SUMMARY
RNA viruses are sensed by RIG-I-like receptors (RLRs), which signal through a mitochondria-associated adaptor molecule, MAVS, resulting in systemic antiviral immune responses. Although RLR signaling is essential for limiting RNA virus replication, it must be stringently controlled to prevent damage from inflammation. We demonstrate here that among all tested UBX-domain-containing protein family members, UBXN1 exhibits the strongest inhibitory effect on RNA-virus-induced type I interferon response. UBXN1 potently inhibits RLR- and MAVS-induced, but not TLR3-, TLR4-, or DNA-virus-induced innate immune responses. Depletion of UBXN1 enhances virus-induced innate immune responses, including those resulting from RNA viruses such as vesicular stomatitis, Sendai, West Nile, and dengue virus infection, repressing viral replication. Following viral infection, UBXN1 is induced, binds to MAVS, interferes with intracellular MAVS oligomerization, and disrupts the MAVS/TRAF3/TRAF6 signalosome. These findings underscore a critical role of UBXN1 in the modulation of a major antiviral signaling pathway.
INTRODUCTION
The success of host defense against viral infections depends upon the ability of the innate immune system to sense and respond to invading pathogens. Intracellular detection of the viral pathogen-associated molecular patterns (PAMPs) of viral RNA is mediated predominantly by cytoplasmic sensors, the RIG-I-like receptors (RLRs) RIG-I (also known as DDX58) and MDA5 (also known as IFIH1) (Honda et al., 2006; Kawai and Akira, 2008; Loo and Gale, 2011; Meylan and Tschopp, 2006; Onoguchi et al., 2011; Wilkins and Gale, 2010; Yoneyama et al., 2004). Upon engagement of viral RNA ligands, both RIG-I and MDA5 trigger a signaling cascade that culminates in the production of cytokines and chemokines (Hornung et al., 2006; Kawai and Akira, 2008; Meylan and Tschopp, 2006; Pichlmair et al., 2006; Wilkins and Gale, 2010; Yoneyama et al., 2004). A central event of RLR signaling is the rapid induction of type I interferons (IFNs) to establish an antiviral state, via transcription factors such as nuclear factor-κB (NF-κB), activator protein 1, and interferon regulatory factor (IRF) (Hiscott et al., 2006; Maniatis et al., 1998; Taniguchi et al., 2001). The synergistic effect of these trans-activators on the promoters of type I interferon genes induces an immediate-early interferon response, mainly IFN-β and IFN-α1 (Honda et al., 2006). Secreted interferons then activate the Jak and STAT signaling to induce the expression of hundreds of interferon-stimulated genes (ISGs) (Liu et al., 2011; Matsumiya and Stafforini, 2010; Sadler and Williams, 2008; van Boxel-Dezaire et al., 2006).
Viral RNA binding enables the amino-terminal (N-terminal) caspase-recruitment domain (CARD) of RLRs to interact with the adaptor molecule MAVS (alternatively known as IPS-1/CARDIF/VISA), a mitochondrial outer membrane protein (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS then recruits E3 ligases TRAF3 and TRAF6 via its TRAF-interacting motif (TIM) (Paz et al., 2011; Xu et al., 2005), leading to the activation of canonical IKKs and the IKK-related kinases TBK1 and IKKε, and eventually to the production of type I interferons and proinflammatory cytokines (Hornung et al., 2006; Kumagai et al., 2008; Nakhaei et al., 2009; Pichlmair et al., 2006). MAVS has been recently found to localize to peroxisomes as well (Dixit et al., 2010). Upon viral infection, peroxisomal MAVS rapidly induces IFN-independent expression of defense factors that provide short-term protection, while mitochondrial MAVS activates an IFN-dependent signaling pathway with delayed kinetics, which amplifies and stabilizes the antiviral response. Although activation of RLR signaling is necessary to limit the spread of a broad spectrum of viral infections, it must be tightly regulated to prevent excessive responses that may result in deleterious effects on the host (Loo and Gale, 2011; Matsumiya and Stafforini, 2010). Recently, accumulating evidence has suggested that aberrant IFN induction due to RLR signaling is associated with autoimmune diseases (Loo and Gale, 2011; Matsumiya and Stafforini, 2010). For example, genetic studies have demonstrated that decreased or defective MDA5-mediated IFN signaling is associated with resistance to type 1 diabetes (Loo and Gale, 2011). As a central molecule of the RLR signaling, MAVS is regulated by host factors and is also the target of viral proteins (Liu and Gu, 2011). The NLR family member X1 (NLRX1) and poly(C)-binding protein 2 (PCBP2) have been reported to be involved in the negative regulation of MAVS-mediated signaling through different molecular mechanisms (Moore et al., 2008; You et al., 2009). NLRX1 is a member of nucleotide-binding domain and leucine-rich-repeat-containing (NLR) protein family, which is a ubiquitously expressed protein that resides on the outer mitochondrial membrane where MAVS is located. The amino terminus of NLRX1 contains a mitochondria-targeting sequence. In resting cells, NLRX1 is associated with MAVS, and expression of positive factors of RLR signaling or IFN-β treatment is unable to alter the mRNA and protein levels of NLRX1 (Moore et al., 2008). NLRX1 is therefore thought of as an inhibitor of steady-state antiviral signaling rather than as a negative-feedback inhibitor (Komuro et al., 2008; Moore et al., 2008). PCBP2 belongs to the hnRNP E family of RNA-binding proteins, which is retained mainly in the nuclei of resting cells. In contrast to NLRX1, PCBP2 is highly inducible by viral infection or IFN-α treatment. The induced PCBP2 translocates to cytosol and binds to MAVS, which leads to the recruitment of a HECT-domain-containing E3 ligase AIP4 to polyubiquitinate and degrade MAVS (You et al., 2009). In addition to NLRX1 and PCBP2, other host factors are also involved in the temporal-spatial regulation of MAVS-induced signaling complex (Komuro et al., 2008), although the molecular mechanisms remain elusive.
The human genome encodes at least 13 ubiquitin-regulatory-X (UBX)-domain-containing proteins, designated UBXN (Alexandru et al., 2008). The UBX domain shares weak amino acid homology with ubiquitin, and it adopts the same 3D fold as ubiquitin (Alexandru et al., 2008; Schuberth and Buchberger, 2008). One of the best understood UBXN proteins is p47, a cofactor of the AAA ATPase p97 (Schuberth and Buchberger, 2008; Yeung et al., 2008). p97 is highly conserved across species and involved in diverse cellular processes, including ubiquitin-dependent protein degradation, vesicle fusion, and cell-cycle control (Chapman et al., 2011; Yeung et al., 2008). It has been recently shown that several of the UBXN proteins bind to p97 and a large number of E3 ubiquitin ligases, suggesting that the UBXN proteins participate in the global regulation of protein turnover, most likely including proteins of immune function (Alexandru et al., 2008; Besche et al., 2009; Chapman et al., 2011; Schuberth and Buchberger, 2008; Yeung et al., 2008). UBXN proteins may determine the substrate specificity of p97, but for the majority of UBXN proteins the cellular function is largely unknown. By using small interfering (si) RNA screening, we demonstrate here that among all family members UBXN1 (also known as SAKS1/2B28) exhibits the strongest inhibitory effect on RNA-virus-induced interferon promoter activity. Further studies show that UBXN1 binds to MAVS, disrupting the MAVS-TRAF3/6 signaling complex and downstream antiviral immune responses.
