Abstract
Innate immune gene repertoires are restricted primarily to germline variation. Adaptive immunity, by comparison, relies on somatic variation of germline-encoded genes to generate extraordinarily large numbers of non-heritable antigen recognition motifs. Invertebrates lack the key features of vertebrate adaptive immunity, but have evolved a variety of alternative mechanisms to successfully protect the integrity of “self”; in many cases, these appear to be taxon-specific innovations. In the protochordate Branchiostoma floridae (amphioxus), the variable region-containing chitin-binding proteins (VCBPs) constitute a multigene family (comprised of VCBPs 1–5), which possesses features that are consistent with innate immune-type function. A large number of VCBP alleles and haplotypes are shown to exhibit levels of polymorphism exceeding the elevated overall levels determined for the whole amphioxus genome (JGI). VCBP genes of the 2 and 5 types are distinguished further by a highly polymorphic segment (exon 2) in the N-terminal immunoglobulin domain, defined previously as a “hyper-variable region” or a “hotspot.” Genomic deoxyribonucleic acid (DNA) and complementary DNA (cDNA) sequences from large numbers of animals representing different populations reveal further significant differences in sequence complexity within and across VCBP2/5 haplotypes that arise through overlapping mechanisms of genetic exchange, gene copy number variation as well as mutation and give rise to distinct allelic lineages. The collective observations suggest that mechanisms were in place at the time of divergence of the cephalochordates that could selectively hyperdiversify immune-type receptors within a multigene family.
Keywords: Variable region-containing chitin-binding protein, Evolution, Innate immunity, Variable immunoglobulin polymorphism
Introduction
The mechanisms by which immune recognition molecules are diversified are of fundamental biological interest as examples of adaptive genetic changes that can shape both population structure and the dynamics of host–pathogen interactions. In jawed vertebrates, the mechanisms that effect somatic change in the variable (V) regions of immunoglobulin (Ig) and T-cell antigen receptor (TCR) genes during the development and maturation of individual lymphocytes are well characterized (Dudley et al. 2005). The basic process of somatic recombination signal sequence-mediated gene rearrangement has remained intact in all jawed vertebrates, although many species-specific alternatives have been observed (Cannon et al. 2004a). Recent evidence suggests that an alternative (i.e., parallel) mechanism, based on diversified leucine-rich repeats, effects somatic variation and accounts for related immune effector functions in jawless vertebrates (Nagawa et al. 2007; Pancer et al. 2004). The cellular basis for adaptive immunity in vertebrates presents a strong case for evolutionary convergence (Guo et al. 2009).
The origins of the somatic and germline mechanisms that diversify V regions of immune receptors in species that are more phylogenetically removed from contemporary vertebrates is of considerable interest (Litman and Cooper 2007). A particularly wide range of mechanisms, including specialized forms of replication-dependent template switching, deamination-based hypermutation, spontaneous rearrangements, and differential RNA processing, give rise to somatic variation in other families of immune receptors in vertebrates and invertebrates (Dheilly et al. 2009; Dong et al. 2006; Kurtz and Armitage 2006; Litman et al. 2007; Schulenburg et al. 2007; Tasumi et al. 2009).
Variable region-containing chitin-binding proteins (VCBPs) are a family of immune-type molecules that have been characterized at the cellular, genetic, and structural levels (Cannon et al. 2002, 2004b; Hernandez Prada et al. 2006) in amphioxus (Branchiostoma floridae), a protochordate that occupies a critical position in the evolution of the vertebrate forms (Delsuc et al. 2006; Holland et al. 2004, 2008; Putnam et al. 2008). VCBPs also have been identified in Ciona (Cannon et al. 2004b), a solitary tunicate. Of the different VCBP families in amphioxus, the VCBP2 and VCBP5 gene cluster exhibits the highest degree of polymorphism. In order to characterize the genetic mechanisms underlying this complex variation, a comprehensive analysis of VCBPs has been carried out. The evolution of these tightly clustered genes is not restricted to point mutations of static haplotypic units, but rather is a dynamic, multifactorial process that under selection has stabilized a variety of discrete allelic groups.
Materials and methods
Animals and nucleic acid isolation
Specimens of the Florida lancelet, B. floridae, were collected along the coastline of Tampa Bay. Genomic deoxyribonucleic acid (DNA) was isolated via standard proteinase K digestion and phenol extraction-based methods. Total RNA was isolated with the one-step RNA isolation reagent, RNAbee (Tel-Test., Friendswood, TX). Complementary cDNA (cDNA) was synthesized with Superscript III (Invitrogen, Carlsbad, CA) and Oligo-dT anchored primers according to manufacturer' recommended protocol. Bacterial artificial chromosome (BAC) and P1 artificial chromosome (PAC) clones were handled as previously described (Dishaw et al. 2008).
