Abstract
Understanding commonalities and differences of how symbiotic and parasitic microbes interact with plants will improve advantageous interactions and allow pathogen control strategies in crops. Recently established systems enable studies of root pathogenic and symbiotic interactions in the same plant species.
Interactions between organisms shape ecological communities [1]. It is fascinating how plants fine-tune defense to prevent detrimental interactions while supporting development towards advantageous interactions [2]. As plants are unable to escape parasite attacks, they needed to evolve strong defense mechanisms to effectively ward off pathogens [3]. But plants also engage in symbiosis with advantageous microorganisms such as root-associated bacteria or fungi to gain extended nutrient access [4]. Knowing how plants control their interactions has a direct impact on our crop plants and influence on our agricultural strategies, and is thus a very important field of research.
Driven by the impact of diseases on agriculture, plant-pathogen research has resulted in extensive knowledge on how plants defend themselves against above-ground pathogens. Also, how plants engage in beneficial root symbiosis is a field of intense research [5]. However, there is much less known on the overlap between the two types of interaction. One reason is that historically research into plant-pathogen interactions and symbiosis research were motivated by different aims. Economically relevant pathogens such as the fungus-like oomycete Phytophthora infestans, trigger of the Irish potato famine, continue to cause dramatic yield losses in crops such as potato and tomato [6]. Given these economic and societal impacts, plant pathology research has focused on disease resistance, and has therefore been dominated by the study of pathogen modulation of plant immunity [7].
In contrast, research into beneficial effects of plant microbes is mainly guided by nutritional aspects [5] with much less focus on immunity and compatibility aspects. A well-studied example for beneficial symbiosis is the association of plant roots with fungi [8]. This mycorrhiza can be found in 80% of all land plants. Arbuscular mycorrhiza (AM) relies on an evolutionarily ancient program dating back to early land plants and was key when plants conquered the land. It is conceivable that pathogens take advantage of this symbiosis program to gain access to the host plant's resources.
We have extensive evidence for commonalities between pathogenic and symbiotic lifestyles. Both interaction types follow similar developmental processes of identification, plant cell penetration and re-differentiation of the host cells to establish intracellular interfaces for nutrient and information exchange (Figure 1) [9]. Undecorated chitin oligomers of microbe origin, known to be potent inducers of plant immunity [10], were found recently to also activate symbiosis-related signaling [11]. Furthermore, effector proteins, hallmarks of animal and plant pathogens and which suppress defense and reprogram the host, were also described recently in mycorrhizal fungi [12,13]. Considering these similarities, it is surprising that very few parallels have been made between modes of pathogen and symbiotic colonization. Since symbiotic mycorrhiza occur only below ground, we are bound to study both types of interactions in roots. This will enable us to generate pathogen-resistant crop plants without affecting beneficial symbiosis. To do so, we need dual research systems that enable these comparative studies.
Figure 1.
Phytophthora species and arbuscular mycorrhizal (AM) fungi follow analogous steps to establish a root interaction. Following chemical cross-talk, the microbe germinates and forms attachment and penetration structures, termed appressoria and hyphopodia, respectively. Penetration occurs through or between cells, and in the case of AM fungi intracellular hyphae are supported by a plant-derived pre-penetration apparatus [17]. Specialized intracellular interfaces, termed haustoria and arbuscules, form within plant root cells. Phytophthora infections eventually result in cell death of the infected tissue, while obligate biotrophic AM fungi continuously reside in living plant roots.
Our ability to compare principles of colonization is hampered by the traditional separation of plant pathology systems and symbiosis systems on different plant species.
