Abstract
Cocaine-Amphetamine Regulated Transcript (CART) peptides are implicated in a wide range of behaviors including in the reinforcing properties of psychostimulants, feeding and energy balance and stress and anxiety responses. We conducted a complex trait analysis to examine natural variation in the regulation of CART transcript abundance (CARTta) in the hypothalamus. CART transcript abundance was measured in total hypothalamic RNA from 26 BxD recombinant inbred (RI) mouse strains and in the C57BL/6 (B6) and DBA/2J (D2) progenitor strains. The strain distribution pattern for CARTta was continuous across the RI panel, which is consistent with this being a quantitative trait. Marker regression and interval mapping revealed significant quantitative trait loci (QTL) on mouse chromosome 4 (around 58.2cM) and chromosome 11 (between 20–36cM) that influence CARTta and account for 31% of the between strain variance in this phenotype. There are numerous candidate genes and QTL in these chromosomal regions that may indicate shared genetic regulation between CART expression and other neurobiological processes referable to known actions of this neuropeptide.
Keywords: Cocaine and amphetamine-regulated transcript, BxD, recombinant inbred mice, hypothalamus, quantitative trait locus, RT-PCR
1. INTRODUCTION
Cocaine and amphetamine-regulated transcript (CART) peptides are neuropeptides that are highly implicated in physiological processes related to addiction and reward. ICV and intra-accumbal CART administration modulates mesolimbic dopaminergic (DA) activity (Yang et al., 2004; Shieh, 2003; Kuhar et al., 2005) and DA-mediated behaviors. For example, intra-ventral tegmental area (VTA) CART injections induce locomotor activation (which is blocked by DA antagonists), attenuates cocaine-induced spontaneous locomotion, and promotes conditioned placed preference (Jaworski et al., 2003; 2007; Couceyro et al., 2005; Kimmel et al., 2000). In addition, CART peptides have been shown to be potent anorectic and anxiogenic agents. The relationship between these disparate actions of this peptide remains obscure, but suggests that a fundamental reward mechanism may be involved in both drug addiction and feeding and satiety behaviors.
CART peptides and mRNA are widely distributed in the brain, mainly concentrated in key brain areas implicated in reward and reinforcement, such as the VTA, nucleus accumbens (NA) and substantia nigra (Douglas et al., 1995, Koylu et al., 1997, 1998; Dallvechia-Adams et al., 2002; Mattson and Morrell, 2005) and are associated with DA-synthesizing and containing neurons in most of these areas (Couceyro et al., 1997; Koylu et al., 1997; Hubert and Kuhar, 2006). CART peptide is also highly expressed in the pituitary, amygdala, and arcuate, lateral, paraventricular and supraoptic nuclei of the hypothalmus (Dominguez et al., 2004a; Van Vugt et al., 2006, Jaworski et al., 2003; Couceyro et al., 1997; Balkan et al., 2006; Sànchez et al., 2007), areas important in stress reactivity.
There is a significant interaction between CART expression and the activity of the hypothalamic-pituitary-adrenal (HPA) axis, with complex negative and positive regulatory interactions, suggesting that CART has a role in stress responsiveness. Corticosterone increases CART mRNA in the NA and in plasma (Hunter et al., 2005; Vicentic et al., 2005). Conversely, ICV CART stimulates corticosterone as well as ACTH in plasma and also c-fos activity in hypothalamic corticotropin-releasing hormone neurons (Vrang et al. 1999; Stanley et al., 2001; Larsen et al., 2003). Thus CART expression appears to be hormonally regulated by glucocorticoids. The CART gene promoter is at least partially under regulation of transcription factors such as cAMP response binding protein (CREB). Second messenger pathways utilizing cAMP are major targets for the actions of drugs of abuse, because they can regulate gene expression by binding to cAMP response elements (CRE) in the promoters of their target genes. Indeed, CREB is in close proximity to the CART proximal gene promoter and mutations introduced to the cAMP response element decrease CART promoter activity, and likely CART mRNA and peptide levels (Dominguez et al., 2002; Dominguez and Kuhar, 2004b; Lakatos, et al., 2002).
While CART is implicated in many physiological functions, the fundamental regulatory mechanisms of CART expression remain relatively elusive. Quantitative trait analysis, using panels of BxD recombinant inbred (RI) mice, continues to be one of the leading methods for the study of genes involved in complex neurobehavioral traits. The BxD series has been used to nominate and map QTL that contribute to a wide variety of complex traits including behavioral, pharmacological, neurochemical and neuroanatomical phenotypes (Erwin et al., 1997; Garlow et al., 2005, 2006; Jones et al., 1999, 2003; Gora-Masalak et al., 1991; Belknap et al., 1992; Zhang and Gershenfeld 2003). The major goal of this study is to conduct quantitative trait analysis of CART expression in hypothalamus, by utilizing the BxD series of RI mice.