Genomic haplotyping and allele surveys
Genomic haplotype polymerase chain reaction (PCR), as well as cDNA analysis, was conducted from a single sample of 100 animals collected from four locations in the Tampa Bay area. Analysis of allelic variation that includes intronic flanking regions was characterized using a PCR genotyping approach that targets exon 2 (EX2) from VCBP2 and VCBP5 genes (n=60 animals), hereafter referred to as 5a, 2b and 5b genes or loci. Conserved positions in EX1 and EX3 were used as priming sites to anchor the amplification of products that include EX2 as well as its upstream and downstream flanking intronic regions. This process is referred to as 5a–2b–5b genotyping. Amplification of EX2 sequences (hotspot/hypervariable regions) were performed as previously described (Cannon et al. 2004b). VCBP2 and 5-type cDNAs were analyzed by reverse transcriptase PCR (RT-PCR; n=30 animals) with primers anchored at the 5′ end of the first Ig domain and at the 3′ end of the second Ig domain or at the 3′-most conserved region of the chitin-binding domain.
Sequencing and sequence analysis
RT-PCR products and deduced polypeptide sequences were analyzed with Sequence Manipulation Suite (http://bioinformatics.org/sms2/); (Stothard 2000), the European Molecular Biology Open Software Suite (EMBOSS) package of sequence tools (Rice et al. 2000) via the web portal (http://liv.bmc.uu.se/emboss/) and Basic Local Alignment Search Tool (BLAST) searches (Altschul et al. 1990) of locally maintained databases. Subsequent alignments (genomic DNA, cDNA, or polypeptides) were constructed with either ClustalX (Thompson et al. 1997), the Needleman–Wunsch algorithm (EMBOSS package), or the CHAOS +DIALIGN package (http://dialign.gobics.de/chaos-dialign-submission; Brudno et al. 2004). Routine sequence comparisons were performed with local iterations of the BLAST package against local databases possessing compiled VCBP alleles as well as complete BAC and PAC assemblies.
Genetic diversity and polymorphism
The proportions of synonymous (Ps) and nonsynonymous (Pn) differences and/or the numbers of synonymous (Ds) and nonsynonymous (Dn) substitutions per site were estimated by the Nei and Gojobori (1986) method implemented in the MEGA4 program (Tamura et al. 2007). Complete deletion option was used while estimating their averages and standard errors were obtained by the bootstrap method.
Results
Allelic variation of the VCBP2/5 gene clusters
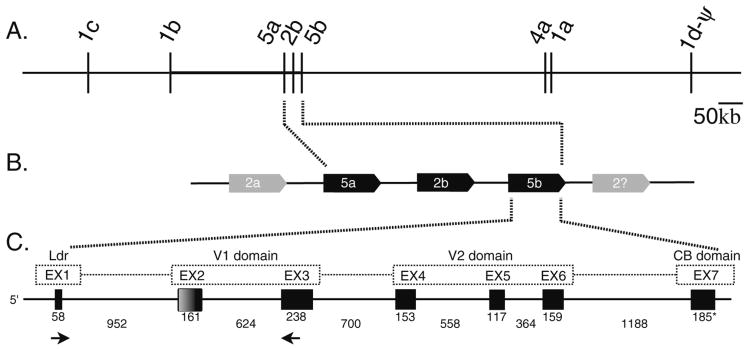
The VCBP2-type and VCBP5-type genes map to a single, tightly grouped allelic cluster (∼40 kb) within a larger chromosomal segment encoding VCBP-encoding DNA (Dishaw et al. 2008). Sequence mapping of BAC clone 62d19 and BAC contig 63n5-43b24 from the current reference genome animal (Brafl1: JGI version 1; Dishaw et al. 2008; Putnam et al. 2008) and PAC 37d17 suggests that a haplotypic unit consists of a single 2b gene flanked by 5a and 5b genes which are paralogous (Fig. 1). The most polymorphic component of the VCBP2/5 genes is referred to as a hypervariable or hotspot region and is localized to EX2 (Fig. 1c; Dishaw et al. 2008).
Fig. 1.
