Introduction
Ataxin 2 binding protein 1 (A2BP1 aka FOX1, RBFOX1) is an RNA binding protein found on chromosome 16p13.2 responsible for regulation of pre-mRNA splicing events in a number of critical developmental genes. It is expressed in muscle, heart and neuronal cells and is known to bind to the (U)GCAUG element found in mRNA precursors [Jin et al., 2003; Shibata et al., 2000; Underwood et al., 2005].
A2BP1 exists in many alternatively spliced isoforms that exhibit a tissue specific expression pattern [Nakahata et al., 2005]. It plays a significant role in alternative splicing of critical neuronal developmental genes and over expression has been shown to increase splicing activation of exons specific for neuronal activation [Jin et al., 2003; Nakahata and Kawamoto 2005; Underwood et al. 2005; Kuroyanagi et al., 2006; Li et al., 2007; Zhou et al., 2007]. For example, A2BP1 is responsible for inclusion of exon five in the neuronal form of glutamate receptor NMDA receptor 1 (GRIN1) [Brudno et al., 2001; Minovitsky et al., 2005; Nakahata and Kawamoto 2005; Underwood et al., 2005; Zhang et al., 2008; Yeo et al., 2009].
Rare copy number abnormalities of A2BP1 have been previously associated with cognitive impairment, attention deficit disorder and autism [Elia et al., 2010; Martin et al., 2007]. Additionally, A2BP1 has been under investigation in autism due to its potential functional relevance as well as its candidate status in positional mapping [Barnby et al., 2005] and association studies [Martin et al., 2007]. Recently, A2BP1 has been identified as a hub in the top expression module in the autistic brain [Voienagu et al., 2011]. Here we provide an in depth case study of a family in which a paternally transmitted A2BP1 deletion was identified in a proband with autism and developmental left hemiparesis.
Molecular Methods
Illumina 1M-Duo DNA Analysis BeadChip
Whole blood DNA from the proband was genotyped for 1.2 million markers using the Human 1M-Duo DNA Analysis BeadChip [Illumina Inc., San Diego, CA, USA] according to the manufactures protocol. Briefly, 200ng of DNA (4uL at 50ng/uL) was amplified, biotin labeled, and hybridized to the microarray. The array was then scanned with default settings using the Illumina BeadArray Reader (BAR) at The Center for Applied Genomics genotyping core [Toronto, CA, http://www.tcag.ca/]. Genotype calls were generated using the Illumina-provided genotype cluster definitions file (Human 1M Duo, generated using HapMap project DNA samples) with a Gencall cutoff of 0.15.
SYBR Green Quantitative Real-Time PCR
SYBR green qPCR was performed on the proband, mother and father to 1) validate the deletion detected by the microarray and 2) determine if the deletion was inherited or de novo. All qPCR primers were picked from hg18 build of genomic DNA sequence obtained from the UCSC Genome Browser (table 1). The primers were designed using Primer3 [Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html. Rozen and Skaletsky, 1998]. Specificity was evaluated in-silico using the UCSC PCR and BLAT tools, and determined empirically by evaluation of the melt curve data. The qPCR reactions contained 4.7µl of 2× Roche SYBR Green PCR Master Mix (Roche), 4.8µl genomic DNA (1ng/µl) and 0.25µl of each primer (10pmol/µl) in a total volume of 10µl. Realtime PCR was run using a Roche 480 Light Cycler Real-Time PCR System. Each sample was amplified in quadruplicate with primers designed to two assay control genes, GUSB and HMBS as well as the putative CNV. qPCR results were analyzed using the ΔΔCt method and the data was normalized by setting a pooled genomic DNA reference (Promega) to a fold change of 1.0.
Table 1.
