Abstract
Piwi-interacting RNAs (piRNAs), a newly identified class of small non-coding RNAs, direct the Piwi-dependent transposon silencing, heterochromatin modification and germ cell maintenance. Owing to our limited knowledge regarding their biogenesis, piRNAs are considered as the most mysterious class of small regulatory RNAs, particularly in pathogenesis such as tumorigenesis. Recently, several lines of evidence have emerged to suggest that piRNAs may be dis-regulated and play crucial roles in tumorigenesis in previously unsuspected ways. In this prospective piece, we will discuss the emerging insights into the potential novel roles of piRNAs in carcinogenesis and highlight their potential implications in cancer detection, classification and therapy.
Keywords: PIWI-interacting RNAs, cancer, diagnosis, therapy
1. Introduction
It has been recently noted that the genomes of all studied eukaryotes are transcribed almost entirely. In fact, >90% of the human genome is likely to be transcribed; however, only ~2% of this is subsequently translated [1,2]. The remaining regulatory noncoding RNAs (ncRNAs) can be roughly categorized into large and small ncRNAs, which mainly include small interfering RNA (siRNA), microRNA (miRNA), small nucleolar RNA (snoRNA) and PIWI-interacting RNA (piRNA). Among these small regulatory ncRNAs, piRNAs are the newest class and the least investigated.
The first hint indicating that piRNAs exist was the study that Stellate protein-coding gene repeats were silenced in the Drosophila melanogaster germ line [3], and subsequently, repeat-associated small interfering RNAs (rasiRNAs) derived from repetitive genomic elements were found in the D. melanogaster testes and early embryos [4]. However, it was not known, at that time, that these small non-coding RNAs were, in fact, piRNAs. In 2006, a class of small RNAs named piRNAs binding to Argonaute proteins in mammalian germ line cells, was purified [5–8]. piRNAs have been shown to act mainly in the Piwi-dependent transposon silencing, heterochromatin modification, and in germ cell maintenance [9–12]. Piwi proteins play a key role in spermatogenesis. For example, Miwi, a Piwi subfamily protein, null mice displayed spermatogenic arrest [13], and piRNAs were found to accumulate at the onset of meiosis or during spermatogenesis [5, 7]; it is easy to understand that piRNAs demonstrated critical regulation in reproduction and fertility [11, 14–18]. However, the expression status and roles of piRNAs as well as other factors involved in functions and mechanisms of piRNAs remain poorly understood in tumorigenesis.
Despite the common assumption that piRNAs play a key role in germline development, recent evidences have suggested that piRNAs had important function in tumorigenesis. For instance, independent studies have shown that aberrantly expressed piRNAs suggested their roles in the process of cancer development [19–21]. Furthermore, one study observed that piRNAs might function as a tumor suppressor in human gastric cancer [22]. Here, we will discuss the emerging insights into the potential roles of piRNAs in carcinogenesis, and the possible applications for cancer diagnosis and treatment.
2. Biogenesis of piRNAs
piRNAs are produced independently of Dicer ribonuclease, which is required for double-stranded precursors to generate 21 to 24 nt small regulatory ncRNAs such as siRNAs and miRNA [23–25]. Unlike siRNAs, piRNAs neither show any phasing within a cluster sequence nor do they overlap with each other [26]; Unlike miRNAs, no significant secondary structures of the stem-loop structures in their precursors have been detected in regions surrounding piRNAs [26, 27]. However, like these extensively studied small regulatory ncRNAs, precursors of piRNAs need further post transcriptional processing to become fully matured.
