Abstract
Purpose
Approximately 1/3 of the patients with advanced non-small cell lung cancer (NSCLC) will initially respond to platinum-based chemotherapy, but virtually all tumors will progress (acquired resistance). The remainder will progress during initial treatment (primary resistance). In this study, we test whether the treatment can be improved by inhibiting hepatoma-derived growth factor (HDGF).
Methods
Thirteen primary NSCLC hetero-transplant models were used to test four treatment regimens including platinum-based chemotherapy with and without bevacizumab (VEGF neutralizing antibody) or HDGF-H3 (HDGF neutralizing antibody) and chemotherapy with bevacizumab and HDGF-H3. Expression of stem-cell related genes was measured using quantitative RT-PCR and immunohistochemistry.
Results
Among 13 primary NSCLC hetero-transplant models, 3 (23%) responded to chemotherapy but all relapsed within 20 days. The residual tumors after response to the chemotherapy exhibited an increased expression in 51 (61%) of 84 genes related with stem-cell proliferation and maintenance, particularly those in Notch and Wnt pathways, suggesting an enrichment for stem-cell populations in the residual tumors. Interestingly, tumors from 2 of the 3 models treated with HDGF-H3, bevacizumab, and chemotherapy combination did not relapse during 6 months of post treatment observation. Importantly, this treatment combination substantially down-regulated expression levels in 57 (68%) of the 84 stem-cell related genes, including 34 (67%) of the 51 genes up-regulated after the chemotherapy.
Conclusion
These data support the hypothesis that cancer stem-cells (CSCs) are a mechanism for chemotherapy resistance and suggest HDGF may be a target for repressing CSCs to prevent relapse of NSCLC sensitive to chemotherapy.
Keywords: HDGF, antibody, NSCLC, cancer stem-cell, therapy
INTRODUCTION
Lung cancer is the leading cause of cancer mortality in the industrialized world (1). 80–90% of lung cancer is the non-small cell (NSCLC) type. The median survival of patients with advanced NSCLC in randomized trials is approximately 12 months with very few survivors beyond three years (2, 3).
Hepatoma-derived growth factor (HDGF) is a heparin-binding growth factor and has been implicated in angiogenesis (4–6). Although the mechanisms how HDGF functions at the molecular level, its binding to a candidate cell surface receptor as well as its re-entering cells are thought critical for its cellular functions (7–9). We have shown that HDGF is overexpressed in NSCLC and higher expression is strongly associated with poorer clinical outcomes (10). We further demonstrated that HDGF promotes tumorigenicity of NSCLC cells and targeting HDGF inhibits growth of these cancer cells both in vitro and in vivo (11). To test HDGF as a therapeutic target, we developed monoclonal antibodies against HDGF and evaluated them in NSCLC xenograft models and selected HDGF-H3 for further evaluation (12).
In this study, we utilized a panel of NSCLC hetero-transplant models developed directly from primary NSCLC to test a potential impact of inhibiting HDGF in treating NSCLC using a treatment design similar to randomized phase II clinical trials.
MATERIALS AND METHODS
NSCLC hetero-transplant tumor models
Primary NSCLC tissues from patients underwent surgical resection were obtained from Departments of Pathology, the University of Texas M. D. Anderson Cancer Center and the University of Maryland School of Medicine according to protocols approved by the Institutional Review Boards. Fresh tumor tissues were cut into pieces of 1–2 mm3 in sterile culture medium. Three to four pieces of the tissues were inoculated into the lower back and anterior chest of 6–8 weeks age female Nu/Nu mice according to the protocols approved by Institutional Animal Care and Use Committees. Two to three mice were used for each tumor. Tumors grown and reached at least 10 mm in diameter were considered established. Each hetero-transplant tumor model established was confirmed by pathology examination. Among the 38 primary NSCLC tissues, 17 hetero-transplant tumor models were established (45% take rate). Tumors can be re-established in nude mice in a reasonable time frame to allow drug testing in 13 of the 17 models. The 3rd or 4th generations of the 13 models were selected for this study (Table 1). The tumor generation is defined as the number of passages in animals starting from the implant of the primary tumors directly obtained from patients.
Table 1.
