Abstract
Background
Vitamin D deficiency is common in chronic liver disease particularly in those with severe liver fibrosis. Aims: To determine the effect of 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3) on the human α1(I) collagen promoter and collagen formation by human stellate LX-2 cells and the mechanism of the effect of the vitamin D receptor (VDR) on the promoter.
Methods
Type I collagen was assessed by measurements of collagen mRNA and collagen protein and by transfection experiments. Binding of VDR to the α1(I) collagen promoter was determined by EMSA and ChIP assays.
Results
1,25-(OH)2D3 decreased human α1(I) collagen mRNA and protein and the secretion of type I collagen by stellate cells after exposure to TGFβ1. Furthermore, 1,25-(OH)2D3 inhibited TGFβ1–induced activation of the α1(I) collagen promoter in transfected LX-2 cells. The effect of 1, 25-(OH)2D3 is mediated by the VDR, which binds at a proximal Sp1 site and also at a newly identified distal site on the collagen promoter. A VDR expression vector reduced the activities of the collagen promoter in transfected LX-2 cells.
Conclusions
1,25-(OH)2D3 inhibits type I collagen formation in human stellate cells. The effect of 1,25-(OH)2D3 is mediated by its receptor which binds at a proximal Sp1.1 site and at a newly identified distal site on the collagen promoter. Correction of vitamin D deficiency in patients with chronic liver disease is a potential therapy to inhibit progression of fibrosis.
Keywords: 1,25-dihydroxyvitamin D3; Sp1; stellate cells; transforming growth factor β1; vitamin D receptor; α1(I) collagen
Hepatic fibrosis and cirrhosis result from the excessive deposition of predominantly type I collagen, which is composed of two α1 and one α2 chains.
1,25-dihydroxyvitamin D3 (1,25-(OH)2D3)2, the active form of vitamin D, is formed by sequential 25 and 1a hydroxylation in the liver and kidney respectively (1). Low serum levels of 25-hydroxyvitamin D are common in chronic cholestatic and non-cholestatic liver disease (2–4) with the lowest levels observed in patients with severe hepatic fibrosis (4).
1,25-(OH)2D3 was shown to reduce type I collagen mRNA and collagen synthesis in osteoclasts of rat fetal calvaria (5), the activity of the rat α1(I) collagen promoter in transfected rat osteosarcoma cells (6) and to inhibit transforming growth factor beta 1 (TGFβ1)-induced fibroblast proliferation and type I collagen expression (7). More recently 1,25-(OH)2D3 was shown to decrease rat α1(I) collagen promoter activity in transfected rat stellate cells, decrease α1(I) collagen mRNA and collagen content in the rat stellate cells and to diminish thioacetamide-induced liver fibrosis in rats (8).
1,25-(OH)2D3 is the ligand of the vitamin D receptor (VDR) which mediates its biological effects. Most of the actions of VDR are mediated by heterodimers of VDR with retinoid X receptor (VDR-RXR heterodimers) which bind to vitamin D response elements (VDREs) in promoters of vitamin D responsive genes (9). VDREs usually consist of two core motifs in the form of Pug (G/T)TCA (X)3 PuG(G/T)TCA (10). However, several variations of these motifs can be tolerated (11). VDRE has been identified in the murine α1(I) collagen promoter (12), however, we found no reports of identification of VDREs in the human α1(I) collagen promoter. VDR was also shown to modulate the expression of genes that lack VDRE via Sp1 binding sites by forming a complex with Sp1 (13). Sp1 binds to the human α1(I) collagen promoter ( − 164 to −142), forms a complex with Smads and is a powerful mediator of the activating effect of TGFβ on the collagen promoter (14, 15).
The aim of this study was to determine the effect of 1,25-(OH)2D3 on the human α1(I) collagen promoter, collagen formation by human stellate LX-2 cells and the mechanism of the effect of VDR on the promoter.
Experimental procedures
Materials
Dulbecco’s modified Eagle’s medium (DMEM), and Schneider’s Drosophila medium were purchased from Invitrogen (Gibco, Carlsbad, CA, USA). Fetal bovine serum (FBS), bovine serum albumin (BSA) and 25-dihydroxyvitamin D3 (1,25-(OH)2D3) were obtained from Sigma (St. Louis, MO, USA). Human TGFβ1 was purchased from R&D Systems (Minneapolis, MN, USA). Trichostatin A was obtained from Biomol (Plymouth Meeting, PA, USA). Protease Inhibitor Cocktail was obtained from Roche (Indianapolis, IN, USA). Poly (dIdC) was from GE Healthcare (Piscataway, NJ, USA). [α-32 P]dATP and [α-32 P]dCTP were purchased from ICN Biochemicals, Inc. (Irvine, CA, USA).
Cell culture
LX-2, a human stellate cell line, was a gift of Dr Scott L. Friedman from the Mount Sinai School of Medicine (New York, NY, USA). The LX-2 cells were cultured in 75-cm2 tissue culture flasks and maintained in DMEM containing 10% FBS, penicillin G (100 U/ml), streptomycin (100 mg/ml) and fungizone (2.5 mg/ml) at 37°C with a humidified atmosphere of 5% CO2 and 95% air. Drosophila Schneider L2 cells were obtained from American Type Culture Collection (Manassas, VA, USA). The Drosophila cells were maintained at room temperature in Schneider’s Drosophila medium, supplemented with 10% FBS, penicillin G (100 U/ml) and streptomycin (100 mg/ml).
Plasmids
The −2.3 kb to +42 CAT (p2.3k α1CAT) and the −174 to +42 CAT (p174 α1CAT) constructs of the human α (I) collagen promoter (14) were provided by Dr Sergio A. Jimenez from Thomas Jefferson University, Philadelphia, PA, USA. The luciferase construct of pGL3-2.3k α1 was made as described previously (16). The pGL-174 α1 construct was made by cutting p174 α1CAT with HinfI and KpnI filling the 5′-end overhang caused by the HinfI cut and inserting it by blunt ligation into the KpnI site of the pGL3-Enhancer vector. pGL3-Enhancer, pGL3-Promoter and phRL-CMV were purchased from Promega (Madison, WI, USA). The expression vector pPacSp1 was obtained from Dr Robert Tijan from the University of California (Berkeley, CA, USA). The VDR expression vector pcDNA-hVDR was provided by Dr J. Wesley Pike from the Department of Biochemistry, University of Wisconsin (Madison, WI, USA).
Nuclear protein extraction
LX-2 stellate cells were harvested for nuclear protein extraction as described previously (17). Protein concentration was determined by the method of Lowry et al. (18). The nuclear protein extracts were aliquoted and stored at −80°C.