Primers used in SYBR green assays and exon sequencing assay. All SYBR green assays were run in quadruplicate at an annealing temperature of 55 degrees on the Roche 480 Light Cycler. The exon sequencing reaction was run using standard conditions at an annealing temperature of 58 degrees.
| Primer ID | Application | Gene Name |
Amplicon Size |
Primer Sequence |
|---|---|---|---|---|
| A2BP1_1F | SYBR target gene | A2BP1 | 115 | TGTTTCTTACACAGCACAGG |
| A2BP1_1R | SYBR target gene | A2BP1 | 115 | CTTGTTTCCTGATTACACAGC |
| A2BP1_2F | SYBR target gene | A2BP1 | 125 | AATACATCCATGTCTTTGATCC |
| A2BP1_2R | SYBR target gene | A2BP1 | 125 | TTCTCATCTCTGATACAATAGGG |
| GUSB_1F | SYBR reference gene | GUSB | 90 | CTGAATGCAGCCTTGACCTA |
| GUSB_1R | SYBR reference gene | GUSB | 90 | GGGCATGGTTATGAGTGCTT |
| HMBS_1F | SYBR reference gene | HMBS | 90 | GCAGCTCATAGGTGGGTTTT |
| HMBS_1R | SYBR reference gene | HMBS | 90 | CCCAGCCATTCTTGCAGT |
| A2BP1_set1f | Exon Sequencing | A2BP1 | 334 | CTTTGCCAGGATCTGAGAGG |
| A2BP1_set1r | Exon Sequencing | A2BP1 | 334 | GAGAACAGGATCAAAGACATGG |
Sequencing
An assay was developed to sequence the 2nd exon of the 4th isoform of A2BP1, which is located within the deletion region. Primers were selected using the hg18 assembly on the UCSC browser (table 1). The sequencing PCR was run using standard conditions at 58° in triplicate on an ABI 9700 thermocycler. The resultant product was sequenced using an ABI BigDye reaction, which was subsequently analyzed with an ABI 3730 Sequencer and visualized using the Sequencher program.
Copy Number Detection
We used CNVision, a program developed by Stephan Sanders and Christopher Mason at Yale University, to detect copy number variations in our Illumina 1M array data [http://www.softpedia.com/get/Science-CAD/CNVision.shtml]. CNVision utilizes PennCNV [Wang et al 2007], QuantiSNP [Collela et al., 2007] and Gnosis (unpublished) in a bundled program designed to identify and merge overlapping CNV calls for high confidence CNV detection.
Phenotypic Characterization
Behavioral Assessment
The family was recruited and assessed by the Autism Center of Excellence (ACE) at the University of Illinois at Chicago according to an approved IRB protocol. The proband, at the time of testing, was twelve years old and in fourth grade. He was in a general education classroom, receiving occupational, speech, and vision therapy while at school. His family indicated that he is right-hand dominant.
A full diagnostic evaluation was performed including the Autism Diagnostic Interview-Revised (ADI-R) [Le Couteur et al., 1989], completed by the proband’s mother and module 3 of the Autism Diagnostic Observation Schedule (ADOS-WPS) [Lord, Rutter, et al., 1999]. Additional measures including the Repetitive Behavior Scale-Revised (RBS-R)[Lam and Aman, 2007], the Aberrant Behavior Checklist-Community Version (ABC-CV) [Aman, Singh, et al., 1985] and the Childhood Routines Inventory (CRI) [Evans, Leckman et al., 1997] were completed to gather information on repetitive behaviors. The Social Communication Questionnaire (SCQ) [Rutter, Bailey et al., 2003] and the Social Responsiveness Scale (SRS) [Constantino, Davis et al., 2003] assessed his current level and quality of social interaction. The Clinical Evaluation of Language Fundamentals (CELF) [Semel, Wiig, et al., 2003] was administered to evaluate language ability and the Wechsler Abbreviated Scale of Intelligence (WASI) [Wechsler, 1999] was used to evaluate current levels of verbal and nonverbal abilities. Finally, the Vineland Adaptive Behavior Scales (VABS-II) [Sparrow, Cicchetti et al., 2005] were administered to assess adaptive functioning. Results from these tests are summarized and presented in table 2.
Table 2.