2. RESULTS
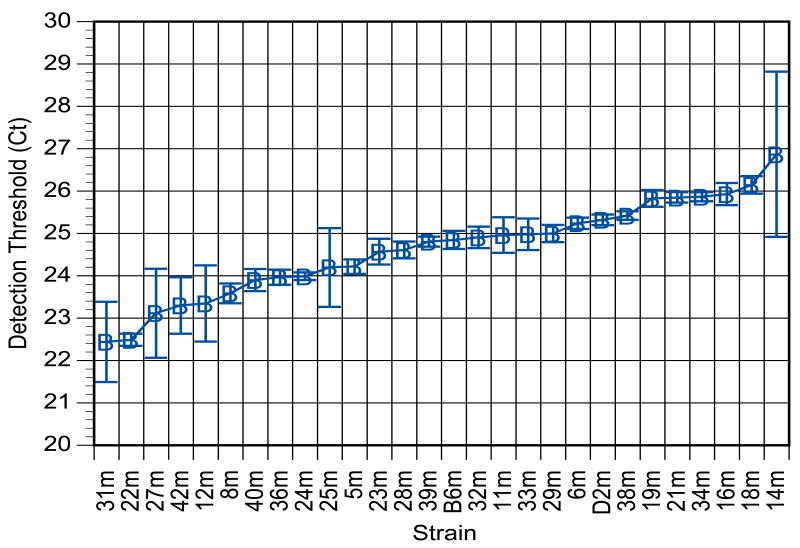
Quantitative trait analysis was used for a genome-wide screen for associations between hypothalamic cocaine and amphetamine-regulated transcript abundance (CARTta) and 975 genetic markers in 26 BxD and C57BL/6 (B6) and DBA/2J (D2) parent strains. The strain distribution pattern (SDP) for mean CARTta, presented as detection threshold cycle (Ct), across the panel of BxD lines is displayed in Figure 1. As detection cycle is the output measure, higher Ct values correspond to lower transcript abundance, as more PCR cycles are required to amplify the target above the detection threshold. CARTta was continuously and quantitatively distributed across strains, a pattern consistent with polygenic inheritance for the trait. The mean Ct for CARTta was 24.53±0.112 and ranged from 22.44 (BxD31) to 26.87 (BxD14). This amounts to a 1.2-fold difference in CARTta expression. CARTta for B6 and D2 progenitors fell within the distribution of CARTta of the RI strains. The observation of both higher and lower trait values for CARTta in some RI strains than in either progenitor (i.e., B6 and D2) is consistent with multifactorial inheritance for the trait (Neumann 1992). This distribution pattern is also suggestive of gene-gene interaction.
Figure 1. Strain distribution pattern (SDP) for hypothalamic CART transcript abundance (CARTta) in C57/BL6 and DBA/2J inbred mice.
SDP are expressed as mean detection threshold cycle (Ct), where higher Ct values correspond to lower transcript abundances. Results are the mean ± SEM from 8–10 per strain. There was a significant difference among strains on Ct (F=3.856; df=27, 140; p<0.0001).
ANOVA revealed significant strain differences in mean CART transcript abundance (F=3.856; df=27, 140; p<0.0001) (Table 1). Strain accounted for 31% of the total variance in CARTta (estimated by ω2), which was virtually identical to the adjusted R2. For CART, the B6 parent strain had a lower Ct (i.e., higher transcript abundance) than the D2 strain. We have observed similar SDP amongst B6 and D2 strains in previous studies of hypothalamic peptide transcripts in our laboratory (Garlow et al., 2005, 2006). Comparison of the SDP for hypothalamic CART and corticotrophin-releasing factor (Garlow et al., 2005) transcription abundance expression reveals a significant correlation in strain mean transcript abundance between the targets (Pearson’s r=0.46, p<0.0192).
Table 1.
Statistical analysis of CART system transcript abundance (CARTta) abundance in B6 and D2 inbred mice.
| Source | Sum of Squares | df | Mean Square | F | Significance | Ω2 | Adjusted R2 |
|---|---|---|---|---|---|---|---|
| Strain | 200.597 | 27 | 7.430 | 3.586 | 0.0001 | 0.314 | 0.317 |
| Error | 268.239 | 139 | 1.930 | ||||
| Total | 168.836 | 166 |
In order to examine any possible relationships between genetic markers and transcript abundance, marker regression analysis was carried out for CARTta at increasing point significance thresholds of P<0.001 through p<10−6 (Table 2, p<0.0001). Critical LRS values that correspond to genome-wide significance levels [highly significant p<0.001 (25.4), significant p<0.05 (16.8), suggestive p<0.63 (9.8)] were calculated with the permutation test run through 10,000 iterations. Results showed two prominent spikes in the LRS profile for CARTta. Potentially significant associations were detected on chromosomes Chr 4 and 11, while suggestive level associations were also detected on Chr 3 and 13 (data not shown).
Table 2.