Electrophoretic mobility shift assay to identify VDR binding sites
Oligonucleotides with potential VDR binding motifs identified in the α1(I) collagen promoter were synthesized and tested by electrophoretic mobility shift assay (EMSA) for the formation of protein–DNA complexes with supershift using VDR antibody. One oligonucleotide was found to bind VDR. Further EMSA were then done with this VDR oligonucleotide: 5′-TCACACCT TGGAGGTTTCAACT-3′ (−2240 to −2219) and with the Sp1.1 oligonucleotide 5′-CTTCCCTCCTCCTCCC CCTCTCC-3′ (−164 to −142) (15). Mutated VDR oligonucleotides contained 2 bp nucleotide substitutions as shown in Fig. 3B. Complementary strands of oligonucleotide were annealed and the double-stranded oligonucleotides were labelled with [α-32 P]dATP and [α-32 P]dCTP using Klenow enzyme according to the method of Feinberg and Vogelstein (19). DNA–protein binding reactions were performed following the previously described EMSA procedure (17). For ‘supershift’ EMSA experiments, rabbit polyclonal antibodies to VDR and to Sp1 were obtained from Abcam Inc. (Cambridge, MA, USA) and to RARα, RXRα and N-CoR were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) were used.
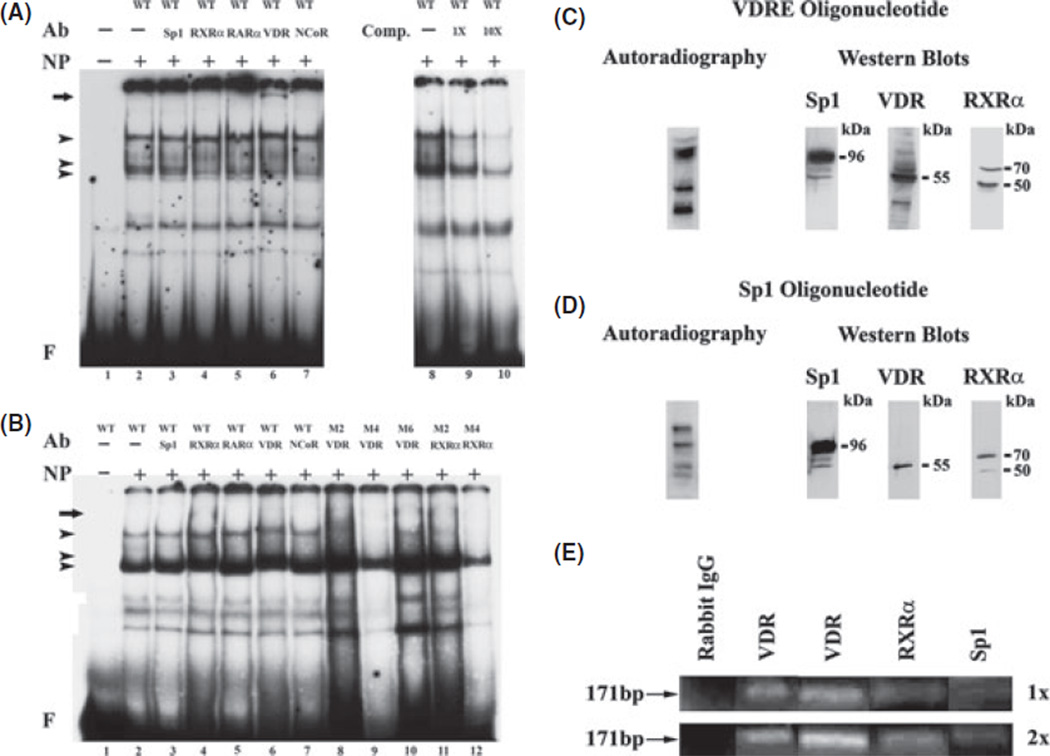
Fig. 3.
(A) EMSA with supershift showing binding of nuclear extracts from LX-2 stellate cells to the VDR oligonucleotide. The EMSAswere performed with 8 mg of nuclear protein extract (NP) and the labelled wild type (WT) or the mutated oligonucleotides (M2, M4, M6).Competition was performed with increasing amounts (0, 1Xand 10X) of the unlabeled nucleotide. For supershifts the antibodies (Ab) to Sp1, RXRα, RARα, VDR and NCoR (1:200 dilution) or pre-immune serum were added to the reaction mixture. The arrowheads indicate the protein–DNA complexes formed. The arrows indicate the location of the supershifted complexes. F indicates the position of the free probe. The labels across the top of panel B indicate either the wild type oligonucleotide −2240 to −2219 or the oligonucleotide with two base pair mutations with the mutations in bold. WT: 5′-TCACACCTTGGAGGTTTCAACT −3′, M2: 5’-TCACATTTTGGAGGTTTCAACT −3’, M4: 5′-TCACACCTTTTAGGTTTCAACT −3’, M6: 5’-TCACACCTTGGAGAATTCAACT −3′ (C) Autoradiography and western blot showing the presence of VDR, Sp1 and RXRα in nuclear extract from 1,25-(OH)2D3-treated LX-2 cells UV crosslinked to the labelled VDR (C) and Sp1.1 (D) oligonucleotides. The cells were treated with 1,25-(OH)2D3 (10 ng/ml) for 24 h. The blots were exposed to film to determine the UV cross-inking of the proteins and incubated with antibodies to VDR, Sp1 and RXRα for the western blots. The location of the bound proteins and their molecular mass (kDa) obtained from protein standards is shown. (E). Chromatin immunoprecipitation (ChIP) assay of VDR binding. One-(1x) and two-step (2x) cross-linked DNA–protein complexes were immunoprecipitated with antibodies to VDR, RXRα, and Sp1. Negative control was performed with purified rabbit IgG. The covalent linkage was reversed and precipitated double-stranded DNA was amplified to the region −2302 to −2131 (171 bp)of the α1(I) collagen promoter.
Type I collagen
Human Type I collagen secreted by the stellate cells into the culture media was determined with the Human Collagen Type I ELISA kit of Cosmo Bio Co., LTD (Tokyo, Japan).
Determination of messenger RNA by real-time quantitative polymerase chain reaction (RT-qPCR)
Total cellular RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA, USA). The concentration of the isolated RNA was determined from the optical density at 260 nm and its purity from the 260 nm/280 nm OD. Equal amounts of RNA were used for the determinations. Superscript III First Strand synthesis from Invitrogen was used to synthesize the first strand cDNA from the purified RNA. The 7900 HT (Applied Biosystems, Foster City, CA, USA) and the SDS 2.2.1 software were used to perform RT-qPCR at the Johns Hopkins DNA Analysis Facility. RT-qPCR for α1(I) collagen, TGFβ mRNA and β-actin mRNA, a housekeeping endogenous control, were performed in triplicate using sequence-specific probes from TaqMan gene expression assays of Applied Biosystems. Variations in the amount of mRNA transcripts were corrected by the level of expression of the β-actin gene in each individual sample.