clinical and pathological characteristics of the hetero-transplant models
| Model | Age(year) | Sex | Histology | Differentiation | Stage | Prior Treatment | Response |
|---|---|---|---|---|---|---|---|
| MDA2131-1 | 76 | M | Adeno | Moderate | IA | None | NA |
| MDA2131-4 | 65 | F | Squamous | Moderate | IIA | 3 cycles: Cisplatin/docetaxel | PR |
| MDA2131-5 | 78 | F | Pleomorphic | Poor | IV (Adrenal metastasis) | None | NA |
| MDA2131-7 | 55 | M | Adeno | Poor | IIIA | None | NA |
| MDA2131-8 | 84 | M | Adeno | Poor | IIA | None | NA |
| MDA2131-11 | 65 | F | Squamous | Poor | IIIA | 3 cycles: docetaxel/carboplatin | SD |
| MDA2131-15 | 64 | M | Squamous | Poor | IIA | None | NA |
| MDA2131-19 | 77 | F | Adeno | Poor | IA | None | NA |
| MDA513 | 77 | M | Squamous | Poor | IB | 1 cycle: cisplatin/docetaxel | SD |
| MDA889 | 75 | M | deno | Poor | IV (Brain metastasis) | None | NA |
| MDA132 | 66 | M | Adeno | Moderate | IIIA | 2 cycles: Cisplatine/docetaxel | SD |
| UMB410 | NA | NA | Squamous | Poor | IIIA | None | NA |
| UMB710 | NA | NA | Squamous | Poor | IA | None | NA |
AD: Adenocarcinoma; SQ: Squamous cell carcinoma; M: Male; F: Female; PR: Partial response; SD: Stable disease; NA: Not available
Treatment and assessment of treatment responses
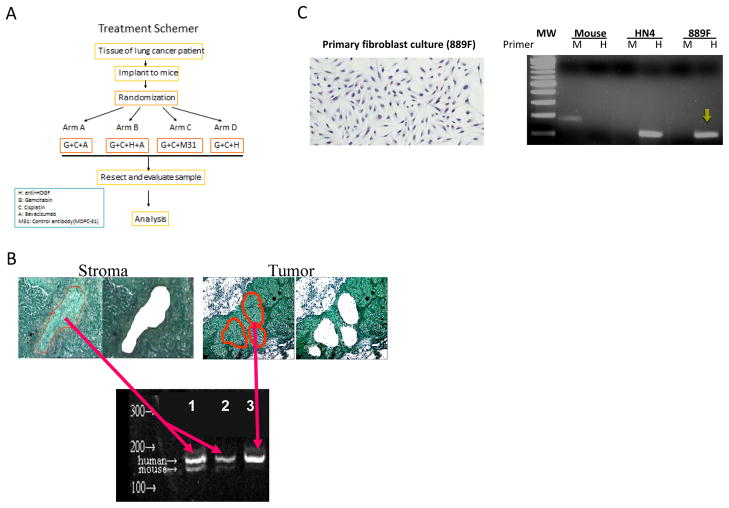
For each tumor model, 5 mice were inoculated with tumor pieces (3–4 pieces each mouse) subcutaneously under the lower back. Tumors were allowed to grow to at least 10 mm in diameter before being randomly selected for one of the four treatment arms: Arm A, cisplatin (CDDP) + Gemcitabine + Bevacizumab + control monoclonal antibody (M31); Arm B, CDDP + Gemcitabine + Bevacizumab + Anti-HDGF monoclonal antibody (HDGF-H3); Arm C, CDDP + Gemcitabine + M31; and Arm D, CDDP + Gemcitabine + HDGF-H3 (Fig. 1A). All the drugs were given intra-peritoneal. CDDP was given weekly at 1mg/kg based on previous report (13) and the dosing toxicity test; gemcitabine, weekly at 125mg/kg; bevacizumab, twice weekly at 5mg/kg; HDGF-H3, twice weekly at 10mg/kg; M31, twice weekly at 10mg/kg. The tumors in the fifth mouse from each model were used as treatment naïve controls in molecular analyses. Four cycles of treatment were given to each animal with each cycle last for one week. Tumor sizes were measured twice a week and the tumor volumes were calculated using the following formula: length (mm) × width (mm) × height (mm) = mm3. RECIST (Response Evaluation Criteria in Solid Tumors) criteria (14) were used to evaluate tumor responses after four cycles of treatment. Animals were weighted twice a week during treatment. The weight loss during chemotherapy was not detectable in some tumor models but observed in other models. The loss was generally less than 20% and no significant differences were observed among the four treatment arms. For the tumor models responded to chemotherapy and other combinations, the experiments were repeated with 2 animals for each treatment arm in order to verify earlier observations and to obtain sufficient tumor tissues for molecular analyses. One of the animals in each arm was scarified at the end of treatment (28 days) to obtain tumor tissues for molecular analyses whereas other animals were further observed to verify early observations. For molecular analyses, resected tumors were divided into two parts, one fresh frozen and the other formalin fixed. These tissues were subsequently used for hematoxylin and eosin (H&E) staining, immunohistochemistry staining, and DNA/RNA extraction.
Figure 1.
NSCLC models used for the experiments. A, treatment schema where the strategy and the agents used in each of the 4 treatment arms are outlined. B, tissue sections from the third generation of MDA2131-8 showing before and after stroma and tumor nests were micro-dissected by Arctures® LCM system. DNA from the dissected cells was amplified using PCR primers which amplifies both human (153-bp) and mouse (144-bp) HDGF gene sequences. The 9-bp difference in sizes allows distinction between mouse and human origins. Only human sequences were detected when DNA extracted from dissected tumor cells was used as a PCR template whereas both human and mouse sequences were detected with predominantly human sequences when DNA extracted from dissected stromal cells was used as the PCR template (2 different amounts of the PCR product were loaded in the lanes 2 and 3. C, Fibroblasts were cultures from a third generation heterotransplant tumor (MDA889) termed as 889F (left panel). A human-specific primer set to amplify a 122-bp fragment of human chromosome 7p15 region and a mouse-specific primer set to amplify a181-bp DNA fragment inserted into our transgenic mouse (3B4DG). The right panel shows PCR products from DNA extracted from tissues of 3B4DG (transgentic mouse), HN4 (a human oral cancer cell line), and 889F. MW, molecular weight marker; M, mouse-specific primer set; H, human-specific primer set. The yellow arrow indicates the amplified human DNA fragment from 889F.