RESULTS
UBXN1 Is a Negative Regulator of RNA-Virus-Induced Innate Immune Responses
To explore the potential role of UBXN proteins in modulating antiviral response, we performed a limited small interfering RNA (siRNA) screen in combination with a dual-luciferase reporter assay, to identify either positive or negative regulators of type I IFN signaling. All UBXN mRNA expression was readily detectable by quantitative RT-PCR, and, except for FAF2, all other UBXNs were upregulated upon Sendai virus (SeV) infection of HEK293 cells (Figure S1A). We then attempted to silence each of the UBXNs using a mix of three individual siRNA and examined type I IFN response using a luciferase reporter. Results demonstrate that of all tested family members silencing of UBXN1 (SAKS1/2B28) led to the greatest induction of ISRE reporter luciferase activity after RNA virus infection, including vesicular stomatitis virus (VSV)-GFP and SeV, suggesting that UBXN1 may be a negative regulator of virus-induced IFN signaling (Figure 1A). To confirm this, we generated a cell line expressing UBXN1-small hairpin RNA (shRNA) and measured IFN-β production in UBXN1-silenced cells infected with VSV-GFP. In agreement with the siRNA screening findings, these cells secreted greater amounts of IFN-β protein than scrambled shRNA-treated cells (Figure 1B). Consequently, VSV titers were decreased 6-fold when UBXN1 was silenced (Figure 1C). These data suggest that UBXN1 is potentially involved in IFN regulation. This was further strengthened by the observations that UBXN1 protein expression was induced by not only viral infection but also IFN-β (Figure S1B).
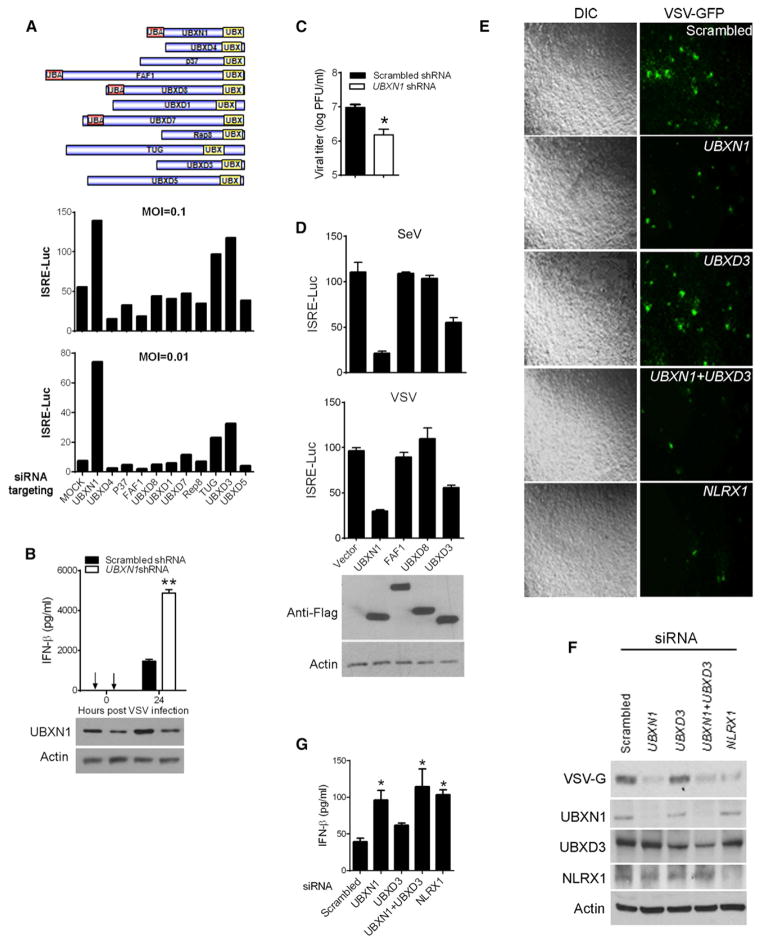
Figure 1. UBXN1 Negatively Regulates RNA-Virus-Induced Type I IFNs.
(A) Schematic illustration of UBX family members for siRNA screening and quantification of ISRE promoter activity by dual-luciferase assay in HEK293 cells transfected with scrambled (mock) or UBX-family-member-specific siRNAs and infected with VSV for 16 hr at a moi of 0.1 or 0.01.
(B) ELISA assay for IFN-β protein in VSV-infected HEK293 cells transfected with scrambled (mock) or UBXN1-specific shRNAs. Bottom: immunoblots of UBXN1 and actin from the whole-cell lysate.
(C) VSV titers from the cell culture medium of (B) at 24 hr after infection.
(D) Dual-luciferase assays of ISRE promoter activity in either VSV or SeV-infected HEK293 cells overexpressing Flag (vector) or Flag-tagged UBXN1, FAF1, UBXD8, or UBXD3. Bottom: immunoblots of indicated proteins from the relevant whole-cell lysates.
(E) Microscopic images of VSV-GFP-infected HEK293 cells. Cells were transfected with indicated siRNA and 48 hr later infected with VSV (moi = 0.01) for 16 hr. Images were acquired with a Zeiss fluorescence microscope (objective 5×).
(F) Immunoblots of indicated proteins from the whole-cell lysates of (E).
(G) Quantification of secreted IFN-β concentrations from HEK293 cells transfected with indicated siRNA and subsequently infected with VSV by ELISA. Luciferase values in (A) and D) are expressed as fold induction over uninfected control.
Data represent the mean ± SEM (n = 3), *p < 0.05; **p < 0.01. See also Figure S1.
Based on their protein domain composition, the human UBXNs are subclassified into two main groups: the UBA-UBX proteins and UBX-only proteins (Alexandru et al., 2008). To validate our siRNA screening results, we assessed the effect of UBXN overexpression on virus-induced ISRE promoter activation. We included two of the UBA-UBX proteins FAF1 and UBXD8, and one of the UBX-only proteins, UBXD3. Consistent with siRNA results, overexpression of UBXN1 dramatically repressed VSV-GFP-induced ISRE activation compared to the vector control, while overexpression of FAF1 and UBXD8 had no effect on ISRE activity. In agreement with the siRNA screening results, UBXD3 also demonstrated an inhibitory effect on VSV-GFP or SV-induced ISRE promoter activation, albeit to a much lesser extent than UBXN1 (Figure 1D). We next assessed the relative importance of UBXN1, UBXD3, and a previously established negative regulator NLRX1 (Moore et al., 2008), in controlling VSV-GFP infection by siRNA knockdown in parallel. Silencing either UBXN1 or NLRX1, but not UBXD3 greatly reduced VSV replication in terms of GFP intensity (Figure 1E) and amount of VSV glycoprotein (VSV-G), as assessed by immunoblotting (Figure 1F). Type I IFN response was increased in UBXN1 and NLRX1, but only modestly in UBXD3 knockdown cells (Figure 1G). In addition, SV-induced ISRE, NF-κB-, IFNβ-, and IFNα4-promoter activation were also inhibited by UBXN1 in a dose-dependent manner (Figures 2A and 2B). In parallel, SV-induced IFN-β protein production was strikingly decreased when UBXN1 was overexpressed (Figure 2C). Taken together, these results demonstrate that UBXN1 inhibits virus-induced IFN and NF-κB signaling.
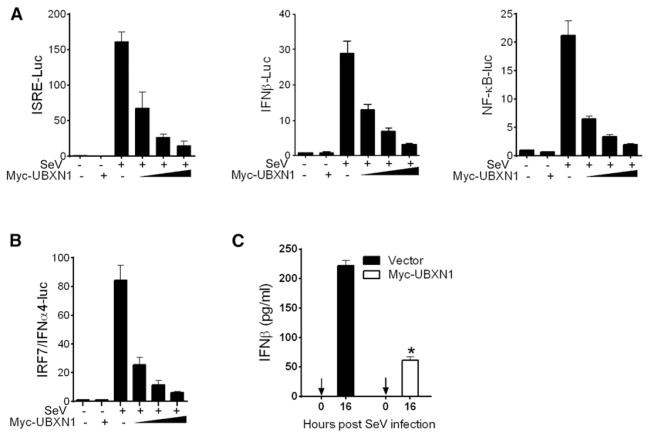
Figure 2. UBXN1 Inhibits SV-Induced IFN Signaling.