Western blot analysis
LX-2 cells from culture were harvested and homogenized in 50 mm Tris-HCl buffer pH 7.6 containing 150 mm NaCl, 10 mm CaCl2, 0.25% Triton-X, 0.1 µm phenylmethanesulfonyl fluoride (PMSF), 10 µm leupetin, 10 µm pepstatin, 0.1 mm iodoacetamide and 25 µg aprotonin and then centrifuged at 3000g for 10 min at 4°C. The cytosol protein in the supernatant was initially stored at −80°C. Equal amounts of protein were loaded and separated on mini-SDS gels at 100 V for 1 h and electrotransferred to nitrocellulose transblot membranes (Bio-Rad, Hercules, CA, USA). The membranes were washed in PBS, pH 7.6, containing 0.1% Tween 20 (PBS-T), blocked with 5% (w/v) dry non-fat milk in PBS-T for 1 h, rinsed with PBS-T and then incubated with mouse antibodies to α1(I) collagen or rabbit anti-human antibodies to MMP-9, TIMP-1, TIMP-2 and α actin, obtained from Santa Cruz Biotechnology, Inc. or to VDR obtained from Abcam (Cambridge, MA, USA). After repeated washing, the membranes were incubated with either horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (1:10 000 dilution; Amersham Biosciences, Piscataway, NJ, USA) at room temperature for 1 h. The membranes were then washed again and visualized by enhanced chemiluminescence reaction (ECL Plus; Amersham Biosciences). Densitometry was determined using Image J v 1.30 obtained from NIH (Bethesda, MD, USA).
Apoptosis
Apoptosis of stellate cells was determined by measurements of DNA fragmentation. DNA fragmentation was evaluated using the Cell Death Detection ELISA kit from Roche Diagnostic. This assay measures cytoplasmic histone-associated DNA fragments. The cells were harvested and homogenized in the ice-cold lysis buffer and incubated for 30 min at 25°C. The lysate was centrifuged at 12 000g for 10 min, and the ELISA assay performed in the supernatant. The Vmax values of absorbance were divided by protein concentration determined by the method of Lowry et al. (18) and expressed as a per cent of control.
Caspase activity
The activity of caspase 3 was determined in liver homogenates by measuring proteolytic cleavage of the specific fluorogenic substrate, DEVD-AFC (AFC:7-amino-4-trifluoromethyl coumarin) (BioVision, Milpitas, CA, USA). The results are expressed as relative units per mg of protein. The concentration of protein was determined by the method of Lowry et al. (18).
Transient transfections and luciferase assay
LX-2 stellate cells were grown as described until they reached 60% confluence. Each cell culture plate was transfected with 3 µg of the pGL3- α1 collagen promoters and either alone or with either 2 µg of pPacSp1, pcDNA-hVDR, or their combination and 2 µg of pGL3-Enhancer and pGL3-Promoter vectors as controls, using calcium phosphate precipitation (20). Transfection efficiency was determined by co-transfection of 0.2 µg of the Renilla luciferase vector phRL-CMV (Promega). Four hours after transfecting, the cells were washed twice with PBS and then shocked with 10% DMSO. A similar procedure (no shock) was followed for transfection in Drosophila cells. The cells were harvested 24 h after transfection. The harvested cells were exposed to two freeze-thaw cycles in Reporter Lysis +Buffer (Promega). Firefly luciferase activity was determined using the Dual luciferase assay system (Promega) and normalized to total cell protein (18).
Ultraviolet cross-linking of nuclear protein to oligonucleotides
The binding reactions were carried out as in EMSA for 30 min at room temperature followed by 15 min on ice using nuclear extracts (16 µg of the protein content) and radioactively labelled oligonucleotides (50 fmol) specifying the Sp1.1 binding site. After completion of the reaction, the samples were kept on ice followed by exposure to UV radiation at 120 000 mJ for 10 min in a UV Crosslinker 1800 (Stratagene Cloning Systems, La Jolla, CA, USA). The cross-linked protein–DNA complexes were subsequently resolved on 4–15% gradient denaturing SDS-polyacrylamide gels. The resolved protein–DNA complexes were electrophoretically transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham Life Science., Inc., Arlington Heights, IL, USA) in a Trans-Blot Cell at 40 V/0.34 A overnight, according to the manufacturer’s protocol (Bio-Rad Laboratories, Richmond, CA, USA). The membranes were washed in Tris-buffered saline pH 7.6 containing 0.1% Tween 20 (TBS-T) and subsequently blocked with 5% dried non-fat milk and 0.5% FBS in TBS-T for 1 h at room temperature. The proteins of interest were then analysed as described for Western blot.
Chromatin immunoprecipitation (ChIP) assay
One and two-step cross-link procedures were performed. For the two-step procedure (21, 22) protein-protein cross-linking, utilizing disuccinimidyl glutarate to facilitate n-hydroxysuccinimide-mediated protein– protein esterification, was done prior to protein–DNA cross-linking with formaldehyde. Briefly, T75 flasks containing LX2 cells in monolayer were washed three times with PBS, after which PBS containing 1 mm MgCl2, pH 8.0 was added to each flask. Disuccinimidyl glutarate was then rapidly added, mixed to a final concentration of 2 mm, and the cells incubated at room temperature for 45 min. The cells were then washed three times with PBS following which they were incubated at room temperature for 15 min in 10 ml of a fresh 1% formaldehyde solution in PBS containing 1 mm MgCl2, pH 8.0. The flasks were again washed three times with PBS, and the cells scraped and transferred to 2 ml microfuge tubes. Following the method outlined in the Magna ChIP procedure manual (Upstate, Charlottesville, VA, USA), nuclei were prepared from the cells and lysed. The nuclear lysates were sonicated to produce soluble chromatin with an average length between 200 and 500 bp. The protein–DNA complexes were immunoprecipitated with the respective antibodies, the protein–DNA cross-links reversed and the DNA purified using spin columns (Qiagen). Anti-Acetyl Histone H3 was used for the positive control and normal rabbit IgG for the negative control. A one-step ChiP assay (23) was also done by incubating the cultured LX-2 cells after washing them with PBS with 10 ml of fresh 1% formaldehyde solution in PBS. The formaldehyde was then quenched by adding 1 m glycine to a final concentration of 145 mm following which the flasks incubated at room temperature for 5 min. The flasks were washed three times with PBS, scraped and the DNA recovered as described above (22). The purified DNA was used as a template for PCR amplification. Primers specific for the detection of the VDR binding site region −2302 to 2131 were as follows: 5′-CAAAGCCAGGGATCCCCAAAT AT-3′ and 5′-TTCGTAGTAAGAAGGGCTCCTG-3′. Because of the high GC content of the region of the collagen gene to be amplified, the PCR was optimized by titrating enhancer solution (Invitrogen) in the reaction and performing 30 cycles of PCR amplification. A 1x enhancer solution concentration proved optimal.