Results of marker regression analysis of CART transcript abundance (CARTta) data.
| Target | Chr (cM) (position) | Marker | LRS | 95% CI (cM) | Significance (p<0.0001) | Additive Reg. Coeff. |
|---|---|---|---|---|---|---|
| CARTta | 4 (58.2) | D4Mit249 | 16.4 | 30 | 0.00005 | 0.65 |
| 11 (20) | D11Mit20 | 16.9 | 28 | 0.00004 | 0.68 | |
| 11 (27.5) | D11Mit154 | 21.1 | 19 | 0.00000 | 0.69 | |
| 11 (31) | S11Gnf059.515 | 20.5 | 20 | 0.00001 | 0.68 | |
| 11 (33) | D11Mit208 | 19.8 | 21 | 0.00001 | 0.65 | |
| 11 (34) | D11Mit318 | 20.3 | 20 | 0.00001 | 0.66 | |
| 11 (36) | D11Mit177 | 17.9 | 25 | 0.00002 | 0.67 |
QTLs associated with CARTta expression in the hypothalamus. Chr, the chromosome containing associated QTL; LRS, likelihood ratio statistic. The criteria for LRS values for regressed CARTta are 25.4 (highly significant) 16.8 (significant) and 9.8 (suggestive) at the genome-wide 0.001 level. A positive additive regression coefficient indicates that DBA/2J alleles increase CARTta trait values.
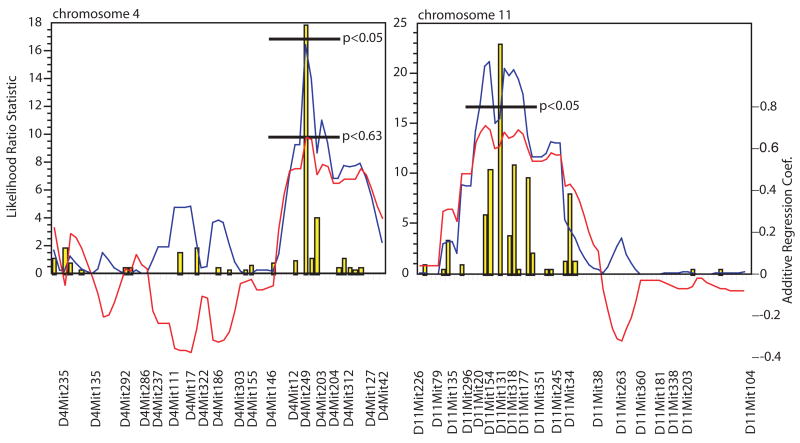
Due to our finding of a considerable (and possibly shared) genetic component to CARTta, simple interval mapping was used to localize the significant linkage signals detected in the initial marker regression onto the respective chromosomes (Figure 2). A cluster of significant associations for CARTta was detected on the proximal region of Chr 11. The strongest linkage was centered on marker D11Mit154 between D1Mit20 and D11Mit17 (LRS 21.1; p<0.05; 20cM–36cM). This marker was added to the background and the marker regression was recalculated. No additional associations were detected. At this interval, the additive effect is positive, thus indicating that the D2 allele increased transcript abundance expression. Scrutiny of the known genes on chromosome 11 around D11Mit154 reveals a number of intriguing possibilities for candidate genes including the QTL dopamine uptake transporter binding 3 (Dautb3) at 25 cM, a number of potential transcription factors, structural genes for GABA-A subunit beta 2 (Gabrb2) at 28.6 cM, glycine receptor alpha 1 (Glra1) at 30 cM and glutamate receptor, ionotropic, AMPA alpha 1 (Gria1) at 31 cM.
Figure 2.
Results of simple interval mapping data for CART system transcript abundance (CARTta) on chromosomes 4 and 11. Critical likelihood ratio statistic (LRS) values, represented here as a solid blue line, were determined with permutation test run through 10,000 iterations for the CARTta target. Confidence intervals for mapping were calculated by a bootstrap-resampling algorithm and are displayed as yellow bars in the chromosome-mapping figure. The Y-axis represents the LRS scores (as determined by marker regression) at each marker located on X-axis. Red line represents additive regression coefficient (scale represented on right-hand margin). A positive additive regression coefficient indicates that DBA/2J alleles increase CARTta trait values, whereas negative coefficient values indicate C57BL/6 alleles increase CARTta expression.
Interval mapping of CARTta on Chr 4 also revealed a highly suggestive-LRS peak (16.4) at D4Mit249, just at the edge of genome-wide significance (16.8). The Chr 4 LRS peak is a gene dense region with many potential candidate genes, including a large number of potential transcription factors and the structural gene for the hypocretin (orexin) receptor 1 (Hcrtr1). Clearly additional investigations will have to be completed to further localize these QTL and specify the potential causal genes and variants.
3. DISCUSSION
We conducted this experiment, using the BxD RI strains, to detect putative genetic loci regulating cocaine and amphetamine transcript abundance (CARTta). The distribution of CARTta is continuous across the BxD strains, with the average CARTta Ct value ranging from 22.44 (BxD31) to 26.87 (BxD14), consistent with this being a quantitative trait. We found that CARTta is a multifactorial trait with a substantial genetic component. The heritability of hypothalamic CARTta was ~31%. Genome-wide analysis revealed significant evidence of linkage to regions of mouse Chr 11 and chromosome 4.