Immunohistochemistry staining
Part of the resected tumor tissues were fixed in formalin for 24 h and then embedded in paraffin. For immunohistochemistry staining, the formalin-fixed and paraffin-embedded tissues were sectioned at 5 μM thickness sections and placed on glass pathology slides. The slides were de-paraffinized and rehydrated in graded concentrations of alcohol using standard techniques. The slides were incubated with 1:50 anti-cleaved Notch1 (Val1744) (D3B8) Rabbit mAb (Cell Signaling) for 24 h. The slides were then incubated with biotinylated secondary antibody (sheep anti-rabbit) in PBS with 0.05% tween20 for 30 min at room temperature followed by sequentially in ABC-Peroxidase Solution for 30 min, substrate-chromagen solution (DAKO Liquid DAB+ Substrate Chromogen System) for 5 min. Cells with nuclear staining were considered positive of Notch intracellular domain (NICD) expression. For HDGF, the slides were blocked with 5% normal horse serum and Mouse-on Mouse Blocking Reagent (M.O.M, Vector Laboratories, Burlingame, CA) for 30 min followed by incubating with anti-HDGF H3 mouse monoclonal antibody (1:500) at 4 °C overnight. After three washes with PBS containing 0.05% tween-20, the slides were incubated with biotinylated horse anti-mouse IgG (1:200, Vector Laboratories) for 30 min. The expression was detected using Vectastain Elite ABC Reagent (Vector Laboratories) and DAB reagent (Vector Laboratories) according to manufactures instruction.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) Assay
Slides were deparaffinized and rehydrated in graded concentrations of alcohol and then incubated with proteinase K for 15 min. Each slide was incubated with 200μl reaction mixture (Terminal deoxynucleotidyl Transferase, TdT, and biotin dUTP) for 1 h at room temperature, followed by TB buffer for 15 min and then washed with ddH2O. The slides were then incubated with 2% BSA for 10 min at room temperature followed by incubation in ABC-Peroxidase Solution for 30 min, substrate-chromagen solution (DAKO Liquid DAB + Substrate Chromogen System) for 5 min, methyl green staining for 2 min, and dehydration. TUNEL positive cells were accounted in the tissue sections.
DNA/RNA extraction
Frozen tissues were lysed and homogenized in a highly denaturing guanidine isothiocyanate–containing buffer. The lysate was then passed through an AllPrep DNA spin column. This column, in combination with the high-salt buffer, allows selective and efficient binding of genomic DNA. The column was washed and pure, ready-to-use DNA was then eluted with water. Ethanol was then added to the flow-through from the AllPrep DNA spin column to provide appropriate binding conditions for RNA, and the sample was then applied to an RNeasy spin column, where total RNA binds to the membrane and contaminants were efficiently washed away. High-quality RNA was then eluted in 30 μl water.
Expression analysis of stem-cell related genes
Approximately 120ng total RNA was used for cDNA synthesis using the RT2 First Strand kit (SA Biosciences, Frederick, MD, USA) following the manufacturer’s instructions. Gene expression was quantitatively analyzed using Human Stem Cell RT2 Profiler PCR Array and RT2 SYBR Green Master Mix (SA Biosciences, Frederick, MD, USA) per the manufacturer’s instructions. PCR was performed on ABI 7900HT Fast Real-time PCR system (Applied Biosystems). For data analysis, ΔΔCt values were calculated online at the manufacturer’s Web site (http://www.sabiosciences.com/pcr/arrayanalysis.php). Fold-changes were calculated as the differences in gene expression levels before and after treatment in each model and treatment arm. A positive value indicates gene up-regulation and a negative value indicates gene down-regulation. Genes that showed >2-fold differences in the expression levels were highlighted in the data presentation.
Primary culture of tumor-derived fibroblasts
Tissues for primary culture of fibroblasts were obtained from a lung cancer hetero-transplant model. These specimens were dissected with scissors into small pieces (after washing with PBS) and transferred into flasks containing collagenase IV (200U/ml) in DMEM with 10% FBS for 6 h followed by exhaustively rinsed (8 times) with 40ml of PBS under vigorous agitation. After disaggregation, the suspension of cells was washed by settling and then centrifuged. Cells were then put into a flask with DMEM for 15 min, aspirate the suspension for cells. The fibroblasts were cultured in DMEM and used for experiments.
Micro-dissection of individual cells
Ten micron tissue sections were mounted on glass slides. Tumor nests and stromal parts were micro-dissected using Arcturus® Laser Capture Micro-dissection (LCM), respectively. The isolated cells were put into 1.5ml plastic tubes for DNA extraction.
DNA amplification and PCR product visualization
DNA from micro-dissected tissues was amplified by PCR using the primer set: 5′-gaggtggaaaagaatagcaccc-3′ (forward) and 5′-ctacaggctctcatgatctctgat-3′ (reverse) to amplify a fragment in HDGF gene. Human DNA would result in a 153-bp PCR fragment whereas mouse DNA would result in a 144-bp fragment. For tumor-derived fibroblasts and mouse tissues, DNA was amplified by PCR using either Human (Hu)-specific primers (forward: 5′-ctgttttgtggcttgttcag-3′ and reverse: 5′-aggaaaccttccctcctcta-3′) to amplify a 122-bp fragment of a region located in 7p15–p12 or mouse (Mo)-specific primers (forward: 5′-ttggttgagaagcagaaaca-3′ and reverse: 5′-cacacagtcaagttcccaaa-3′) to amplify a 181-bp fragment of a region located in B2m gene. Amplified PCR products were separated in 2% agarose gels and visualized using ethidium bromide.