Scores on clinical measures administered as a part of the UIC ACE diagnostic and clinical battery of tests
| Proband | Father | Mother | ||
|---|---|---|---|---|
| Autism Diagnostic Interview - Revised | ||||
| Impairments in Reciprocal Social Interaction | 23 | -- | -- | |
| Qualitative Abnormalities in Communication | 20 | -- | -- | |
| Restricted And Repetitive Patterns of Behavior | 6 | -- | -- | |
| Abnormality Of Development Evident At Or Before 36 Months | 3 | -- | -- | |
| Autism Diagnostic Observation Schedule | ||||
| Communication | 4 | -- | -- | |
| Social | 7 | -- | -- | |
| Social Affect | 7 | -- | -- | |
| Restricted Repetitive Behaviors | 3 | -- | -- | |
| Calibrated Severity Score | 6 | -- | -- | |
| Additional Clinical Measures | ||||
| Vineland Adaptive Behavior Scale | 98 (45) | -- | -- | |
| Clinical Evaluation of Language Fundamentals (CELF-4) | 109 (73) | -- | -- | |
| Broader Autism Phenotype Questionnaire | ||||
| Rigid | -- | 2.92 | 3.08 | |
| Aloof | -- | 3.75 | 1.58 | |
| Pragmatic Language | -- | 2.67 | 2.00 | |
| Additional Research Measures | ||||
| Social Communication Questionnaire | 26 | -- | -- | |
| Repetitive Behavior Scale - Revised | 6 | -- | -- | |
| Aberrant Behavior Checklist-Community | 27 | -- | -- | |
| Childhood Routines Inventory, version 1.2 | 44 | -- | -- | |
At evaluation, his language was fluent, and conversational, but could be repetitive. His use of gestures was limited and he primarily communicated verbally. Per his evaluation, the proband used overly formal words in conversation and his tone and prosody were atypical. He also showed impairments in nonverbal communication. The proband exhibited multiple sensory-seeking behaviors, and was also sensitive to certain stimuli. For example, he often touched objects to his lips, neck, and face to see how they felt. He was sensitive to loud noises, like train horns, and covered his ears in response to these sounds.
Notable behavior patterns included avoidance of eye contact, staring at hands, preference for certain foods and routines, and sensitivity to particular pieces of clothing. While he appeared to want to be around others and enjoyed physical contact, he indicated that other people irritated him. He primarily had conversations about a circumscribed range of topics, preferred solitary activities, and talked to himself loudly. He consistently engaged in reciprocal social smiling, but was described as often having a flat affect. During his evaluation, he exhibited limited insight into others’ experiences and showed a narrow understanding of the nature of interpersonal relationships.
Developmental and Family History
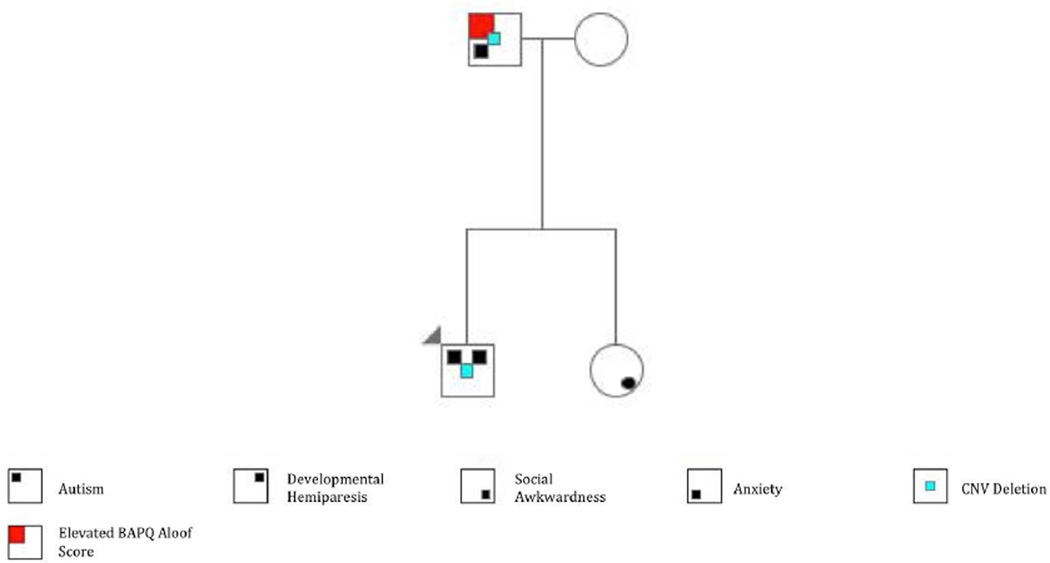
Though the family is considered simplex, the pedigree is notable for anxiety (sister) according to maternal report (Figure 1) and social isolation (father) (Table 1). The father previously had a dermatofibrosarcoma protuberans removed.