CART is implicated in many neurobiological processes including drug addiction, dopamine system function, stress responses and feeding and satiety, thus QTL impacting these types of behaviors should be considered as potential candidate genes. While proposing candidate genes for any QTL detected in this study is premature, there were a number of interesting genes that mapped in the same locations as CART system transcript abundance QTL. The phenotypes to which these known QTL were associated could be alternative manifestations of the same fundamental genetic process detected with CART transcript abundance. The QTL Dautb3 (dopamine transporter binding 3), which impacts abundance of the dopamine transporter (DAT) in the caudate-putamen in both sexes maps at 25 cM on Chr 11 (Jones et al., 1999). This is an intriguing QTL as the DAT is one of the main targets for psychostimulant action. Another QTL potentially related to dopamine system function, Vmbic10 (ventral midbrain iron content) also maps at 24 cM on Chr 11 (Jones et al., 2003). A series of QTL related to body weight, food intake, energy balance and other metabolic parameters map to Chr 4 and 11 in the exact same regions as the CARTta QTL (Bevova et al., 2006; Moody et al., 1999; Plum et al., 2000; Ueda et al., 1999; Allan et al., 2005; Rocha et al., 2004).
Our study also revealed a significant association between D11Mit20 (20cM) and CARTta. The QTL Desp2 (despair 2), defined by responses in the forced swim test and tail suspension tests, maps at 21 cM on Chr 11, suggests a possible relationship between CART expression and these measures of stress response (Yoshikawa et al., 2002). This location is in close proximity to the GABAA receptor gene and a series of QTL associated with ethanol and pentobarbital dependence and withdrawal severity in BxD mice (Buck et al. 1998, 1999). This region is also implicated in initial sensitivity to ethanol (Kirstein et al. 2002). Comparison of the CARTta data to the WebQTL phenotypes database revealed modest correlations of CARTta-Ct values to a number of drug-related phenotypes such as ethanol sensitivity on test of loss of righting reflex, sleep time and hypothermia (r=−0.625, P<0.005;r=−0.5569 P<0.016;r=−0.5880, P<0.03 respectively), ethanol intake (r=0.683, P<0.003) and preference (r=−0.535, P<0.03) (Table 3).
Table 3.
Genetic correlation analysis of CARTta and published phenotypes using the BXD/Ty recombinant inbred mouse.
| PHENOTYPE | Correlation | p Values | Authors |
|---|---|---|---|
| Drd2 expression in nucleus accumbens | −0.8376 | 5.75e-05 | Jones et al., 1999 |
| Drd1 expression in caudate putamen | −0.8486 | 7.61e-05 | Jones et al., 1999 |
| Ethanol-duration of LORR | 0.6829 | 0.00244 | Rodriguez et al., 1995 |
| DA transporter expression in caudate-putamen | −0.6428 | 0.01140 | Jones et al., 1999 |
| Preference for 10% ETOH | −0.5349 | 0.03138 | Phillips et al., 1994 |
| Tolerance to 4g/kg ETOH-induced hypothermia | 0.5498 | 0.03171 | Crabbe et al., 1994 |
| Corticotropin-releasing factor transcript abundance in total hypothalamic RNA | 0.4614 | 0.01923 | Garlow et al., 2005 |
Selected phenotypes significantly associated with CARTa expression in the hypothalamus.
A suggestive linkage was also located within the mid-distal region of Chr 4 (at marker D4Mit249; cM 58, LRS 16.4, p<0.0001). This region is in very close proximity to Txlna, a ubiquitous cell membrane tethering protein, which has a role in intracellular trafficking of hormones, ions, etc. This suggestive QTL may be identical to an alcohol withdrawal QTL that has been mapped to this same region of Chr 4 (Buck et al. 1997, etc.). Thus, suggestive linkage on Chr 4 near D4Mit249 and Chr 11 near D11Mit20 suggests CARTta may be involved in liability for sedative-hypnotic drug dependence and withdrawal and should be considered as a plausible candidate involved in the addiction process.
There are several limitations to this study. The first is that it was conducted in one set of segregating animals, which was the BxD recombinant inbred set, so the linkage findings need to be confirmed in a separate, independent analysis. These experiments are ongoing, but the statistical significance of some of the associations, in particular for CARTta on chromosome 11, argues against these being false-positive findings. Another limitation is that all of the animals in this experiment were male, so no conclusions can be drawn as to the genetic regulation of the CART system in females. The resolution of the mapping algorithms employed, and the density of recombination events in the strain set are not sufficient to engage in fine mapping, so linkages in smaller candidate intervals cannot be detected, thus limiting the resolution of the mapping results. Fine scale mapping of these QTL will be dependent on additional analysis of other sets of animals (segregating F2 cross, advanced intercross recombinant inbred set, etc.) and only with high resolution fine mapping will genuine candidate genes become apparent as the target interval shrinks (Complex Trait Consortium, 2003).
