Abstract
Formation of the periodontium begins following onset of tooth-root formation in a coordinated manner after birth. Dental follicle progenitor cells are thought to form the cementum, alveolar bone and Sharpey's fibers of the periodontal ligament (PDL). However, little is known about the regulatory morphogens that control differentiation and function of these progenitor cells, as well as the progenitor cells involved in crown and root formation. We investigated the role of bone morphogenetic protein-2 (Bmp2) in these processes by the conditional removal of the Bmp2 gene using the Sp7-Cre-EGFP mouse model. Sp7-Cre-EGFP first becomes active at E18 in the first molar, with robust Cre activity at postnatal day 0 (P0), followed by Cre activity in the second molar, which occurs after P0. There is robust Cre activity in the periodontium and third molars by 2 weeks of age. When the Bmp2 gene is removed from Sp7+ (Osterix+) cells, major defects are noted in root, cellular cementum and periodontium formation. First, there are major cell autonomous defects in root-odontoblast terminal differentiation. Second, there are major alterations in formation of the PDLs and cellular cementum, correlated with decreased nuclear factor IC (Nfic), periostin and α-SMA+ cells. Third, there is a failure to produce vascular endothelial growth factor A (VEGF-A) in the periodontium and the pulp leading to decreased formation of the microvascular and associated candidate stem cells in the Bmp2-cKOSp7-Cre-EGFP. Fourth, ameloblast function and enamel formation are indirectly altered in the Bmp2-cKOSp7-Cre-EGFP. These data demonstrate that the Bmp2 gene has complex roles in postnatal tooth development and periodontium formation.
Keywords: Bmp2 gene, cementum, dentinogenesis, periodontium development, root formation
Introduction
In order to regenerate a tooth and its supporting structures, it is necessary to have in depth knowledge of the critical events that regulate normal tooth and periodontium development. The process of cementogenesis is dependent on tooth-root formation. As the formation of the tooth-root begins, the inner and outer enamel epithelium proliferates and forms the bilayered Hertwig's epithelial root sheath.1,2 After root dentin formation, Hertwig's epithelial root sheath becomes perforated and the newly formed root dentin comes in contact with dental follicle cells.3 It is speculated that epithelial signals from the Hertwig's epithelial root sheath are responsible for the differentiation of dental follicle precursor cells into cementoblasts4 to form cementum. Moreover, bone morphogenetic protein-2 (Bmp2), has been shown to promote the differentiation of immortalized dental follicle cells towards an osteoblast/cementoblast phenotype.5 However, the role of Bmp2 in exerting its effects on the development of tooth-root, and its supportive tissues such as the periodontal ligament (PDL) and cementum as well as on pulp vasculogenesis in vivo has not been tested.
We previously showed that Bmp2 plays a critical role in postnatal tooth development and cytodifferentiation when deleted in mature odontoblasts.6 We also presented a model in which terminal odontoblast differentiation was required to set up a feedback loop to control vascularization and candidate stem cells for the odontoblasts. To test components of this model, we deleted the Bmp2 gene (Bmp2-cKOSp7-Cre-EGFP) by using the Sp7-Cre-EGFP mouse model that expresses Cre in a subset of dental pulp cells, very early osteoblasts, pre-odontoblasts, and in a subset of dental follicle cells. We now report the link between the Bmp2 gene in Sp7+ cells to the critical roles in cementogenesis, as well as formation and organization of the periodontal ligaments, and coordination with tooth-root formation. Our findings should provide fundamental knowledge in understanding the factors and cells regulating the formation of the tooth-root and the periodontium. It will also lay a profound foundation for designing new and effective therapeutic approaches for the repair and regeneration of the tooth root and periodontium.
Materials and methods
Mice
All mice studies were conducted under the proper guidelines for use of experimental animals as defined in our protocols and IACUC-UTHSCSA policies on use of experimental animals. The Bmp2 floxed mice are as described in Yang et al.7 The Sp7-Cre-EGFP mice were obtained from Andy McMahon at Harvard University.8 Bmp2 fx/fx mice were crossed with Sp7-Cre-EGFP;Bmp2fx/+ to obtain wild-type (WT), heterozygotes (Het, Sp7-Cre-EGFP;Bmp2fx/+), and Bmp2-cKOSp7-Cre-EGFP (Sp7-Cre-EGFP;Bmp2fx/Bmp2fx). The Rosa26-loxP-stop-loxP-tdTomato (Rosa-LSL-tdTomato) mice were obtained from Jackson Laboratories (Maine).
Mapping Sp7-Cre-EGFP+ cells and Cre events with the Rosa-LSL-tdTomato reporter
Sp7-Cre-EGFP mice were crossed with Rosa-LSL-tdTomato reporter mice, cryostat sections on a tape system were then visualized with confocal microscopy.7 The green nuclear fluorescence marks cells that express Cre, and the cytoplasmic red marks the Cre event and progeny of the Sp7-Cre-EGFP+ cells in the mice. First, second and third molars were mapped at postnatal day 0 (P0) and 2 weeks of age.
Histology, histomorphometric evaluation and acid etching of plastic embedded mandibles
Mandible tissues were collected from Bmp2-cKOSp7-Cre-EGFP and their littermate controls at several postnatal stages. Mandibles from the left side were prepared for radiology study, and the right mandibles were prepared for bone and teeth histological evaluation. Mandible samples were fixed in RNase free 4% formaldehyde, demineralized in RNase free 15% ethylene diaminetetraacetic acid (EDTA) for 6 weeks (changed once per week) and embedded in paraffin.6 Eight-micrometer sections were then prepared for toluidine blue,9 hematoxylin and eosin with orange G, picrosirius ed10 and van Gieson stain11 for collagen bundles. To assess the maturation status of calcification and mineralization, mandible samples were fixed in 70% ethanol for 3 days followed by plastic resin embedding without demineralization and cut at 8-μm sections. In some experiments, the surface was polished and thin sections were prepared for acid etching and scanning electron microscopy evaluation.9
Radiography
Male mice aged at 2 weeks, 1 month and 3 months were used and mandible samples were processed and stored in 70% ethanol for high-resolution X-rays. Digital X-ray images were captured using a Faxitron radiographic inspection unit (Model MX-2 Faxitron; Field Emission Corporation, Inc., Tucson, AZ, USA).