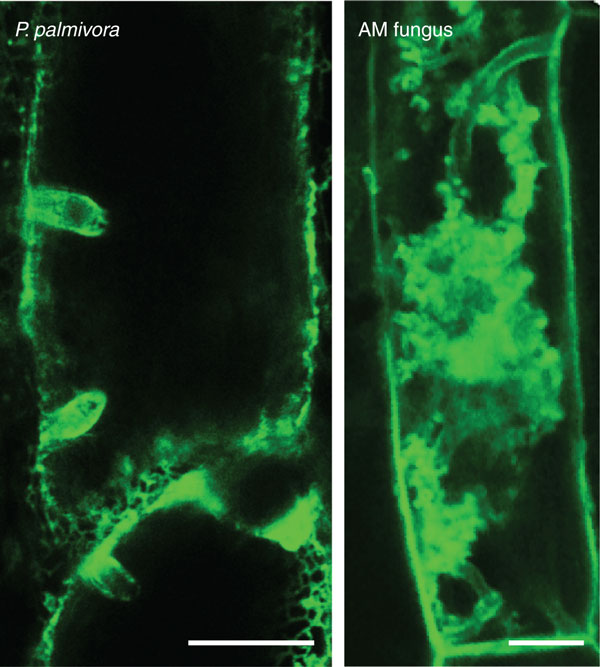
Arabidopsis thaliana, the plant system of choice for numerous plant-pathogen interactions, does not support feeding structure formation by endomycorrhizal fungi, and thus is limited to studies of non-host interactions [14]. Notably, separate research of Phytophthora pathogens in its host plants potato and tomato, and beneficial AM fungi in legumes and rice, has shown that both follow analogous steps to establish an interaction (Figure 1). Moreover, both form specialized accommodation structures within plant cells (Figure 2). Thus, it would be good to have a single plant species that allows direct comparison between pathogenic and symbiotic interactions.
Figure 2.
Accommodation structures formed by filamentous microbes in Nicotiana benthamiana roots. Phytophthora palmivora projects digit-like haustoria into root cells that are surrounded by plant endoplasmic reticulum (labeled using green fluorescent protein, GFP). Arbuscular mycorrhizal (AM) fungi form arbuscules, visualized using a plant membrane-associated GFP fusion protein.
Dual systems enable the study of root colonization by filamentous pathogens and symbionts
Dual systems are crop plants whose roots are colonized by filamentous symbiotic microbes (for example, the widely used AM fungus Glomus irregularis) but in addition can also be infected by other biotrophic pathogens. The legume species Medicago truncatula and Lotus japonicus have served as genetic model plants for symbiosis research [4,8] and a huge genetic resource has been established by the community, rendering these plants prime candidates for systems to study similarities and differences between symbiosis and pathogenicity. Another established monocot system for symbiosis research is rice [15]. It is surprising, however, that not many root pathogen infection systems with clearly distinguishable biotrophic stages exist for these plant species.
In AM fungi colonized roots, the ideal microbial pathogen partner to compare with is a naturally root colonizing filamentous organism with a broad host range. Other than G. irregularis it should be cultivatable, transformable and efficiently traceable in living tissues - for example, by fluorescent proteins. Several filamentous microbes have been employed to unravel the mechanisms involved in root colonization (Table 1). Historically, most research has been carried out using Aphanomyces euteiches [16], Colletotrichum trifolii [17] and Verticillium species [18], and to a major extent using Magnaporthe oryzae [19]. Piriformospora indica colonization of roots and its growth-promoting effects have also been studied in the economically relevant barley [20], a monocot plant that also establishes interactions with AM fungi [21].
Table 1.
