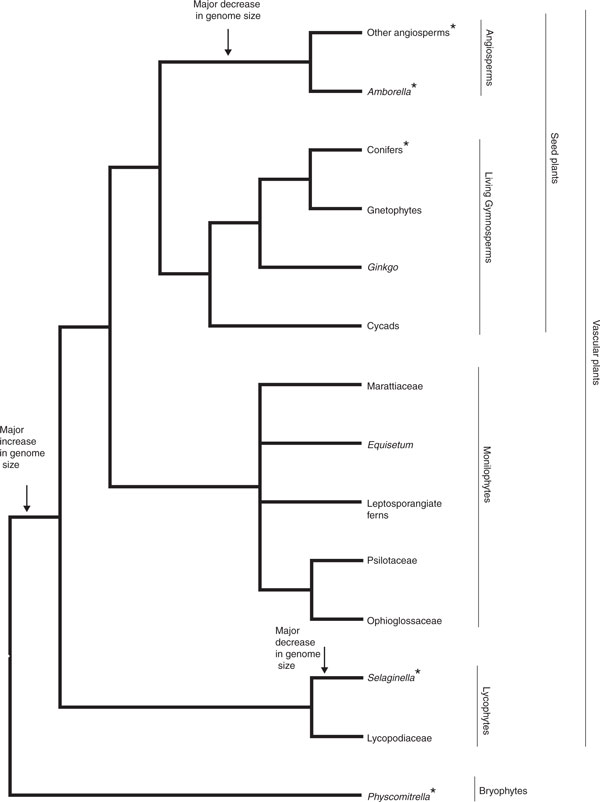
In the past year a burst of plant genome sequences have been published, providing enhanced phylogenetic coverage of green plants (Figure 1) and inclusion of new agricultural, ecological, and evolutionary models. Collectively, these sequences are revealing some extraordinary structural and evolutionary attributes in plant genomes. Perhaps most surprising is the exceptionally high frequency of whole-genome duplication (WGD): nearly every genome that has been analyzed has borne the signature of one or more WGDs, with particularly notable events having occurred in the common ancestors of seed plants, of angiosperms, and of core eudicots (the latter 'WGD' represents two WGDs in close succession) [1,2]. Given this tendency for plant genomes to duplicate and then return to an essentially diploid genetic system (an example is the cotton genomes, which have accumulated the effects of perhaps 15 WGDs [3]), the conservation of genomes in terms of gene number, chromosomal organization, and gene content is astonishing. From the publication of the first plant genome, Arabidopsis thaliana [4], the number of inferred genes has been between 25,000 and 30,000, with many gene families shared across all land plants, although the number of members and patterns of expansion and contraction vary. Furthermore, conserved synteny has been detected across the genomes of diverse angiosperms, despite WGDs, diploidization, and millions of years of evolution.
Figure 1.
Simplified phylogeny of land plants, showing major clades and their component lineages. Asterisks indicate species (or lineage) for which whole-genome sequence (or sequences) is (are) available. Increases and decreases in genome size are shown by arrows.
Despite the proliferation of genome sequences available for angiosperms, genome-level data for both ferns (and their relatives, collectively termed monilophytes; Figure 1) and gymnosperms have been conspicuously lacking - until recently, with the publication of the genome sequence of the gymnosperm Norway spruce (Picea abies) [5]. The large genome sizes for both monilophytes and gymnosperms have discouraged attempts at genome sequencing and assembly, whereas the smaller genome size of angiosperms has resulted in more genome sequences being available (Table 1) [6]. Because of this limited phylogenetic sample, our understanding of the timing and phylogenetic positions of WGDs, the core number of plant genes, possible conserved syntenic regions, and patterns of expansion and contraction of gene families across both tracheophytes (vascular plants) and across all land plants is imperfect. This sampling problem is particularly acute in analyses of the genes and genomes of seed plants; many hundreds of genes are present in angiosperms that are not present in mosses or lycophytes, but whether these genes arose in the common ancestor of seed plants or of angiosperms cannot be determined without a gymnosperm genome sequence. The Norway spruce genome therefore offers tremendous power, not only for understanding the structure and evolution of conifer genomes, but also as a reference for interpreting gene and genome evolution in angiosperms.