Micro computed tomography analysis
Quantitative micro computed tomography (μCT) analysis of mandibles was carried out on 1-month-, 2-month- and 3-month-old animals. Total volume of dentin, radicular (root) dentin, enamel and total pulp volume, as well as periodontal volume, was quantitated in the first and second molars in 1 month, 2 months and 3 months of age, with one mouse of each genotype at each age. A total of 12 molars were evaluated for the indicated parameters, combining the data from the three ages for statistical evaluation, and as described in detail in Yang et al.6
In situ hybridization
Tissues for in situ hybridization were processed RNase free with diethyl pyrocarbonate (DEPC) water in all reagents. The tissues were fixed in 4% formaldehyde overnight, decalcified in buffered 15% EDTA at 4 °C for 4–6 weeks, embedded in paraffin, mounted on ProbeON Plus slides (Fisher Scientific, Pittsburgh, PA, USA) and kept at 4 °C. All RNA probes for in situ hybridization were transcribed in vitro in the presence of Digoxigenin for production of antisense and sense probes with T3, T7 or SP6 polymerases, where appropriate. Sections were first deparaffinized and treated with proteinase K. Hybridization was performed at 58 °C overnight with a probe concentration of 1 µg⋅mL−1. After overnight hybridization, sections were treated with RNase, washed with 5XSSC and with 50% formamide in 2XSSC. Detection of hybridization signal was done by adding alkaline phosphatase substrate (NTB/BCIP; Roche Diagnostic Corp., Dallas, TX, USA) in detection buffer (10% polyvinyl alcohol 70–100 kD, 100 mmol⋅L−1 Tris, pH 9, 100 mmol⋅L−1 NaCl, 2 mmol⋅L−1 Levamisole; Sigma-Aldrich, Inc., St Louis, MO, USA). The duration of hybridization signal development was from 1 h to overnight at 30 °C, depending on the probe and abundance of the transcript in the tissue. The sections were then lightly stained with methyl green. Procedures for in situ hybridization and numerical estimation of mRNA expression were performed as previously described.6,11
Immunohistochemistry
Immunohistochemistry was performed as previously described.6,11 Primary antibodies included polyclonal rabbit anti-mouse vascular endothelial growth factor A (VEGF-A), Ab9571 (1∶200), rabbit anti-mouse CD146, Ab75769 (1∶200) and rabbit anti-mouse α-SMA, Ab5694 (1∶200), were obtained from Abcam Limited (Boston, MA, USA). Polyclonal rabbit anti-mouse nuclear factor IC (Nfic), LS-B4029 (1∶200) was obtained from LifeSpan BioSciences, Inc. (Seattle, WA, USA), rabbit anti-mouse Smad1/5/8 (1∶200) was obtained from Cell Signaling Technology, Inc. (Boston, MA, USA), and rabbit anti-mouse Periostin (1∶100) was obtained from Innovative Research (Novi, MI, USA).
Immunofluorescence
Mouse mandibles from control (WT and Het) and Bmp2-cKOSp7-Cre-EGFP were fixed in RNase free 4% formaldehyde, demineralized in RNase free 15% EDTA for 1 week (changed every day), rinsed twice in phosphate-buffered saline (PBS) for 30 min and placed in 30% sucrose overnight at 4 °C. The mandibles were serially sectioned in a longitudinal plane at 8 µm with a cryostat. Tissue sections were placed onto Superfrost glass slides (Fisher Scientific, Pittsburgh, PA, USA). Samples were then incubated in blocking solution consisting of 4% normal goat serum (Sigma-Aldrich, St Louis, MO, USA), 2% bovine gamma-globulin (Sigma-Aldrich, St Louis, MO, USA) and 0.3% Triton X-100 (Fisher Scientific, Pittsburgh, PA, USA) in PBS for 60 min before incubation for 16 h in primary antibodies rabbit anti-mouse Ki67, Ab15580 (1∶50), rabbit anti-mouse α-SMA, Ab5694 (1∶50) and rat anti-mouse CD31, Ab7388 (1∶50) antibodies. These antibodies were obtained from Abcam Limited. Rabbit anti-mouse cleaved caspase 3 (1∶50) was obtained from Cell Signaling Technology. Slides were rinsed in PBS and then incubated in Alexa Fluor 568 goat anti-rabbit and Alexa Fluor 488 goat anti-rat secondary antibodies (Molecular Probes, Eugene, OR, USA) at a 1∶100 dilution. Primary and secondary antibodies were both diluted in blocking solution. Slides were rinsed in PBS, water, air dried and coverslipped with fluoroshield with 4′, 6-diamidino-2-phenylindole (DAPI) (Sigma, St Louis, MO, USA) and immunoreactivity was visualized with a confocal microscope in our Core Optical Imaging Facility (Olympus FV-1000 with four lasers and DIC; Olympus, Center Valley, PA, USA). Control preparations consisted of adjacent sections stained exactly like the experimental sections except that they lacked primary antibodies, and some slides lacked primary and secondary antibodies.
Terminal deoxynucleotidyl transferase dUTP nick end labeling assay
In situ cell death detection kit, TMR red (Roche Applied Science, IIndianapolis, IN, USA) was used. Eight-micrometer paraffin-embedded sections of control and Bmp2-cKOSp7-Cre-EGFP were deparaffinized in xylene and ethanol (absolute, 95%, 90%, 80% and 70% diluted in double distilled water), washed in PBS, treated with proteinase K, nuclease free (20 µg⋅mL−1 in 10 mmol⋅L−1 Tris-HCl, pH 7.5) for 30 min, and rinsed with PBS. Sections were then incubated in terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) reaction mixture (mixture of label solution and enzyme solution) for 60 min at 37 °C in a humidified atmosphere in the dark. Slides were then rinsed in PBS (three times, 5 min each), coverslipped with fluoroshield with DAPI, and evaluated under a confocal microscope. For negative controls, sections were treated with label solution instead of TUNEL reaction mixture. For positive controls, sections were treated with DNase 1, grade 1 (3 000 U⋅mL−1 in 50 mmol⋅L−1 Tris-HCl, pH 7.5, 1 mg⋅mL−1 bovine serum albumin) for 10 min prior to treatment with TUNEL reaction mixture. TUNEL reaction was visualized with a confocal microscope.
Statistical analysis
Student's t-test and analysis of variance were performed to compare data between Bmp2-cKOSp7-Cre-EGFP and their littermate control (WT and Het) mice using GraphPad Prism software and Microsoft Excel. P<0.05 was considered statistically significant. For immunocytochemistry, at least two independent litters were evaluated for each antibody with two to three sections per mandible from a given animal.