Figure 1. Pedigree.
The proband presented with autism and mild paresis on his left side. The father was diagnosed with anxiety and had an elevated BAPQ aloof score. Both the proband and the father carried a deletion in the A2BP1 gene (chr16:6356187-6369513). The proband’s sibling was unaffected, but was described as socially awkward. The arrowhead denotes the proband.
The proband was the 7lb 15 oz product of a 38-week gestation. The mother reported six miscarriages and one ectopic pregnancy prior to conception. The parents participated in a recurrent miscarriage study between the fifth and twelfth weeks of pregnancy, which involved maternal subcutaneous injection of paternal white blood cells. No apparent complications arose as a result of participation in the study.
During infancy the proband demonstrated feeding difficulties, generalized hypotonia and weakness of the left leg and arm. At approximately 19 months his mother noticed loss of previously normal language development, including eight different words. Language loss persisted for 11 months. At the age of 22 months, the proband underwent a neurological examination, which was notable for macrocephaly. Tests included EEG, MRI of the brain, serum amino acids, and Fragile X testing, all of which were negative or normal. At the age of 43 months, the proband was evaluated and given a diagnosis of autistic disorder. Repeat EEG at 5 years showed left temporal and frontal epileptiform waves, but to date there has been no report of clinical seizure.
Neuropsychiatric Assessment
During a neurological examination, his parents reported that he had achieved gross motor milestones within normal limits, but that he dragged his left leg while learning to crawl. At present, he shows a pattern of asymmetric physical development consistent with mild left hemiparesis. His left arm is shorter (56.5 vs 60 cm) and has reduced circumference relative to his right arm. Stretch reflexes were 2+ and symmetrical throughout. Additionally there were no asymmetries evident in his legs. The left side of his face was less mobile than the right side and his head circumference was in the macrocephalic range (56.5 cm, 98th percentile).
Neuropsychological testing was repeated after one year to ensure that results that appeared much poorer than his functional level were not confounded by inattention or fatigue (Table 3). He was right-hand dominant for the majority of tasks (16/23) but also was ambidexterous for 7 tasks [Oldfield, 1971]. The proband’s performance was in the average range on tests of visuomotor planning (Trails A) [Spreen and Gaddes, 1969], cognitive flexibility (Trails B) and bilateral gross motor speed (finger tapping) [Baron, 2003]. Overall grip strength was greater with his dominant relative to his non-dominant hand (32 vs. 23 kg). During a test of fine-motor planning and speed (grooved pegboard), his performance was relatively impaired for his dominant compared to his non-dominant hand (dominant hand standard score (SS); time 1: <50; time 2: 80; non-dominant hand SS; time 1: 87; time 2; 107) [Knights and Norwood, 1980; Heaton, Grant, et al., 1986]. An isometric grip force test was administered in which the proband was instructed to sustain a steady force contraction while a line moved vertically to provide online visual feedback about his performance [Vaillancourt, Mayka, et al., 2006]. He showed a 38% increase in error variability for his dominant (root mean squared error = 10.45 (7.50)) compared to his non-dominant hand (RMSE=7.57 (5.56)) across different force levels.
Table 3.