2.1. Transcription mechanism
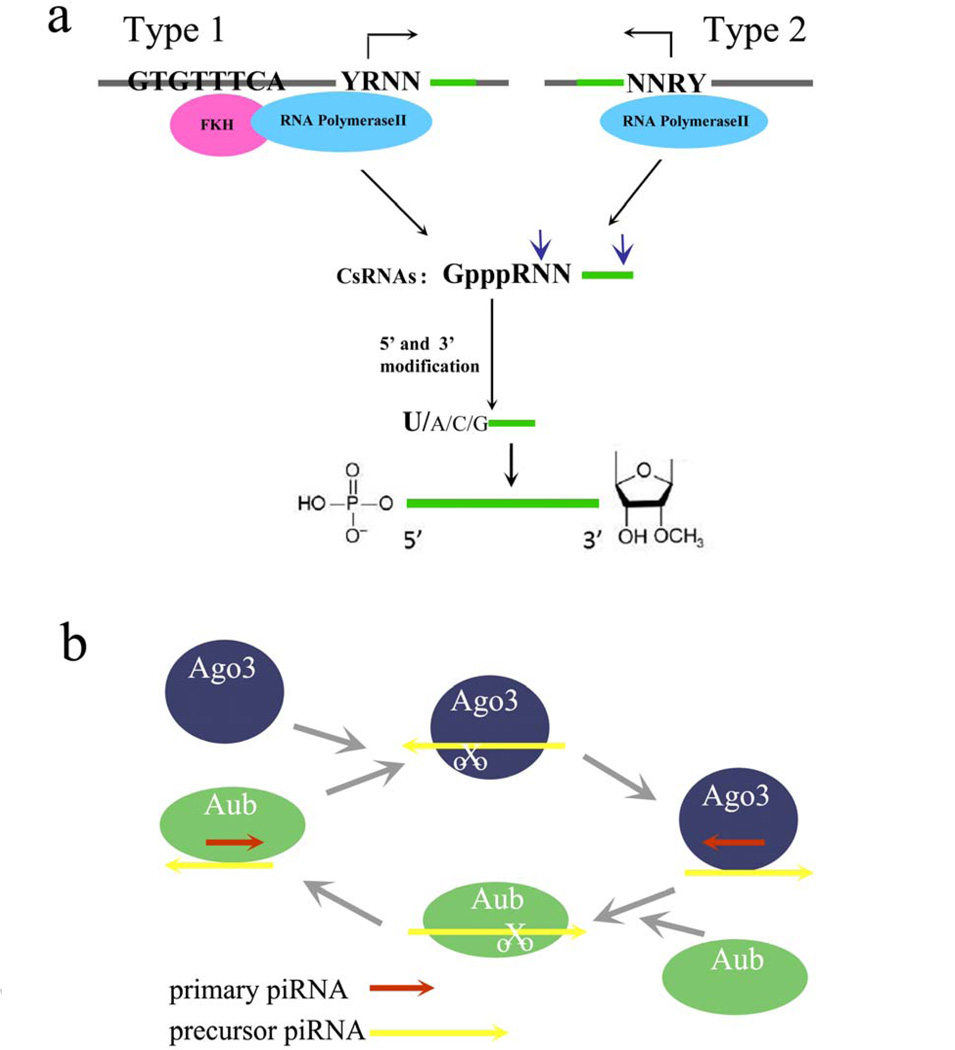
Initial analysis of genomic sequences revealed extremely high diversity of piRNAs which have been mapped to a relatively small number of genomic regions called piRNA clusters [5, 6, 10]. Recently, in a series of well-executed compelling experiments, the specific mechanism involved in matured 21U-RNA piRNAs transcription in C. elegans was logically and persuasively explained by observing that capped small RNAs (csRNAs) were precursors of piRNAs [28]. There are two types of loci producing csRNAs: type 1 has a feature of CTGTTTCA motif upstream of transcription start site [28, 29], and type 2 locus has no typical motif governing tendency to produce csRNAs. The precursors of csRNAs share some characteristics: (1) ~26nt in length; (2) initiate precisely 2nd nt upstream of mature piRNAs; (3) exhibit a strong bias for a 2 nt motif of pyrimidine (Y) purine (R), or YR; and (4) a U at the csRNA +3 position for piRNA processing or stability [28]. These precursors are then processed into matured piRNAs (Fig.1a), of which with 1U bias in 5’ ends and 2'-O-methylated at their 3' ends.
Figure 1.
Biogenesis of piRNAs. (a) Model for transcriptional biogenesis of piRNAs. There are two types of loci producing csRNAs, which are modified at 5’ and 3’ to be matured ones. A strong bias for a U at 5’ end of matured piRNA is indicated in bold letter. (b) Amplification ping-pong mechanism of piRNAs. It involves two distinct Piwi proteins. The number of piRNAs is increased after cycle.
Consistent with previous observations [9, 10, 30], these csRNAs were mapped to both genomic strands, suggesting bidirectional transcription. Also, single-stranded RNAs were indeed transcribed from piRNA clusters [9, 10, 28, 30]. Different from other observations, these studies did not show that long RNA molecules spanning the whole piRNA cluster were processed into mature piRNAs [10, 31]. One of the possibilities for the difference was that the transcriptional tendency was dependent on specific species. However, additional biochemical experiments are needed to determine the structure and processing of csRNAs caps, and how these caps could be formed or added to 5’ end of the original transcripts. Additionally, it is possible that other motif of loci could govern the tendency to produce precursors other than pol II-based transcription.