Genomic organization of the VCBP2/5 gene cluster. A In Brafl1, the reference genome animal, two VCBP5 paralogs are separated by a VCBP2 gene, which lies within a larger chromosomal region encoding other VCBP genes (Dishaw et al. 2008). Chromosomal distances in (A) are to scale. B Copy number variants (CNVs) observed in different VCBP2/5 haplotypes are depicted in grey shading. C VCBP2 and VCBP5 genes consist of seven exons (EX1-7) that encode two V-type Ig domains and one chitin-binding domain as illustrated for the 5b gene locus. A region of elevated, localized, polymorphism in exon 2 (EX2, hotspot motif) is shaded. Nucleotide sequence length in the reference BAC contig 63n5-43b24 is indicated below introns and exons. Arrows below EX1 and EX3 depict the location of priming sites that were used for PCR genotyping of 5a, 2b, and 5b genes
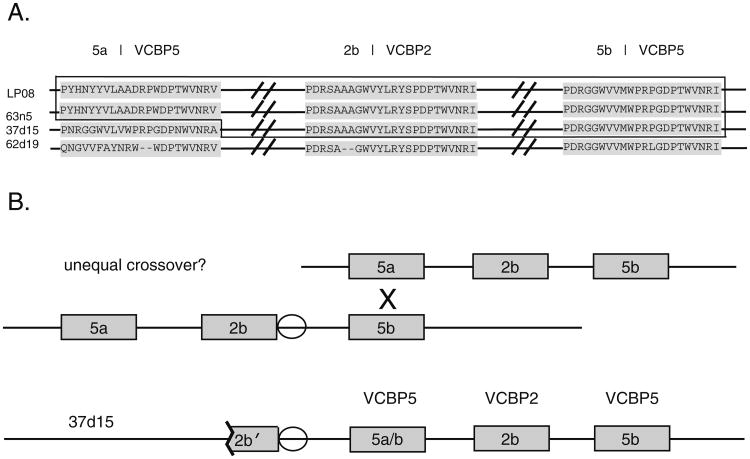
Using a molecular genetic approach, a preliminary screen identified many of the haplotypes as conforming to the VCBP5a/2b/5b arrangement. However, other animals possess allele types (spanning both coding and non-coding regions) that cannot be classified readily with the positional 5a–2b–5b genetic designation (see below). The most definitive comparisons of alleles can be made to the haplotypes defined by the genome reference BAC and other PAC clones (Dishaw et al. 2008; Fig. 2a). For example, animal LP08 shares nearly identical alleles to BAC contig 63n5-43b24 across the VCBP 5a–2b–5b loci (see below), indicating the conservation of a haplotype termed “A” (Dishaw et al. 2008). However, the haplotype from reference PAC clone 37d15, which is nearly identical to the A haplotype across VCBP 2b and 5b genetic regions, possesses a unique 5a allele (identified most readily by the translated coding sequence of EX2, see Fig. 2a). The translated product of EX2 in the 37d15 gene 5a is more typical of 5b-type alleles than of the known 5a types. Approximately five to six putative 5a types can be distinguished in our current accumulation of comparable sequences. In addition, this haplotype termed “C” (Dishaw et al. 2008) possesses an additional VCBP2-like coding region (locus 2a) upstream of the 5a locus. The upstream intron (5′ to EX1 of the 5a locus) of 37d15 and an intron between some 2b and 5b loci (Fig. 2b) reveal additional similarity, suggesting that unequal crossover has modified some 5a loci and explains further the upstream copy number variation (CNV). Other atypical and/or mosaic alleles that reflect exchanges between haplotypes of the VCBP2/5 gene cluster have been identified (see below).
Fig. 2.
Evidence for recombination involving different 5a–2b–5b haplotypes. A The representative EX2 hotspot motifs for each locus are compared in reference haplotypes: BAC clone 62d19, BAC contig 63n5-43b24, and PAC clone 37d15. Haplotypes recovered from animal LP08 and BAC 63n5 are conserved. Haplotype 37d15 shares homology with LP08 and 63n5 across the 2b and 5b loci, but not at the 5a locus. B PAC 37d15 possesses a segment of an upstream VCBP2-like CNV that is interrupted by cloning. The 5a allele of 37d15 shows evidence for mosaicism with a 5b-type allele and is designated 5a/b. A related sequence flanking 2b-type alleles is evident upstream of the 5a (5a/b) locus in 37d15. The 37d15 haplotype may have originated through unequal crossover between 5a and 5b genes. Rectangular boxes in (B) imply full-length gene loci
In order to further address the issue of allelic complexity, amphioxus (n=30) were screened by RT-PCR in order to recover full-length cDNAs corresponding to the 5a–2b–5b loci. Nearly all animals possessed at least one 5b-type allele that is highly similar (93–99% sequence identity) to alleles of the above-referenced BAC and PAC reference clones; several exceptions have been noted that can be accounted for by primer incompatibilities. Only four animals in this group possess alleles that are equally similar (in the 90% range) at the 5a locus.
Despite the extensive polymorphism observed in the cDNA sequences, EX2-coding regions of 5a-, 2b-, and 5b-type genes can be recovered readily from genomic DNA. Many of the translated products of EX2 are homologous to those of the reference BAC and PAC clones (Fig. 2a) and serve as reference alleles. Among the 100 animals sampled, 39 animals possess EX2-coding regions that are related closely to those of the BAC and PAC reference 5a loci, 44 animals possess EX2 coding regions that are related closely to the reference 2b loci, and 93 animals possess EX2-coding regions that are nearly identical to the corresponding 5b reference loci. The stability of distinct allelic lineages within large natural populations is evident from the recovery of several near-identical full-length sequences from individual animals recovered from four locations or populations (Tampa Bay), over a 4-year sampling period.
VCBP hotspot sequences identify specific allelic lineages
Allele frequencies and haplotype content were examined further by PCR genotyping across EX1–EX3 of the 5a, 2b, and 5b loci (n=60 animals, same animal set). When compared to the three reference sequences, 12–28% (range due to three unrelated alleles present at 5a) of the recovered alleles were found to be related closely (90–99% identity) to one of the three reference 5a-locus genes (i.e., of the same peptide type; Fig. 2a), as many as 30% of the alleles are related closely (95–98%) to the 2b-locus genes, and as many as 87% of the alleles are related closely (93–99%) to the 5b-locus genes.