Statistical analysis
Data were analysed with Student’s t-test when appropriate or by two-way analysis of variance when comparing means of more than two groups.
Results
1,25-(OH)2D3 inhibits collagen formation
Exposure of cultured LX 2 cells to 1,25-(OH)2D3 (10 nm) increased VDR protein in the absence or presence of TGFβ 1 (10 ng/ml) (Fig. 1A).
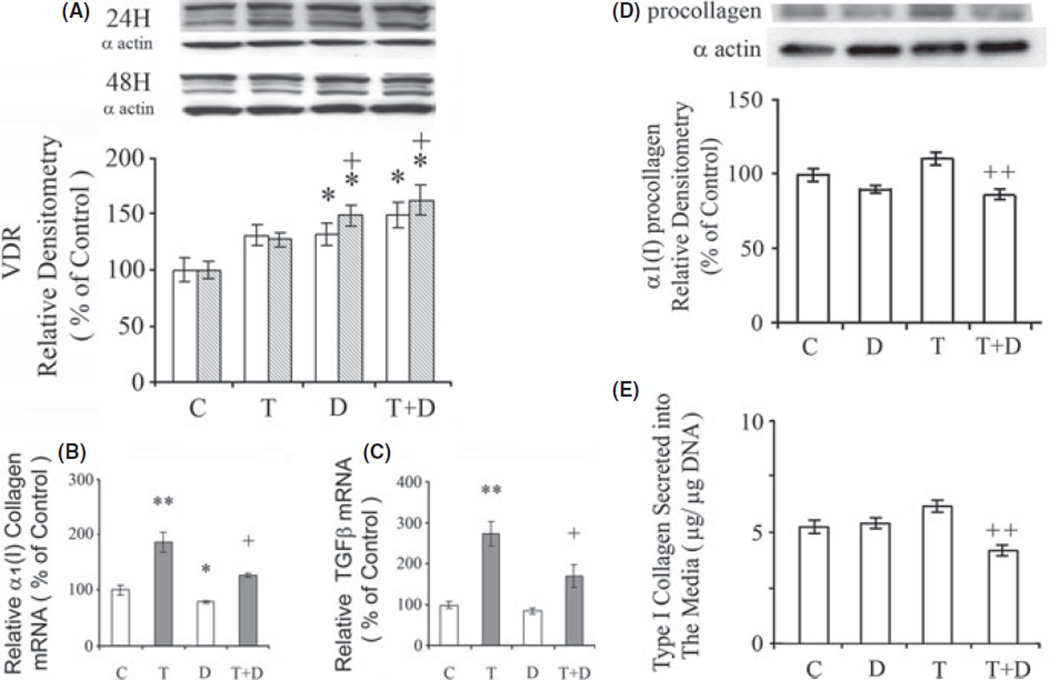
Fig. 1.
Effects of 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3 and TGFβ1 on (A) vitamin D receptor (VDR) protein. The cells were exposed to 1,25-(OH)2D3 (D) (10 nm), TGFβ1 (T) (10 ng/ml) or their combination for 24 h (clear bars) and/or 48 h (shaded bars). VDR protein was determined by western blot with α-actin as a control on the same blots, The amounts of VDR determined by densitometry were normalized to α-actin. Effects of 1,25-(OH)2D3 and TGFβ1 on (B) aα1(I) collagen mRNA and (C) TGFβ mRNA. The cells were exposed to either 1,25-(OH)2D3 (D) (10 nm), TGFβ1 (T) (10 ng/ml) or their combination for 24 h. The mRNAs were determined by real-time polymerase chain reaction. The relative expressions of the cDNAs were normalized against β-actin DNA in the same samples. Effects of 1,25-(OH)2D3 and TGFβ1 on (D) α1(I) collagen protein in LX-2 cells and (E) Type I collagen accumulation in the media secreted by LX-2 stellate cells in culture. The cells were cultured for 24 with or without 1,25-(OH)2D3 (D) (10 nm) and TGFβ1 (T) (10 ng/ml). α1(I) collagen protein was determined by western blot with α- actin as a control on the same blots, The amounts of α1(I) collagen determined by densitometry were normalized to α-actin. Type I collagen in the culture media was determined by ELISA. The data are expressed as means ± of 6 determinations. *P < 0.05 vs. respective control. **P < 0.01 vs. respective control. +P < 0.05 vs. TGFβ1. ++P < 0.01 vs. TGFβ1.
1,25-(OH)2D3 (10 nm) decreased α1(I) collagen mRNA in LX-2 cells (P < 0.05) (Fig. 1B). TGFβ 1 (10 ng/ml) enhanced α1(I) collagen mRNA (P < 0.01) and this effect was inhibited by 1,25-(OH)2D3 (P < 0.05). TGFβ1 enhanced TGFβ mRNA (P < 0.01) and this effect was inhibited by 1,25-(OH)2D3 (P < 0.05) (Fig. 1C).
1,25-(OH)2D3 (10 nm) decreased α1(I) collagen protein in the LX-2 cells (Fig. 1D) and Type I collagen secretion (Fig. 1E) by the cultured LX-2 cells in the presence of TGFβ1 (10 ng/ml) after 24 h of culture (P < 0.01).