2.2. Amplification mechanism
After a primary piRNA is generated, piRNA’s accumulation requires amplification by the so-called ping-pong mechanism involving at least two distinct Piwi proteins, a process which occurs in the cytoplasm (Fig. 1b) [10]. This mechanism is similar to secondary siRNA generation [32] in which primary piRNAs recognize their complementary targets: antisense piRNA associates with PIWI/AUB complex, while sense piRNA associates with AGO3 protein. Cleavage of transcript targeted by AUB-bound 1U primary piRNA leads to the generation of the 5’ end of new secondary piRNA, which has a 10A bias and then accordingly can be loaded onto AGO3. Finally, piRNA-AGO3 complex binds to the retro-transposon transcript, creating another set of anti-sense piRNA [10, 26, 33–36]. Ping-pong signatures have been identified in zebrafish and D. melanogaster as well as in very primitive animals such as sponges, suggesting the existence of the ping-pong cycle a somewhat early stage evolutionarily [37].
piRNAs biogenesis during adult spermatogenesis in mice, however, is independent of the ping-pong mechanism [38]. Only Piwi proteins were detectable in the adult and post natal testes and no interaction was observed between Mili and Miwi or piRNA sequence features for the ping-pong mechanism. Additionally, both populations of piRNAs were biased for 5’ Uracil but not for Adenine on the 10th nucleotide position, and displayed no complementarity. Furthermore, in Miwi mutants, Mili-associated piRNAs were upregulated, but not downregulated. Together, these results showed that piRNAs biogenesis during adult spermatogenesis in mice did not use ping-pong ball cycle at all.
3. Involvement of piRNAs in cancer development
Cancer cells and germ cells, as well as stem cells, share important biological features such as infinite self-renewal and rapid proliferation. Therefore, it is reasonable to assume that the newly developed cancer cells appropriate the self-renewing division machineries used factors specific for germ cell regulation [39,40]. In fact, studies have shown that many of these transiently expressed genes in developing germcells are proto-oncogenes or oncogenes. For example, cancer/testis antigens (CTAs), a category of tumor antigens whose normal expression was restricted to male germ cells in the testis, were also expressed in various human tumor types including melanoma, breast cancer, bladder cancer, gastric cancer, lung cancer, sarcoma, ovariancancer, prostate cancer and hepatocellular carcinoma, among others [41–44]. Furthermore, CTAs profiles in cancers have been linked to neoplastic phenotype including immortality, invasiveness, immune evasion, and metastatic capacity, thus, CTAs may sever as biomarkers, new targets, and predictors of biochemical recurrence for these cancers [41–44]. These studies are consistent with the concept that tumorigenesis adapts pathways utilized in germ-cell development.
As a class of germline-expressed small regulatory ncRNAs, piRNAs are mainly linked to epigenetic programming, which might confer some of the central characteristics of malignancy.
3.1. piRNAs-associated Piwi proteins in cancer
Piwi proteins have evolutionarily conserved essential roles in germline development, as their mutations in mice, Drosophila, and Zebrafish can cause defects in gametogenesis [14–17,45].
There are four Piwi proteins expressed in humans: PiwiL1/Hiwi, PiwiL2/Hili, PiwiL3, and PiwiL4/Hiwi2 [46]. The first report of Piwi expression in cancer was in testicular seminomas-tumors originating from embryonic germ cells, which retained a germ cell phenotype [47]. Hiwi was detected in seminomas, but not in non-seminomas, spermatocytic seminomas, or testicular tumors originating from somatic cells, indicating Hiwi was specifically expressed in both normal and malignant spermatogenic cells in a maturation stage-dependent pattern and might function in cell proliferation [47]. The report on seminomas was followed by reports on a wide variety of cancers, and Piwi protein expression profiles have recently received much attention for their potential functional involvement in a wide variety of human cancers (Table 1), suggesting Piwi proteins may play a role in oncogenesis and may serve as diagnostic and prognostic biomarkers.
Table 1.