RESULTS
Rodent-based randomized phase II treatment trial
To recapitulate treatment strategies used in treating patients with advanced NSCLC in animal models, we selected the commonly utilized regimen of cisplatin plus gemcitabine in treating patients with advanced NSCLC. We also tested this regimen with bevacizumab based upon data demonstrating prolonged progression free survival (3). We included HDGF-H3 in each of the regimens to determine a role of targeting HDGF in treating NSCLC (Fig. 1A). Thirteen NSCLC hetero-transplant models (15) which are derived directly from primary NSCLC without in vitro passage (Table 1) were evaluated. For each model, 5 animals were implanted with 3rd or 4th generation hetero-transplant tumors with an average tumor size of 448mm3 at the time treatment started. The large size of the tumors mimics the actual presentation of patients with advanced NSCLC. Four of the 5 animals were then randomly selected to be treated with one of the four treatment arms (Fig. 1A) with 13 animals in each arm. Together with repeated experiments in two of the models, a total of 62 animals were used in the animal trial. The identical genetic background of all the animals minimizes the impact of host factors. Additionally, each treatment arm utilized the same tumor panel, which minimizes the impact of tumor heterogeneity. Together, this experimental strategy recapitulates an initial biomarker-integrated randomized phase II human therapeutic trial.
NSCLC hetero-transplants contain human-origin stroma
Tumor stroma is increasingly recognized as an important component of tumor biology, contributing to the malignant phenotypes and treatment resistance (16, 17). In the NSCLC hetero-transplant tumors which reflect properties of their clinical counterparts (18, 19), stromal cells of human origin were observed in the third and fourth generation tumors using assays which are sensitive and specific to differentiate the genetic origins of these cells (Fig. 1B and 1C).
Responses to treatment regimens
Tumor response was assessed utilizing the response criteria in solid tumors (RECIST) after 4 treatment cycles with each cycle last for one week. The chemotherapy only arm (Arm C) resulted in 3 partial responses (PR), 6 stable diseases (SD), and 3 progressive diseases (PD) (Table 2) for a 23% response rate (RR). This degree of response is comparable to previous reports using similar models and clinical trials (18, 20). We analyzed HDGF expression levels in the tumor models and found no direct relationship between the expression levels and tumor responses (Table 2). The 3 models with PR also showed PR or complete response (CR) in other treatment arms (Table 2). Consistent responses were also observed in other tumor models with SD and PD with limited variation (Table 2).
Table 2.
Responses to treatment in the hetero-transplant models
| Model | HDGF level | Arm A | Arm B | Arm C | Arm D |
|---|---|---|---|---|---|
| MDA2131-1 | +++ | PD | SD | SD | PD |
| MDA2131-4 | + | SD | PR | SD | SD |
| MDA2131-5 | +++ | PR | PR | PR | PR |
| MDA2131-7 | +++ | SD | SD | SD | SD |
| MDA2131-8 | ++ | CR | CR | PR | PR |
| MDA2131-11 | + | SD | PD | PD | PD |
| MDA2131-15 | ++ | PD | SD | SD | SD |
| MDA2131-19 | + | PD | SD | SD | SD |
| MDA513 | +++ | SD | SD | SD | SD |
| MDA889 | + | PD | SD | PD | PD |
| MDA132 | ++ | SD | PD | PD | PD |
| UMB410 | +++ | PD | PD | PD | PD |
| UMB710 | +++ | PR | PR | PR | PR |
Impact of chemotherapy to expression of stem-cell related genes
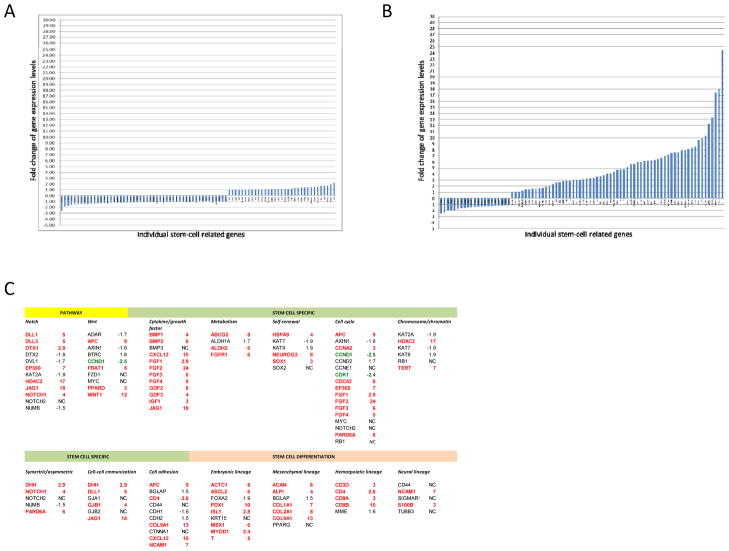
Expression patterns of 84 genes representing putative stem-cell specific markers, stem-cell differentiation markers, and stem-cell maintenance signaling pathways were quantitatively analyzed in 6 tumor models, 3 (MDA2131-1, MDA889, and UMB410) with primary chemotherapy resistance and 3 (MDA2131-5, MDA2131-8, and UMB710) with acquired resistance, before and after chemotherapy. There was minimal change in the expression patterns of these 84 genes in the tumor models with primary resistance (Fig. 2A). Only 1 of the genes was up-regulated >2-fold (TERT, 2.2-fold) and 1 down-regulated (FGFR1, -2.6-fold). In contrast, 51 (61%) of the 84 genes were up-regulated >2-fold in the tumor models with acquired resistance but only 4 (5%) of the genes down-regulated (Fig. 2B). As shown in Figure 2C, the up-regulated genes include 7 of the 12 critical for Notch pathway, 4 of the 10 for Wnt pathway as well as stem-cell specific molecules including 11 of the 12 cytokines/growth factors, 3 of the 4 metabolic markers, 3 of the 6 self-renewal markers, 9 of the 17 cell-cycle regulators, 2 of the 6 chromosome/chromatin modulator, 3 of the 5 symmetric regulators, 4 of the 6 cell-cell communication regulators, and 5 of the 10 cell adhesion regulators. Up-regulated genes also included 5 of the 7 mesenchemal lineage markers, 7 of the 9 embryonic lineage markers, and 4 of the 5 hematopoietic lineage markers, and 2 of the 5 neural lineage markers (Fig. 2C).