Standard scores and percentile ranks (in parentheses) for the proband, father and mother on a battery of neuropsychological tests (mean=100, sd=15). The trails tasks, the grooved pegboard task and the finger tapping task were administered to the proband at two different time points, one year apart in order to confirm a persistent asymmetric pattern of motor performance. The Edinburgh Handedness (Oldfield, 1971) was administered to quantitatively assess the proband’s lateralization of functions of his hands, feet, and eyes. Scores reflect number of behaviors for which the patient preferred to use his right or left hand, or had no lateral preference.
| Proband | Father | Mother | ||
|---|---|---|---|---|
| Time 1 | Time 2 | |||
| Trails A | 98 (45) | 106 (66) | 105 (63) | 101 (53) |
| Trails B | 98 (45) | 109 (73) | 115 (84) | 119 (90) |
| Grooved Pegboard | ||||
| Left Hand | 87 (19) | 107 (68) | 113 (81) | 93 (32) |
| Right Hand | <50 (<.1) | 80 (9) | 101 (53) | 77 (6) |
| Finger Tapping | ||||
| Left Hand | 115 (84) | 102 (55) | 100 (50) | 87 (19) |
| Right Hand | 112 (79) | 97 (27) | 85 (16) | 74 (4) |
| Wechsler Abbreviated Scale of Intelligence | ||||
| Full Scale IQ | 126 (96) | 120 (91) | 111 (77) | |
| Verbal IQ | 129 (97) | 122 (93) | 119 (90) | |
| Performance IQ | 117 (87) | 112 (73) | 101 (53) | |
| Edinburgh Handedness | ||||
| Left Hand | 0 | |||
| Right Hand | 16 | |||
| Ambidexterity | 7 | |||
Molecular Characterization of A2BP1 Deletion
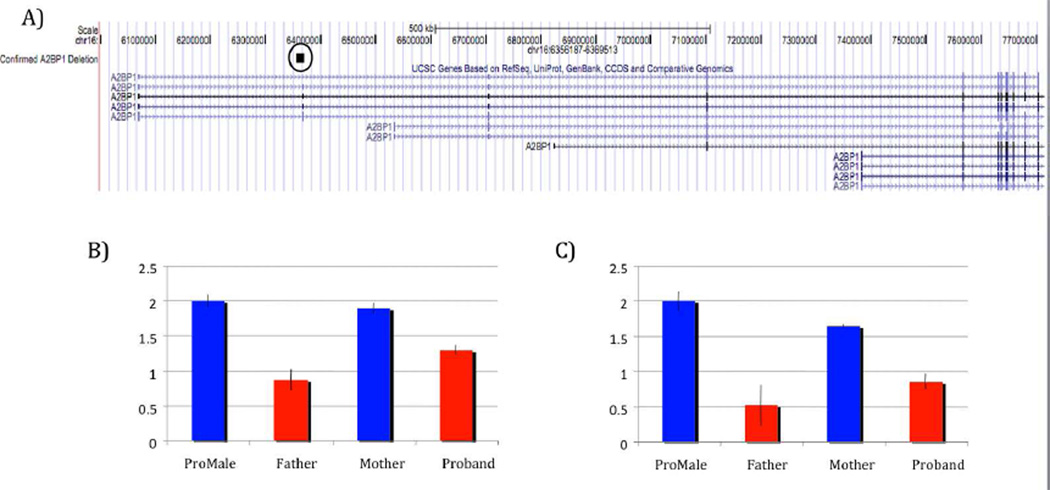
The 1.3 kb deletion was detected in the proband by all three CNV algorithms in CNVision based on a loss of signal intensity for 16 consecutive probes on the 1M array. The deletion (chr16:6356187-6369513, hg19) removes the second exon (63 bps) of the fourth (i.e., the longest) isoform of A2BP1 along with flanking intronic sequence. The deletion was validated by real-time PCR in the proband and found to be paternally inherited (Figure 2). The exon residing within the deleted region was sequenced to rule out the possibility of a compound heterozygous mutation. The sequencing was negative for any variation within the deleted region.
Figure 2. Validated Deletion of A2BP1.