Results
Characterization of the Sp7-Cre-EGFP model and expression in the mandible
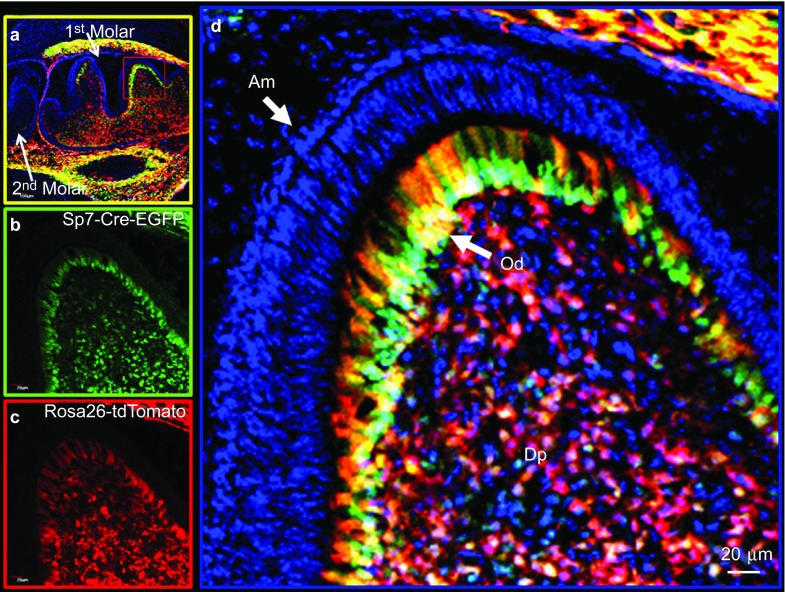
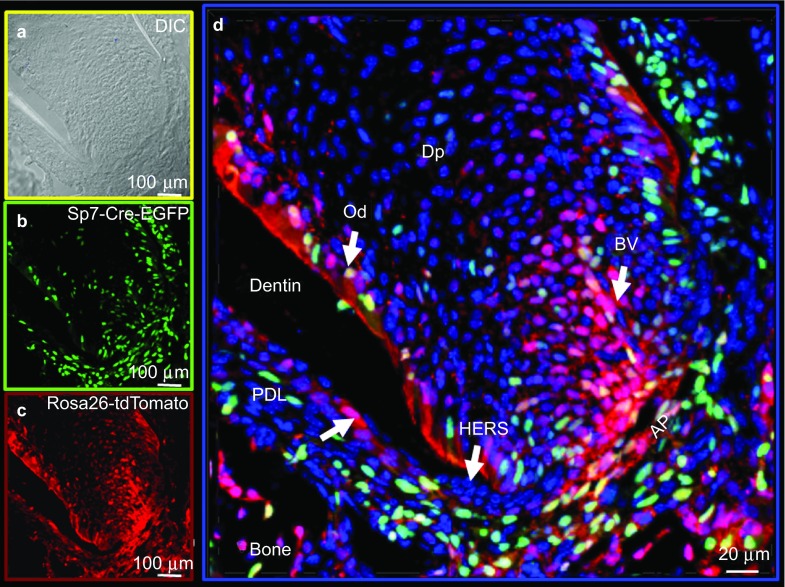
Using a Sp7-Cre-EGFP mouse model and a Rosa26-loxP-stop-loxP-tdTomato reporter, we mapped the cells in and around the tooth structures that underwent Cre recombination and progressed to new lineages (marked by the Red tdTomato expression). Therefore, we could follow their potential lineage progression. In our initial experiments, we used the Sp7-Cre-EGFP mouse model and a Rosa26-LSL-tdTomato reporter to demonstrate that there are a large number of Sp7+ (green) cells in the pulp, odontoblasts and alveolar bone osteoblasts at postnatal day 0 (P0), with a large number of Cre events (red) in the pulp and in the odontoblast layer of the frst molar. At P0, there is little Sp7+ cells (Cre events) observed in the second molar (Figure 1a, Supplementary Figure S1d and S1h). Supplementary Figure S1h also shows no activity in the third molar. At 2 weeks of age, there are a large number of Sp7+ cells in the pulp, periodontium and apical papilla region (green). Many of the progeny (red) most likely go on to become root odontoblasts, cementoblasts, alveolar bone osteoblasts and/or PDL fibroblasts (Figure 2). Supplementary Figure S2 shows a similar pattern in the crown region at this 2-week age in the second molar. Supplementary Figure 3 shows the Sp7+ cells and progeny (red) in the third molar at 2 weeks of age. Of note, there were no Sp7+ cells or Cre events in any of the epithelial components, including all stages of amelogenesis at either P0 or 2 weeks of age in the first, second or third molars. When we crossed this Sp7-Cre-EGFP model with the Bmp2-floxed model, we uncovered a complex phenotype that includes disruption of the formation of tooth root and associated periodontium as well as amelogenesis.12
Figure 1.
Lineage studies by mapping Cre activity in the first molar of a postnatal day 0 mouse by confocal microscopy using the Olympus FV 1000. Combined DAPI-stained nuclei (blue), Sp7-Cre-EGFP+ (green) and tdTomato+ (red) (Cre event) cells are shown. (a) Low magnification of the first and second molars with strong Cre activity in the alveolar bone and the first molar, but no detectable Cre activity in the second molar. (b) Sp7+ green signal from EGFP. (c) tdTomato signal representing Cre events and cells derived from Cre events (red box). (d) Shown at high magnification (×400) of the first molar crown region. Am, ameloblasts; DAPI, 4′,6-diamidino-2-phenylindole; DP, dental-pulp chamber; Od, odontoblasts.
Figure 2.
Lineage studies by mapping Cre activity in the root region of a second molar in a 2-week-old mouse by confocal microscopy using the Olympus FV 1000. Combined DAPI-stained nuclei (blue), Sp7-Cre-EGFP+ (green) and tdTomato+ (red) cells are shown. (a) DIC image. (b) Sp7+ EGFP signal. (c) tdTomato signal representing Cre event and cells derived from the Cre events. AP, apical papilla; BV, blood vessel; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; Dp, dental-pulp chamber; Od, odontoblasts; PDL, periodontal ligament region.
Phenotype of the Bmp2-cKOSp7-Cre-EGFP mice in the tooth and supporting structures
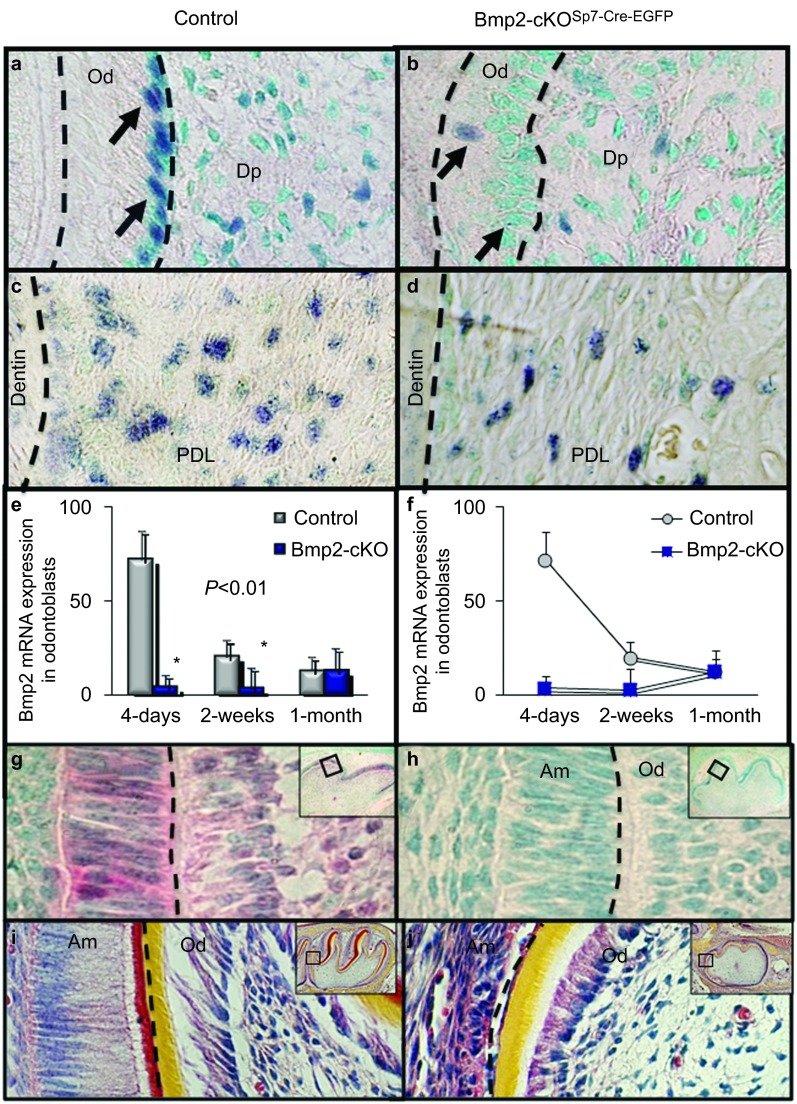
In Figure 3a and 3b, Bmp2 Exon 3 expression is reduced by over 85% in the odontoblasts and in a subset of pulp cells in the molars. Arrows point to the blue hybridization signal. The green color is the methylene green counterstain. Note the reduced number of dental pulp cells hybridizing to the Bmp2 Exon 3 probe, indicating Bmp2 deletion in a subset of Sp7+ cells in the pulp. The Sp7-Cre-EGFP model is effective at deleting Bmp2 Exon 3 expression, as shown and numerically estimated.6 Bmp2 Exon 3 is also deleted in a subset of periodontal cells (Figure 3c and 3d). Expression is very high in early odontoblasts in the control mice and is reduced as dentinogenesis is completing by 1 month (Figure 3e and 3f). While the expression of Bmp2 is reduced at 4 days and 2 weeks compared to the controls, the Bmp2 expression levels off at 1 month (n=2). Phospho-Smad1/5/8 levels, a read-out of Bmp signaling, is reduced as early as E18 in both the ameloblasts and in the odontoblasts of the first molar (Figure 3g and 3h). This indicates Cre activity as early as E18, and that Bmp2 in developing odontoblasts is indirectly affecting Bmp signaling in the epithelial ameloblast.12 Hematoxylin and eosin-stained sections from mouse molars show a strong tooth phenotype as early as 4 days in the first molar (Figure 3i and 3j). Both odontoblasts and ameloblasts are dysmorphic and fail to differentiate into highly polarized structures.