Figure 2.
Responses of the NSCLC models to a combination of chemotherapy. A, Fold changes of the 84 stem-cell related genes quantified using real-time RT-PCR after treated with chemotherapy (Arm C) in the tumor models not responding to the treatment. The genes are arranged based on fold changes from the most down-regulated to the most up-regulated from left to right horizontally. B, Fold changes of the 84 stem-cell related genes after treated with chemotherapy (Arm C) in the tumor models (MDA2131-5, MDA2131-8, and UMB710) significantly responded to the treatment. The genes are arranged based on fold changes from the most down-regulated to the most up-regulated from left to right horizontally. C, List of the 84 stem-cell related genes arranged by pathways or functionalities involved (some of the genes involved in multiple pathways or functionalities). The data is based on results exhibited in Fig. 2B. Gene names in red for those up-regulated (>2 fold) and in green for those down-regulated (fold changes are rounded to closest full digits except those <3 fold).
HDGF-H3 and NSCLC relapse
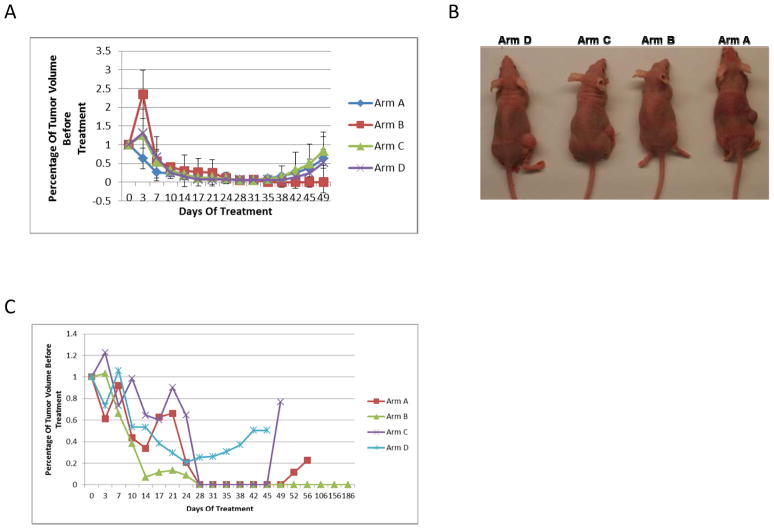
To test whether an anti-HDGF based combination can disrupt tumor microenvironment and inhibit cancer-associated stromal stem cells to result in preventing or delaying NSCLC relapse, we extended the observation time of the 3 chemotherapy sensitive tumor models. Ten to 20 days after treatment completion, all but 2 (MDA2131-5 and MDA2131-8) tumors in Arm B (HDFD-H3, bevacizumab, and chemotherapy) relapsed (Fig. 3A–C), the experiment was repeated once for MDA2131-5 tumor model with similar results (Fig. 3A). In the MDA2131-5 case, the residual tumor in the treatment arm completely regressed one week after the completion of treatment (Fig. 3A). The mice remained relapse-free for >6 months (at which time they were sacrificed) demonstrating a role for the combination in preventing or delaying relapse of the tumors.
Figure 3.
A, tumor growth curves of model MDA2131-5 (third generation including results from the repeated experiment) treated in four arms. B, mice one month after treatment stopped. C, Tumor growth curves of model MDA2131-8 (fourth generation) treated in four arms.
HDGF-H3 and expression of stem-cell related genes
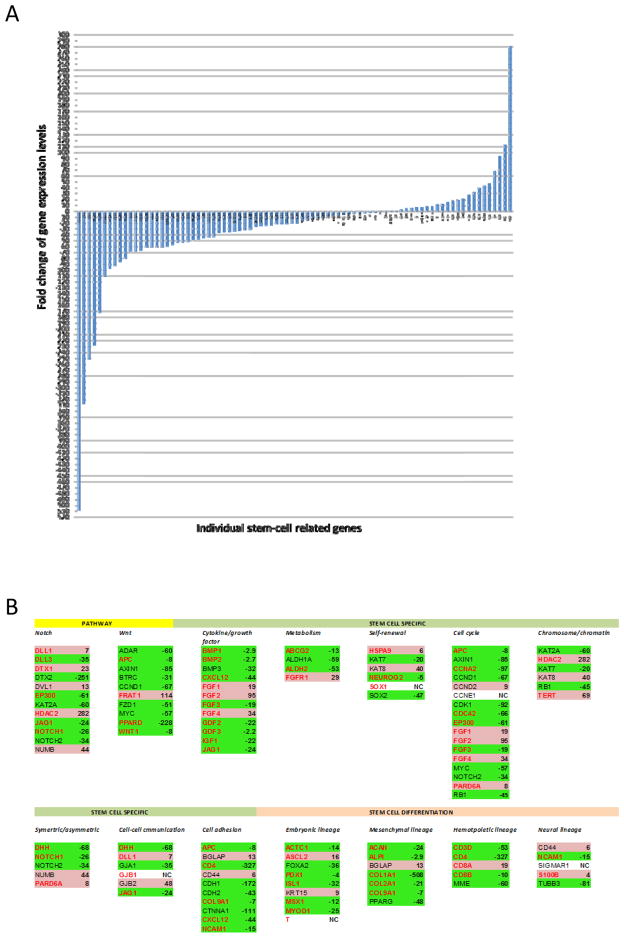
To determine whether this effect is due to inhibition of stem-cells enriched by chemotherapy, we compared expression patterns of the 84 stem-cell gene panel using real-time PCR array system (http://www.sabiosciences.com/rt_pcr_product/HTML/PAHS-405Z.html) in tumors after each treatment regimen with the treatment naïve tumors in the 2 models (MDA2131-5 and MDA2131-8). In Arm B, 57 (68%) of the genes, including 34(67%) of the 51 up-regulated in the chemotherapy arm (Arm C), were down-regulated >2-fold (Fig. 4A and 4B). There were 22 genes, including 14 up-regulated by chemotherapy, were up-regulated >2-fold (Fig. 4A and 4B). In contrast, only modest impacts were observed when either bevacizumab (Arm A) or HDGF-H3 (Arm D) was added to chemotherapy (Supplemental Fig. 1A and 1B).