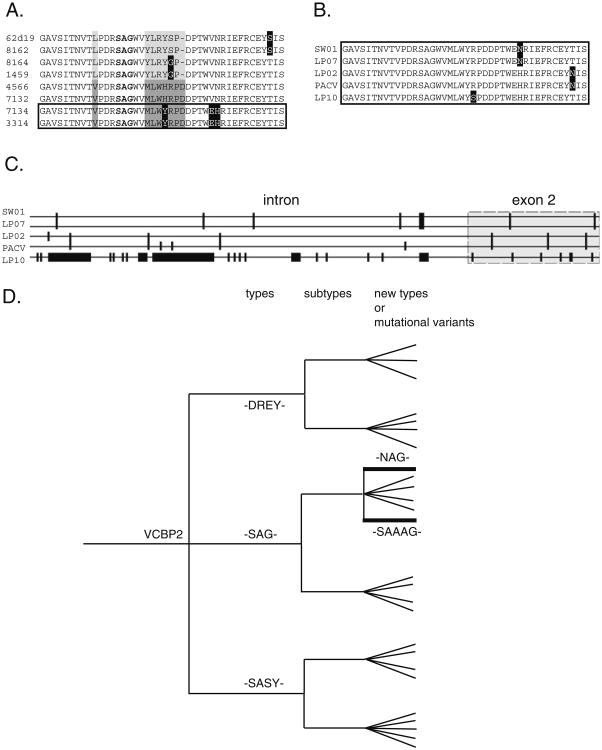
The alleles recovered for each of the loci, as defined provisionally by the translated products of EX2 at 5a, 2b, and 5b loci (Fig. 2), can be categorized into distinct groups sharing specific hotspot motifs, which are referred to collectively as hotspot-type variants, and can specify lineages of diversifying alleles at each locus. Mutational variants of these hotspot types, which distinguish subtypes, are encountered frequently, and can define discrete lineages of alleles (Figs. 3 and S1). Related haplotypes, such as those defined by the reference BAC and PAC sequences, all share alleles of the same type (Fig. 2). VCBP2b (VCBP2 at 2b locus) hotspot-type alleles always include the -SAG- or -SAAAG- (predicted sequence) motifs; other VCBP2-type alleles possess -NAG-, -SASY-, and -DREY- motifs (some of these may represent CNV, see below). The reference haplotypes define at least three different motif types at the 5a locus (Fig. 2a). Other VCBP5a-type candidate sequences are difficult to assign without additional genomic data. A variety of hotspot types are associated with the 2b and 5a loci, suggesting allelic complexity; however, only a single predominant allele type is seen at the 5b locus. Allelic variation at the 5b locus is consistent with mutational differences.
Fig. 3.
Exon 2 motif type and subtype variation generates diversified allelic lineages. A Comparison of two subtypes of -SAG-type 2b alleles (-SAG-motif in bold; defined in mixed animal surveys are distinguished by grey shading). Polymorphisms representing point mutations (reverse image) account for additional variation. B Alleles from additional animals exhibit subtype variation (reverse image) from (A). C Shared polymorphisms in EX2 (boxed) extend to adjacent introns (e.g., SW01 and LP07) or intron variation in otherwise conserved alleles (e.g., LP02 and PACV). Sequence differences can be extensive (e.g., LP10). D Schematic illustration of EX2 polymorphism and relatedness; EX2 types and subtype variants give rise to diverse allelic lineages. In this example, three types of VCBP2 alleles [defined by core EX2 peptide motifs (i.e., “types”): DREY, SAG and SASY] can be classified into subtypes, exhibiting further polymorphic variation (terminal branches; subtype variants) that can result in additional subtype lineages (e.g., NAG and SAAAG). Some VCBP2 alleles (i.e., alternative EX2 types) represent paralogous genes from haplotype-specific CNVs. Bold lines for -NAG- and -SAAG- imply novel lineages of allele types. PacV=Bf Pac 47J9. Numbers in (A) refer to individual sequence clones. Figure in (C) is not meant to depict phylogenetic relationships
Unequal crossover and CNV
The classification criteria applied here were derived initially from the reference BAC/PAC sequences, but generally can be applied to a large group of the animals that have been haplotyped. Intron similarity, in addition to motif type, is important for accurately assigning alleles presumably derived from the 5a, 2b, or 5b loci; however, a significant number of alleles fail to meet these criteria (Figs. 4 and S2). Analyses of near full-length transcripts of VCBPs by RT-PCR identified multiple divergent forms of VCBP2-type genes that differ at the amino acid level by as much as 30% (Fig. S3). Many of these alleles represent examples of CNV (<70% identity and/or unrelated introns in genes; Figs. 4, S2, and S3). VCBP2-type alleles with an overall >70% amino acid identity (e.g., of the -DREY- or -SASY- type hotspot, Figs. S1–3) that exhibit very limited sequence similarity across flanking genomic introns have been recovered frequently. A VCBP2-like CNV is evident in 37d15 (i.e., locus 2a); the previously described VCBP2 allele (AF520473) does not share the same 2b-motif type and may represent a haplo-specific variant or CNV (see below). Most CNVs thus far characterized are of the VCBP2-type and are encountered frequently. Some of the CNV haplotypes have been preserved in the population, presumably due to selected functional specificities.