In conclusion, the results from this study suggest that CART regulation in the hypothalamus is a complex trait (i.e., it is under influence of several genes) with a substantial genetic component. To our knowledge, this study is the first to identify and map QTL related to regulation of the CART system. The results from this study provide strong evidence for a number of QTL on chromosomes 4 and 11 that regulate the abundance of hypothalamic CART transcript.
4. EXPERIMENTAL PROCEDURES
4.1. Animals and Tissue Preparation
Foundation stock for 26 BxD lines and the C57BL/6 and DBA/2J parent lines were acquired from the Jackson Laboratory (Bar Harbor, ME) and used to establish a breeding colony. For a complete description of housing and colony conditions, please see Garlow et al., (2005, 2006). Animals were sacrificed by cervical dislocation at PND 75–90, under low stress conditions. Brains were rapidly dissected on ice into constituent anatomical regions and stored at −80°C until utilized.
4.2. RNA Quantification
Total hypothalamic RNA was isolated with the RNeasy Mini Kit (Quiagen Inc, Valencia, CA) by manufacturer’s method. See Garlow et al., (2005 2006) for complete RNA quantification methods. Amplification and detection primers for RT-PCR assays were from ABI “Assays-On-Demand” and included CART (ABI assay # Mm00489086; Accession # NM_013732.3), which generated an amplicon of 71 base pairs. This is a “relative abundance assay”, and target transcript abundance is expressed as the “detection threshold cycle” (Ct), which is in the middle of the exponential phase of the PCR amplification. As detection cycle is the readout parameter, a higher Ct value corresponds to lower transcript abundance, as more PCR cycles were required for target amplification. A dilution series positive control and no transcript and no reverse transcriptase negative controls were included in each assay. All assays were performed in duplicate and showed identical results.
4.3. Statistical Analysis and Genetic Mapping
Strain mean transcript abundances were determined from 6 individual male animals per line (Figure 1). Statistical analysis of transcript abundance data was conducted with the JMP-5 computer program (SAS Institute Inc, Cary, NC) on Macintosh G-4 computers. Descriptive statistics were calculated for each line, comparison of strain mean values was by ANOVA, and correlation of strain mean transcript abundance values was with Pearson product-moment analysis as implemented by JMP-5 (SAS Institute, Cary, NC). Effect size was estimated by ω2.
Quantitative trait genetic mapping was performed with the Windows version of QTX (http://www.mapmanager.org/mmQTX.html) (Manly et al., 2001). The BxD genetic marker set used with QTX was from Dr. Robert Williams at the University of Tennessee at Memphis (Williams, et al., 2001). This genetic marker set consisted of 975 error-checked and non-redundant loci. Results of genetic mapping with QTX are expressed as Likelihood Ratio Statistic (LRS). Statistical significance is the probability of a Type I (false positive) error, or the probability of obtaining by chance an LRS value as great as that observed (Haley and Knott, 1992). Marker regression was used to detect potential QTL and these calculations were carried out at increasingly stringent point (X2) significance levels, from p<0.001 through p<10−6. During the marker regression operation, QTX calculates a 95% confidence interval (CI) for the LRS peaks, expressed in centiMorgans (cM), and the width of the CI is inversely proportional to the strength of a QTL at that location (Darvasi and Soller, 1997). To control for the possible influence of other QTL impacting expression of the transcript abundance phenotype, the marker with the highest LRS value from the marker regression analysis was added to the trait background. The marker regression was then repeated to control for possible background loci effects. Simple interval mapping was used to localize QTL detected with marker regression on the relevant chromosomes, independent of the effects of other QTL (Lander and Botstein, 1989; Zeng, 1993, 1994). Significance levels (critical LRS) for interval mapping for each target transcript were determined empirically by permutation testing (10,000 iterations at 1 cM intervals) (Churchill and Doerge, 1994). The critical LRS values are defined as suggestive (corresponding to the 37th percentile or p<0.63), significant (at the 95th percentile or p<0.05) and highly significant (at the 99.9th percentile or p<0.001) (Lander and Kruglyak, 1995). Chromosomal locations of candidate and target structural genes are from the Mouse Genome Database (MGD), Mouse Genome Informatics (v3.1) Web Site [http://www.informatics.jax.org], at the Jackson Laboratory queried on (1/20/2005) (Blake et al., 2003).
This research project has been reviewed and approved by the Emory University Institutional Animal Care and Use Committee (IACUC) and complied with all relevant regulations.
Acknowledgments
This work was supported by National Institute of Health Grants K23 RR15531 and MH60745 (SJG) and by a United Negro College Fund/Merck Postdoctoral fellowship (EB). We thank Dr Robert Williams of the University of Tennessee at Memphis for advice and guidance in conducting these experiments.