Figure 3.
The Bmp2 gene is deleted in both the odontoblast and in the periodontal ligament region and Bmp2 signaling is reduced in both odontoblasts and ameloblasts. (a–d) Bmp2 Exon 3 in situ hybridization at 2 weeks in the dental pulp, odontoblast and PDL. (e, f) Numerical estimation6 at 4 days, 2 weeks and 1 month of the first mandibular molars (a–f). Bmp2 Exon 3 is reduced at 4 days and 2 weeks compared to the control (>90%). Bmp2 expression normally decreases as terminal differentiation is completed and is noted with little change in Bmp2 Exon 3 expression at 1 month in the first mandibular molars. (g, h) Phospho-Smad1/5/8 immunohistochemistry in control and Bmp2-cKOSp7Cre-EGFP of the first mandibular molars in E18 embryos. (i, j) H&E staining of the first mandibular molars of 4-day-old control and Bmp2-cKOSp7-Cre-EGFP mice. Note the beginning of a severe tooth phenotype in both ameloblasts and odontoblasts. Am, ameloblasts; Bmp2, bone morphogenetic protein-2; Dp, dental-pulp chamber; H&E, hematoxylin and eosin; Od, odontoblasts; PDL, periodontal ligament.
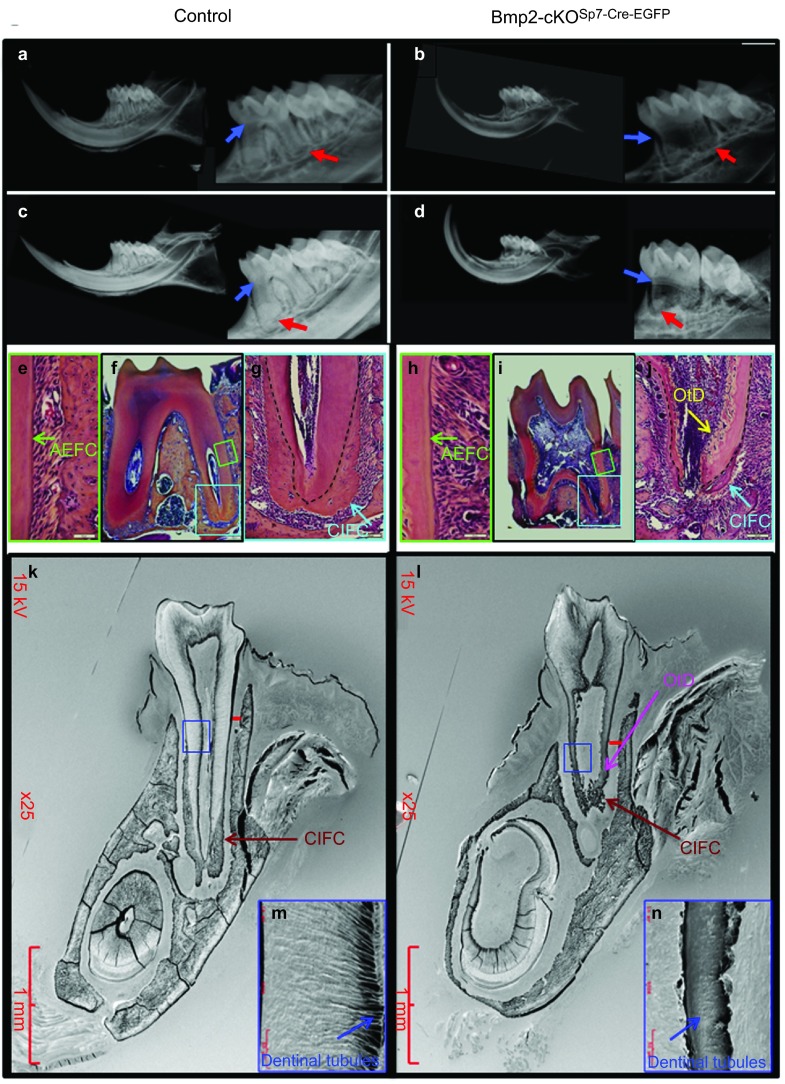
Figure 4 shows μCT analysis of control HetSp7-Cre-EGFP;Bmp2fx/+ (4A) and Bmp2-cKOSp7-Cre-EGFP (Figure 4b) mice. We quantified the total dentin volume (Figure 4c), root dentin volume (Figure 4d), root pulp volume (Figure 4e), enamel volume (Figure 4f) and periodontal volume (Figure 4g) in the first and second molars of control and Bmp2-cKOSp7-Cre-EGFP mice at 1 month, 2 months and 3 months of age, combining the data for all three ages for statistical evaluation The total dentin volume was reduced 30%–40% in both the first and second molars (Figure 4c), and root dentin is reduced 50%–80% (Figure 4d). Corresponding root pulp volume is increased approximately threefold (Figure 4e). As noted in Figure 4f, the enamel volume is reduced 25 % in the first molar with a similar trend in the second molar. The periodontal volume is increased significantly to 30% in the second molars, with a similar trend in the first molars (Figure 4g). Supplementary Figure. S4 shows similar results at 6 weeks of age in both first and second molars. In Figure 5a–5d, X-ray analysis demonstrates enlarged pulp chamber (blue arrow in Figure 5b and 5d) with a failure to form proper roots (Figure 5b and 5d) and thinner root dentin in the Bmp2-cKOSp7-Cre-EGFP mice at 1 (Figure 5a and 5b) and 3 months of age (Figure 5c and 5d) compared to control animals. We also noticed ectopic mineralized regions within the pulp near the apical region of the root in the Bmp2-cKOSp7-Cre-EGFP mice that was verified by histology in Figure 5j (blue arrow in Figure 5j) within the mandibles of 1.5-month-old mice. The material appears to be abnormal dentin with cells inside similar to bone, or osteodentin (OtD). In addition, in the Bmp2-cKOSp7-Cre-EGFP mice, the cellular intrinsic fiber cementum (CIFC) is greatly reduced as shown (blue arrow) in Figure 5j compared to Figure 5g. However, no obvious changes were seen in the acellular extrinsic fiber cementum (green arrow in Figure 5e and 5h). Using 3-month-old plastic embedded mouse mandibles, combined with acid etching and scanning electron microscopy, we noted the roots were not well developed and the CIFC was greatly reduced, as shown in Figure 5k and 5l (red arrow). We also confirmed the presence of osteodenin (pink arrow in Figure 5l and labeled OtD). In addition, the alveolar bone height was reduced (red bar in Figure 5k and 5l) and the dentinal tubules were defective in the absence of the Bmp2 gene (blue arrow in Figure 5m and 5n).