Piwi proteins profiles in human cancers
| Piwi types | Disease | Material | Reference |
|---|---|---|---|
| PiwiL1/Hiwi | Seminomas | Tissue | [47] Qiao,et al. 2002 |
| Gastric cancer | Tissue and cell lines | [55] Wang,et al. 2012 | |
| Sarcoma | Tissue | [48]Taubert,et al. 2007 | |
| Pancreatic cancer | Tissue | [54] Grochola, et al.2008 | |
| Livwe cancer | Tissue and cell lines | [58] Zhao,et al. 2012 | |
| Glioma | Tissue and cell lines | [57] Sun, et al.2011 | |
| Esophageal cancer | Tissue and cell lines | [60] He,et al. 2009 | |
| Colorectal cancer | Tissue and cell lines | [59] Zeng, et al. 2011; | |
| PiwiL2/Hili | Breast cancer | Tissue and cell lines | [61] Liu, et al. 2010 |
| Cervical | Tissue and cell lines | [52] He, et al. 2010 | |
| Seminoma cancer | Tissue and cell lines | [49] Lee,et al. 2006 | |
| Colon cancer | Tissue | [53] Li, et al. 2012 | |
| PiwiL3 | Gastric cancer | Tissue | [55] Wang, et al. 2012 |
| PiwiL4/Hiwi2 | Gastric cancer | Tissue | [55] Wang, et al. 2012 |
3.1.1. Piwi proteins are involved in cancer cell proliferation andapoptosis
Qiao et al. first reported that Piwi protein was overexpressed in testicular seminomas-tumors originating from embryonic germ cells with retention of germ cell phenotype [47]. PiwiL2 silencing could significantly reduce tumor cell proliferation, colony formation but increased apoptosis in vitro, and inhibited tumor growth in vivo [48]. PiwiL2 was reported to be widely expressed in tumors and acted as an oncogene by inhibiting apoptosis and promoting cell proliferation via Stat3/Bcl-X(L) signaling pathway [49], because induction of high-level expression of the anti-apoptotic gene Bcl-X(L) was observed in cells expressing PiwiL2, whereas an increased Bcl-X(L) expression correlated with increase of signal transducer and activator of transcription 3 (STAT3) expression. Additionally, there was a link between PiwiL2 overexpression and reduced apoptosis, enhanced proliferation and induced transformation of fibroblast cells. Furthermore, PiwiL2 inhibition suppressed STAT3 and Bcl-X(L) expression and induced apoptosis [49]. One more insight into the role of Piwil2 in tumorigenesis was that PiwiL2 might reduce apoptosis in tumor cells possessing P53, which was a positive regulator of STAT3 signaling pathway [50]. PiwiL2 can directly bind to STAT3 protein via its PAZ domain and form a PiwiL2/STAT3/c-Src triple protein-protein complex. STAT3 was phosphorylated by c-Src and translocated to nucleus, then bound to P53 promoter and repressed its transcription [50].
3.1.2. Piwi proteins are involved in virus infection, cancer cell metastasis and invasion
The expression of both Hiwi and Hili has been linked to human papilloma virus infection [51, 52]. The expression rate of PiwiL1 was 75% in cervical squamous cell carcinoma (CSCC) and had a statistically positive correlation with HPV16, and the elevated expression of PiwiL1 has been associated with invasion of CSCC, which supports the interaction between HPV16 and host cells the carcinogenesis of CSCC [51].
PiwiL2 can be detected in various stages of human CSCC and adenocarcinomas, and was also detected in some metaplastic epithelial cells, histologically "normal" appearing tissues adjacent to malignant lesions, a typical glandular cells, low-grade and high-grade squamous intra epithelial lesions [52]. Additionally, significantly higher expression levels of PiwiL2 were observed in primary colon cancer tissue and in lymph node metastasis in comparison with normal colon mucosa. Interestingly, its expression significantly correlated with more aggressive clinical and pathological parameters, such as five-year metastasis-free survival and overall survival [53]. PiwiL2 mediated increase of matrix metallopeptidase 9 (MMP9) transcriptional activity has been suggested to cause an increased migration and invasion of cancer cells [53].
3.1.3. Piwi proteins have prognostic value in human cancers
Taubert et al. first showed that an increased expression of Hiwi mRNA was a significant negative prognostic factor for patients with soft-tissue sarcomas (STS), and a high level of its mRNA identified STS patients at high risk of tumor-related death [48]. Another study showed that Hiwi alterations in mRNA expression were associated with an increased risk of tumor-related death in male patients with ductal adenocarcinoma of the pancreas (PDAC) [54].
Increasing evidence showed that the specific prognostic value of a subfamily Piwi protein was variable for different cancers or different stages of cancers. PiwiL1 has been shown as an independent prognostic factor in gastric cancer, and both PiwiL1 and PiwiL2 expression in gastric cancer tissue predicted poorer overall survival [55]. PiwiL2 expression was relatively higher in colorectal carcinomas and has been correlated with various clinic-pathologic parameters and a poor prognosis [56]. Hiwi could be used as a potential molecular marker for pathological diagnosis and prognostic indicator for malignant gliomas [57], whereas in hepato-cellular carcinoma (HCC), Hiwi played a key role in proliferation and metastasis, and could be a potential prognostic factor for HCC after curative resection, particularly with well-differentiated HCC [58]. In colorectal cancer, Hiwi was a potential prognostic biomarker especially for those at early stages or without lymph node metastasis [59].