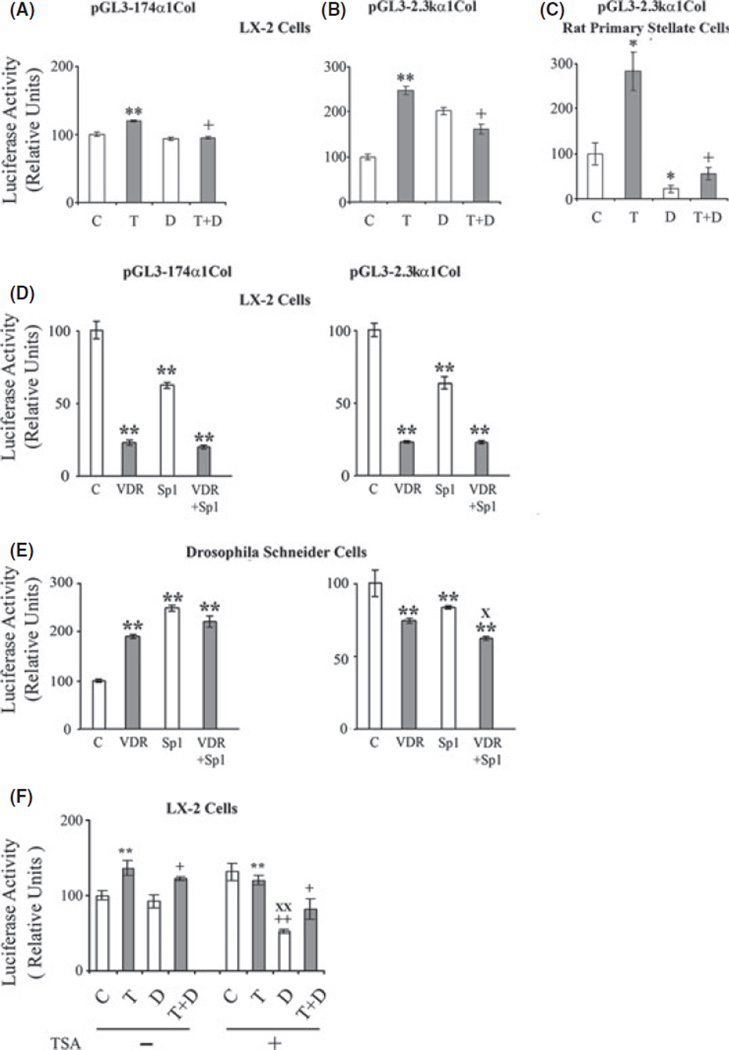
In transient transfection experiments in LX-2 cells, exposure to TGFβ1 (10 ng/ml) enhanced the activities of pGL3–174 α1 and pGL3-2.3k α1 (P < 0.01) (Fig. 2A, B) and 1,25-(OH)2D3 (10 nm) inhibited the TGFβ1-induced activation of both promoters (P < 0.01). Also in transient transfection studies in primary rat hepatic stellate cells, TGFβ1 (10 ng/ml) enhanced the activity of pGL3-2.3k α1 (P < 0.05) (Fig. 2C) and 1,25-(OH)2D3 (10 nm) inhibited the promoter both in the absence (P < 0.05) and presence of TGFβ1 (P < 0.01). cis-Retinoic acid (10 µM), the ligand of RXRα, had no significant effect on the activity of pGL3-2.3k α1, the activation of pGL3-2.3k α1 by TGFβ1 (10 ng/ml) or the inhibitory effect of 1,25-(OH)2D3 (10 nm) (data not shown).
Fig. 2.
Effect of 1,25-(OH)2D3 (D) (10 nm) and TGFβS1 (T) (10 ng/ml) on the activity of (A) pGL-174 α1 and (B) pGL3-2.3k α1 in transfected LX-2 cells and (C) on the activity pGL3-2.3k α1 in transfected rat primary stellate cells. Effects of VDR (pcDNA-hVDR) and Sp1 (pPacSp1) expression vectors on the activities of (D) pGL-174 α1 and pGL3-2.3k α1 collagen promoters in transfected LX-2 cells and (E) in transfected Drosophila Schneider cells. (F) Effect of trichostatin on the activity of the pGL3-2.3k α1 collagen promoter in the presence of 1,25-(OH)2D3 and or TGFβ1. The cells were cultured for 24 h with or without trichostatin A (TSA) (100 nm), 1,25-(OH)2D3 (D) (10 nm) and TGFβ1 (T) (10 ng/ml). Luciferase activities are shown as percentages of the control value. The data are expressed as means ± of 6−8 determinations. *P < 0.05 vs. respective control. **P < 0.01 vs. respective control. +P < 0.05 vs. TGFβ1. ++P < 0.01 vs. TGFβ1. XP< 0.01 vs. VDR. XXP< 0.01 vs. TSA alone.
The VDR expression vector (pcDNA-hVDR) caused a marked reduction in the activities of pGL3–174 a and pGL3-2.3k α1 in transfected LX-2 cells (Fig. 2D) (P < 0.01), and this reduction in activity was similar when VDR was co-transfected with pPacSp1. Because pPacSp1 alone decreased the activity of the transfected collagen promoters, the transfections were also done in Drosophila Schneider L2 cells which lack endogenous Sp1. In the Drosophila cells VDR reduced the activity of pGL3-2.3k α1 in the presence or absence of pPacSp1 as seen in LX-2 cells (P < 0.01) and Sp1 by itself also decreased the activity of pGL3-2.3k α1 (P < 0.01) (Fig. 2E). By contrast, VDR, pPacSp1 and the co-transfection of VDR and pPacSp1 all increased the activity of pGL3–174 α1 in the transfected Drosophila cells (P < 0.01).
Histone acetylation leads to changes in chromatin structure that promote accessibility of DNA to transcription factors (24). Trichostatin A (100 nm), a histone deacetylase inhibitor, increased activation of pGL3-2.3k α1 (P < 0.01), and allowed for marked inhibition of the promoter by 1,25-(OH)2D3 (10 nm) (P < 0.01) (Fig. 2F). Trichostatin A did not significantly alter the inhibitory effect of 1,25-(OH)2D3 on the TGFβ1-activated promoter.
Binding of nuclear proteins to the α1(I) collagen promoter
EMSA of the VDR oligonucleotide with nuclear proteins from the LX-2 cells revealed three DNA–protein complexes (Fig. 3A). Competition experiments with the unlabeled VDR oligonucleotide show that the uppermost two complexes are competed away, and the lowest complex is decreased, indicating that uppermost complexes are specific for protein binding to the VDR oligonucleotide. Antibody to VRD did not decrease or eliminate any of the DNA–protein complexes but resulted in the appearance of a supershift (Fig. 3A, lane 6). EMSA with protein–DNA complexes that were mutated by two base pairs between −2240 and −2219 reveals that one mutation (M4) eliminates the uppermost DNA–protein complex and the supershift with VDR antibody (Fig. 3B, lane 9). By contrast, the upper DNA–protein complexes with the appearance of a supershift with VDR antibody are retained with the M2 and M6 mutations (Fig. 3B, lanes 8 and 10). Antibodies to Sp1, RXRα, RARβ or have no effect on the DNA–protein complexes and do not result in supershifts.
EMSA of the Sp1.1 oligonucleotide with nuclear proteins from the LX-2 cells revealed four major DNA–protein complexes with supershifts observed with Sp1 antibody as demonstrated previously (15) (data not shown). However, no changes in the DNA–protein complexes and no appearance of supershifts were obtained with antibodies to VDR, RXRα, RARα or n-CoR (data not shown).
To further evaluate the binding of VDR, Sp1 and RXRα to the α1(I) collagen promoter nuclear extracts of LX-2 cells previously exposed to 1,25-(OH)2D3 (10 nm) for 24 h were cross-linked with the VDR and Sp1 oligonucleotides followed by resolution of the protein–DNA complexes on a 4–15% gradient denaturing SDS-poly-acrylamide gel. Figure 3C shows that VDR, and Sp1 bind to the VDR oligonucleotide with kDa of 55 and 96, respectively, while RXRα binds with kDa of 70 and 50. VDR also binds to the Sp1 oligonucleotide but the binding is much weaker than seen with the VDR oligonucleotide (Fig. 3D).