(A) Quantification of ISRE, IFNβ, and NFκB promoter activities in HEK293 cells transfected with either empty vector or increasing amounts of Myc-UBXN1 plasmid 16 hr after SeV infection.
(B) Quantification of IFN-α4 promoter activity of HEK293 cells transfected with either empty vector or increasing amounts of Myc-UBXN1 plasmid and a constant amount of IRF7 plasmid 16 hr after SV infection.
(C) ELISA of IFN-β secreted from HEK293 cells transfected with either control vector or Myc-UBXN1 expression plasmid 16 hr after SV infection.
Shown are mean ± SEM (n = 3). *p < 0.05.
UBXN1 Negatively Regulates West Nile Virus- and Dengue Virus-Induced Innate Immune Response
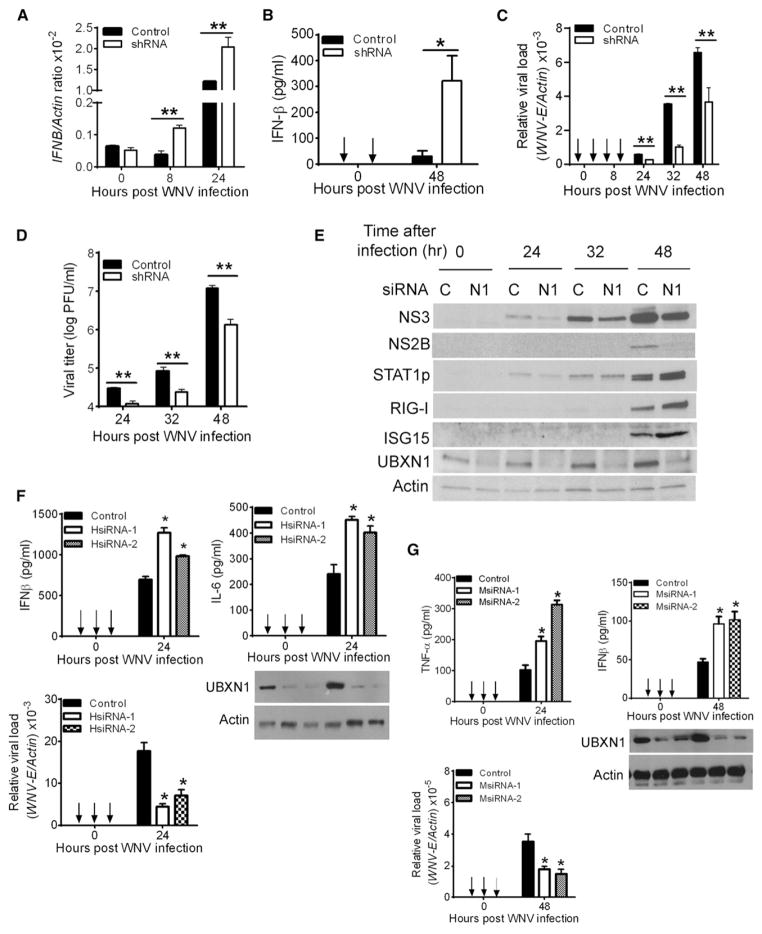
West Nile virus (WNV) is an enveloped, single-stranded, positive-sense RNA virus of the Flaviviridae family and is endemic in the Middle East and parts of North America (Fredericksen et al., 2008; Suthar et al., 2010). There have been over 1,400 lethal human cases of WNV since 1999 and over 240 deaths in 2012 alone in the United States. To expand our findings, we employed WNV to test whether endogenous UBXN1 suppresses WNV-induced host antiviral responses. We first generated a stable UBXN1-silenced A549 cell line by using plasmid-mediated shRNA interference. These UBXN1-silenced A549 cells produced more IFN-β protein and mRNA than that of control shRNA-treated cells, which is concomitant with decreased intra-cellular viral replication and yield of extracellular infectious viral particles (Figures 3A–3D and S2A–S2C). In parallel, IFN-stimulated gene (ISG) expression in response to WNV infection was also monitored by immunoblotting assays. Phosphorylation of STAT1 and the expression levels of ISG15 and RIG-I were enhanced in UBXN1-silenced cells, as compared to control cells (Figure 3E). Moreover, the expression levels of NF-κB-responsive genes, TNFα and IL6, were elevated in UBXN1-silenced cells (Figure S2C). In line with these results, siRNA directed against human and mouse UBXN1 resulted in similar effects in A549 cells and mouse primary bone-marrow-derived macrophages, respectively (Figures 3F, 3G, and S2D).
Figure 3. RNA Interference of UBXN1 Results in Enhanced Antiviral Response.
(A and B) Quantification of (A) IFNβ mRNA levels by Taqman quantitative real-time PCR (q-PCR) normalized to β-actin, and (B) secreted IFN-β levels by ELISA in A549 cells stably expressing either scrambled shRNA (control) or UBXN1-targeting shRNA, after WNV infection (moi = 1).
(C and D) Quantification of (C) intracellular WNV loads by q-PCR or (D) infectious viral particles in the culture medium by plaque assay from A549 cells stably expressing either scrambled shRNA (control) or UBXN1-targeting shRNA.
(E) Immunoblots of viral proteins (NS3, NS2B), select ISGs, and UBXN1, using specific antibodies in A549 cells stably expressing either scrambled shRNA (control) or UBXN1-targeting shRNA at various times (hr) after WNV infection.
(F and G) Quantification of indicated cytokine levels by ELISA, and WNV loads by q-PCR from (F) A549 cells or (G) bone-marrow-derived mouse macrophages in the presence of control or UBXN1-targeting siRNA. The immunoblots indicate the knockdown efficiency.
Data represent mean ± SEM (n = 3). *p < 0.05, **p < 0.01. See also Figures S2 and S3.
We further extended our study to another medically important flavivirus, type II dengue virus (DENV), which is endemic throughout the tropics and South Asia (Loo et al., 2008). We measured protein production of IFN-β, the NF-κB-responsive proinflammatory cytokine tumor necrosis factor α (TNF-α), and DENV replication, as measured by real-time RT-PCR and plaque assay in UBXN1-silenced cells. As expected, these cells exhibited enhanced protein production of IFN-β and TNF-α and reduced DENV replication, compared to control siRNA-treated cells (Figure S3). Thus, we show that UBXN1 plays an important role in controlling the innate antiviral response to virus infection.