A)The circled block highlights the confirmed deletion found in the A2BP1 gene of the proband and father. This deletion consist of a loss of signal across 16 consecutive probes covering chr16:6296188-6309514. B) Quantitative PCR performed to confirm the deletion and establish inheritance in the proband utilizing HMBS as the reference gene. C) Quantitative PCR performed to confirm the deletion and establish inheritance in the proband utilizing GUSB as the reference gene (SD shown).
Discussion
A2BP1 is one of the largest genes in the genome (1.7 Mb) and plays an important role as a master regulator of splicing activity in the brain and muscle. Interestingly, the protein serves, in a tissue specific manner, to repress splicing when bound to an intron located upstream of the target alternative exon and as a splicing enhancer when bound to a downstream element of the alternative exon [Black, 1992; Huh and Hynes, 1994; Modafferi and Black, 1997; Jin et al., 2003; Underwood et al., 2005; Zhang et al., 2008; Yeo et al., 2009]. For example, muscle specific isoforms of A2BP1 have been shown to repress inclusion of muscle specific exons in nonmuscle tissues in mice while neuronal isoforms of A2BP1 are responsible for inclusion of neuron-specific exons in brain [Nakahata and Kawamoto, 2005; Underwood et al., 2005]. While the mRNA recognition element for A2BP1 has been known for some time, the target genes are less well characterized. Zhang et al. [2008] attempted to identify and characterize all targets of A2BP1. Eleven thousand and three target transcripts were predicted and significant enrichment of neuromuscular GO terms and disease annotations were found in this target set. Notable within the list of targets were NLGN3 and NLGN4X, which have previously been implicated in autism [Jamain et al., 2003].
There is a growing body of evidence to suggest that A2BP1 may play a role in the etiology of developmental delay and autism. Previous studies of autism, ADHD, intellectual disability and epilepsy have identified rare copy number variants in A2BP1 [Elia et al., 2010; Martin et al, 2007; Bhalla et al., 2004]. One such study found translocations in two unrelated patients with cognitive impairment and epilepsy, both of which disrupted the A2BP1 gene [Bhalla et al., 2004]. Martin et al. [2007] identified a de novo translocation with a subsequent loss of A2BP1 at one of the breakpoints. Additionally Elia et al., [2010] reported three copy number variants in their sample of children with ADHD including one duplication and two deletions, all of which were inherited and two of which were inherited from affected parents. The deletion presented in this paper along with previously reported translocations and CNVs associated with psychiatric phenotypes are presented in Figure 3. More recently, Voingeau et al. [2011] also identified differential alternative splicing events in the brain tissue of donors with autism and reduced expression of A2BP1, when compared to controls. GRIN1, and MEF2C are notable genes in this list of A2BP1 mediated splicing events unique to the autism transcriptome.
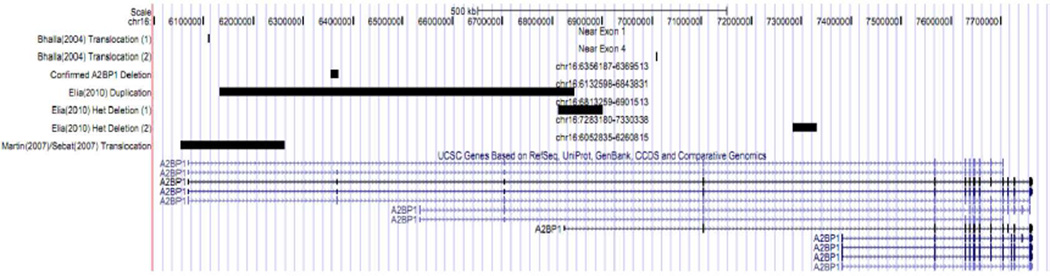
Figure 3. Confirmed A2BP1 Deletion and Previously Confirmed Deletions in A2BP1.