Figure 4.
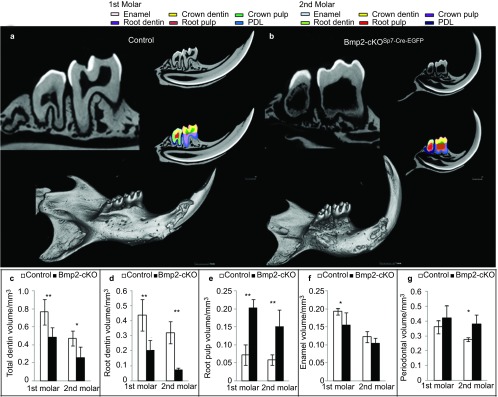
μCT analysis of mandibles from 1-month-, 2-month- and 3-month-old control Het and Bmp2-cKOSp7CreEGFP mice. (a, b) Images of overall mandible and sample sections through the middle showing examples with this 3-month data of various components that were quantified at all ages in both the first and second molars. The first molar, enamel, pink; crown dentin, yellow; crown pulp, green; root dentin, purple; root pulp, rust; periodontal volume, blue. The second molar, enamel, light blue; crown dentin, yellow; crown pulp, purple; root dentin, light green; root pulp, red; periodontium volume, dark blue. (c, d) Quantitation of total dentin volume and root dentin volume in the first and second molars. (e) Quantiation of root pulp volume (non-mineralized) in the first and second molars. (f) Quantitation of the enamel volume in the first and second molars. (g) Quantitation of the periodontal volume in the first and second molars. **P<0.01, *P<0.05. Total of 12 molars were quantified, six control molars and six Bmp2-cKO Sp7-Cre-EGFP molars, the first and second. Bmp2, bone morphogenetic protein-2.
Figure 5.
Bmp2 gene deletion in Sp7+ (Osterix+) cells results in loss of root formation, odontoblast dysmorphic differentiation, and a failure to form cellular cementum. (a–d) Digital X-rays of control and Bmp2-cKOSp7-Cre-EGFP mouse mandibles at 1 month (a and b) and 3 months (c and d) using Faxitron high-resolution X-ray resolution. X-ray analysis demonstrates enlarged pulp chamber (blue arrows) with a failure to form proper roots with thinner root dentin (red arrows) at 1 and 3 months of age of Bmp2-cKOSp7-Cre-EGFP compared to control heterozygous animals. (e–j) Histology of 1.5-month-old control Het and Bmp2-cKOSp7-Cre-EGFP mice. In e and h, green arrows point to AEFC. g and j show CIFC, blue arrow and yellow arrow points to abnormal mineralization or OtD in the root pulp chamber, j–l shows abnormal tooth root development and cementum formation, as revealed by acid etching of plastic embedded and SEM of 3-month-old third mandibular molars from Bmp2-cKOSp7-Cre-EGFP mice compared to control WT. Red bars denote level of formation of the alveolar bone between the teeth and the width of the periodontium. Low magnification, red arrow and label indicate well-formed cellular cementum in the control WT (k), but little organized cellular cementum is visible in the Bmp2- cKOSp7-Cre-EGFP (l). Note the OtD in the Bmp2- cKOSp7-Cre-EGFP (pink arrow in l) near the base of the roots. (m, n) High magnification of root region showing normal dentinal tubules in the WT (m) and highly dysmorphic dentinal tubules in Bmp2-cKOSp7-Cre-EGFP (n). Green box, area shown in e and h at high magnification. Blue box, area shown in g and h at high magnification. Blue box, area shown in m and n at high magnification. AEFC, acellular extrinsic fiber cementum; Bmp2, bone morphogenetic protein-2; CIFC, cellular intrinsic fiber cementum; OtD, osteodentin; SEM, scanning electron microscope.
Sp7 is the key transcription factor for CIFC formation9 and root formation and directly regulates collagen, type 1 and alpha 1 (Col1a1) at the osteoblast enhancer, and possibly directly regulates dentin sialophosphoprotein and dentin matrix protein 1. Sp7 also directly regulates VEGF through a proximal promoter region.13 As shown in Figure 6, dentin matrix protein 1 (Figure 6a and 6b), dentin sialophosphoprotein (Figure 6c and 6d), Col1a1 (Figure 6e and 6f) and Sp7 (Figure 6g and 6h) are all greatly reduced in mRNA expression by 80%–95% as estimated by numerical estimation in the Bmp2-cKOSp7-Cre-EGFP animals (Supplementary Figure S6).6 There is suggestive data that Sp7 may in fact be part of a set of transcription factors that regulates nuclear factor IC (Nfic) and part of a feed forward transcription gene network. Since Nfic is a key transcription factor for tooth-root formation and periodontium formation,14,15,16,17,18 we carried out in situ hybridization for Nfic expression and Nfic protein expression by immunocytochemistry (Figure 7i and 7j). As shown in Figure 6i and 6j, Nfic mRNA is reduced by 85% (Supplementary Figure S6) in root odontoblasts of these first molars at 2 weeks of age.
Figure 6.
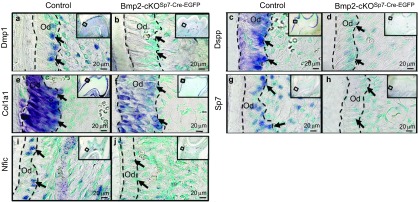
In situ hybridizations. (a, b) Dmp1; (c, d) Dspp; (e, f) Col1a1; (g, h) Sp7; (i, j) Nfic in control and Bmp2-cKOSp7Cre-EGFP mice of the first mandibular molars, 2 weeks of age. Bar=20 µm. Numerical estimation of the changes in levels of these key transcription factors involved in root formation and key markers for terminal differentiation of odontoblasts are given in Supplementary Fig. S6. Col1a1, collagen, type 1, alpha 1; Dmp1, dentin matrix protein 1; Dspp, dentin sialophosphoprotein; Nfic, nuclear factor IC; Od, odontoblasts.
Figure 7.