The binding of VDR to the VDR binding region was also determined by ChIP assay after 24 h exposure of the LX-2 cells in culture to 1,25-(OH)2D3 (10 nm). The ChIP assay demonstrates the binding of VDR to the promoter region between −2302 and −2131 (Fig. 3E). The ChIP assay also shows that binding of Sp1 and RXR occurs in association with VDR.
Collagen degradation
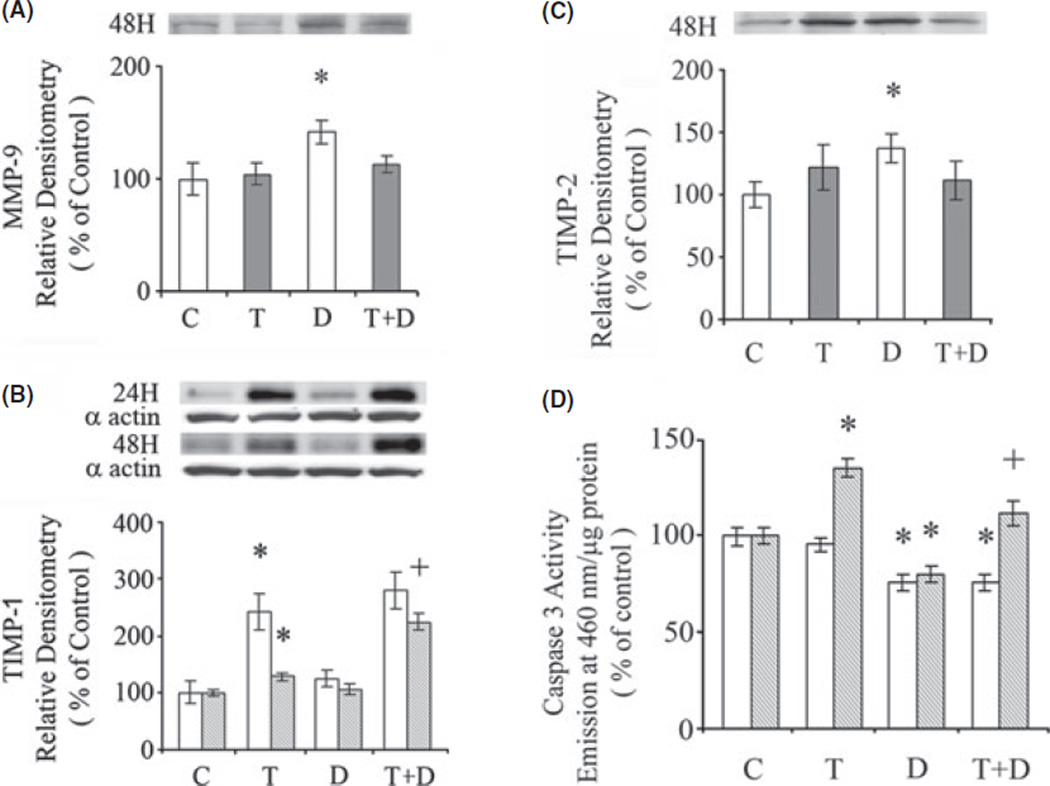
1,25-(OH)2D3 (10 nm) increased MMP-9 protein (P < 0.05) in the absence but not in the presence of TGFβ1 (10 ng/ml) (Fig. 4A). 1,25-(OH)2D3 (10 nm) had no effect on TIMP-1 in the absence of TGFβ1, but increased TIMP-1 in the presence of TGFβ1 (P < 0.05) (Fig. 4B). By contrast, 1,25-(OH)2D3 (10 nm) increased TIMP-2 in the absence (P < 0.05), but not in the presence of TGFβ1 (Fig. 4C).
Fig. 4.
Effect of 1,25-(OH)2D3 on (A) MMP-9, (B) TIMP1 and (C) TIMP2 protein. The cells were exposed to either 1,25-(OH)2D3 (D), (10 nm), TGFβ1 (T) (10 ng/ml) or their combination for 24 or 48 h. The proteins were determined by western blot with α-actin as a control on the same blots. The amounts of MMP-9, TIMP1 and TIMP2 determined by densitometry were normalized to α-actin. (D). Effects of 1,25-(OH)2D3 and TGFβ1 on caspase 3 activity. The cells were exposed to 1,25-(OH)2D3 (D) (10 nm), TGFβ1 (T) (10 ng/ml) or their combination for 24 h (clear bars) and 48 h (shaded bars). The data are expressed as means ± of 6 determinations. *P < 0.05 vs. respective control. +P < 0.05 vs. TGFβ1.
1,25-(OH)2D3 inhibits apoptosis
Apoptosis determined in the LX-2 cells by DNA fragmentation was increased by TGFβ1 after 48 h exposure (P < 0.05), and this effect was inhibited by 1,25-(OH)2D3 (P < 0.01) (Fig. 4, Table 1). Exposure of LX-2 cells to 1,25-(OH)2D3 (10 nm) for 24 and 48 h, in the absence of TGFβ1, resulted in decreased caspase 3 activity (P < 0.05) (Fig. 4D). TGFβ1 (10 ng/ml) increased caspase activity at 48 h and this effect was inhibited by 1,25-(OH)2D3 (P < 0.05).
Table 1.
Effects of TGFβ1, and 1,25-(OH)2D3 on DNA fragmentation in cultured LX-2 cells
| DMA fragmentation (% of control) | |
|---|---|
| Control | 100 ± 8.9 |
| TGFβ1 | 218 ± 36.5* |
| 1,25-(OH)2D3 | 94.5 ± 13.7 |
| TGFβ1 + 1,25-(OH)2D3 | 88.8 ± 7.4† |
The cells were cultured for 48 h with or without TGFβ1 (10 ng/ml)or 1,25-(OH)2D3 (10 nm). DNA fragmentation per mg of LX-2 cell protein is expressed as a per cent of control cells. The values are shown as means ± SE of 6 determinations.
P< 0.05 vs. control.
P< 0.01 vs. TGFβ1.
Discussion
Low levels of serum 25-hydroxyvitamin D are common in patients with chronic liver disease (2–4) with the lowest level observed in patients with severe fibrosis (4). This study shows that 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3), the active form of vitamin D, decreases human α1(I) collagen mRNA and collagen protein in human LX-2 stellate cells, and also decreases the secretion of Type I collagen after exposure to TGFβ1. This confirms a recent study showing that 1,25-(OH)2D3 reduced rat α1(I) mRNA and collagen content in primary rat stellate cells (8). Furthermore, 1,25-(OH)2D3 in vivo diminished thioacetamide-induced liver fibrosis in rats (8). 1,25-(OH)2D3 is the ligand of the VDR which mediates its biological effects. In this study, VDR was demonstrated in the LX-2 cells and shown to be enhanced by 1,25-(OH)2D3.