Abbreviations
- CART
Cocaine and amphetamine-regulated transcript
- QTL
quantitative trait locus
- RT-PCR
Real-time PCR
- mRNA
- CARTta
CART transcript abundance
- ICV
Intracerebroventricular
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allan MF, Eisen EJ, Pomp D. Genomic Mapping of Direct and Correlated Responses to Long-Term Selection for Rapid Growth Rate in MIce. Genetics. 2005;170:1863–1877. doi: 10.1534/genetics.105.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkan B, Gozen O, Yararbas G, Koylu EO, Akinturk S, Kuhar MJ, Pogun S. CART expression in limbic regions of rat brain following forced swim stress: sex differences. Neuropeptides. 2006;40:185–193. doi: 10.1016/j.npep.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Phillips TJ, O’Toole LA. Quantitative trait loci associated with brain weight in BxD/Ty recombinant inbred mouse strains. Brain Res Bulletin. 1992;29:337–344. doi: 10.1016/0361-9230(92)90065-6. [DOI] [PubMed] [Google Scholar]
- Bevova MR, Aulchenko YS, Aksu S, Renne U, Brockman GA. Chromosome-Wise Dissection of the Genome of the Extremely Big Mouse Line DU61. Genetics. 2006;172:401–410. doi: 10.1534/genetics.104.040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Richardson JE, Bult CJ, Kadin JA, Eppig JT Group atmotMGD. The Mouse Genome Database. Nuc Acid Res. 2003;31:193–195. doi: 10.1093/nar/gkg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck KJ, Hood HM. Genetic association of a GABA(A) receptor gamma2 subunit variant with severity of acute physiological dependence on alcohol. Mamm Genome. 1998;9:975–978. doi: 10.1007/s003359900909. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TheComplexTraitConsortium. The nature and identification of quantitative trait loci: a community’s view. Nat Reviews Genetics. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couceyro PR, Evans C, McKinzie A, Mitchell D, Dube M, Hagshenas L, White FJ, Douglass J, Richards WG, Bannon AW. Cocaine and amphetamine-regulated transcript (CART) peptides modulate the locomotor and motivational properties of psychostimulants. Journal of Pharmacology and Experimental Therapeutics. 2005;315:1091–1000. doi: 10.1124/jpet.105.091678. [DOI] [PubMed] [Google Scholar]
- Couceyro PR, Koylu EO, Kuhar MJ. Further studies on the anatomical distribution of CART by in situ hybridization. J Chem Neuroanat. 1997;12:229–241. doi: 10.1016/s0891-0618(97)00212-3. [DOI] [PubMed] [Google Scholar]
- Dallvechia-Adams S, Kuhar MJ, Smith Y. Cocaine- and amphetamine-regulated transcript peptide projections in the ventral midbrain: Co-localization with gamma-aminobutyric acid, melanin-concentrating hormone, dynorphin, and synaptic interactions with dopamine neurons. J Comp Neurol. 2002;448:360–372. doi: 10.1002/cne.10268. [DOI] [PubMed] [Google Scholar]
- Darvasi A, Soller M. A Simple Method to Calculate Resolving Power and Confidence Interval of QTL Map Location. Behav Genetics. 1997;27:125–132. doi: 10.1023/a:1025685324830. [DOI] [PubMed] [Google Scholar]
- Dominguez G, Vicentic A, Del Giudice EM, Jaworski J, Hunter RG, Kuhar MJ. CART peptides: modulators of mesolimbic dopamine, feeding and stress. Ann NY Acad Sci. 2004;1025:363–369. doi: 10.1196/annals.1316.044. [DOI] [PubMed] [Google Scholar]
- Dominguez G, Kuhar MJ. Transcriptional regulation of the CART promoter in CATH. a cells. Brain Res Mol Brain Res. 2004;126:22–9. doi: 10.1016/j.molbrainres.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Dominguez G, Lakatos A, Kuhar MJ. Characterization of the CART peptide gene promoter and its activation by a cyclic AMP-dependent signaling pathway in GH2 cells. J Neurochem. 2002;80:885–893. doi: 10.1046/j.0022-3042.2002.00775.x. [DOI] [PubMed] [Google Scholar]
- Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J Neurosci. 1995;15:2471–81. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin VG, Markel PD, Johnson TE, Gehle VM, Jones BC. Common quantitative trait loci for alcohol-related behaviors and central nervous system neurotensin measures: hypnotic and hypothermic effects. J Pharm Exp Therapeutics. 1997;280:911–918. [PubMed] [Google Scholar]
- Garlow SJ, Boone E, Kinkead B, Nemeroff CB. Genetic analysis of the hypothalamic neurotensin system. Neuropsychopharmacology. 2006;31:535–543. doi: 10.1038/sj.npp.1300870. [DOI] [PubMed] [Google Scholar]
- Garlow SJ, Boone E, Li W, Owens MJ, Nemeroff CB. Genetic Analysis of the Hypothalamic Corticotropin Releasing Factor (CRF) System. Endocrinology. 2005;146:2362–2368. doi: 10.1210/en.2004-1450. [DOI] [PubMed] [Google Scholar]