Table 1.
Genome sizes in land plants
| Lineage | Range (1C; pg) | Mean |
|---|---|---|
| Gymnosperms | ||
| Conifers | ||
| Pinaceae | 9.5-36.0 | 23.7 |
| Cupressaceae | 8.3-32.1 | 12.8 |
| Sciadopitys | 20.8 | n/a |
| Gnetales | ||
| Ephedraceae | 8.9-15.7 | 8.9 |
| Gnetaceae | 2.3-4.0 | 2.3 |
| Cycadaceae | 12.6-14.8 | 13.4 |
| Ginkgo biloba | 11.75 | n/a |
| Monilophytes | ||
| Ophioglossaceae | 10.2-65.6 | 31.05 |
| Equisetaceae | 12.9-304 | 22.0 |
| Psilotum | 72.7 | n/a |
| Leptosporangiate ferns | ||
| Polypodiaceae | 7.5-19.7 | 7.5 |
| Aspleniaceae | 4.1-9.1 | 6.2 |
| Athyriaceae | 6.3-9.3 | 7.6 |
| Dryopteridaceae | 6.8-23.6 | 11.7 |
| Water ferns | ||
| Azolla | 0.77 | n/a |
| Angiosperms | ||
| Oryza sativa | 0.50 | n/a |
| Amborella trichopoda | 0.89 | n/a |
| Arabidopsis thaliana | 0.16 | n/a |
| Zea mays | 2.73 | n/a |
n/a, not applicable. Data based on [6].
An overview of the P. abies genome
Like the genomes of other conifers, the Norway spruce genome is immense [5], despite a typical conifer chromosome number of 2N = 24: the genome size for species of Picea ranges from 15.75 to 19.125 pg, and the draft genome for P. abies is 20 Gb (or about 20.5 pg). Based on analyses using synonymous substitution rates for inferring ancient WGDs, P. abies lacks evidence of WGDs other than the one that predated all extant seed plants [1]. The large genome of Picea and other conifers has occurred through mechanisms other than WGD: proliferation of long terminal repeat retrotransposons (LTR-RTs), including both the well known Ty3-gypsy and Ty1-copia superfamilies of transposable elements, accounts for its genomic 'obesity' [5] and, by extension, that of other conifers [7].
Furthermore, the two fundamental differences between angiosperms and other seed plants relate to reproduction and water-conducting ability, and the genes found in the Picea genome provide information on these systems. P. abies, as expected, lacks FLOWERING LOCUS T, a set of key activation genes for flowering in angiosperms, and contains an expanded set of FT/TFL1-like genes, which probably act as repressors of flowering [5]. In contrast, the genetic control of water conduction is not as clear. Water transport in conifers is accomplished by cells called tracheids, but most angiosperms have more efficient conducting cells (vessels). Angiosperm-specific innovations in water conduction are controlled by a gene family (VASCULAR NAC DOMAIN, VND) that may have originated in gymnosperms, or possibly earlier - two VND genes were detected in P. abies, compared with seven in Arabidopsis.
Significance of the P. abies genome for understanding plant genomes
The sequencing of the Norway spruce genome [5] is a landmark development in our understanding of plant genomes. The gene space of conifers is not substantially different from that of angiosperms - despite the much larger conifer genomes. In fact, the number of predicted genes for essentially all sequenced plant genomes is approximately 25,000 regardless of genome size and the number of WGDs. Even the bladderwort Utricularia gibba (an angiosperm), with a genome size of only 77 Mb, has an estimated 28,500 genes [8], nearly the same as that predicted for P. abies (28,345). In contrast, the sacred lotus Nelumbo nucifera (also an angiosperm), with a genome size more than 10-fold greater than that of U. gibba and 20 times smaller than that of P. abies at 929 Mb, contains approximately 26,685 genes [9]. The consistency of these three estimates is striking, especially as the three papers [5,8,9] appeared within a month of one another and followed community standards for gene annotation. Furthermore, despite the much larger and more complex genomes of plants compared to those of most animals, the number of genes in their genomes is similar and does not seem to be proportional to genome size. Finally, the Norway spruce genome has expanded by the slow and steady accumulation of LTR-RTs, a phenomenon also observed in pine genomes [7]; this may reflect the lack of an efficient mechanism for eliminating these transposable elements.