The effect of 1,25-(OH)2D3 in inhibiting the α1(I) collagen promoter in the LX-2 stellate cells was demonstrated in transient transfection experiments. Most of the actions of VDR are mediated by heterodimers of VDR with retinoid X receptor (VDR-RXR heterodimers) which bind to vitamin D response elements (VDREs) in promoters. In this study, cis-retinoic acid the ligand of RXR had no effect on the α1(I) collagen promoter.
In Drosophila Schneider L2 cells, which lack endogenous Sp1, VDR reduced the activity of the full length pGL-2.3kα1 promoter, but resulted in an enhancement of the shorter pGL3–174 α1 promoter indicating that VDR can act at two separate sites, one proximally and another distally, to −174 with different actions independently of Sp1. Co-transfection of Sp1 with VDR did enhance the inhibitory effect of VDR on the full length promoter. The effect of VDR in increasing the activity of the short pGL3–174 α1 promoter in the absence or presence of co-transfected Sp1 is unexplained, but could be because of another not yet identified VDR binding site or to lack of other transacting factors in the Drosophila cells that usually interact with VDR or Sp1 and their binding to the promoter.
We show that VDR binds at least two sites on the α1(I) collagen promoter, proximally at a previously identified Sp1-1 site and at a newly identified distal site (−2240 to −2219). This newly identified site has a TGGA sequence previously shown to be part of a VDR responsive element (25). We found that the nucleotides GG in this distal site of the α1(I) collagen promoter are essential for the binding of VDR. Sp1 and RXRα also bind to this distal VDR site, while VDR binds to the Sp1.1 site albeit with a weaker binding than observed at the distal VDR site. As regards the binding of VDR to the Sp1.1 site, studies with other promoters show that VDR binds indirectly to Sp1 sites by forming a complex with Sp1 and that Sp1 is required for the binding of Sp1 to the Sp1 sites (14, 26).
The histone deacetylase inhibitor trichostatin A increased activation of pGL3-2.3k α1 and allowed for marked inhibition of the promoter by 1,25-(OH)2D3 indicating that chromatin reorganization enhances the association of transcription factors such as VDR and Sp1 and/or enhances their binding to their cis-acting sites of the collagen promoter. Prior studies with the 5-lipoxygenase promoter showed that trichostatin A potentiated the effects of transfected VDR and RXR on enhancing this promoter (27). In other studies trichostatin A increased 5-lipoxygenase promoter activity by enhanced recruitment of Sp1 and Sp3 to the promoter (28).
There is no evidence in this study for an effect of 1,25-(OH)2D3 on enhancing collagen degradation. Matrix metalloproteinases (MMPs) which degrade collagen and other matrix proteins are modulated by tissue inhibitor metalloproteinases (TIMPs). A number of studies indicate that the net accumulation of collagen in chronic liver disease is mainly because of decreased TIMP expression (29). Although 1,25-(OH)2D3 increased MMP-9 protein, this only occurred in the absence of TGFβ1, while TIMP-1 was increased in the presence of TGFβ1.
TGFβ1 is known to increase reactive oxygen species and apoptosis in stellate cells (30–32). Iredale et al. (29) showed that apoptosis of stellate cells contributes to resolution of fibrosis after discontinuation of chronic CCl4 administration or bile duct ligation in rats. The effects of TGFβ1 on stimulating fibrosis and also causing apoptosis, which is associated with decreased fibrosis, are important opposing factors on collagen deposition which are regulated independently (32). It is possible that apoptosis limits the amount of new collagen formation during enhanced fibrogenesis. In this study 1,25-(OH)2D3 inhibited TGFβ1-induced apoptosis of stellate cells, indicating that the effect of 1,25-(OH)2D3 on TGFβ1-induced type I collagen formation is most likely because of inhibition of collagen synthesis and not because of stellate cell death (apoptosis). The mechanism of the effect 1,25-(OH)2D3 on apoptosis is unknown, but other studies showed that vitamin D attenuates apoptosis induced by oxidative stress or toxic agents (33, 34).
In conclusion, this study shows that 1,25-(OH)2D3 inhibits type I collagen formation in human LX-2 stellate cells. The effect of 1,25-(OH)2D3 is mediated by its receptor which binds at a proximal Sp1.1 site and also at a newly identified distal site on the collagen promoter. There is no evidence in this study for an effect of 1,25-(OH)2D3 on enhancing collagen degradation. Correction of vitamin D deficiency in patients with chronic liver disease is a potential therapy to inhibit progression of fibrosis that deserves further study.
Acknowledgements
This study was supported by Grant AA000626 from the United States Public Health Service.