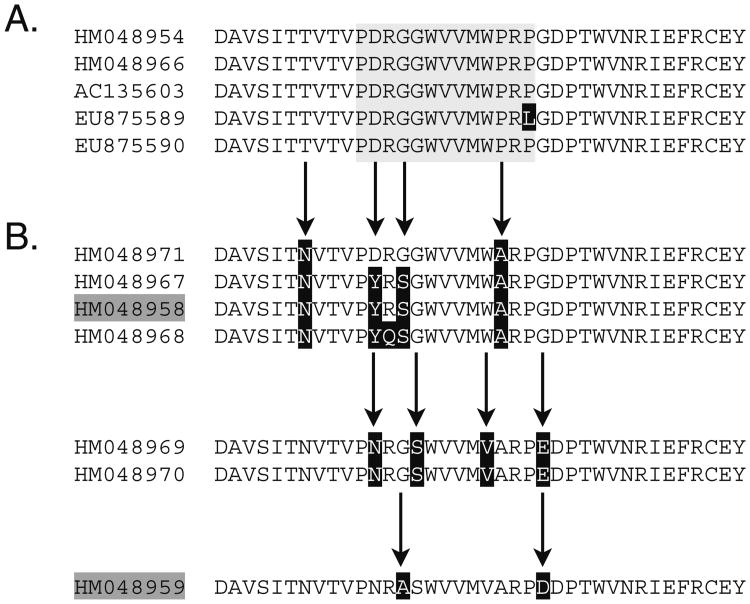
Fig. 4.
VCBP2 genes with VCBP5b-type EX2 motifs; exceptions to the EX2 allele-type classification rules confound accurate placement of VCBP2/5-type alleles. A 5b-type EX2 polypeptide sequences exhibit subtype variation (reverse image). B VCBP2 cDNAs (shaded, dark grey) have been identified with similar EX2 peptides. Conserved VCBP5b-type EX2 motif is shaded (light grey), intended for reference purposes only
Some coding regions exhibit features consistent with unequal crossover and/or gene conversion (Figs. 4 and 5, Table S1; see below), whereas other coding regions reflect equal crossovers (e.g., Fig. S4). Within specific coding regions, some VCBP5-like EX2 variants were identified in VCBP2-type cDNAs (Fig. 4). Extensive variation across haplotypes has been reported and suggests the exchange of unrelated DNA into and out of the VCBP regions (Dishaw et al. 2008). Deletions of EX2 have been observed in several 5a- and 2b-related alleles and may represent byproducts of these recombination events.
Fig. 5.
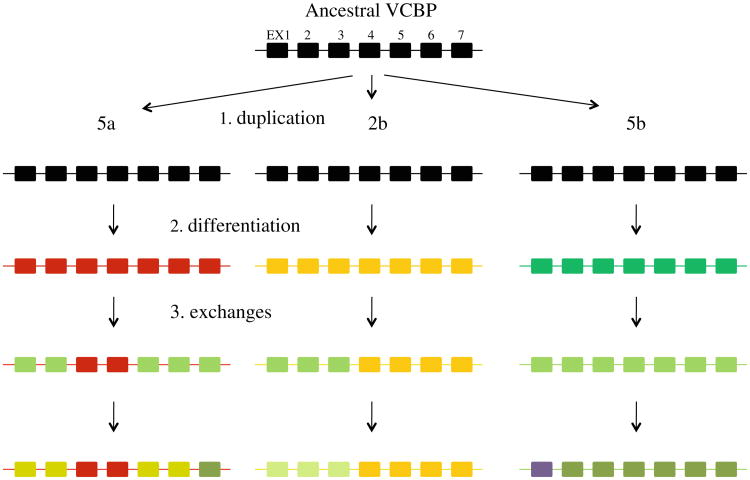
A hypothetical evolutionary scenario for VCBP2/5 haplotype diversification via sequence change: (1) duplication generated at least three VCBP2/5 genes, each consisting of seven exons; (2) differentiation of gene loci; (3) exchange has occurred between genes 5a and 5b in EX1–2 as well as in EX5–7. EX1–EX3 have undergone genetic exchange between genes 2b and 5b. EX1 of the 5b gene was replaced by unrelated or distantly related genes. All or part of EX7 was exchanged between 5a and 5b. The VCBP 5b locus is used as a reference (except for the unusual EX1), likely owing to more functional constraint resulting in less polymorphism at this locus
Nucleotide polymorphism
In order to further characterize patterns of nucleotide polymorphism, both the proportion of synonymous substitutions (Ps) and number of synonymous differences per site (Ds) were estimated within and compared among the coding regions of the VCBP2/5 gene clusters in the reference BAC/PAC haplotypes as well as in unrelated alleles. The overall averaged distance values were found to be higher in the VCBP2/5 genes than in unrelated genes flanking the VCBP gene cluster, including MSDS2, GAPDH, and the Big Defensin gene, that were analyzed in parallel (Tables S1 and S2). Of particular note, VCBP3 alleles, which are encoded in a separate genetic locus, exhibit similar distance values to those seen for the VCBP2/5 alleles.