Because the P. abies genome is the sole representative of the four extant lineages of gymnosperms (conifers, cycads, Ginkgo, and gnetophytes; Figure 1), it is difficult to infer which features are common to other conifers and which are unique to P. abies. However, taken together with data for other land plants, including genomic resources available for other gymnosperms, we will be able to start to assemble an understanding of the features that are unique to seed plants as a whole and those that are restricted to angiosperms. Further assembly and annotation are needed to understand genome structure and gene content in Norway spruce; genome sequences for additional conifers or other lineages of gymnosperms, despite their generally large size, should help clarify the uniqueness of the P. abies genome and provide the information needed for comparative studies that will enable the application of genomic data to forestry, breeding, and analysis of seed plant traits.
Genome and organismal evolution in a phylogenetic context
Despite the relatively small size of most plant genomes sequenced so far, extensive genetic and physical map resources have typically been required for the organization of the sequencing effort and the genome assembly. Therefore, for both scientific and practical reasons, sequencing efforts have focused mostly on genetic models with small genomes. However, as efforts to understand the evolution of plant genes and genomes expand, species will be sequenced solely because of their pivotal phylogenetic position - and these species will probably lack genetic and genomic resources.
Fortunately, the emergence of next-generation sequencing technologies, as well as new strategies for genome assembly, now make it possible to generate and assemble high-quality, cost-effective genome sequences for evolutionary models lacking genetic resources. For example, fluorescent in situ hybridization of bacterial artificial chromosomes or other probes, coupled with whole-genome mapping (optical mapping), can be used to guide and validate de novo genome assembly based on next-generation sequencing data. This strategy should be widely applicable to non-model plants with poor genomic resources, thus facilitating whole-genome sequencing and assembly for many other plant species.
Technical and analytical breakthroughs provide unprecedented opportunities for gaining new and fundamental insight into genome and organismal evolution across land plants. The Picea genome, at 20 Gb, takes us in a bold new direction. But what genomes to sequence next? A phylogenetic perspective can help identify future targets.
Viewing genome size across land-plant phylogeny reveals a dynamic pattern of genome size evolution, with an increase in genome size coincident with the origin of vascular plants, subsequent independent genome reductions in Selaginella and angiosperms, and further increases within some groups of angiosperms (such as monocots; Figure 1). Genome sizes are labile even within gymnosperms; from a large ancestral gymnosperm genome, independent increases occurred in Ephedra (a gnetophyte), Pinaceae, Pinus, and two non-Pinaceae conifers (Sciadopitys and Sequoia, the latter the only known polyploid conifer), and reduction in Gnetum (a gnetophyte), Ginkgo, and most non-Pinaceae conifers [10]. Monilophytes typically have very large genomes and high chromosome numbers, features typically attributed to ancient WGD. Although recent episodes of polyploidy have been documented in many fern genera, there is no compelling evidence for ancient WGD in any monilophyte lineage.