Sequenced filamentous microorganisms with biotrophic stages that enable dual root interaction studies
| Species | Organism | Interaction | Mainly used plant systems | Root invasion described | Host range | Intracellular interface | Axenic growth | Transform-able | Genome |
|---|---|---|---|---|---|---|---|---|---|
| Aphanomyces euteiches | Oomycota, saprolegniales | Pathogen | Medicago truncatula, Pisum sativum | Yes | Narrow | None | Yes | No | In progress |
| Colletotrichum trifolii | Fungi, Ascomycota | Pathogen | M. truncatula | No | Narrow | Specialized hyphae | Yes | Yes | Other species [42] |
| Glomus irregularis | Fungi, glomeromycota | Mutualist | M. truncatula, Lotus japonicus, Oryza sativa, P. sativum, Hordeum vulgare | Yes | Broad | Arbuscules | No | No | In progress |
| Laccaria bicolor | Fungi, basidiomycota | Mutualist | Populus trichocarpa | Yes | Broad | None | Yes | Yes | Yes [43] |
| Magnaporthe oryzae | Fungi, ascomycota | Pathogen | O. sativa | No | Narrow | Specialized hyphae | Yes | Yes | Yes [44] |
| Phytophthora palmivora | Oomycota, peronosporales | Pathogen | M. truncatula, Nicotinia benthamiana | Yes | Broad | Haustoria | Yes | Yes | In progress |
| Piriformospora indica | Fungi, basidiomycota | Mutualist | H. vulgare | Yes | Broad | Sporadic coils | Yes | Yes | Yes [45] |
| Verticillium sp. | Fungi, ascomycota | Pathogen | M. trunctatula | Yes | Broad | None | Yes | Yes | Yes [46] |
Notably, C. trifolii and M. oryzae are major leaf colonizers in nature; however, they can be employed for root infection under laboratory conditions [17,22]. C. trifolii experiments have helped to extend the role of the DMI3 (DOESN'T MAKE INFECTIONS 3) calcium/calmodulin kinase, a classical symbiosis signaling element, from symbiotic to pathogenic interactions [17]. Infections with C. trifolii showed differential responses between plants that carried either a DMI3 wild-type or a mutated allele.
While C. trifolii and M. oryzae were reported to establish biotrophic stages inside the root, others such as apoplast-colonizing A. euteiches [16] and Verticillium albo-atrum [18] lack the potential to form intracellular structures such as the arbuscular feeding structures of AM fungi, thereby complicating the delimitation of their biotrophic stages. Nevertheless, A. euteiches has been successfully employed to identify signaling elements that are shared between symbiotic and pathogen perception mechanisms. An example is NFP (NOD FACTOR PERCEPTION), a lysin motif receptor-like kinase (LysM-RLK) that is integral to perception of lipochitooligosaccharidic symbiosis factors from AM fungi by the plant. Recently, NFP was shown to also affect colonization by the pathogen A. euteiches [16]. Larger sets of LysM-RLK receptor variants can be found in root nodule and AM-forming legumes [23,24] and non-nodulating but AM-forming rice [25] compared with non-mycorrhized and non-nodulated Arabidopsis. This enlarged receptor repertoire could correlate with a requirement for higher resolution signal discrimination between pathogens, mycorrhizal fungi and symbiotic bacteria. Further research is required to clarify how specificity in signal perception is achieved.
Phytophthora palmivora provides new opportunities for comparative studies in roots
A key element of beneficial AM fungi is nutrient exchange through tree-like arbuscules in the inner cortical cells of the host root (Figure 2) [9]. Thus, it is of particular interest to compare arbuscules with other morphologically distinguishable intracellular interfaces formed by pathogens in roots. M. oryzae and C. trifolii do form specialized intracellular hyphae and P. indica infrequently forms coils in root cells [26]. However, abundant haustoria of pathogenic fungi and oomycetes such as Phytophthora most strikingly resemble dedicated interfaces as observed in arbuscules [9].
Similar to the arbuscules of AM fungi, which are surrounded by a periarbuscular membrane spiked with an exclusively positioned phosphate transporter [27,28], Phytophthora haustoria are enveloped by an extrahaustorial membrane with distinct features to normal plant plasma membranes [29]. We still do not fully understand why Phytophthora forms these intricate structures in plant cells. So far, haustoria are known to facilitate the deployment of pathogen-encoded effector proteins [30]. Currently, cellular research on Phytophthora haustoria is mainly focused on its interaction with above-ground plant parts [31]. However, the majority of Phytophthora species are prominent root pathogens [32], such as P. palmivora, which forms haustoria in roots and thus allows us to comparatively study arbuscules and haustoria in the same tissue (Figure 2). Indeed, the P. palmivora-M. truncatula system has recently helped to identify an overlap in the chemical language spoken between microbe and plant. A cutin-derived signal produced by M. truncatula roots was found to be required for the interaction of both beneficial AM fungi and pathogenic oomycetes [33].