Additionally, cellular distribution of a Piwi protein influenced its function. Expression level of Hiwi in cytoplasm of esophageal cancer cells was significantly associated with histological grade, clinical stage and poorer clinical outcome. However, there is no correlation between the nuclear Hiwi expression and clinic-pathological features [60]. Interestingly, PiwiL2 expression patterns were different in different stages of breast cancers: the cytoplasm, nucleus or both cytoplasm and nucleus expression patterns were observed in invasive and metastatic breast cancers, while nucleus pattern was less common in breast pre-cancers, indicating that PiwiL2 was expressed in various stages of breast cancers and has the potential to be used a novel biomarker [61].
3.2. piRNAs play a key role in epigenetic regulation for genome programming
All organisms balance the need to maintain genetic variation against the danger of accumulating potentially deleterious genesor pathogenic sequences [62,63]. In C. elegans and Drosophila, the piRNAs have been shown to be necessary for silencing transposons during germline development by RNA degradation at the post transcriptional level [9, 26, 31, 64]. piRNAs function in silencing transposons by DNA methylation also leaded to transcriptional gene silencing. For example, piRNAs expressed in the central nervous system (CNS) and other somatic tissues in Aplysia could mediate CpG methylation and transcriptional silencing of a key plasticity-related gene, CREB2 [65]. These observations indicate that piRNAs have function in epigenetic regulation, as transposons are ubiquitous genome pathogens that can mobilize and induce mutations that alter gene expression, cause disease, and drive evolution [64, 66, 67]. In Oxytricha, piRNAs can protect against loss during genome rearrangement [68], showing that piRNAs are powerful trans-generational carriers of epigenetic information for genome programming. The same observation of the piRNAs’ function has been further confirmed in multi generational epigenetic memory in C. elegans [69], in which piRNAs were found to initiate an epigenetic memory of nonself RNA [70]. Furthermore, piRNAs guide both Piwi and Piwi-associated epigenetic factors to program the genome by binding to numerous piRNA-complementary sequences, which explained that how functions of specific genomic sites were regulated by epigenetic factors [71].
3.3. piRNA regulates its host genes and its interacting proteins
In contrast to the mechanism known for miRNA-mediated modulation of gene expression, piRNA can regulate its host gene expression. The human melatonin receptor 1A gene (MTNR1A) gene, expression of which has been shown altered in prostate cancer cells, is located on the chromosomalregion 4q35.2 with two exons. piR_015520 is in intron 1of MTNR1A gene, 14 kb downstream of the ATG start codon. This piRNA negatively regulated MTNR1A gene expression by binding to its genomic region, as overexpression of piR_015520 resulted in a repression of MTNR1A expression in a concentration-dependent manner [65]. This finding suggests that changes in individual piRNA levels could influence the expression of the gene in which the piRNA is located, offering a new perspective for piRNAs functioning as gene regulators in humans.
Also, piRNAs have function in regulating its interacting proteins stability. For example, during mouse sperm maturation, its Piwi protein Miwi ubiqutination was regulated by piRNAs through enhancing Miwi interaction with an APC/C substrate-binding subunit [18]. This observation not only identified how piRNAs-interacting protein was regulated by its binding piRNAs, but also expanded our knowledge in piRNAs regulation processes [72].
3.4. piRNAs have oncogenic or tumor suppressor roles in cancer development
Recently, several independent lines of evidence have indicated that the piRNAs were involved in cancer development (Table 2). The relationship between piRNAs and carcinogenesis has been suggested by results of microarray screening, next generation sequencing, and real-time quantitative reverse transcription-polymerase chain reaction analyses. The up-regulated expression of piR-651, piR-4987, piR-20365, piR-20485 and piR-20582 was observed in cancer cell lines and in primary tumors compared to matched non-cancerous tissues [20, 21]. piR-Hep1 was found to be up-regulated in hepatocellular carcinoma tumors compared to their corresponding adjacent non-cancerous liver tissues [73]. On the other hand, the level of piR-823 in gastric cancer tissues was found significantly lower than that in the non-cancerous tissues [21]. These phenomena of dys-regulated piRNA expression implied that piRNA could act as an oncogene or a tumor suppressor in carcinogenesis and these piRNAs might be involved in regulating cancer cell activities in the following ways:
Table 2.