Abbreviations
- 1,25-(OH)2D3
1,25-dihydroxyvitamin D3
- ELISA
Enzyme-Linked Immuno-Sorbent Assay
- MMP
liver matrix metalloproteinase
- RXR
retinoid X receptor
- TGFβ
transforming growth factor β
- TIMP
tissue inhibitor metalloproteinase
- VDR
vitamin D receptor
References
- 1.Christakos S, Ajibade DV, Dhawan P, Fechner AJ, Mady LJ. Vitamin D: metabolism. Endocrinol Metab Clin N Am. 2010;39:243–253. doi: 10.1016/j.ecl.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arteh J, Narra S, Nair S. Prevalence of vitamin D deficiency in chronic liver disease. Dig Dis Sci. 2010;55:2624–2628. doi: 10.1007/s10620-009-1069-9. [DOI] [PubMed] [Google Scholar]
- 3.Targher G, Bertolini L, Scala L, et al. Associations between serum 25-hydroxyvitamin D3 concentrations and liver histology in patients with non-alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2007;17:517–524. doi: 10.1016/j.numecd.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Petta S, Camma C, Scazzone C, et al. Low vitamin D serum level is related to severe fibrosis and low responsiveness to INF-based therapy in genotype 1 chronic hepatitis C. Hepatology. 2010;51:1158–1167. doi: 10.1002/hep.23489. [DOI] [PubMed] [Google Scholar]
- 5.Genovese C, Rowe D, Kream B. Construction of DNA sequences complementary to rat α1 and α2 collagen mRNA and their use in studying the regulation of Type I collagen synthesis by 1,25-dihydroxyvitamin D. Biochemistry. 1984;23:6210–6216. doi: 10.1021/bi00320a049. [DOI] [PubMed] [Google Scholar]
- 6.Lichtler A, Stover ML, Angilly J, Kream B, Rowe DW. Isolation and characterization of the rat α1(I) collagen promoter. Regulation by 1,25-hydroxyvitamin D. J Biol Chem. 1989;264:3072–3077. [PubMed] [Google Scholar]
- 7.Ramirez AM, Wongtrakool C, Welch T, et al. Vitamin D inhibition of pro-fibrotic effects of transforming growth factor b1 in lung fibroblasts and epithelial cells. J Steroid Biochem Mol Biol. 2010;118:142–150. doi: 10.1016/j.jsbmb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abramovitch S, Dahan-Bachar L, Sharvit E, et al. Vitamin D inhibits proliferation and profibrotic marker expression in hepatic stellate cells and decreases thiocetamide-induced fibrosis in rats. Gut. 2011;60:1728–1739. doi: 10.1136/gut.2010.234666. [DOI] [PubMed] [Google Scholar]
- 9.Haussler MR, Haussler CA, Jurutka PW, et al. The vitamin D hormone and its nuclear receptor: molecular actions and disease states. J Endocrinol. 1997;154:S57–S73. [PubMed] [Google Scholar]
- 10.Jimenez-Lara A, Aranda A. Interaction of vitamin D and retinoic receptors on regulation of gene expression. Horm Res. 2000;54:301–305. doi: 10.1159/000053276. [DOI] [PubMed] [Google Scholar]
- 11.Carlberg C. Mechanisms of nuclear signaling by vitamin D3. Interplay with retinoid and thyroid hormone signaling. Eur J Biochem. 1995;231:517–527. [PubMed] [Google Scholar]
- 12.Kuroki Y, Shiozawa S, Kano J, Chihara K. Competition between c-fos and 1,25 (OH)2 vitamin D3 in the transcriptional control of type I collagen synthesis in MC3T3-E1 osteoblastic cells. J Cell Physiol. 1995;164:459–464. doi: 10.1002/jcp.1041640303. [DOI] [PubMed] [Google Scholar]
- 13.Huang YC, Chen JY, Hung WC. Vitamin D3 receptor/Sp1 complex is required for the induction of p27kip1 expression of vitamin D3. Oncogene. 2004;23:4856–4861. doi: 10.1038/sj.onc.1207621. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez SA, Varga J, Olsen A, et al. Functional analysis of human α1(I) procollagen gene promoter. Differential activity in collagen-producing and–nonproducing cells and response to transforming growth factor β 1. J Biol Chem. 1994;269:12684–12691. [PubMed] [Google Scholar]
- 15.Sysa P, Potter JJ, Liu X, Mezey E. Transforming growth factor-beta1 up-regulation of human α1(I) collagen is mediated by Sp1 and Smad2 transacting factors. DNA Cell Biol. 2009;28:425–434. doi: 10.1089/dna.2009.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang M, Potter JJ, Mezey E. Activation of the human a2(I) collagen promoter by leptin in not mediated by transforming growth factor β responsive elements. Biochem Biophys Res Commun. 2003;312:629–633. doi: 10.1016/j.bbrc.2003.10.167. [DOI] [PubMed] [Google Scholar]
- 17.Potter JJ, Mezey E, Christy RJ, et al. CCAAT/enhancer binding protein binds and activates the promoter to the rat class I alcohol dehydrogenase gene. Arch Biochem Bio-phys. 1991;285:246–251. doi: 10.1016/0003-9861(91)90356-n. [DOI] [PubMed] [Google Scholar]
- 18.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Di Nocera PP, Dawid IB. Transient expression of genes introduced into cultured cells in Drosophila . Proc Natl Acad Sci U.S.A. 1983;80:7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowak DE, Tian B, Brasier AR. Two-step crosslinking method for identification of NF-KappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–724. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 22.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by P300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 23.Aparicio O, Geisberg JV, Sekinger E, et al. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Pro-toc Mol Biol. 2005;21:21.3.1–21.3.33. doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- 24.Schubeler D, MacAlpine DM, Scalzo D, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorg BL, Klan N, Seuter S, et al. Analysis of the 5-lipoxygenase promoter and characterization of the vitamin D receptor binding site. Biochim Biophys Acta. 2006;1761:686–697. doi: 10.1016/j.bbalip.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Huang YC, Hung WC. 1,25-dihyroxy vitamin D3 transcriptionally represses p45skp2 expression via Sp1 sites in human prostate cancer cells. J Cell Physiol. 2006;209:363–369. doi: 10.1002/jcp.20741. [DOI] [PubMed] [Google Scholar]
- 27.Klan N, Seuter S, Schnur N, Jung M, Steinhilber D. Trichostatin A and structurally related histone deacetylase inhibitors induce 5-lipoxygenase promoter activity. Biol Chem. 2003;384:777–785. doi: 10.1515/BC.2003.086. [DOI] [PubMed] [Google Scholar]
- 28.Schnur N, Seuter S, Katryniok C, Radmark O, Steinhilber D. The histone deacetylase inhibitor trichostatin A mediates upregulation of 5-lipoxygenase promoter activity by recruitment of Sp1 to distinct GC boxes. Biochim Biophys Acta. 2007;1771:1271–1282. doi: 10.1016/j.bbalip.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Iredale JP, Benyon RC, Pickering J, et al. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J Clin Invest. 1998;102:538–549. doi: 10.1172/JCI1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Blesser PJ, Xu G, Rombouts K, Rogiers V, Geerts A. Glutathione levels discriminate between oxidative stress and transforming growth factor-b signaling activated rat hepatic stellate cells. J Biol Chem. 1999;274:33881–33887. doi: 10.1074/jbc.274.48.33881. [DOI] [PubMed] [Google Scholar]
- 31.Herrera B, Murillo MM, Alvarez-Barrientos A, et al. Source of early reactive species in the apoptosis induced by transforming growth factor-b in fetal rat hepatocytes. Free Radic Biol Med. 2004;36:16–26. doi: 10.1016/j.freeradbiomed.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Issa R, Williams E, Trim N, et al. Apoptosis of hepatic stellate cells involvement in resolution of biliary fibrosis and regulation by soluble growth factors. Gut. 2001;48:548–557. doi: 10.1136/gut.48.4.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diker-Cohen T, Koren R, Liberman A, Ravid A. Vitamin D protects keratinocytes from apoptosis induced by osmotic shock, oxidative stress, and tumor necrosis factor. Ann NY Acad Sci. 2003;1010:350–353. doi: 10.1196/annals.1299.064. [DOI] [PubMed] [Google Scholar]
- 34.Regulska M, Leskiewics M, Budziszewska B, et al. Inhibitory effect of 1,25 hydroxyvitamin D3 and its low calcemic analogues on staurosporin-induced apoptosis. Pharmacol Rev. 2007;59:393–401. [PubMed] [Google Scholar]