P. palmivora has a broad host range infecting numerous monocot and dicot cash crops such as oil palms, rubber trees, cocoa and coconut. As with other pathogens, its colonization potential resembles that of widespread AM fungi (Table 1). The broad host range of P. palmivora also enables it to infect the workhorse species of time-efficient plant cell biology and fast-forward genetics: Nicotiana benthamiana [34], a species that has also been studied in the context of AM fungal symbiosis [35] (Figure 2). Transformation of Phytophthora facilitates the generation of tracer lines and allows highlighting pathogen-formed haustoria components [31]. Transformability and also knowledge of the P. palmivora genome will enable us to functionally assess pathogen-encoded factors that contribute to colonization such as secreted effector proteins.
We need to know more about effectors
Historically, microbe-encoded effector proteins with functionality inside the host were a hallmark of bona fide pathogens [36]. Cytoplasmic effectors enter the host cells and modify plant processes to suppress immunity and support pathogen colonization [7]. Notably, effectors that target plant root cell processes have recently also been reported from Laccaria bicolor and G. irregularis, which are both beneficial mycorrhizal fungi [12,13]. Evidently, also in beneficial plant-microbe interactions the microbe needs to suppress plant immunity to establish an interaction and prove its usefulness to the plant. The growing repertoire of characterized effectors and availability of numerous pathogen genomes (Table 1) will provide inroads to study common and contrasting effector-targeted processes of mutualistic and parasitic interactions. Furthermore, effector functions have been studied mainly in leaves, but transformable filamentous microbes that colonize roots now give us the tools to study effector functions below ground, where principles might diverge from the known.
We have the tools and we will use them
The described systems (Table 1), which are based on dual plant infections utilizing AM fungi and pathogens, will allow us to challenge findings obtained previously in either pathogen or symbiosis research. This will give insights into the commonalities and differences of both types of interaction outcomes. Moreover, several biotrophic pathogens (M. oryzae, P. palmivora), endophytes (P. indica) and beneficial AM fungi (G. irregularis) are directly applicable on rice, barley and other dicot crops, thereby bypassing the step to transfer knowledge of mechanisms from model plants into application.
Systems that form specialized intracellular structures will be integral in solving open questions on filamentous microbe interactions with plants. We need to elucidate how similar arbuscules and pathogen interfaces are [9], and whether they generally also serve as devices for nutrient uptake [37]. Knowledge on general and specific transport mechanisms between microbe and plant may be decisive for our ability to protect plants from pathogens while maintaining symbiosis.
Common sets of regulated genes point to a large overlap in development processes during beneficial and detrimental interactions [19]; however, we still need to clarify whether observed structural features, such as the pre-penetration apparatus formed during root colonization by AM fungi [38] or the typical tree-like branching of AM fungal arbuscules, are defined by the microbe species or the plant, or both.
Root-colonizing microbes are guided by chemical plant signals but we do not know the extent to which these signals overlap. For example, plant flavonoids act as attractants for mobile oomycete zoospores and beneficial nitrogen-fixing root bacteria, while symbiotic fungi but also parasitic Striga hermontica plants perceive strigolactones released by the plant [39,40]. Whether some filamentous pathogens are consistently responsive to strigolactones remains to be clarified [41].
Furthermore, it is important to identify contrasting principles in effector-mediated reprogramming and immune suppression between symbiotic microbes and pathogens. This might enable us to engineer the host processes they target in order to direct the outcome of an interaction towards the beneficial side.
Abbreviations
AM: arbuscular mycorrhiza; LysM-RLK: lysin motif receptor-like kinase; NFP: nod factor perception.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Thomas Rey, Email: thomas.rey@slcu.cam.ac.uk.