Distance estimates comparing the entire VCBP2/5 coding regions reveal that the overall average distance between loci 5a and 5b is smaller than the distance to locus 2b. A similar relationship is seen for exons 5–7, despite the large variance caused by the relatively small numbers of nucleotides that were examined. This observation is consistent with the VCBP 2b locus, being more distantly related to VCBP 5a or 5b. However, the average Ps between loci 5a and 5b for EX7 is smaller than the values observed for EX5 or EX6 and is consistent with recent gene exchanges between loci 5a and 5b at EX7.
Further inspection of sequence variation at other exons reveals that the estimated distances are inconsistent with this overall pattern of relatedness. Specifically, in EX1 (encoding the secretion signal sequence), the average Ps (0.274) or average Ds (0.360) between loci 5a and 2b is much smaller than between loci 5a and 5b. The predicted peptide sequence in EX1 is distinct in locus 5b and may not be homologous (or at least distantly related) to EX1 from 5a or 2b. In EX2 (hotspot exon, V1 domain), the average values for Ps (0.217, 0.216, 0.229) or Ds (0.258, 0.276, 0.263) among different loci are similar to or slightly higher than the individual values estimated for locus 5a (Ps 0.199 and Ds 0.234). As noted previously, an unequal crossover event may have resulted in a 5a allele of 37D15 acquiring the sequence features of 5b and resulted in low Ps (0.100) between loci 5a and 5b of 37d15. Such past gene exchanges would be predicted to partially lower the average distance between 5a and 5b loci by mixing paralogous loci and to increase the average distance within a locus by introducing distantly related paralogous sequences. For EX3 (second exon of V1 domain), the average Ps (0.195) and/or average Ds (0.227) of loci 2b and 5b are smaller than between 5a– 2b or 5a–5b; this is consistent with a role for genetic exchange between the VCBP 2b and the VCBP 5b loci. Collectively, these observations suggest that the VCBP2/5 cluster was derived by unequal crossover, gene conversion and/or other mechanisms of gene exchange rather than by simple gene duplication. The processes that gave rise to the diversified VCBP2/5 gene family can be represented as a series of discrete events (Fig. 5).
Discussion
Mounting evidence suggests that certain polymorphisms can benefit recognition receptors of invertebrate innate defense systems by increasing recognition potential in the face of shifting pathogen threats (Kurata 2006; Litman et al. 2005; Messier-Solek et al. 2010). In amphioxus and sea urchin, innate receptors have undergone dramatic expansions in gene copy number and predicted functional complexity (Holland et al. 2008; Messier-Solek et al. 2010; Rast et al. 2006). Such adaptations appear to reflect specialization that may better serve to discriminate pathogenic challenges or refine specificities in commensal relationships.
Amphioxus VCBPs exhibit a particularly high level of genetic polymorphism (Cannon et al. 2004b; Dishaw et al. 2008), even when considered within the context of an overall genome rate variation of 0.0562 in amphioxus (Putnam et al. 2008). While this number is quite high, it is consistent with the variation calculated for non-VCBP regions reported here and the site variation described previously for the expanded family of amphioxus toll-like receptors (Yuan et al. 2009), which are related to the smaller families of these genes found in mammals and other vertebrates.
High degrees of polymorphism generally are explained by high rate of mutation and/or large effective population size, a feature common to many invertebrate species. Systematic phylogenetic analyses of several gene families in amphioxus thus far have failed to provide evidence for accelerated rates of mutation; the source of high site variation in this species likely relates at least in part to its large effective population size (Delsuc et al. 2006; Putnam et al. 2008). However, the population size effect itself is insufficient to account for the high degree of polymorphism observed at VCBP loci (Tables 1 and 2). A multifactorial biological process, such as immune recognition, could be consistent with this difference.
VCBP2/5 alleles can be grouped by characteristic EX2 (hotspot) motifs (i.e., types). Whereas some of the hotspot types (i.e., allelic lineages) exhibit strong conservation, others are highly diverse and account for a large number of assignable subtype variants. In some animals, as many as 8–10 VCBP2/5-type alleles have been described (Cannon et al. 2002, 2004b). Without exception, all of the genomic sequencing and transcriptional analyses conducted confirm extraordinary levels of complexity (e.g., Fig. S3) and define haplotypic CNVs as the source of additional alleles that are encountered in some genotypes.