piRNAs expression in human cancers
| SpeificpiRNA | Expression Level |
Disease | Refence |
|---|---|---|---|
| piR-651 | up-regulated | gastric, colon, lung, breast, cervical mesothelium, and liver cancer | [20] Cheng, et al. 2011 |
| piR-823 | downregulated | gastric cancer | [22] Cheng, et al. 2012 |
| piR-4987 | upregulated | ductal carcinoma of breast | [21] Huang, et al.2012 |
| piR-20365 | upregulated | ductal carcinoma of breast | [21] Huang, et al.2012 |
| piR-20485 | upregulated | ductal carcinoma of breast | [21] Huang, et al.2012 |
| piR-20582 | upregulated | ductal carcinoma of breast | [21] Huang, et al.2012 |
| piR-19825 | upregulated | ductal carcinoma of breast | [21] Huang, et al.2012 |
| pi-17458 | downregulated | ductal carcinoma of breast | [21] Huang, et al.2012 |
| piR-Hep1 | upregulated | liver cancer | [73] Law, et al. 2013 |
| piR-015520 | upregulated | Tissues and HEK293 | [65] Esposito, et al. 2011 |
3.4.1. Cell proliferation and viability
The growth of gastric cancer cells such as MGC-803 and SGC-7901was significantly inhibited by a piR-651 inhibitor in a dose-dependent manner [20]. The mechanism of the decreased proliferation ability by the piR-651 inhibitor was that the cells transfected with the piR-651 inhibitor were arrested at G(2)/M phase [20]. However, cell growth was significantly inhibited by piR-823 overexpression in these two cell lines [22], and the effect of growth inhibition by ectopic piR-823 expression was further confirmed by in vivo experiments. Both tumor volume and weight were decreased significantly in cells treated with more ectopic piR-823 level, indicating that elevation of piR-823 level resulted in inhibition of tumor formation. Also, in hepatocellular carcinoma cell line HKCI-8, cells transfected with piR-Hep1 inhibitor displayed reduced cell viability by knockdown piR-Hep1 when compared with the control inhibitor [73].
3.4.2. Cell invasion and transwell motility
Cell invasion and trans-well motility decreased when inhibiting piR-Hep1in Hepatocellular carcinoma HKCI-8 cells, whereas overexpression of piR-Hep1 cells could result in profound augmentations on cell migrationin immortalized hepatocyte line (MIHA) cells [73]. These observations suggested that piR-Hep1played a key role in enhancing cells’ capability to invade and migrate [73]. Although overexpression of piR-Hep1 in MIHA cells showed no apparent effect on cell viability, the authors thought that functional effects on motility and invasiveness might be more evident in non-tumorigenic cells than in carcinoma cells, as pre-malignant cells displayed a more modest genetic background [73].
3.5. Other possibilities of piRNAs contributing to tumorigenesis
One of the most important functions of piRNAs is to repress transposon, a kind of transposable elements or “jumping genes” which are pieces of DNA that insert themselves into other locations in the genome. These transposable elements of somatic insertions were present in multiple tumor types [74]. There were transposons insertion occurred in genes that were frequently mutated in cancer, suggesting that transposon actively contributed to the development of cancer by introducing mutations that disrupted gene expression [74–76].
4. piRNAs are potential biomarkers for cancer diagnosis and prognosis
Given the important role of piRNAs and their interacting proteins in diverse cellular processes, defining different expression profiles of cancer type-specific piRNAs should allow development of novel cancer biomarkers. Indeed, we have seen the emergence of certain piRNAs as potential regulators of cancer cell development in the past two years, suggesting that piRNAs can serve as potential markers for cancer diagnosis. For example, the level of piR-651 was associated with tumor-node-metastasis (TNM) stage in gastric cancer [20]; and up-regulated piR-4987 expression was associated with lymph node positivity in breast cancer [21]. These findings suggested that a fraction of piRNAs would be expressed in a highly cancer-specific way, and the quantitative differences in the expression level of these piRNAs may allow determine tissue-origin of cancers. Furthermore, the possible molecular mechanisms by which piRNAs may carry out the regulatory functions are also partially explained: the growth of gastric cancer cells was inhibited by a piR-651 inhibitor and arrested at the G(2)/M phase [20], and Silencing of piRHep1 inhibited cell viability, motility and invasiveness with a concomitant reduction in the level of active AKT phosphorylation in hepatocellular carcinoma [73]. Although the proposed mechanisms require further testing and may not provide a complete explanation for piRNAs as biomarkers for cancer, the emerging evidences demonstrated that piRNAs might serve as potential biomarkers for both diagnosis and prognosis of malignancies. In addition, piRNAs are 26–32 nt in length, and such a short fragment of RNA means that piRNAs are not so easily degraded as other long RNAs, and also means piRNAs can pass through cell membrane easily. These characteristics imply that piRNAs can be detected in patient samples, and potentially isolated from easily obtained bodily fluids, such as blood plasma and serum, saliva, sputum, and urine. Therefore, piRNAs have attracted a great deal of attention to serve as a potential noninvasive approach to improve diagnosis of human cancers, and the exploitation of this knowledge is already presenting potential opportunities for advances in cancer diagnosis. Thus, elucidating the relationship between piRNAs and cancer is a promising new field of cancer research.