- Gora-Masalak G, McClearn GE, Crabbe JC, Phillips TJ, Belknap JK, Plomin R. Use of recombinant inbred strains to identify quantitative trait loci in psychopharmacology. Psychopharm. 1991;104:413–424. doi: 10.1007/BF02245643. [DOI] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hubert GW, Kuhar MJ. Colocalization of CART peptide with prodynorphin and dopamine D1 receptors in the rat nucleus accumbens. Neuropeptides. 2006;40:409–415. doi: 10.1016/j.npep.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hunter RG, Vicentic A, Rogge G, Kuhar MJ. The effects of cocaine on CART expression in the rat nucleus accumbens: a possible role for corticosterone. Eur J Pharmacol. 2005;517:45–50. doi: 10.1016/j.ejphar.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Jaworski JN, Kimmel HL, Mitrano DA, Tallarida RJ, Kuhar MJ. Intra-VTA CART 55–102 reduces the locomotor effect os systemic cocaine in reats: An isobolographic analysis. Neuropeptides. 2007;41:65–72. doi: 10.1016/j.npep.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski JN, Kozel MA, Philpot KB, Kuhar MJ. Intra-accumbal injection of CART (cocaine and amphetamine-regulated transcript) peptide reduces cocaine-induced locomotor activity. Journal of Pharmacology and Experimental Therapeutics. 2003;307:1038–1044. doi: 10.1124/jpet.103.052332. [DOI] [PubMed] [Google Scholar]
- Jones BC, Reed CL, Hitzemann R, Wiesinger JA, McCarthy KA, Buwen JP, Beard JL. Quantitative genetic analysis of ventral midbrain and liver iron in BxD recombinant inbred mice. Nutr Neurosci. 2003;6:369–377. doi: 10.1080/10284150310001624192. [DOI] [PubMed] [Google Scholar]
- Jones BC, Tarantino LM, Rodriguez LA, Reed CL, McClearn GE, Plomin R, Erwin VG. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9:607–617. [PubMed] [Google Scholar]
- JMP Statistics and Graphics Guide, Version 5. Cary, N.C.: SAS Institute Inc; 2002. [Google Scholar]
- Kimmel HL, Gong W, Vechia SD, Hunter RG, Kuhar MJ. Intra-ventral tegmental area injection of rat cocaine and amphetamine-regulated transcript peptide 55–102 induces locomotor activity and promotes conditioned place preference. Journal of Pharm and Exp Ther. 2000;294:784–792. [PubMed] [Google Scholar]
- Kirstein SL, Davidson KL, Ehringer MA, Sikela JM, Erwin VG, Tabakoff B. Quantitative trait loci affecting initial sensitivity and acute functional tolerance to ethanol-induced ataxia and brain cAMP signaling in BxD recombinant inbred mice. J Pharmacol Exp Ther. 2002;302:1238–1245. doi: 10.1124/jpet.302.3.1238. [DOI] [PubMed] [Google Scholar]
- Koylu EO, Couceyro PR, Lambert PD, Kuhar MJ. Cocaine- and amphetamine-regulated transcript peptide immunohistochemical localization in the rat brain. J Comp Neurol. 1998;391:115–132. [PubMed] [Google Scholar]
- Koylu EO, Couceyro PR, Lambert PD, Ling NC, DeSouza EB, Kuhar MJ. mmunohistochemical localization of novel CART peptides in rat hypothalamus, pituitary and adrenal gland. J Neuroendocrinol. 1997;9:823–833. doi: 10.1046/j.1365-2826.1997.00651.x. [DOI] [PubMed] [Google Scholar]
- Kuhar MJ, Jaworski JN, Hubert GW, Philpot KB, Dominguez G. Cocaine and amphetamine regulated transcript peptides play a role in drug abuse and are potential therapeutic targets. AAPS Journal. 2005;7:E259–E265. doi: 10.1208/aapsj070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos A, Dominguez G, Kuhar MJ. CART promoter CRE site binds phosphorylated CREB. Brain Res Mol Brain Res. 2002;104:81–85. doi: 10.1016/s0169-328x(02)00321-2. [DOI] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L. Genetic dissection of complex traits - Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PJ, Seier V, Fink-Jensen A, Holst J, Warberg J, Vrang N. Cocaine- and amphetamine-regulated transcript is present in hypothalamic neuroendocrine neurones and is released to the hypothalamic-pituitary portal circuit. J Neuroendocrinol. 2003;15:219–226. doi: 10.1046/j.1365-2826.2003.00960.x. [DOI] [PubMed] [Google Scholar]
- Manly K, Cudmore JRH, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mammal Gen. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- Mattson BJ, Morrell JI. Preference for cocaine versus pup-associated cues differentially activates neurons expressing either Fos or CART in lactating, maternal rodents. Neuroscience. 2005;135:315–328. doi: 10.1016/j.neuroscience.2005.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DE, Pomp D, Nielsen MK, Van Vleck LD. Identification of Quantitative Trait Loci Influencing Traits Related to Energy Balance in Selection and Inbred LInes of Mice. Genetics. 1999;152:699–711. doi: 10.1093/genetics/152.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann PE. Infrences in linkage analysis of multifactorial traits using recombinant inbred strains of mice. Behavior Genetics. 1992;22:665–676. doi: 10.1007/BF01066637. [DOI] [PubMed] [Google Scholar]