Sebastian Schornack, Email: sebastian.schornack@slcu.cam.ac.uk.
Acknowledgements
We apologize to those authors whose work was not emphasized due to space limitations. We are indebted to Siobhan Braybrook and Sophien Kamoun for critical reading and commenting on an earlier draft of this manuscript. TR and SS acknowledge funding from the Gatsby Charitable Foundation. SS also acknowledges funding by the Royal Society and motivational support by Dr Féi māo.
References
- Van Der Heijden MGA, Bardgett RD, Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Bonneau L, Huguet S, Wipf D, Pauly N, Truong HN. Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol. 2013. doi: 10.1111/nph.12234. [DOI] [PubMed]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, Palmer TM, West SA, Vandenkoornhuyse P, Jansa J, Bücking H. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333:880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- Fry W. Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Pathol. 2008;9:385–402. doi: 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease?. Curr Opin Plant Biol. 2000;3:320–328. doi: 10.1016/s1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pages V, Novero M, Rey T, Fournier J, Rochange S, Becard G, Bonfante P, Barker DG. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca(2+) spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- Plett JM, Kemppainen M, Kale SD, Kohler A, Legue V, Brun A, Tyler BM, Pardo AG, Martin F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol. 2011;21:1197–1203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21:1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- Veiga RS, Faccio A, Genre A, Pieterse CM, Bonfante P, van der Heijden MG. Arbuscular mycorrhizal fungi reduce growth and infect roots of the non-host plant Arabidopsis thaliana. Plant Cell Environ. 2013. doi: 10.1111/pce.12102. [DOI] [PubMed]
- Sawers RJ, Gutjahr C, Paszkowski U. Cereal mycorrhiza: an ancient symbiosis in modern agriculture. Trends Plant Sci. 2008;13:93–97. doi: 10.1016/j.tplants.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Rey T, Nars A, Bonhomme M, Bottin A, Huguet S, Balzergue S, Jardinaud MF, Bono JJ, Cullimore J, Dumas B, Gough C, Jacquet C. NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 2013;198:875–886. doi: 10.1111/nph.12198. [DOI] [PubMed] [Google Scholar]
- Genre A, Ortu G, Bertoldo C, Martino E, Bonfante P. Biotic and abiotic stimulation of root epidermal cells reveals common and specific responses to arbuscular mycorrhizal fungi. Plant Physiol. 2009;149:1424–1434. doi: 10.1104/pp.108.132225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben C, Toueni M, Montanari S, Tardin MC, Fervel M, Negahi A, Saint-Pierre L, Mathieu G, Gras MC, Noël D, Prospéri JM, Pilet-Nayel ML, Baranger A, Huguet T, Julier B, Rickauer M, Gentzbittel L. Natural diversity in the model legume Medicago truncatula allows identifying distinct genetic mechanisms conferring partial resistance to Verticillium wilt. J Exp Bot. 2013;64:317–332. doi: 10.1093/jxb/ers337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güimil S, Chang H-S, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, Briggs SP, Paszkowski U. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA. 2005;102:8066–8070. doi: 10.1073/pnas.0502999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Huckelhoven R, Neumann C, von Wettstein D, Franken P, Kogel KH. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Maier W, Miersch O, Kramell R, Strack D. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol. 2002;130:1213–1220. doi: 10.1104/pp.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesma A, Osbourn AE. The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature. 2004;431:582–586. doi: 10.1038/nature02880. [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, Geurts R, Dénarié J, Rougé P, Gough C. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann GV, Shimoda Y, Nielsen MW, Jorgensen FG, Grossmann C, Sandal N, Sorensen K, Thirup S, Madsen LH, Tabata S, Sato S, Stougaard J, Radutoiu S. Evolution and regulation of the Lotus japonicus LysM receptor gene family. Mol Plant Microbe Interact. 