5. Potential use of piRNAs in cancer therapies and future directions
Although our understanding of the molecular mechanisms of piRNA function in tumorigenesis is still very limited, some features of piRNAs would make them ideal candidates for therapeutic intervention. For instance, piRNAs functioning in RNA degradation at the post transcriptional level could be of therapeutic benefit, or its function in DNA methylation leading to transcriptional gene silencing could be used to inhibit certain oncogenes expression. Furthermore, RNAi-mediated gene silencing for cancer treatment could be applied to selectively silence oncogenic piRNAs. Moreover, although piRNAs are discovered to be restricted to germ cells and germline tissues, recent findings have showed that piRNAs are also expressed in the central nervous system (CNS) and other somatic tissues as well as cancer cells and tumor tissues [20–22, 65, 73, 77]. This indicates that there are undiscovered piRNAs-interacting proteins depending on tissue- or cancer- specific manner; because the advantage that piRNAs indeed have protein-binding ability provides a means of therapeutic intervention. Although piRNAs are discovered to be restricted to bind to Piwi proteins, there are observations have showed that piRNAs also bind to other proteins. For example, two complexes other than PIWI proteins could bind to piR_015520 in cytoplasmic extract from HEK 293 by electrophoretic mobility shift assay, suggesting that other RNA-binding proteins, which could bind piRNAs, not yet identified [65].
The fields of piRNAs and their interacting proteins in cancer have yet to be fully explored, and this area has the potential to yield many promising leads. Future use of comprehensive and high throughput techniques could identify more informative piRNA-biomarkers and their interacting proteins specific to different type or stage of cancers. If the function and mechanism were explored for all of them, piRNAs and their interacting proteins would attract a great deal of attention as effective biomarkers for diagnosis as well as novel targets for capable therapeutic manipulation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
None
Reference
- 1.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 5.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 6.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 7.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 9.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 11.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 14.Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 16.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 17.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Gou LT, Zhang M, Zu LD, Hua MM, Hua Y, Shi HJ, Li Y, Li J, Li D, Wang ED, Liu MF. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24:13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Li C, Zhang K, Sun H, Tao D, Liu Y, Zhang S, Ma Y. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010;43:635–641. doi: 10.5483/BMBRep.2010.43.9.635. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new noncoding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2012 Nov 15; doi: 10.1007/s12094-012-0966-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 24.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 25.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 26.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr, Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cecere G, Zheng GX, Mansisidor AR, Klymko KE, Grishok A. Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol Cell. 2012;47:734–745. doi: 10.1016/j.molcel.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 33.Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med. 2012;10:103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samji T. PIWI, piRNAs, and germline stem cells: what's the link? Yale J Biol Med. 2009;82:121–124. [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 37.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beyret E, Liu N, Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012;22:1429–1439. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki R, Honda S, Kirino Y. PIWI Expression and Function in Cancer. Front Genet. 2012;3:204. doi: 10.3389/fgene.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 41.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 42.Costa FF, Le Blanc K, Brodin B. Concise review: cancer/testis antigens, stem cells, and cancer. Stem Cells. 2007;25:707–711. doi: 10.1634/stemcells.2006-0469. [DOI] [PubMed] [Google Scholar]
- 43.He C, Zuo Z, Chen H, Zhang L, Zhou F, Cheng H, Zhou R. Genome-wide detection of testis- and testicular cancer-specific alternative splicing. Carcinogenesis. 2007;28:2484–2490. doi: 10.1093/carcin/bgm194. [DOI] [PubMed] [Google Scholar]