UBXN1 Blocks RLR-Dependent Antiviral Signaling
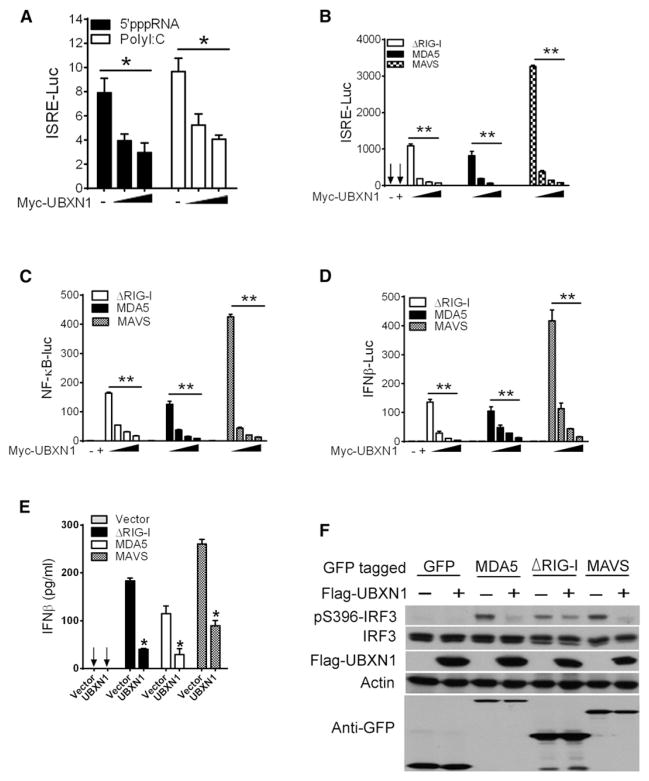
Given that WNV, DENV, VSV, and SV are potent agonists of the RLR signaling pathway (Cui et al., 2010; Kato et al., 2008; Loo et al., 2008; Saha et al., 2006), we next evaluated the impact of UBXN1 overexpression on ISRE promoter activity using synthetic analogs of viral RNAs, 5′-triphosphate RNA (a potent stimulator of RIG-I) (Hornung et al., 2006; Pichlmair et al., 2006), and high-molecular-weight poly(I:C) (an agonist of MDA5 [Kato et al., 2008]). UBXN1 significantly inhibited RLR-agonist-induced ISRE promoter activation in a dose-dependent manner (Figure 4A). To confirm the inhibitory role of UBXN1 in the RLR signaling pathway, we ectopically expressed the cytosolic receptor MDA5, the constitutively active form of RIG-I (ΔRIG-I), or the adaptor molecule MAVS along with increasing amounts of UBXN1. ΔRIG-I-, MDA5-, and MAVS-induced ISRE, NF-κB, and IFNβ promoter activation were significantly reduced by UBXN1 in a dose-dependent manner (Figures 4B–4D). The UBXN1-mediated regulation of the IFN pathway was also confirmed by ELISA, demonstrating that UBXN1 inhibited ΔRIG-I-, MDA5-, and MAVS-induced IFN-β protein production (Figure 4E). Furthermore, UBXN1 significantly inhibited ΔRIG-I-, MDA5-, and MAVS-mediated IRF3 phosphorylation, a hallmark of IFN activation (Figure 4F). These data collectively demonstrate that UBXN1 is a potent inhibitor of RLR signaling and inhibits the IFN antiviral response downstream of MAVS. To determine whether the immunoregulatory function of UBXN1 is conserved across species, we generated an expression plasmid encoding the mouse Ubxn1 cDNA. The amino acid sequences of UBXN1 are highly homologous between human, mouse, and rat (Ishibashi et al., 2005). Both human and mouse UBXN1 proteins are 297 amino acids and share 91.3% homology, implying conserved function (Figures S4A and S4B). Overexpression of mouse Ubxn1 markedly blocked ΔRIG-I-, MDA5-, and MAVS-induced ISRE and IFN-β promoter activation, suggesting that its function as a negative regulator of RLR signaling is evolutionarily conserved across species (Figures S4C–S4E).
Figure 4. UBXN1 Blocks RLR Signaling.
(A) Quantification of ISRE promoter activity of A549 cells transfected with either an empty vector or increasing amounts of UBXN1 plasmid and treated with 5′-triphosphate RNA or polyI:C. Data are expressed as fold induction over nontreated cells.
(B–D) Quantification of (B) ISRE-, (C) NFκB-, and (D) IFNβ-promoter activity of HEK293 cells expressing GFP-ΔRIG-I, GFP-MDA5, or GFP-MAVS, together with increasing amounts of Myc-UBXN1. Data are presented as fold induction over empty vector.
(E) ELISA of IFN-β secreted from HEK293 cells transfected with indicated expression plasmids and either an empty vector or Myc-UBXN1 expression plasmid.
(F) Immunoblotting analyses of whole-cell lysates of HEK293 cells transfected with plasmids as described in (E).
Data represent mean ± SEM (n = 3). *p < 0.05; **p < 0.01. See also Figures S4 and S5.
MAVS Is the Target of UBXN1
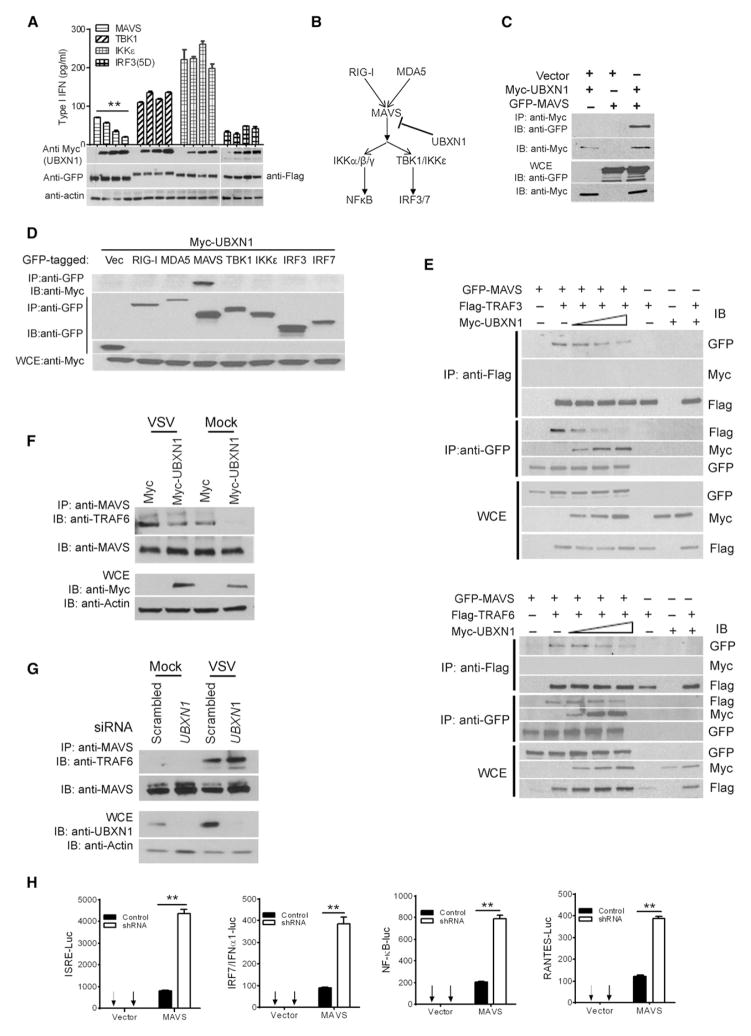
To identify the potential target of UBXN1 in RLR signaling, we expressed MAVS, the noncanonical kinase IKKε-, TBK1, or the constitutively active form of IRF3 [IRF3(5D)], along with increasing amounts of UBXN1. UBXN1 strongly inhibited the type I IFN production induced by MAVS but not by TBK1, IKKε, or IRF3 (5D) (Figure 5A), though UBXN1 modestly inhibited TBK1- or IKKε-mediated ISRE promoter activity (Figure S5I). Similar results were also observed with mouse Ubxn1 (Figures S4F–S4H). These data therefore suggest that UBXN1 targets the MAVS signaling complex (Figure 5B). Consistent with this, UBXN1 interacted only with MAVS, but not with other known components of the RLR signaling pathway, including RIG-I, MDA5, TBK1, IKKε, IRF3, or IRF7, indicating the specificity of UBXN1-MAVS physical and functional interactions (Figures 5C and 5D). Given that MAVS contains TRAF-interacting motifs that allow MAVS to recruit TRAF3 and TRAF6 and assemble the MAVS signaling complex (Paz et al., 2011; Saha et al., 2006; Xu et al., 2005), we investigated whether UBXN1 can interfere with the assembly of MAVS/TRAF3/TRAF6 signalosome. Competitive coimmunoprecipitation assays demonstrate that both MAVS-TRAF3 and MAVS-TRAF6 interaction were disrupted by UBXN1 in a dose-dependent manner, and there was no detectable interaction of UBXN1 with either TRAF3 or TRAF6 (Figure 5E). Overexpression of UBXN1 also disrupted endogenous MAVS-TRAF6 interaction in the presence or absence of VSV-GFP infection (Figure 5F). In contrast, depletion of UBXN1 by siRNA enhanced endogenous MAVS-TRAF6 interaction following VSV-GFP infection (Figure 5G). Furthermore, MAVS-induced activation of ISRE, NF-κB, RANTES, and IFNα1 promoters were enhanced in UBXN1-silenced cells (Figure 5H). To assess the relative importance of UBXN1 on MAVS regulation, we compared it with a well-known MAVS inhibitor, NLRX1. Overexpression of UBXN1 resulted in a similar fold inhibition on MAVS-induced type I IFN response as NLRX1 (Figure S5A). We next asked if UBXN1 interferes with TLR-mediated or DNA-virus-induced type I IFN response. Overexpression of UBXN1 had no effect on TLR3-mediated, TLR4-mediated, or herpes-simplex-virus (HSV)-induced IFNβ promoter activation (Figures S5B–S5D). Taken together, these observations strongly indicate that UBXN1 specifically interferes with MAVS function.