The genotype screening reported here permits comparisons across extended sequence stretches and provides evidence that certain haplotypes include mosaic and hybrid alleles. However, accurate placement of discrete crossover sites and/or conversion tracts is confounded by “layers” of sequence polymorphism, reflecting not only gene conversion and crossover (e.g., homologous and nonhomologous) events but also a relatively high frequency of point mutations. The dataset includes additional alleles that are comprised of haplotypically diverse introns and unrelated intergeneic regions (Dishaw et al. 2008) yet share identical (or nearly identical) coding regions. Notwithstanding this multilayered and multileveled complexity, both genomic and cDNA sequencing of VCBP 5a, 2b, and 5b alleles from individual animals, representing different populations, underscore the preservation of distinct allelic lineages. Some allele types have diverged into distinct lineages reflected in single amino acid changes within the hotspot (EX2) exon (e.g., Fig. S1). Functional subspecialization of each lineage or subpopulation structuring can account for such strong patterns of evolutionary conservation.
Different evolutionary and selective forces influence genetic events, such as point mutation and gene exchange, which include unequal crossover and CNV, that contribute to the high levels of polymorphism in the VCBP2/5 genes (e.g., Fig. S3). Frequent gene exchanges solely among paralogous loci theoretically result in concerted evolution (in effect homogenizing paralogous genes). Individuals that are homozygous at the VCBP loci have not been identified, and it would appear as if some form of selection (including nonhomologous gene exchange) is acting to maintain variation in VCBP loci. Related effects described here have been demonstrated previously in the immune genes encoding plant nucleotide-binding site leucine-rich repeat proteins (R-genes), receptor-like kinases, and receptor-like proteins. In several cases, clustered and/or tandem loci associated with functional immune restriction have been shown to reach a balance between intergenic polymorphism and the degree of diversifying sequence exchange within coding regions (Mondragon-Palomino and Gaut 2005; Parniske et al. 1997). In the absence of such compensatory balancing processes, allelic diversity would diminish in time and compromise a broad-based function such as immunity.
Whereas the function(s) of VCBPs only now are being elucidated, increasing evidence suggests they may play a role in some immune processes mediated at the gut epithelial surfaces (Cannon et al. 2002). The diversity that is inherent in VCBPs is effected at the germline level through haplotypic complexity within a large interbreeding population. Polymorphisms of innate receptors can alter population structures to meet the demands of reciprocal waves from pathogen challenge. Patterns of genetic polymorphism remarkably similar to those reported here for VCBPs have been described in primate killer immunoglobulin receptors (KIRs) in which allelic polymorphism and CNV give rise to diverse and functionally relevant individual genotypes influenced by variable modes of selection (Hsu et al. 2002; Khakoo et al. 2000; Norman et al. 2009; Shilling et al. 2002). Notably, plant immune genes, KIRs, and VCBPs are all organized as relatively small gene clusters. Immune selection may be particularly suited to gene families that are organized in clusters; this configuration appears to create an optimal matrix for the generation of diversity in immune receptors (Friedman and Baker 2007). This cluster organization itself may be driven by variable selective pressure. Irrespective of the precise genetic basis for such effects, complex genetic histories molded by variable selective forces, which preserve discrete allelic lineages and/or haplotypes, are emerging as a general feature of eukaryotic innate immune receptor gene families.
Supplementary Material
Acknowledgments
We thank Barbara Pryor for editorial assistance and Dr. Michelle M. Roux for constructive criticisms on earlier versions of the manuscript. This work was supported by the National Institutes of Health Grant AI23338 to G.W.L. L.J.D. is supported by a fellowship from the H. Lee Moffitt Cancer Center and Research Institute.
Sequences have been deposited into GenBank, http://www.ncbi.nlm.nih.gov/ (accession # HM048928–HM048976).
Abbreviations
- BAC
Bacterial artificial chromosome
- CNV
Copy number variation
- KIR
Killer immunoglobulin receptor
- PAC
P1 artificial chromosome
- TCR
T-cell antigen receptor
- V
Variable region
- VCBP
Variable region-containing chitin-binding protein
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s00251-010-0464-x) contains supplementary material, which is available to authorized users.
Contributor Information
Larry J. Dishaw, Department of Molecular Genetics, All Children' Hospital, St. Petersburg, FL, USA; H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA
Tatsuya Ota, Department of Evolutionary Studies of Biosystems, The Graduate University for Advanced Studies, Hayama, Japan.