These patterns of genome size change raise intriguing questions about the evolution of plant genomes. Through analysis of the P. abies genome, we now know that conifer genomes have expanded through proliferation of LTR-RTs, but does that mechanism apply to other large genomes, such as those of other gymnosperms and monilophytes, especially Psilotaceae and Ophioglossaceae, which have even larger genomes than conifers? Are the large monilophyte genomes comparable in structure and function to the P. abies genome? Do other large plant genomes also have long introns, similar to P. abies? What are the ancestral features of vascular plant genomes? Is genomic obesity the result of LTR-RTs retained from the ancestral vascular plants? What is the structural and functional role of the large chromosomes associated with large genomes? What role, if any, has ancient WGD played in the evolution of large monilophyte genomes? Reductions in genome size have occurred independently in gymnosperms (for example Gnetum), monilophytes (for example, Azolla, a water fern), and angiosperms: did genome downsizing occur by the same mechanism, for example, by repression of transposable element expansion coupled with loss of genetic material?
With the P. abies genome as a reference, analysis of genomes from a cycad, Ginkgo, gnetophyte, and other conifers could reveal how many features of the P. abies genome are actually shared, ancestral features of all gymnosperms and which are unique to conifers. Inclusion of a leptosporangiate fern (Ceratopteris) and a member of Marattiales (Angiopteris), both of which have some genetic resources, and of a lycophyte with a large genome (Lycopodium) would facilitate testing hypotheses of ancestral patterns of genomic change in vascular plants and their underlying mechanisms.
Conclusions
Genome sequences carry the keys to understanding genotype-to-phenotype relationships - for features as diverse as morphological characters, biochemical pathways, transcriptional networks, stress response, and more. Increased strategic sampling of plant genomes from across land plants and their green algal relatives will yield unparalleled information on the genes and gene families responsible for the major transitions in plant evolutionary history - the move onto land and the origins of vascular tissue, seeds, and flowers - as well as the genes controlling traits that could be harnessed for human benefit.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Pamela S Soltis, Email: psoltis@flmnh.ufl.edu.
Douglas E Soltis, Email: dsoltis@ufl.edu.
References
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Leebens-Mack J, Ayyampalayam S, Bowers JE, McKain MR, McNeal J, Rolf M, Ruzicka DR, Wafula E, Wickett NJ, Wu X, Zhang Y, Wang J, Zhang Y, Carpenter EJ, Deyholos MK, Kutchan TM, Chanderbali AS, Soltis PS, Stevenson DW, McCombie R, Pires JC, Wong GK, Soltis DE, Depamphilis CW. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hällman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Käller M, Luthman J, Lysholm F, Niittylä T, Olson Å, Rilakovic N, Ritland C, Rosselló JA, Sena J. et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (embryophyta). Ann Bot. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse AM, Peterson DG, Islam-Faridi MN, Smith KE, Magbanua Z, Garcia SA, Kubisiak TL, Amerson HV, Carlson JE, Nelson CD, Davis JM. Evolution of genome size and complexity in Pinus. PLoS One. 2009;4:e4332. doi: 10.1371/journal.pone.0004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Lyons E, Hernández-Guzmán G, Pérez-Torres CA, Carretero-Paulet L, Chang TH, Lan T, Welch AJ, Juárez MJ, Simpson J, Fernández-Cortés A, Arteaga-Vázquez M, Góngora-Castillo E, Acevedo-Hernández G, Schuster SC, Himmelbauer H, Minoche AE, Xu S, Lynch M, Oropeza-Aburto A, Cervantes-Pérez SA, de Jesús Ortega-Estrada M, Cervantes-Luevano JI, Michael TP, Mockler T, Bryant D, Herrera-Estrella A, Albert VA, Herrera-Estrella L. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Vanburen R, Liu Y, Yang M, Han Y, Li LT, Zhang Q, Kim MJ, Schatz MC, Campbell M, Li J, Bowers JE, Tang H, Lyons E, Ferguson AA, Narzisi G, Nelson DR, Blaby-Haas CE, Gschwend AR, Jiao Y, Der JP, Zeng F, Han J, Min XJ, Hudson KA, Singh R, Grennan AK, Karpowicz SJ, Watling JR, Ito K. et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 2013;14:R41. doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh JG, Barbazuk WB, Davis JM, Morse AM, Soltis PS. Exploring diversification and genome size evolution in gymnosperms through phylogenetic synthesis. J Bot. 2012.