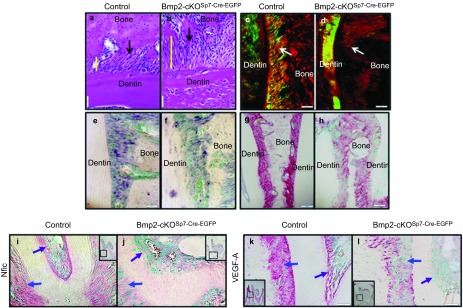
Bmp2 gene deletion in Sp7+ (Osterix+) cells leads to disorganized periodontal ligaments, decreased periostin and Nfic expression, and decrease VegfA expression in the periodontium and root odontoblasts region. (a, b) van Gieson stain for collagen bundles and fibers of control and Bmp2-cKOSp7-Cre-EGFP periodontium region, respectively, at ×400. The yellow line denotes the width of the periodontium. Black arrows, bundles of periodontal ligament collagen type 1-rich fibers in the periodontium, with disorganized Sharpey's fibers in the Bmp2-cKOSp7-Cre-EGFP mice. (c, d) Picrosirius Red stain for collagen shows highly disorganized Sharpey's fibers (white arrows) in the Bmp2-cKOSp7-Cre-EGFPmice (d), compared to the control (c) at ×200. (e, f) In situ hybridization of periostin at ×200. (g, h) Immunohistochemistry of periostin in control and Bmp2-cKOSp7Cre-EGFP of PDL at ×100. a–h are from 1.5-month-old mice. (i, j) Immunohistochemistry of Nfic in control and Bmp2-cKOSp7Cre-EGFP of the second molars at ×200. (k, l) Immunohistochemistry of VEGF-A in control and Bmp2-cKOSp7Cre-EGFP of the second molars at ×200. i–l are from 1-month-old mice, the second molars. Blue arrow points to periodontium and purple arrow points to root odontoblasts. Bmp2, bone morphogenetic protein-2; Nfic, nuclear factor IC; VEGF, vascular endothelial growth factor A.
Cell proliferation and apoptosis assays in the Bmp2-cKOSp7-Cre-EGFP model
Using 2-week-old mandibles in a Ki67 immunofluorescence assay, we note no major change in the proliferation index between WT and Bmp2-cKOSp7-Cre-EGFP mice, as shown in Supplementary Figure S5a and S5b. There appears to be decreased apoptosis, as assayed with cleaved caspase 3 antibody (Supplementary Figure S5c and S5d) and TUNEL assay along the root dentin odontoblast regions (Supplementary Figure S5e and S5f) in the second molars of the Bmp2-cKOSp7-Cre-EGFP. We speculate that this is reflection of decreased cell death of either the microvascular bed that normally disappears after odontogenesis and dentin formation terminates, or that a fraction of the odontoblasts undergo apoptosis, as do osteoblasts, while most odontoblast become resting odontoblasts, or ‘odontocytes' and serve functions in load responses and repair. In the Bmp2-cKO Sp7-Cre-EGFP model, we have a failure or decreased terminal differentiation of odontoblasts, as well as decreased vascularization within the odontoblast layer, both of which could lead to decrease observed apoptosis in the absence of the Bmp2 gene.
Also, the PDLs are dysmorphic in the Bmp2-cKOSp7-Cre-EGFP mice. To further explore and validate these observations on altered periodontium in the Bmp2-cKOSp7-Cre-EGFP mice, we carried out Van Gieson stain for collagen fiber bundles. As shown in Figure 7b from 1.5-month-old mice, the PDLs were highly disorganized and the periodontal space was expanded (yellow bar) in the Bmp2-cKOSp7-Cre-EGFP mice. There is also little alveolar bone between the molar teeth for attachment of the PDLs. Picrosirius Red staining of the periodontium for collagen under polarized light (Figure 7c and 7d) shows highly disorganized Sharpey's fibers in the Bmp2-cKOSp7-Cre-EGFP mice (Figure 7d) compared to the control (Figure 7c). Periostin is an extracellular matrix protein that is highly expressed in the periodontal ligament. As shown in Figure 7e–7h, there is reduced expression of Periostin mRNA (Figure 7f) and protein (Figure 7h) in the in the PDL in the Bmp2-cKOSp7-Cre-EGFP mice.
Possible mechanism of Bmp2 action in tooth and supporting structure development
We had previously observed that removal of the Bmp2 gene in mature odontoblasts resulted in decreased vascularization in the pulp. This decreased vascularization was linked to a reduction in candidate stem cells, at least one niche being the capillaries and associated pericytes within the pulp.6,19 Candidate follicular stem cells within the periodontium have also been shown to be decreased in the Sp7 conditional knock out model.9 One of the indirect mechanisms we propose is reduced VEGF-A production by Bmp induced odontoblasts, pulp cells and/or other cells in the periodontium, and osteoblast precursors, leading to alterations in the stem cell population in and around the vascular beds.6 As shown in Figure 7k and 7l, there is a great reduction of VEGF-A protein levels in the root odontoblasts (Figure 7l, purple arrow), as well as in the periodontium (Figure 7l, blue arrow) of the Bmp2-cKOSp7-Cre-EGFP mice as well.
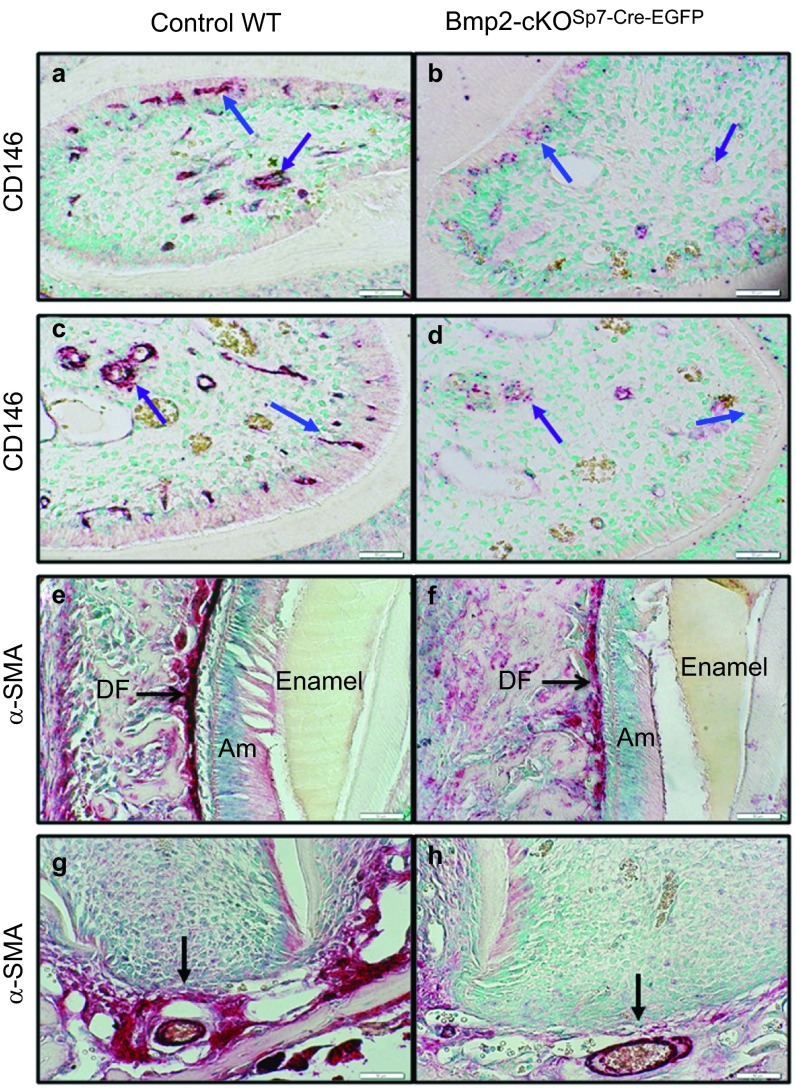
In the human bone marrow, CD146+ cells have been clearly shown to possess all the properties of mesenchymal stem cells (MSCs) that can differentiate into functioning osteoblasts and other differentiated tissues such as cartilage and muscle.20,21,22 Most of these CD146+ cells are found associated with the small blood vessels and have many properties referred to by some as pericytes.22 These CD146+ vascular associated cells were proposed as candidate stem cells for odontoblasts, as well as osteoblasts.23 The hypothesis proposed was that without the formation of proper microvascular structures within teeth, in part driven by Bmp2-induced VEGF-A production, there will be reduced levels of these candidate CD146+ MSCs.6,11 As shown in Figure 8, CD146 is highly expressed in vascular and microvessel structures in the pulp (purple arrow) of both the third molar (Figure 8a) and incisor (Figure 8c) of these 1-month-old animals, undergoing active dentinogenesis. There are also large numbers of CD146+ microvessels in and next to the odontoblast layer as shown in both the third molars and incisors (Figure 8a and 8c, respectively) of the control mice. We show a major reduction in CD146+ in microvascular structures within the odontoblast layer (Figure 8b–8d, blue arrow) and pulp (Figure 8b–8d, purple arrow) in the Bmp2-cKOSp7-Cre-EGFP mice littermates.