Figure 4.
A, Fold changes of the 84 stem-cell related genes after treated with HDGF-H3 + bevacizumab + chemotherapy (Arm B) in the tumor models MDA2131-5 and MDA2131-8. The genes are arranged based on fold changes from the most down-regulated to the most up-regulated from left to right horizontally. B, List of the 84 stem-cell related genes arranged by pathways or functionalities involved in a format as presented in Fig. 1F. The fold changes are based on the data presented in Fig. 4A. Gene names with green highlight are those down-regulated in the treatment arm whereas those with pink highlight are those up-regulated. Gene names in red are those up-regulated in the tumor models sensitive in Arm C.
To verify the inhibition of Notch in tumors from Arm B, we measured Notch intracellular domain (NICD) using immunohistochemistry and found that NICD was highly expressed in nucleus of tumor cells in treatment naïve tumors but undetectable in the residual tumor cells after treatment combinations containing HDGF-H3 (Supplemental Fig. 2A). We further evaluated Wnt signaling molecules because the pathway is important in embryonic stem cell self-renewal (21) and interaction between cancer stem-cells (CSCs) and stromal stem-cells (22, 23). In the residual tumors, 9 of the 10 Wnt pathway genes were significantly down-regulated (Fig. 4B) with 8- and 51-fold reduction of Wnt1 and Frizzled (the key receptor of Wnt), supporting the impairment of Wnt signaling.
HDGF-H3 and apoptosis of tumor stromal cells
To determine whether the modulation of stem-cell related genes is in part due to the death of tumor-associated stromal cells, we used the TUNEL assay to measure apoptotic cells and observed substantially increased TUNEL positive cells in the Arm B treated tumors, predominantly with morphology of stromal cells (Supplemental Fig. 2B).
DISCUSSION
In this study, we used human NSCLC hetero-transplant models in a therapeutic testing similar to a randomized phase II clinical trial. Because the tumors derived directly from primary tumors, the models avoid in vitro adaption and therefore better reflect in vivo tumor conditions. It has been well-recognized that the morphologies of the tumors from heterotransplant models mimic their primary tumor counterparts. Here, we provide evidence indicating the presence of human stromal cells in these tumors. It is well-know that stromal cells in cancer tissues play critical roles in supporting and promoting cancer initiation and progression (24, 25). Somatic mutations of tumor suppressor genes and epigenetic alterations commonly seen in cancer cells can be identified in the stroma of breast cancers (26, 27). Therefore, it is possible that some of the stroma cells in NSCLC are immortal and possess stem cell features. These cells may co-proliferate together with cancer cells in these tumor models and become a critical tumor component for assessing treatments targeting cancer microenvironment. The identical genetic background of the host animals and the tumor source of each model helped the uniform treatment responses observed in the study (Table 2).
CSC hypothesis implies that resistance to treatment occurs as a consequence of the presence of a small number of chemotherapy resistant self-renewing tumor cells, which are primarily based on observation in cell-based models (28–31). Recently, the hypothesis has been validated in several animal models (32–34) but has not been confirmed in any human cancers. Here, the hypothesis was evaluated by examining expression patterns of 84 genes representing putative stem-cell specific markers, stem-cell differentiation markers, and stem-cell maintenance signaling pathways in tumors sensitive and resistant to chemotherapy. Compared to the treatment naïve tumors, the residual tumors from chemo-sensitive models showed a substantial up-regulation of stem-cell related genes, suggesting an enrichment of stem-cell populations, but virtually no expression change of these genes in the chemo-resistant tumors. The data support the hypothesis that the emergence of a CSC population is associated with acquired chemotherapy resistance. However, it is also possible that these cells are resistant before chemotherapy stimulation and are simply enriched when the sensitive cells are eliminated during the treatment course. Notch signaling activation is critical in lung carcinogenesis and stem-cell renewal (35–37). The up-regulation of multiple Notch ligands (DLL1, DLL3, and JAG1), Notch itself, and transcriptional co-activators (EP300 and DTX1) suggests that the Notch pathway is activated in the residual tumor tissues.