Figure 5. UBXN1 Blocks MAVS Function.
(A) Quantification of type I IFN, using a 2fGTH cell line stably expressing ISRE-Luc, from the cell culture medium of HEK293 cells expressing increasing amounts of Myc-UBXN1 together with GFP-tagged MAVS, TBK1, IKK, or Flag-IRF3(5D), respectively.
(B) Schematic graph showing the potential target of UBXN1 in the RLR signaling pathway.
(C) Coimmunoprecipitation (coIP) of MAVS and UBXN1 from HEK293 cells expressing Myc-UBXN1 and GFP-MAVS using an Myc monoclonal antibody, followed by immunoblotting (IB). WCE, whole-cell extract.
(D) CoIP of MAVS and UBXN1 from HEK293 cells expressing Myc-UBXN1 and GFP-tagged proteins using a GFP monoclonal antibody, followed by immunoblotting (IB) using a Myc or GFP antibody.
(E) Displacement of TRAF from MAVS by UBXN1 in a dose-dependent manner. CoIP was performed with HEK293 cells expressing MAVS-GFP, Flag-TRAF, and increasing amounts of UBXN1-Myc, using either an Flag or GFP antibody.
(F) CoIP of endogenous MAVS and TRAF6 from HEK293 cells expressing Myc or Myc-UBXN1 using a rabbit MAVS polyclonal antibody, followed by immunoblotting (IB) using a mouse MAVS or TRAF6 antibody.
(G) CoIP of MAVS and TRAF6 from HEK293 cells transfected with a scrambled or UBXN1 siRNA and immunoblotting (IB) as in (F).
(H) Quantification of ISRE, NFκB, RANTES, and IFNα1 promoter activity by dual-luciferase reporter assay in scrambled (control) shRNA or UBXN1 shRNA-treated HEK293 cells transfected with either empty vector or GFP-MAVS plasmid (MAVS). Luciferase values in are expressed as fold induction over vector control.
Data represent mean ± SEM (n = 3). **p < 0.01. See also Figure S5.
UBXN1 Targets a TRAF3/6-Binding Site of MAVS
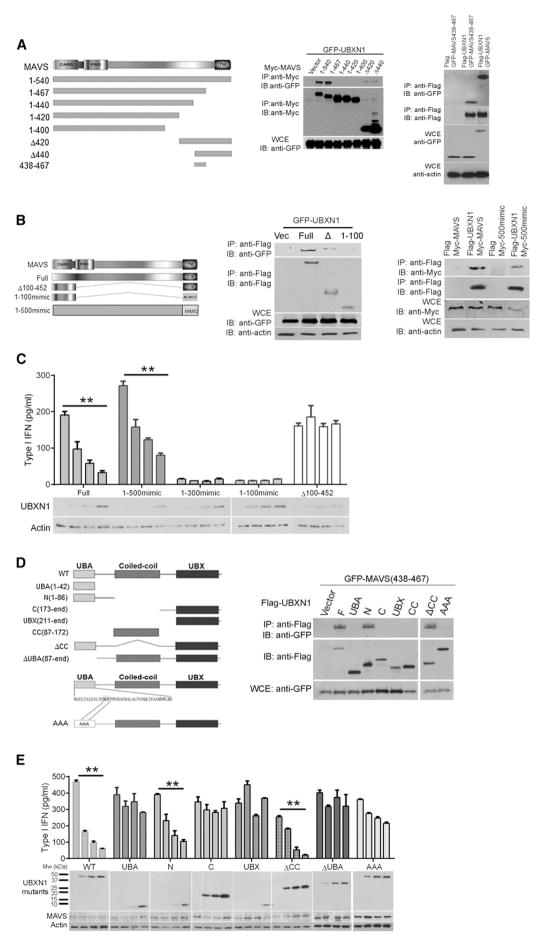
To characterize the UBXN1-binding motif of MAVS, we generated a series of MAVS deletion constructs. We transfected cells with plasmids encoding each MAVS mutant, along with an expression plasmid of UBXN1, and assessed binding to UBXN1 by coimmunoprecipitation. UBXN1 bound strongly to full-length and aa 1–467 MAVS, weakly to the C terminus containing the transmembrane domain (Δ420 and Δ440), but failed to bind to other C-terminally truncated forms of MAVS (1–400, 1–420, and 1–440, Figure 6A, left panel). These results suggest that UBXN1 predominantly binds to the amino acid spanning 440–467 of MAVS. Indeed, this region alone was sufficient for UBXN1 binding (Figure 6A, right panel). MAVS with the trans-membrane domain (TM; required for MAVS subcellular localization; Figure 6A) (Seth et al., 2005) and CARD domain only (MAVSΔ) was able to bind UBXN1, but to a much less extent than full-length protein. Substitution of the TM domain of MAVSΔ with the TM domain of a mitochondrial membrane protein OMP25 (MAVS1–100mimic; Dixit et al., 2010) completely abolished UBXN1 binding (Figure 6B, left panel). However, substitution of the TM domain of full-length MAVS with the TM domain of a mitochondrial membrane protein OMP25 (MAVS1–500mimic) did not appreciably reduce UBXN1 binding (Figure 6B, right panel). Taken together, these results indicate that the residues 440–467 of MAVS are necessary and sufficient for strong UBXN1 binding. Interestingly, it has been previously demonstrated that MAVS has two TRAF3-binding sites (aa 143–147 and 455–460) (Xu et al., 2005; Paz et al., 2011) and two TRAF6-binding sites (aa 153–158 and 455–460) (Xu et al., 2005; Saha et al., 2006). Thus, our results suggest that UBXN1 specifically competes for the TRAF3/6-binding site (aa 455–460) of MAVS to block TRAF3/6 recruitment to MAVS. Consistent with this notion, UBXN1 significantly repressed type IFN production induced only by full-length and 1–500mimic MAVS that contains the TRAF3/6-binding site, but not by 1–100mimic, 1–300mimic, or MAVSΔ100-452 (Figures 6C and S6A).
Figure 6. Characterization of MAVS and UBXN1 Mutual Binding Motifs.
(A) Interaction of MAVS truncation mutants with full-length UBXN1. Left panel: schematic of MAVS deletion mutants. Middle panel: coimmunoprecipitation (IP) of MAVS and UBXN1 from HEK293 cells expressing GFP-UBXN1 together with Myc-tagged MAVS variants using a Myc monoclonal antibody, followed by immunoblotting (IB) using a Myc or GFP antibody. Right panel: binding of UBXN1 to MAVS438-467. WCE, whole-cell extract.
(B) CoIP of MAVS truncation mutants with a mimic transmembrane domain with full-length UBXN1 as in (A).
(C) Quantification of type I IFN, using a 2fGTH cell line stably expressing ISRE-Luc, in the cell culture medium of HEK293 cells expressing increasing amounts of Myc-UBXN1 together with Flag-tagged MAVS full-length, MAVSΔ, or MAVS-mimic. Immunoblots under the chart indicate the intracellular Myc-UBXN1 expression levels.