- Plum L, Kluge R, Giesen K, Altmuller J, Ortlepp JR, Joost HG. Type-2 diabetes-like hyperglycemia in a backcross model of NZO and SJL mice: characterization of susceptibility locus on chromosome 4 and its relationship with obesity. Diabetes. 2000;49:1590–1596. doi: 10.2337/diabetes.49.9.1590. [DOI] [PubMed] [Google Scholar]
- Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: II Body composition. Mamm Genome. 2004;15:100–113. doi: 10.1007/s00335-003-2308-6. [DOI] [PubMed] [Google Scholar]
- Salinas A, Wilde JD, Maldve RE. Ethanol enhancement of CART mRNA and peptide expression in the nucleus accumbens. J Neurochem. 2006;97:408–415. doi: 10.1111/j.1471-4159.2006.03745.x. [DOI] [PubMed] [Google Scholar]
- Sanzchèz E, Fekete C, Lechan RM, Joseph-Bravo P. Cocaine-and amphetamine-regulated transcript (CART) expression is differentially regulated in the hypothalamic paraventricular nucleus of lactating rats exposed to suckling or cold stimulation. Brain Research. 2007;9:120–128. doi: 10.1016/j.brainres.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh KR. Effects of cocaine and amphetamine-regulated transcript peptide on turnover of central dopaminergic neurons. Neuropharmacology. 2003;44:940–8. doi: 10.1016/s0028-3908(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Murphy KG, Bewick GA, Kong WM, Opacka-Juffry J, Gardiner JV, Ghatei M, Small CJ, Bloom SR. Regulation of rat pituitary cocaine-and amphetamine-regulated transcript (CART) by CRH and glucocorticoids. Am J Physiol Endocrinol Metab. 2004;287:E583–90. doi: 10.1152/ajpendo.00576.2003. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Small CJ, Murphy KG, Rayes E, Abbott CR, Seal LJ. Actions of cocaine and amphetamine-regulated transcript peptide on regulation of appetite and hypothalamo-pituitary axes in vitro and in vivo in male rats. Brain Research. 2001;893:186–94. doi: 10.1016/s0006-8993(00)03312-6. [DOI] [PubMed] [Google Scholar]
- Ueda H, Ikegami H, Kawaguchi Y, Fulisawa T, Yamato E, Shibata M, Ogihara T. Genetic analysis of late-onset type 2 diabetes in a mouse model of human complex trait. Diabetes. 1999;48:1168–1174. doi: 10.2337/diabetes.48.5.1168. [DOI] [PubMed] [Google Scholar]
- Van Vugt DA, Lujan ME, Froats M, Krzemien A, Couceyro PR, Reid RL. Effect of fasting on cocaine-amphetamine-regulated transcript, neuropeptide Y, and leptin receptor expression in the non-human primate hypothalamus. Neuroendocrinology. 2006;84:83–93. doi: 10.1159/000097494. [DOI] [PubMed] [Google Scholar]
- Vicentic A, Hunter RG, Kuhar MJ. Effect of corticosterone on CART peptide levels in rat blood. Peptides. 2005;26:531–535. doi: 10.1016/j.peptides.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Vincentic A, Lakatos A, Hunter R, Philpot K, Dominguez G, Kuhar MJ. CART peptide diurnal rhythm in brain and effect of fasting. Brain Res. 2005;1032:111–115. doi: 10.1016/j.brainres.2004.10.053. [DOI] [PubMed] [Google Scholar]
- Vincentic A, Dominguez G, Hunter RG, Philpot K, Wilson M, Kuhar MJ. Cocaine and amphetamine-regulated transcript peptide levels in blood exhibit a diurnal rhythm: regulation by glucocorticoids. Endocrinology. 2004;145:4119–4124. doi: 10.1210/en.2003-1648. [DOI] [PubMed] [Google Scholar]
- Vrang N, Tang-Christiansen M, Larsen LK, Kristensen P. Recombinant CART peptide induces c-Fos expression in central areas involved in control of feeding behaviour. Brain Research. 1999;818:499–509. doi: 10.1016/s0006-8993(98)01349-3. [DOI] [PubMed] [Google Scholar]
- Williams RW, Gu J, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:1–18. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Pan JT, Li HY. CART peptide increases the mesolimbic dopaminergic neuronal activity: a microdialysis study. European Journal of Pharmacology. 2004;494:179–82. doi: 10.1016/j.ejphar.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Watanabe A, Ishitsuka Y, Nakaya A, Nakatani N. Identification of multiple genetic loci linked to the propensity for ‘behavioral despair’ in mice. Genome Res. 2002;12:357–366. doi: 10.1101/gr.222602. [DOI] [PubMed] [Google Scholar]
- Zeng ZB. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. PNAS. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gershenfeld HK. Genetic contributions to body weight in mice: relationship of exploratory behavior to weight. Obes xcRes. 2003;11:828–838. doi: 10.1038/oby.2003.114. [DOI] [PubMed] [Google Scholar]