Figure 8.
Candidate stem cell marker CD146 associated with small blood vessels in the odontoblast layer are reduced in the Bmp2-cKOSp7-Cre-EGFP model, and αSma+ stem cells in the cervical and apical regions of the periodontium are also greatly reduced in the Bmp2-cKOSp7-Cre-EGFP model. (a–d) Candidate stem cell marker CD146 immunocytochemistry localized within the dental pulp (a–d, purple arrows) and in association with blood vessels adjacent to the odontoblast layer (a–d, blue arrows) on capillaries. CD146 protein expression is greatly reduced in the Bmp2-cKOSp7-Cre-EGFP in 1-month-old mice (×200). a and b are from the third molars, and c and d are from incisor cervical loop region. Candidate stem cells, marked with α-SMA immunocytochemistry are noted all within the periodontium of the first and second molars (data shown in Supplementary Figure S7) and in the less developed dental follicle cells of the third molars (e and f, black arrow), and apical papilla region (g and h, black arrow). Expression of α-SMA stem cell marker is greatly reduced in the Bmp2-cKOSp7-Cre-EGFP mice (h) as compared to control (g), example from the second molar, 1 month of age, and shown at ×200. Bmp2, bone morphogenetic protein-2.
Recently, α-SMA+ cells have also been shown to be stem cells for osteoblasts by lineage tracing procedures.24 Α-SMA+ cells have also been shown to be highly expressed in both the dental follicle and apical papilla region.25 Our hypothesis was that α-SMA+ cells in the dental follicle and apical papilla region could be candidate stem cells for cementum, periodontal ligaments and alveolar bone osteoblasts. To begin to explore this idea, we carried out immunocytochemistry with α-SMA antibody, and found that α-SMA+ cells are greatly reduced in the dental follicle in the Bmp2-cKOSp7-Cre-EGFP mice (Figure 8f, black arrow) as compared to control (Figure 8e). We also noted very high levels of α-SMA+ cells in the apical papilla region (Figure 8g, black arrow) as compared to Bmp2-cKOSp7-Cre-EGFP (Figure 8h, black arrow). The apical papilla is thought to harbor root odontoblast stem cells and possibly cementoblast stem cells associated with the microvascular tissue.26,27 We extended these findings with double immunofluorescence assays with CD31-endothelial (green) and α-SMA+ (red), as shown in Supplementary Figure S7. In the periodontium and apical papilla regions, of the control (Supplementary Figure S7a, S7c and S7e), there are a large number of red α-SMA+ cells localizing around the CD31 immunostain, while the levels of these vascular associated α-SMA+ cells are greatly reduced in the Bmp2-cKOSp7-Cre-EGFP mice, in both the peridontium and apical papilla regions and in the vascular structures within the odontoblast layer (Supplementary Figure S7b, S7d and S7f).
Discussion
We discovered new and unique roles of the Bmp2 gene in both development of the periodontium and tooth root that appear coordinated in the postnatal animals. There are direct cell autonomous effects on terminal differentiation of odontoblasts, as well as differentiation of precursor cells to components of the periodontium. Also, the enamel and ameloblast differentiation were altered in an indirect fashion since there was no Sp7-Cre-EGFP expression in any of the ameloblast-epithelial lineages, as shown by lineage analysis. The development of molar roots was permanently altered in the Bmp2-cKOSp7-Cre-EGFP. The mechanism of the tooth root and cementum phenotype is related to a failure of terminal differentiation that indirectly leads to decreased candidate stem cells that are CD146+ in the dental pulp, and also decreased in the periodontium and apical papilla region that are α-SMA+. The nexus of these phenotypes are thought to be linked to decreased vascularization as a result of decreased Bmp2-dependent VEGF-A production and other factors involved in vascularization and angiogenesis in the pulp, odontoblast layer and periodontium.6 Recent data have demonstrated by lineage tracing studies that in the context of bone marrow, the α-SMA+ cells are precursors to osteoblasts, and the α-SMA+ cells in the periodontium have a high capacity to form mineralizing structures similar to bone and cementum.24,25
α-SMA+ cells are found not only on many of the smooth muscle cells of the larger blood vessels, but also on the smaller microvessels or capillary walls, sometimes referred to as pericytes. α-SMA+ cells are also scattered throughout the odontoblast layer and in the pulp region, not necessarily on microvessels, and are highly expressed in the periodontium.25 Recent data using an incisor model and the NG2 marker for pericytes, Feng et al.,19 has shown that a fraction of these pericytes can in fact become odontoblasts by lineage tracing procedures. In addition, a non-pericyte niche for odontoblasts was also identified in the cervical loop region, but still in a highly vascularized region.19 Therefore, there appears to be several niches for stem cells even within the pulp region, however; the localization of these MSC niches has not been explored in the context of developing molars. We propose the α-SMA+ cells, at least a subset, are stem cells for odontoblasts, and for other components of the supporting structures of the teeth, such as cementoblasts, PDL fibroblasts and alveolar bone osteoblasts. Our hypothesis is that the Bmp2 gene is critical for development of the tooth and tooth roots in coordination with formation of the supporting structures of the tooth.
We also noted massive osteodentin in the pulp of the Bmp2-cKOSp7-Cre-EGFP mice, suggesting alterations and cell autonomous dysmorphic differentiation of odontoblasts. Recent data have shown that conditional removal of the Bmp2 induced transcription factor, Sp7, leads to major defects in cementogenesis.9 The Sp7 transcription factor is highly expressed in a subset of dental pulp cells, odontoblasts, cementoblasts and a subset of dental follicle cells in the periodontium, as demonstrated and published using a Sp7 conditional KO mouse model.9 When the Bmp2 gene is deleted in these several cell types or states, we have shown major defects in tooth root formation, as well as cementum, PDL and alveolar bone formation. The underlying mechanism in part seems to be linked to a failure to form the vascular system in the dental pulp and in the periodontium in the absence of the Bmp2 gene in odontoblast and periodontium precursors. Since there are many examples now of the link of vascularization and associated stem cells on the vascular walls, and that there is a strong correlation of the formation of blood vessels and formation of osteoblasts, we put forth the hypothesis that this principle applies to many of the stem cells in the dental pulp and in the periodontium.6,7,28,29 Supporting evidence for this hypothesis is presented with the decrease in the CD146+ candidate stem cells on the small microvessels in the dental pulp, and decreased VEGF-A production when the Bmp2 gene is removed from Sp7+ cells. Overexpression of VEGF-A in the bone microenvironment has been shown by others to cause massive new vascularization and new bone formation.30
Another major candidate marker for MSCs in the periodontium and dental pulp is the α-SMA antigen. These α-SMA+ cells have been shown to have MSC properties by several criteria.24,31 We show major decrease in expression of α-SMA+ cells in both the dental pulp region and in the periodontium in the absence of the Bmp2 gene. We propose that loss of these stem cells in the periodontium leads to the observed defects in cementum formation and in the disordered periodontal ligament fiber bundles observed in the Bmp2-cKOSp7-Cre-EGFP mice. As well, we hypothesize that the failure in formation of these α-SMA+ cells and/or CD146+ cells is linked to the failure to form a vascular niche. Moreover, without sufficient Bmp2 in the early stages of odontoblasts, the root and crown odontoblasts fail to terminally differentiate.