(D) Interaction of UBXN1 deletion mutants with MAVS438-467. Left panel: schematic diagram of UBXN1 deletion mutants. Right panel: coIP of MAVS and UBXN1 from HEK293 cells expressing GFP-MAVS438-467 and Flag-tagged UBXN1 mutants using anti-Flag agarose beads, followed by IB using a Flag or GFP antibody.
(E) Quantification of type I IFNs from the culture medium of HEK293 cells expressing increasing amounts of Myc-tagged UBXN1 truncates and GFP-MAVS, as described in (C). Immunoblots under the chart indicate the intracellular Myc-UBXN1and GFP-MAVS expression levels.
Data in (E) and (G) represent mean ± SEM (n = 3). **p < 0.01. See also Figure S6.
The N Terminus Containing the UBA Domain of UBXN1 Is Required and Sufficient for MAVS Inhibition
We next asked which domain of UBXN1 binds to and inhibits MAVS function. UBXN1 contains an N-terminal UBA domain, a coil-coiled (CC) domain, and a C-terminal UBX domain. To identify the MAVS-binding motif within UBXN1, we generated a series of UBXN1 deletion or mutation constructs tagged with GFP or Myc. Surprisingly, MAVS interacted with all the functional domains of UBXN1 (Figure S6B). A similar interaction mode was also observed between NLRC5 and IKKβ, in which NLRC5 negatively regulates IKKα/β-mediated NF-κB activation (Cui et al., 2010). As it appears that residues 438–467 mediate MAVS binding to UBXN1, we hypothesize that the UBXN1 domain that binds to MAVS 438–467 may be of prime physiological significance. Only the N terminus (1–86) and the ΔCC (aa 1–86 joined with 173-end) retained comparable MAVS438-467-binding as full-length UBXN1 (Figure 6D). Consistent with these findings, only the N terminus and the ΔCC forms of UBXN1 tained the same fold-inhibitory effect on MAVS-induced type I IFN production as full-length UBXN1 (Figure 6E). The C terminus (173-end) and UBX domain (211-end) of UBXN1 were dispensable for its MAVS-inhibiting function. In addition, deletion of the UBX domain did not affect the inhibitory activity of UBXN1 on both MAVS- and SV-induced IFNβ promoter activation, indicating that the UBX domain is not essential (Figures S6C and S6D). Although the minimal UBA domain (1–42) alone was unable to inhibit MAVS function, the UBA-truncated form (ΔUBA) and the UBA domain mutant (AAA) also failed to inhibit MAVS function (Figures 6E, S6C, and S6D). Taken together, these results indicate (1) the UBA domain (1–42) is necessary, but not sufficient, (2) the N terminus (1–86) alone is sufficient, and that (3) physical binding to MAVS is necessary, but not sufficient, for the observed inhibitory effect of UBXN1 on MAVS signaling.
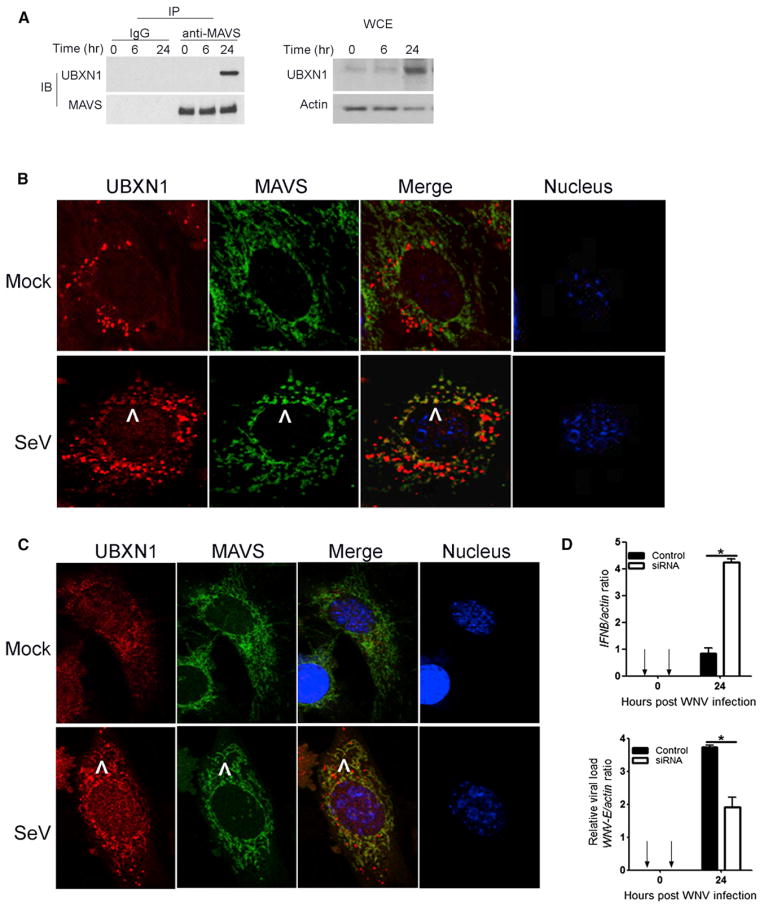
Endogenous UBXN1 Colocalizes with MAVS after Viral Infection
Our previous data demonstrated that UBXN1 interacts with MAVS under the experimental condition of transient transfection and overexpression. To test whether endogenous UBXN1 interacts with MAVS after RNA virus infection, we infected A549 cells with SV at a high multiplicity of infection (moi). Infected cells were collected at varying time points after infection, lysed, and subjected to coimmunoprecipitation. Twenty-four hours after SV infection, endogenous MAVS coimmunoprecipitated with endogenous UBXN1, which itself was induced several-fold (Figure 7A). This result is supported by the observation that after SV infection UBXN1 and MAVS colocalized in primary human foreskin fibroblast cells (HFFs) (Figure 7B) and mouse embryonic cells (MEFs) (Figure 7C), as assessed by confocal immunofluorescence microscopy. In parallel, the negative regulatory effects of Ubxn1 in primary MEFs were also evaluated after WNV infection. Specific silencing of endogenous Ubxn1 expression resulted in upregulation of Ifnβ expression, concomitant with decreased WNV replication (Figure 7D). Taken together, these findings suggest that at a relatively late phase after RNA virus infection endogenous UBXN1 is induced and interacts with MAVS, curtailing the antiviral immune response.
Figure 7. SV Infection Induces Endogenous UBXN1 and MAVS Interaction and Colocalization.
(A) CoIP of MAVS and UBXN1 from A549 cells after SV infection using a MAVS antibody pull-down and IgGs as a control, followed by immunoblotting (IB). WCE, whole-cell extract.
(B and C) Confocal immunofluorescence microscopy of UBXN1 and MAVS after SeV infection in primary (B) HFFs and (C) MEFs. UBXN1/MAVS/nucleus was stained with secondary antibodies conjugated with Alexa Fluor 546/488 and TO-PRO-3, respectively. Images were acquired using a Zeiss LSM 510 Meta (objective 63×). Arrows indicate representative colocalization.
(D) Quantification of IFNβ mRNA levels and intracellular WNV load by Taqman quantitative real-time PCR (q-PCR), normalized to β-actin, in MEFs transfected with scramble (control) or Ubxn1-targeting siRNA and infected with WNV (moi = 1). Data represent mean ± SEM (n=3). **p < 0.05.
See also Figure S6.
UBXN1 Disrupts MAVS-MAVS Interaction
Upon virus infection MAVS is known to oligomerize and form structures that resemble prion fibrils inside cells (Hou et al., 2011). We wished to see if UBXN1 disrupted MAVS-MAVS interactions. To do so, we constructed four N- and C-terminal fusions of MAVS with enhanced yellow fluorescent protein (eYFP) and cyan fluorescent protein (CFP). These constructs were overexpressed by transient transfection in 293T cells, which were then analyzed by flow cytometry for fluorescence resonance energy transfer (FRET). MAVS FRETed with itself (Figure S7A), and the FRET signal was reduced in the presence of Myc-UBXN1 as competitor but not by HIV Gag (Figures S7B and S7C). MAVS-MAVS FRET signal was also reduced in the presence of Flag-MAVS (Figure S7D). As a known positive control, HIV Gag strongly FRETed with itself (Figure S7E), which was reduced by excess competitor Gag (Figure S7F) but not re- by Myc-UBXN1 or Flag-MAVS (Figures S7G and S7H). These results, taken together, suggest that UBXN1 is able to disrupt MAVS-MAVS interaction in intact, living cells.