2010;23:510–521. doi: 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Savita V, Sahay N, Butehorn B, Franken P. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl Environ Microbiol. 1999;65:2741–2744. doi: 10.1128/aem.65.6.2741-2744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 2002;14:2413–2429. doi: 10.1105/tpc.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Zhang X, Noar RD, Harrison MJ. Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci USA. 2012;109:E665–672. doi: 10.1073/pnas.1110215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Schornack S, Spallek T, Geldner N, Chory J, Schellmann S, Schumacher K, Kamoun S, Robatzek S. Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol. 2012;14:682–697. doi: 10.1111/j.1462-5822.2012.01751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PR. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Avrova AO, Boevink PC, Young V, Grenville-Briggs LJ, van West P, Birch PR, Whisson SC. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol. 2008;10:2271–2284. doi: 10.1111/j.1462-5822.2008.01206.x. [DOI] [PubMed] [Google Scholar]
- Erwin D, Ribeiro O. Phytophthora Diseases Worldwide. St Paul, Minnesota: The American Phytopathological Society; 1996. [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol. 2012;22:2242–2246. doi: 10.1016/j.cub.2012.09.043. [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Wilkinson RC, S G-I, Findlay K, Coffey MD, Zipfel C, Rathjen JP, Kamoun S, Schornack S. The receptor-like kinase SERK3/BAK1 is required for basal resistance against the Irish potato famine pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS One. 2011;6:e16608. doi: 10.1371/journal.pone.0016608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Fester T. Molecular and cell biology of arbuscular mycorrhizal symbiosis. Planta. 2005;221:184–196. doi: 10.1007/s00425-004-1436-x. [DOI] [PubMed] [Google Scholar]
- Win J, Chaparro-Garcia A, Belhaj K, Saunders DG, Yoshida K, Dong S, Schornack S, Zipfel C, Robatzek S, Hogenhout SA, Kamoun S. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb Symp Quant Biol. in press . [DOI] [PubMed]
- Voegele RT, Mendgen K. Rust haustoria: nutrient uptake and beyond. New Phytol. 2003;159:93–100. doi: 10.1046/j.1469-8137.2003.00761.x. [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell. 2005;17:3489–3499. doi: 10.1105/tpc.105.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner S, Lendzemo V, Langer I, Schweiger P, Khaosaad T, Toussaint JP, Vierheilig H. Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant-fungus interactions. Molecules. 2007;12:1290–1306. doi: 10.3390/12071290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytol. 2009;183:180–189. doi: 10.1111/j.1469-8137.2009.02840.x. [DOI] [PubMed] [Google Scholar]
- Dor E, Joel DM, Kapulnik Y, Koltai H, Hershenhorn J. The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta. 2011;234:419–427. doi: 10.1007/s00425-011-1452-6. [DOI] [PubMed] [Google Scholar]
- O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, Altmüller J, Alvarado-Balderrama L, Bauser CA, Becker C, Birren BW, Chen Z, Choi J, Crouch JA, Duvick JP, Farman MA, Gan P, Heiman D, Henrissat B, Howard RJ, Kabbage M, Koch C, Kracher B, Kubo Y, Law AD, Lebrun MH. et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet. 2012;44:1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I. et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ. et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- Zuccaro A, Lahrmann U, Guldener U, Langen G, Pfiffi S, Biedenkopf D, Wong P, Samans B, Grimm C, Basiewicz M, Murat C, Martin F, Kogel KH. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 2011;7:e1002290. doi: 10.1371/journal.ppat.1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BP, Chen Z, Henrissat B, Lee YH, Park J, Garcia-Pedrajas MD, Barbara DJ, Anchieta A, de Jonge R, Santhanam P, Maruthachalam K, Atallah Z, Amyotte SG, Paz Z, Inderbitzin P, Hayes RJ, Heiman DI, Young S, Zeng Q, Engels R, Galagan J, Cuomo CA, Dobinson KF, Ma LJ. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011;7:e1002137. doi: 10.1371/journal.ppat.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]