In summary, these studies outline a potential interesting relationship that has been proposed in the past that tooth and root formation are coupled to the formation of the supporting structures of the teeth. Our data support a critical role for Bmp2 gene in formation and coordination of both the tooth root and supporting structures, including alveolar bone, CIFC and the periodontal ligaments within the periodontium.
Acknowledgments
This study was partly supported by research grant funding: NIH-NIAMS R01-AR054616 (SEH), NIH-NIDCR T32-DE14318 (Rakian) and F32-DE018865 (Yang). Images were generated in the Core Optical Imaging Facility, which is supported by UTHSCSA, NIH-NCI P30-CA54174 (CTRC at UTHSCSA) and NIH-NIA P01-AG19316. Publication of this manuscript is supported by Open Fund of State Key Laboratory of Oral Diseases, Sichuan University.
Footnotes
Note: Supplementary information for this article can be found on International Journal of Oral Science's website(http://www.nature.com/ijos).
Supplementary Information
References
- Huang X, Bringas P, Jr, Slavkin HC, et al. Fate of HERS during tooth root development. Dev Biol. 2009;334 1:22–30. doi: 10.1016/j.ydbio.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XF, Chai Y. Molecular regulatory mechanism of tooth root development. Int J Oral Sci. 2013;4 4:177–181. doi: 10.1038/ijos.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekwisch TG. The developmental biology of cementum. Int J Dev Biol. 2001;5 5/6:695–706. [PubMed] [Google Scholar]
- Saygin NE, Giannobile WV, Somerman MJ. Molecular and cell biology of cementum. Periodontology 2000. 2000;24:73–98. doi: 10.1034/j.1600-0757.2000.2240105.x. [DOI] [PubMed] [Google Scholar]
- Zhao M, Xiao G, Berry JE, et al. Bone morphogenetic protein 2 induces dental follicle cells to differentiate toward a cementoblast/osteoblast phenotype. J Bone Miner Res. 2002;17 8:1441–1451. doi: 10.1359/jbmr.2002.17.8.1441. [DOI] [PubMed] [Google Scholar]
- Yang W, Harris MA, Cui Y, et al. Bmp2 is required for odontoblast differentiation and pulp vasculogenesis. J Dent Res. 2012;91 1:58–64. doi: 10.1177/0022034511424409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Guo D, Harris MA, et al. Bmp2 gene controls bone quantity and quality through regulating osteoblast and development and vascular-skeletal stem cell niche J Cell Sci 2013. in press.
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133 16:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Cao Z, Zhang H, Zhou X, et al. Genetic evidence for the vital function of Osterix in cementogenesis. J Bone Miner Res. 2012;27 5:1080–1092. doi: 10.1002/jbmr.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BL. Methods for studying tooth root cementum by light microscopy. Int J Oral Sci. 2012;4 3:119–128. doi: 10.1038/ijos.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluhak-Heinrich J, Guo D, Yang W, et al. New roles and mechanism of action of BMP4 in postnatal tooth cytodifferentiation. Bone. 2010;46 6:1533–1545. doi: 10.1016/j.bone.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yang G, Yuan G, et al. Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs. 2011;194 2/3/4:216–221. doi: 10.1159/000324644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yang F, Li Y, et al. Transcriptional regulation of Vascular Endothelial Growth Factor (VEGF) by osteoblast-specific transcription factor Osterix (Osx) in osteoblasts. J Biol Chem. 2012;287 3:1671–1678. doi: 10.1074/jbc.M111.288472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Xu X, Bringas P, Jr, et al. Smad4–Shh–Nfic signaling cascade-mediated epithelial–mesenchymal interaction is crucial in regulating tooth root development. J Bone Miner Res. 2010;25 5:1167–1178. doi: 10.1359/jbmr.091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Reyna J, Chen LS, et al. Role of the transcription factor NFIC in odontoblast gene expression. J Calif Dent Assoc. 2009;37 12:875–881. [PubMed] [Google Scholar]
- Lee TY, Lee DS, Kim HM, et al. Disruption of Nfic causes dissociation of odontoblasts by interfering with the formation of intercellular junctions and aberrant odontoblast differentiation. J Histochem Cytochem. 2009;57 5:469–476. doi: 10.1369/jhc.2009.952622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JC, Herr Y, Kim HJ, et al. Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J Periodontol. 2007;78 9:1795–1802. doi: 10.1902/jop.2007.060363. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Butz KG, Lyons GE, et al. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol. 2003;23 3:1075–1084. doi: 10.1128/MCB.23.3.1075-1084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Mantesso A, De BC, et al. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proc Natl Acad Sci U S A. 2011;108 16:6503–6508. doi: 10.1073/pnas.1015449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covas DT, Panepucci RA, Fontes AM, et al. Multipotent mesenchymal stromal cells obtained from diverse human tissues share functional properties and gene-expression profile with CD146+ perivascular cells and fibroblasts. Exp Hematol. 2008;36 5:642–654. doi: 10.1016/j.exphem.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3 3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131 2:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sourcestheir biology and role in regenerative medicine. J Dent Res. 2009;88 9:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grcevic D, Pejda S, Matthews BG, et al. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells. 2012;30 2:187–196. doi: 10.1002/stem.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Miguel SM, Fatahi MR, Li H, et al. Defining a visual marker of osteoprogenitor cells within the periodontium. J Periodontal Res. 2010;45 1:60–70. doi: 10.1111/j.1600-0765.2009.01201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Sonoyama W, Liu Y, et al. The hidden treasure in apical papilla: the potential role in pulp/dentin regeneration and bioroot engineering. J Endod. 2008;34 6:645–651. doi: 10.1016/j.joen.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18 4:696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19 2:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipani E, Maes C, Carmeliet G, et al. Regulation of osteogenesis–angiogenesis coupling by HIFs and VEGF. J Bone Miner Res. 2009;24 8:1347–1353. doi: 10.1359/jbmr.090602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C, Goossens S, Bartunkova S, et al. Increased skeletal VEGF enhances beta-catenin activity and results in excessively ossified bones. EMBO J. 2010;29 2:424–441. doi: 10.1038/emboj.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalajzic Z, Li H, Wang LP, et al. Use of an alpha-smooth muscle actin GFP reporter to identify an osteoprogenitor population. Bone. 2008;43 3:501–510. doi: 10.1016/j.bone.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.