UBXN1was recently demonstrated to potentially participate in p97-mediated protein turnover and to modify protein function by binding to polyubiquitin chains (Alexandru et al., 2008; Besche et al., 2009). We tested if overexpression of UBXN1 facilitated MAVS degradation. Total MAVS protein levels remained largely unchanged when UBXN1 was overexpressed in the absence or presence of the proteasome inhibitors Bortezomib and MG132 (Figure S8). This suggests that UBXN1 does not mediate its inhibitory activity by degrading MAVS or otherwise targeting it to the proteasome.
DISCUSSION
The UBXN family members are believed to participate in multiple biological processes. However, the function of most of the family members still remains to be explored. In particular, the involvement of any UBXN genes in the innate immune system has not yet been reported. In this study, we demonstrated by several lines of evidence that UBXN1 functions as a negative regulator of RNA-virus-induced host antiviral response. First, depletion of endogenous UBXN1 expression by small RNA interference enhances RNA-virus-induced IFNβ promoter activation and IFN-β protein secretion. Second, overexpression of UBXN1 strikingly inhibits either RNA-virus-induced or positive factors of RLR-signaling-induced IFN-β production and NF-κB promoter activation. Third, knockdown of UBXN1 by RNA interference promotes West-Nile- and Dengue-virus-induced IFN-β and proinflammatory cytokine production and repressed viral replication. Fourth, overexpression of UBXN1 disrupts the assembly of MAVS/TRAF3/TRAF6 signaling complex. Fifth, SeV infection induces endogenous UBXN1-MAVS interaction and colocalization. Sixth, UBXN1 disrupts MAVS-MAVS interaction inside cells.
Based on its protein domain composition, UBXN1 is considered as one of the UBA-UBX-containing subfamily of human UBXNs. The UBXNs are generally believed to be the adaptor molecules of p97, an AAA ATPase that is highly conserved across species and involved in diverse cellular processes, including ubiquitin-dependent protein degradation, vesicle fusion, and the cell cycle (Alexandru et al., 2008; Chapman et al., 2011; Schuberth and Buchberger, 2008; Yeung et al., 2008). UBXN1 has been recently shown to bind p97 and polyubiquitin, positively (Alexandru et al., 2008; Besche et al., 2009; Ishibashi et al., 2005) or negatively (LaLonde and Bretscher, 2011), regulating protein degradation. However, our data indicate that MAVS protein levels are largely unaffected in the presence of increasing amounts of exogenous UBXN1. Furthermore, the C-terminal UBX domain of UBXN1, which mediates the binding and recruitment of p97 to a target substrate, is dispensable for its inhibitory effect on MAVS function, suggesting that the regulatory effect of UBXN1 on MAVS is independent of p97 action. Last, of the UBA-UBX proteins, the regulatory effect of UBXN1 on MAVS is unique and specific because either knockdown or overexpression of the other two UBA-UBX proteins, FAF1 and FAF2, shows no substantial impact on either VSV- or SV-induced IFN response.
How does UBXN1 regulate MAVS? As a central adaptor of RLRs, MAVS signaling is held in check by negative regulators to prevent excessive immune response. NLRX1, PCBP2, and RNF5 are recently identified cellular factors that limit MAVS signaling through distinct molecular mechanisms (Moore et al., 2008; You et al., 2009; Zhong et al., 2010). NLRX1 resides on the outer mitochondrial membrane where MAVS is located and acts as an inhibitor of steady-state antiviral signaling (Moore et al., 2008). In contrast, PCBP2, which is predominantly localized in the nuclei of unstimulated cells, translocates to the cytosol and binds to MAVS after viral infection, leading to the recruitment of the E3 ubiquitin ligase AIP4 and subsequent proteasomal degradation of MAVS (You et al., 2009). RNF5 is another ligase that targets MAVS for K48-linked ubiquitination and proteasomal degradation (Zhong et al., 2010). Surprisingly, IKKε, a protein kinase that phosphorylates IRF3 to activate IFN transcription, was recently shown to bind to a TRAF3-binding site (aa 468–540) in MAVS via K63 ubiquitination, resulting in downregulation of the IFN response (Paz et al., 2009, 2011). In contrast, Castanier et al. recently showed that TRIM25-mediated proteasomal degradation of MAVS via lysine 7 and 10 after RLR activation is required for release of TBK1 into cytosol, allowing IRF3 phosphorylation and type I IFN expression (Castanier et al., 2012). Our results show that full-length UBXN1 primarily blocks a TRAF3/6-binding site (aa 450–480) that is critical for MAVS-mediated type IFN induction (Paz et al., 2011), suggesting that UBXN1 simply serves as a dominant-negative binder of MAVS. Except for a putative role in ERAD via p97, UBXN1 also likely blocks the function of its target proteins by binding to their polyubiquitin chains (McNeill et al., 2004; Wu-Baer et al., 2010). Of note, the UBA domain of UBXN1 binds K6-linked polyubiquitin chains conjugated to BRCA1 and blocks its E3 ligase activity (Wu-Baer et al., 2010). Our data, however, show that replacement of the unique lysine residue within the UBXN1-binding motif (438–467) with an arginine (K461R) does not affect MAVS activity or the inhibitory capacity of UBXN1 on MAVS signaling (data not shown). These results suggest that the MAVS-inhibiting activity of UBXN1 is likely independent of ubiquitination. Surprisingly all of the UBXN1 functional domains when overexpressed are capable of binding to the full-length MAVS, only the UBA-containing N terminus (aa 1–86) of UBXN1 binds to MAVS 438–467 and is also necessary and sufficient for its MAVS-inhibitory function. This suggests that an additional cellular factor(s) that specifically interacts with the N terminus of UBXN1 among other possibilities may be required. The fact that MAVS-UBXN1 did not FRET with each other (data not shown) suggests a cofactor may be involved in the observed interaction. Importantly, the presence of excess UBXN1 does not result in MAVS degradation; thus, it is more likely that UBXN1 acts as a steric “antagonist:” of MAVS.
In addition to localization on mitochondrial outer membranes, MAVS has recently been shown to reside on peroxisomal membranes, where it induces swift IFN-independent expression of antiviral factors upon virus infection (Dixit et al., 2010). This rapid-early response may provide a critical short-term protection before the initiation of mitochondrial MAVS signaling, which activates, amplifies, and stabilizes a state of IFN-dependent antiviral immunity. Our results suggest that UBXN1 is unlikely to be an early-stage inhibitor of peroxisomal MAVS-dependent signaling but rather UBXN1 modulates the sustained mitochondrial MAVS-dependent antiviral response.
In conclusion, UBXN1 is induced and recruited to MAVS after virus infection, serving as a brake to prevent excessive RLR signaling, most likely at a late stage of infection. This mode of action may help avoid cell death and tissue destruction due to hyperimmune responses, without compromising necessary antiviral immunity. In this sense, UBXN1 may be an ideal target for the development of novel therapies for inflammatory and autoimmune diseases that have been associated with aberrant or protracted RLR signaling (Loo and Gale, 2011). In addition, our findings regarding UBXN1 offer insights into the molecular functions of this highly conserved gene family.