We noticed predominant mesenchymal and hematopoietic features in the residual tumor tissues after responses to chemotherapy, suggesting an enrichment of cancer-related mesenchymal and hematopoietic stem cells, a finding consistent with the increased tumor stroma observed in NSCLC patients after neoadjuvant chemotherapy (38). Consistent with this notion, almost all the cytokines measured were up-regulated in these tissues. Together with the evidence supporting the critical role of cancer-associated fibroblasts in tumorigenesis and progression (39, 40), these data suggest that the stromal stem-cells may facilitate cancer relapse. The results are also consistent with a recent report demonstrating the existence of distinct types of CSCs in a colon cancer model (41). Thus, it is possible that the anti-HDGF based treatment eliminated stem-cells capable of regenerating whereas the remaining “stem-cells” had limited or no self-renewal capability. Additionally, we observed significantly reduced expression levels of genes encoding cytokines, cell adhesion molecules, and stem-cell differentiation markers. The striking down-regulation of COL1A1 (down >500-fold) (Fig. 4B), together with the down-regulation of COL2A1 and COL9A1, may impact collagen structures and result in a loss of extracellular support for proliferation and differentiation of CSCs.
It should be noted that an increased rate of hemoptysis was observed in patients with lung squamous cell carcinoma (LSCC) treated with bevacizumab in combination with chemotherapeutic agents (2), which limits the use of bevacizumab only for patients with non-squamous type NSCLC. In our earlier study (12), we showed that certain chemotherapeutic agent, such as gemcitabine, could substantially enlarge the vessel size in an experimental NSCLC model. Although bevacizumab could reduce the vessel number but not the vessel size in the model system. In contrast, the anti-HDGF antibody substantially reduced the vessel size but not the vessel number. When bevacizumab was used together with the anti-HDGF antibody, both the vessel size and numbers were substantially reduced. Therefore, combining anti-VEGF and anti-HDGF agents may reduce the possibility of hemoptysis seen in patients with LSCC treated with bevacizumab. More experimental and clinical studies will be needed to test this potential.
In summary, our data provide evidence supporting the CSC hypothesis and may have potential clinical implications. We show that chemotherapy sensitive human lung cancer explants have enriched stem-cell-like populations after chemotherapy. Importantly, a regimen combining chemotherapy/bevacizumab with an anti-HDGF antibody might have impaired the stem-cell populations and prevented relapse. It is possible that this effect is due to a blockade of signaling and other factors required for the proliferation and/or maintenance of CSCs and cancer-related stem cells. Thus, our study suggests that simultaneously targeting CSCs and stromal stem cells in chemo-sensitive NSCLC may prevent relapse and prolong patient survival.
Supplementary Material
Translational Relevance.
Patients with advanced non-small cell lung cancer (NSCLC) are not curable by current treatment strategies. A major obstacle to this poor outcome is due to treatment resistance. In this study, we used NSCLC hetero-transplant models which resemble human primary tumors to test a novel target, hepatoma-derived growth factor (HDGF). We tested platinum-based chemotherapeutic regimens with and without bevacizumab or HDGF-H3 (HDGF neutralizing antibody) and chemotherapy with bevacizumab and HDGF-H3. The control regimens are part of standard treatment for patients with advanced NSCLC and the testing design is similar to a randomized phase II trial. The findings that the anti-HDGF based regimen may prevent tumor recurrence and such anti-tumor effort may be due to inhibition of cancer stem cell features have potentially significant clinical implications in preventing relapse of NSCLC that is initially sensitive to chemotherapy and developing novel strategies to target cancer-related stem cells.
Acknowledgments
Supported by grants from the National Cancer Institute R01 CA126818 and P30 CA134274.
We are grateful to the individuals who donated tissue toward this study.
Footnotes
Prior presentations: None
Disclaimers: None
Conflict of Interest: None
References
- 1.Siegel R, Naishadham D, Jermal A. Cancer Statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–50. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 3.Reck M, von Pawel J, Zatloukal P, Zatloukal P, Ramlau R, Gorbounova V, et al. Overall survival with cisplatin-gemcitabine and bevacizumab or placebo as first-line therapy for nonsquamous non-small-cell lung cancer: results from a randomised phase III trial (AVAiL) Ann Oncol. 2010;21:1804–9. doi: 10.1093/annonc/mdq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everett AD, Stoops T, McNamara CA. Nuclear targeting is required for hepatoma-derived growth factor-stimulated mitogenesis in vascular smooth muscle cells. J Biol Chem. 2001;276:37564–8. doi: 10.1074/jbc.M105109200. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura H, Izumoto Y, Kambe H, Kuroda T, Tori T, Kawamura K, et al. Molecular cloning of complementary DNA for a novel human hepatoma-derived growth factor. Its homology with high mobility group-1 protein. J Biol Chem. 1994;269:25143–9. [PubMed] [Google Scholar]
- 6.Lepourcelet M, Tou L, Cai L, Sawada J, Lazar AJ, Glickman JN, et al. Insights into developmental mechanisms and cancers in the mammalian intestine derived from serial analysis ofg ene expression and study of the hepatoma-derived growth factor (HDGF) Development. 2005;132:415–27. doi: 10.1242/dev.01579. [DOI] [PubMed] [Google Scholar]