M. Gail Mueller, Department of Molecular Genetics, All Children' Hospital, St. Petersburg, FL, USA.
John P. Cannon, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA; Department of Pediatrics, University of South Florida College, of Medicine, USF/ACH Children' Research Institute, St. Petersburg, FL, USA
Robert N. Haire, Department of Pediatrics, University of South Florida College, of Medicine, USF/ACH Children' Research Institute, St. Petersburg, FL, USA
Natasha R. Gwatney, Department of Pediatrics, University of South Florida College, of Medicine, USF/ACH Children' Research Institute, St. Petersburg, FL, USA
Ronda T. Litman, Department of Pediatrics, University of South Florida College, of Medicine, USF/ACH Children' Research Institute, St. Petersburg, FL, USA
Gary W. Litman, Email: litmang@allkids.org, Department of Molecular Genetics, All Children' Hospital, St. Petersburg, FL, USA; H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA; Department of Pediatrics, University of South Florida College, of Medicine, USF/ACH Children' Research Institute, St. Petersburg, FL, USA.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brudno M, Steinkamp R, Morgenstern B. The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Res. 2004;32:W41–W44. doi: 10.1093/nar/gkh361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Rast JP, Litman GW. The phylogenetic origins of the antigen binding receptors and somatic diversification mechanisms. Immunol Rev. 2004a;200:12–22. doi: 10.1111/j.0105-2896.2004.00166.x. [DOI] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Schnitker N, Mueller MG, Litman GW. Individual protochordates possess unique immune-type receptor repertoires. Curr Biol. 2004b;14:R465–R466. doi: 10.1016/j.cub.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dheilly NM, Nair SV, Smith LC, Raftos DA. Highly variable immune-response proteins (185/333) from the sea urchin, Strongylocentrotus purpuratus: proteomic analysis identifies diversity within and between individuals. J Immunol. 2009;182:2203–2212. doi: 10.4049/jimmunol.07012766. [DOI] [PubMed] [Google Scholar]
- Dishaw LJ, Mueller MG, Gwatney N, Cannon JP, Haire RN, Litman RT, Amemiya CT, Ota T, Rowen L, Glusman G, Litman GW. Genomic complexity of the variable region-containing chitin-binding proteins in amphioxus. BMC Genomics. 2008;9:78. doi: 10.1186/1471-2156-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4:1137–1146. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17:493–499. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Prada JA, Haire RN, Allaire M, Jakoncic J, Stojanovic N, Cannon JP, Litman GW, Ostrov DA. Ancient evolutionary origin of diversified variable regions revealed by crystal structures of an immune-type receptor in amphioxus. Nat Immunol. 2006;7:875. doi: 10.1038/ni1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Laudet V, Schubert M. The chordate amphioxus: an emerging model organism for developmental biology. Cell Mol Life Sci. 2004;61:2290–2308. doi: 10.1007/s00018-004-4075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutierrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, Ferrier DE, Garcia-Fernandez J, Gibson-Brown JJ, Gissi C, Godzik A, Hallbook F, Hirose D, Hosomichi K, Ikuta T, Inoko H, Kasahara M, Kasamatsu J, Kawashima T, Kimura A, Kobayashi M, Kozmik Z, Kubokawa K, Laudet V, Litman GW, McHardy AC, Meulemans D, Nonaka M, Olinski RP, Pancer Z, Pennacchio LA, Pestarino M, Rast JP, Rigoutsos I, Robinson-Rechavi M, Roch G, Saiga H, Sasakura Y, Satake M, Satou Y, Schubert M, Sherwood N, Shiina T, Takatori N, Tello J, Vopalensky P, Wada S, Xu A, Ye Y, Yoshida K, Yoshizaki F, Yu JK, Zhang Q, Zmasek CM, de Jong PJ, Osoegawa K, Putnam NH, Rokhsar DS, Satoh N, Holland PW. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- Kurata S. Recognition and elimination of diversified pathogens in insect defense systems. Mol Divers. 2006;10:599–605. doi: 10.1007/s11030-006-9032-6. [DOI] [PubMed] [Google Scholar]
- Kurtz J, Armitage SA. Alternative adaptive immunity in invertebrates. Trends Immunol. 2006;27:493–496. doi: 10.1016/j.it.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Litman GW, Cooper MD. Commentary: why study the evolution of immunity? Nat Immunol. 2007;8:547–548. doi: 10.1038/ni0607-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nat Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Dishaw LJ, Cannon JP, Haire RN, Rast JP. Alternative mechanisms of immune receptor diversity. Curr Opin Biol. 2007;19:526–534. doi: 10.1016/j.coi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier-Solek C, Buckley KM, Rast JP. Highly diversified innate receptor systems and new forms of animal immunity. Semin Immunol. 2010;22:39–47. doi: 10.1016/j.smim.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Gaut BS. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol. 2005;22:2444–2456. doi: 10.1093/molbev/msi241. [DOI] [PubMed] [Google Scholar]
- Nagawa F, Kishihita N, Shimizu K, Hirose S, Miyoshi M, Nezu J, Nishimura T, Nishizumi H, Takahashi Y, Hashimoto S, Takeuchi M, Miyajima A, Takemori T, Otsuka AJ, Sakano H. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8:206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, Graef T, McQueen KL, Guethlein LA, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Tyan D, Verity DH, Vaughan RW, Davis RW, Fraser PA, Riley EM, Ronaghi M, Parham P. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt RA, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin I, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Boehnisch C, Michiels NK. How do invertebrates generate a highly specific innate immune response? Mol Immunol. 2007;44:3338–3344. doi: 10.1016/j.molimm.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tasumi S, Velikovsky CA, Xu G, Gai SA, Wittrup KD, Flajnik MF, Mariuzza RA, Pancer Z. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA. 2009;106:12891–12896. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Huang S, Zhang W, Wu T, Dong M, Yu Y, Liu T, Wu K, Liu H, Yang M, Zhang H, Xu A. An amphioxus TLR with dynamic embryonic expression pattern responses to pathogens and activates NF-kappaB pathway via MyD88. Mol Immunol. 2009;46:2348–2356. doi: 10.1016/j.molimm.2009.03.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.