EXPERIMENTAL PROCEDURES
RNA Interference
For siRNA screening, 1 × 105 HEK293 were plated into polyornithine-coated 24-well cell culture plates. Sixteen hours later, cells were transfected with 40 pmol of siRNA mix directed against human UBXNs (three 27-mer siRNAs from Origene) or NLRX1 (#EHU111901, Sigma-Aldrich) using Lipofectamine 2000 (Invitrogen). Cells were transfected again with luciferase reporter plasmids (100 ng ISRE-luc and 50 ng pRL-TK per well) 36 hr after siRNA transfection. Cells were infected with GFP-vesicular stomatitis virus 48 hr after siRNA transfection (Zhao et al., 2007). Cells were washed gently with PBS and lysed for Dual Glow Luciferase assay following the manufacturer’s instructions (Promega) 16 hr after infection. For siRNA knockdown in bone-marrow-derived mouse macrophages or A549 cells, 1–2 × 106 cells were transfected by electroporation (Lonza) with 100 pmol of mouse Ubxn1 siRNA (Dharmacon #J-051867-09 and J-051867-10) or human UBXN1 siRNA (Origene #SR309485, vial number 451335 and 451337), respectively. Cells were ready for other manipulations (virus infection or luciferase reporter assay) 48 hr after transfection.
For generating stable UBXN1 knockdown cell lines, a UBXN1-specific shRNA (#TF306979 OriGene Technologies) was retrovirally transfected (Zhao et al., 2007) into HEK293 and A549 cells. Stably transduced bulk cultures were selected in media containing puromycin (2 mg/ml; Sigma), and individual cell clones were isolated and confirmed by immunoblotting.
Analysis of Protein-Protein Interactions
HEK293 cells were transfected with expression plasmids using Lipofect-amine 2000. Whole-cell extracts (300 μg) were prepared from transfected cells and were incubated for 1 hr at 4°C with 1 μg of mouse monoclonal Myc (Sigma-Aldrich), GFP (Roche), or Flag (Sigma-Aldrich) antibody cross-linked to protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology). For coimmunoprecipitation of endogenous proteins, A549 cells were infected with SeV at a concentration of 80 hemagglutination units per milliliter. Whole-cell extracts (10 mg) were prepared at the indicated time points by using RIPA buffer and incubating for 4 hr at 4°C with 10 μg of either MAVS (Millipore) or IgG antibody crosslinked to protein A/G Agarose beads (Pierce). Beads were washed five times and proteins eluted by boiling for 3 min in SDS sample lysis buffer.
Virus Infection
NY99 strain of WNV, CT2741, was used to infect A549 or MEFs at a moi of 1 or 3, mouse bone-marrow-derived macrophages at a moi of 5 unless specified. Cantell strain of SeV (ATCC, #VR907) was added to cells at a concentration of 40 hemagglutination units per milliliter, unless specified. DENV-2 (DENV New Guinea C strain) was used at a moi = 1, unless specified.
Enzyme-Linked Immunosorbent Assay, Type I IFN Bioavailability, and Plaque-Forming Assays
ELISA of human or mouse TNF-α, IL-6 (R&D Systems), and IFN-β (PBL Interferon Sources) in the cell culture medium was performed following the manufacturers’ instructions. Type I IFNs in the cell culture medium were quantified using a 2fGTH cell line stably expressing an ISRE-Luc reporter (You et al., 2009). In brief, 200 μl of culture medium was incubated with confluent 2fGTH-ISRE-Luc cells (24-well plate) for 6 hr. Cells were lysed in passive lysis buffer and subjected to luciferase quantification (Promega). A serial dilution of human IFN-β was included as standards. Viral titers from the cell culture medium were determined by plaque-forming assays as previously described (Wang et al., 2008; Cheng et al., 2010). Briefly, virus-containing medium was serially diluted and then added to confluent Vero cells. Plaques were visualized with Neutral Red (Sigma-Aldrich) 3 days postinfection.
Statistical Analysis
Statistical significances were calculated with an unpaired two-tailed Student’s t test using Prism 5 software (GraphPad).
For methods on plasmid construction and site-directed mutagenesis, luciferase reporter assays, cell culture and transfection, quantitative RT-PCR, immunoblotting analyses, immunofluorescence microscopy, and FRET analysis, please refer to Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to Dr. John F. Anderson at The Connecticut Agricultural Experiment Station for providing West Nile virus. We thank Dr. Yingqun Huang for use of equipment. This work is supported by NIH grants AI055749, AI050031, AI067034, AI079348, HHSN272201100019C, and AI099625 and the Canadian Institutes of Health Research grant MOP42562. E.F. is Investigator of the Howard Hughes Medical Institute. G.C. and G.Y. made significant contributions to experimentation. L.Y., P.W., E.F., R.L., and R.E.S. conceived ideas, designed the experiments, and interpreted the results; P.W., L.Y., G.C., G.Y., Z.X., F.Y., and Q.S. conducted the experiments; L.Y., P.W., and R.E.S. wrote the manuscript.
Footnotes
Supplemental Information includes eight figures and Extended Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.02.027.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Zemirli N, Portier A, Garcin D, Bidère N, Vazquez A, Arnoult D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012;10:44. doi: 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E, Fry AN, Kang M. The complexities of p97 function in health and disease. Mol Biosyst. 2011;7:700–710. doi: 10.1039/c0mb00176g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Cox J, Wang P, Krishnan MN, Dai J, Qian F, Anderson JF, Fikrig E. A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell. 2010;142:714–725. doi: 10.1016/j.cell.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, Wang RF. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene. 2006;25:6844–6867. doi: 10.1038/sj.onc.1209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Ogawa S, Hashiguchi Y, Inoue Y, Udo H, Ohzono H, Kato A, Minakami R, Sugiyama H. A novel protein specifically interacting with Homer2 regulates ubiquitin-proteasome systems. J Biochem. 2005;137:617–623. doi: 10.1093/jb/mvi074. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Takeuchi O, Akira S. Pathogen recognition by innate receptors. J Infect Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- LaLonde DP, Bretscher A. The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J Biol Chem. 2011;286:4892–4901. doi: 10.1074/jbc.M110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Gu J. Retinoic acid inducible gene-I, more than a virus sensor. Protein Cell. 2011;2:351–357. doi: 10.1007/s13238-011-1045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Sanchez DJ, Cheng G. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol. 2011;23:57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, García-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- Matsumiya T, Stafforini DM. Function and regulation of retinoic acid-inducible gene-I. Crit Rev Immunol. 2010;30:489–513. doi: 10.1615/critrevimmunol.v30.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Knebel A, Arthur JS, Cuenda A, Cohen P. A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs. Biochem J. 2004;384:391–400. doi: 10.1042/BJ20041498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Tschopp J. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell. 2006;22:561–569. doi: 10.1016/j.molcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmer-mann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Yoneyama M, Fujita T. Retinoic acid-inducible gene-I-like receptors. J Interferon Cytokine Res. 2011;31:27–31. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, et al. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol. 2009;29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, Nguyen TL, Sun Q, Meurs EF, Lin R, Hiscott J. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 2011;21:895–910. doi: 10.1038/cr.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25:3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Wang P, Arjona A, Zhang Y, Sultana H, Dai J, Yang L, LeBlanc PM, Doiron K, Saleh M, Fikrig E. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat Immunol. 2010;11:912–919. doi: 10.1038/ni.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Dai J, Bai F, Kong KF, Wong SJ, Montgomery RR, Madri JA, Fikrig E. Matrix metalloproteinase 9 facilitates West Nile virus entry into the brain. J Virol. 2008;82:8978–8985. doi: 10.1128/JVI.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F, Ludwig T, Baer R. The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol Cell Biol. 2010;30:2787–2798. doi: 10.1128/MCB.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yeung HO, Kloppsteck P, Niwa H, Isaacson RL, Matthews S, Zhang X, Freemont PS. Insights into adaptor binding to the AAA protein p97. Biochem Soc Trans. 2008;36:62–67. doi: 10.1042/BST0360062. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol. 2010;184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.