- 7.Abouzied MM, El-Tahir HM, Prenner L, Haberlein H, Gieselmann V, Franken S. Hepatoma-derived growth factor. Significance of amino acid residues 81–100 in cell surface interaction and proliferative activity. J Biol Chem. 2005;280:10945–54. doi: 10.1074/jbc.M414652200. [DOI] [PubMed] [Google Scholar]
- 8.Okuda Y, Nakamura H, Yoshida K, Enomoto H, Uyama H, Hirotani T, et al. Hepatoma-derived growth factor induces tumorigenesis in vivo through both direct angiogenic activity and induction of vascular endothelial growth factor. Cancer Sci. 2003;94:1034–41. doi: 10.1111/j.1349-7006.2003.tb01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett AD, Narron JV, Stoops T, Nakamura H, Tucker A. Hepatoma derived growth factor is a pulmonary endothelial cell-expressed angiogenic factor. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1194–201. doi: 10.1152/ajplung.00427.2003. [DOI] [PubMed] [Google Scholar]
- 10.Ren H, Tang X, Lee JJ, Feng L, Everett AD, Hong WK, et al. Expression of hepatoma-derived growth factor is a strong prognostic predictor for patients with early-stage non-small-cell lung cancer. J Clin Oncol. 2004;22:3230–7. doi: 10.1200/JCO.2004.02.080. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Ren H, Yuan P, Lang W, Zhang L, Mao L. Down-regulation of hepatoma-derived growth factor inhibits anchorage-independent growth and invasion of non-small cell lung cancer cells. Cancer Res. 2006;66:18–23. doi: 10.1158/0008-5472.CAN-04-3905. [DOI] [PubMed] [Google Scholar]
- 12.Ren H, Chu Z, Mao L. Antibodies targeting hepatoma-derived growth factor as a novel strategy in treating lung cancer. Mol Cancer Ther. 2009;8:1106–12. doi: 10.1158/1535-7163.MCT-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Judde JG, Rebucci M, Vogt N, de Gremoux P, Livartowski A, Chapelier A, et al. Gefitinib and chemotherapy combination studies in five novel human non small cell lung cancer xenografts. Evidence linking EGFR signaling to gefitinib antitumor response. Int J Cancer. 2007;120:1579–90. doi: 10.1002/ijc.22364. [DOI] [PubMed] [Google Scholar]
- 14.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 15.Pathak AK, Bhutani M, Saintigny P, Mao L. Heterotransplant mouse model cohorts of human malignancies: a novel platform for systematic preclinical efficacy evaluation of drugs (SPEED) Am J Transl Res. 2009;1:16–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Davis A, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Soler R, Kemp B, Wu QP, Mao L, Gomez J, Zeleniuch-Jacquotte A, et al. Response and determinants of sensitivity to paclitaxel in human non-small cell lung cancer tumors heterotransplanted in nude mice. Clin Cancer Res. 2000;6:4932–8. [PubMed] [Google Scholar]
- 19.John T, Kohler D, Pintilie M, Pham NA, Li M, Panchal D, et al. The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2011;17:134–41. doi: 10.1158/1078-0432.CCR-10-2224. [DOI] [PubMed] [Google Scholar]
- 20.Goffin J, Lacchetti C, Ellis PM, Ung YC, Evans WK. First-line systemic chemotherapy in the treatment of advanced non-small cell lung cancer: a systematic review. J Thorac Oncol. 2010;5:260–74. doi: 10.1097/JTO.0b013e3181c6f035. [DOI] [PubMed] [Google Scholar]
- 21.Miki T, Yasuda SY, Kahn M. Wnt/β-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Rev. 2011;7:836–46. doi: 10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- 22.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delalove JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–9. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 23.Shin K, Lee J, Guo N, Kin J, Lim A, Qu L, et al. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature. 2011;472:110–4. doi: 10.1038/nature09851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maffini MV, Soto AM, Calabro JM, Ucci AA, Sonnenschein C. The stroma as a crucial target in rat mammary gland carcinogenesis. J Cell Sci. 2004;117:1495–1502. doi: 10.1242/jcs.01000. [DOI] [PubMed] [Google Scholar]
- 25.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 26.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32:355–7. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 27.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 28.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–9. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 30.Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–45. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 32.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–30. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 35.Galluzzo P, Bocchetta M. Notch signaling in lung cancer. Expert Rev Anticancer Ther. 2011;11:533–40. doi: 10.1586/era.10.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García Campelo MR, Alonso Curbera G, Aparicio Gallego G, Grande Pulido E, Antón Aparicio LM. Stem cell and lung cancer development: blaming the Wnt, Hh and Notch signalling pathway. Clin Transl Oncol. 2011;13:77–83. doi: 10.1007/s12094-011-0622-0. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 38.Liu-Jarin X, Stoopler MB, Raftopoulos H, Ginsburg M, Gorenstein L, Borczuk AC. Histologic assessment of non-small cell lung carcinoma after neoadjuvant therapy. Mod Pathol. 2003;16:1102–8. doi: 10.1097/01.MP.0000096041.13859.AB. [DOI] [PubMed] [Google Scholar]
- 39.Brennen WN, Isaacs JT, Denmeade SR. Rationale behind targeting fibroblast activation protein-expressing carcinoma-associated fibroblasts as a novel chemotherapeutic strategy. Mol Cancer Ther. 2012;11:257–66. doi: 10.1158/1535-7163.MCT-11-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura K, Semba S, Aoyagi K, Sasaki H, Yokozaki H. Mesenchymal stem cells provide an advantageous tumor microenvironment for the restoration of cancer stem cells. Pathobiology. 2012;79:290–306. doi: 10.1159/000337296. [DOI] [PubMed] [Google Scholar]
- 41.Dieter SM, Ball CR, Hoffmann CM, Nowrouzi A, Herbst F, Zavidij O, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–65. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.