- 44.Cheng YH, Wong EW, Cheng CY. Cancer/testis (CT) antigens, carcinogenesis and spermatogenesis. Spermatogenesis. 2011;1:209–220. doi: 10.4161/spmg.1.3.17990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 47.Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- 48.Taubert H, Greither T, Kaushal D, Wurl P, Bache M, Bartel F, Kehlen A, Lautenschlager C, Harris L, Kraemer K, Meye A, Kappler M, Schmidt H, Holzhausen HJ, Hauptmann S. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene. 2007;26:1098–1100. doi: 10.1038/sj.onc.1209880. [DOI] [PubMed] [Google Scholar]
- 49.Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15:201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y, Zhang K, Li C, Yao Y, Tao D, Liu Y, Zhang S, Ma Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One. 2012;7:e30999. doi: 10.1371/journal.pone.0030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu WK, Jiang XY, Zhang ZX. Expression of PSCA, PIWIL1 and TBX2 and its correlation with HPV16 infection in formalin-fixed, paraffin-embedded cervical squamous cell carcinoma specimens. Arch Virol. 2010;155:657–663. doi: 10.1007/s00705-010-0635-y. [DOI] [PubMed] [Google Scholar]
- 52.He G, Chen L, Ye Y, Xiao Y, Hua K, Jarjoura D, Nakano T, Barsky SH, Shen R, Gao JX. Piwil2 expressed in various stages of cervical neoplasia is a potential complementary marker for p16. Am J Transl Res. 2010;2:156–169. [PMC free article] [PubMed] [Google Scholar]
- 53.Li D, Sun X, Yan D, Huang J, Luo Q, Tang H, Peng Z. Piwil2 modulates the proliferation and metastasis of colon cancer via regulation of matrix metallopeptidase 9 transcriptional activity. Exp Biol Med (Maywood) 2012;237:1231–1240. doi: 10.1258/ebm.2012.011380. [DOI] [PubMed] [Google Scholar]
- 54.Grochola LF, Greither T, Taubert H, Moller P, Knippschild U, Udelnow A, Henne-Bruns D, Wurl P. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer. 2008;99:1083–1088. doi: 10.1038/sj.bjc.6604653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Liu Y, Shen X, Zhang X, Chen X, Yang C, Gao H. The PIWI protein acts as a predictive marker for human gastric cancer. Int J Clin Exp Pathol. 2012;5:315–325. [PMC free article] [PubMed] [Google Scholar]
- 56.Oh SJ, Kim SM, Kim YO, Chang HK. Clinicopathologic Implications of PIWIL2 Expression in Colorectal Cancer. Korean J Pathol. 2012;46:318–323. doi: 10.4132/KoreanJPathol.2012.46.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun G, Wang Y, Sun L, Luo H, Liu N, Fu Z, You Y. Clinical significance of Hiwi gene expression in gliomas. Brain Res. 2011;1373:183–188. doi: 10.1016/j.brainres.2010.11.097. [DOI] [PubMed] [Google Scholar]
- 58.Zhao YM, Zhou JM, Wang LR, He HW, Wang XL, Tao ZH, Sun HC, Wu WZ, Fan J, Tang ZY, Wang L. HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer. 2012;118:2708–2717. doi: 10.1002/cncr.26524. [DOI] [PubMed] [Google Scholar]
- 59.Zeng Y, Qu LK, Meng L, Liu CY, Dong B, Xing XF, Wu J, Shou CC. HIWI expression profile in cancer cells and its prognostic value for patients with colorectal cancer. Chin Med J (Engl) 2011;124:2144–2149. [PubMed] [Google Scholar]
- 60.He W, Wang Z, Wang Q, Fan Q, Shou C, Wang J, Giercksky KE, Nesland JM, Suo Z. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer. 2009;9:426. doi: 10.1186/1471-2407-9-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K, Jarjoura D, Nakano T, Ramesh GK, Shapiro CL, Barsky SH, Gao JX. Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker. Int J Clin Exp Pathol. 2010;3:328–337. [PMC free article] [PubMed] [Google Scholar]
- 62.Antonovics J, Boots M, Abbate J, Baker C, McFrederick Q, Panjeti V. Biology and evolution of sexual transmission. Ann N Y Acad Sci. 2011;1230:12–24. doi: 10.1111/j.1749-6632.2011.06127.x. [DOI] [PubMed] [Google Scholar]
- 63.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esposito T, Magliocca S, Formicola D, Gianfrancesco F. piR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS One. 2011;6:e22727. doi: 10.1371/journal.pone.0022727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci U S A. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hedges DJ, Belancio VP. Restless genomes humans as a model organism for understanding host-retrotransposable element dynamics. Adv Genet. 2011;73:219–262. doi: 10.1016/B978-0-12-380860-8.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A Major Epigenetic Programming Mechanism Guided by piRNAs. Dev Cell. 2013;24:502–516. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sytnikova Y, Lau NC. Does the APC/C Mark MIWI and piRNAs for a final farewell? Dev Cell. 2013;24:119–120. doi: 10.1016/j.devcel.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.01.032. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 74.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'ullivan MG, Matise I, Dupuy AJ, Collier LS, Powers S, Oberg AL, Asmann YW, Thibodeau SN, Tessarollo L, Copeland NG, Jenkins NA, Cormier RT, Largaespada DA. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen L, Stuart L, Ohsumi TK, Burgess S, Varshney GK, Dastur A, Borowsky M, Benes C, Lacy-Hulbert A, Schmidt EV. Transposon activation mutagenesis as a screening tool for identifying resistance to cancer therapeutics. BMC Cancer. 2013 doi: 10.1186/1471-2407-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]