A) Displays the validated CNV found in both the proband and father covering chr16:6296188-6309514, A2BP1 duplication and deletions detected via beadchip and found to be associated with ADHD and autism [Elia et al., 2010], and deletion detected through FISH from Martin et al. [2007] found in an autistic female with severe developmental disability, epilepsy, delayed walking with mild residual ataxia, behavioral regression, fluctuating liver function tests and mild cerebellar atrophy. B) Graphs display two deletion endpoints from Bhalla et al. [2004] determined by FISH. The deletions were detected in two patients, one with epilepsy, the second with intellectual impairment. The exact length of the deletions could not be determined. (Het, heterozygous deletion)
The first three exons of A2BP1 (isoform 4) are not translated and contain extensive regulatory elements. The deletion found in the proband and his father encompasses the second exon and flanking intronic regions of the fourth isoform (chr16:6296188-6309514). The case presented here is remarkable not only for his autism but also for his global hypotonia, his left side developmental hemiparesis and an additional lateralized deficit in motor planning and coordination affecting his dominant hand. There was no central nervous system abnormality found on MRI during a prior evaluation, and no complications were experienced during the pregnancy or during delivery. During early development, the hemiparesis did not interfere with daily functions using his non-dominant (left) hand or arm. He is right hand dominant and he and his parents had not previously noted the physical asymmetry in the size and circumference of his arms but were aware of early developmental left side weakness. At the time of his clinical evaluation, the heimparesis was largely resolved with no difference between lower extremities and only size asymmetry remaining apparent in upper extremities as well as reduced mobility on the left side of the face. Interestingly, the patient described in Martin et al., [2007] suffered from a similar phenotype of hypotonia in the lower extremities. The left-sided developmental hemiparesis and contrasting right-sided dyspraxia may reflect an alteration in the maturation of hemispheric specialization or independent pathologies that conferred distinct abnormalities on his physical and motor development. In order to confirm our original neuropsychological findings of dyspraxia affecting the hand (right) opposite his hemiparetic limb (left), the proband returned for a second testing session. His performance improved during the second testing session, judged clinically to most likely result from increased attentiveness. The relative deficit was still present in his right hand. Similarly, the proband demonstrated decreased force stability with his right compared to his left hand during a test mediated by cortico-cerebellar circuitry [Coombes et al., 2011]. These findings are consistent with previously documented disruptions of motor lateralization in autism suggesting an alteration in the maturation of hemispheric specialization or an abnormality that differentially affects left motor cortices and/or right cerebellum [Escalante-Mead et al., 2003; Mosconi et al., 2010]. It is not clear whether this distinct neurological phenotype is unique to the A2BP1 deletion or more generally associated with autism. We were not able to collect neuropsychological and sensorimotor data on the father, so it remains unclear whether the motor atypicalities observed in the proband are also present in his father. Heterogeneity among individuals carrying the same risk associated structural variant has been well documented in autism [Sanders et al., 2011].
Copy number variants in A2BP1 have been noted in the Database of Genomic Variants (DGV). Intronic losses and gains are not uncommon among unscreened control populations, such as those residing in the DGV. This phenomenon is not surprising considering that A2BP1 is one of the largest genes in the genome (1.7 Mb). Exonic CNVs, while also present, are much less common. One CNV in the DGV overlaps the CNV region presented in this paper. We present in this study an in depth phenotypic analysis of a confirmed deletion in the A2BP1 gene. As with all rare findings, we cannot conclusively state that the A2BP1 deletion in this proband results directly in his autism. However, taken together with previously published studies, our data provides further evidence for a role of A2BP1 in the development of autism and associated motor asymmetries.
References
- Aman MG, Singh NN, Stewart AW, Field CJ. The Aberrant Behavior Checklist: a behavior rating scale for the assessment of treatment effects. American Journal of Mental Deficiency. 1985;89(5):485–491. [PubMed] [Google Scholar]
- Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. American Journal of Human Genetics. 2005;76(6):950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron IS. Domains and Tests. In: Baron IS, editor. Neuropsychological Evaluation of the Child. New York: Oxford; 2003. pp. 93–106. [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. Journal of Human Genetics. 2004;49(6):308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Black DL. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69(5):795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Research. 2001;29(11):2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collela S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Research. 2007;35(6):2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, Metzger LM, Shoushtari CS, Splinter R, Reich W. Validation of a Brief Quantitative Measure of Autistic Traits: Comparison of the Social Responsiveness Scale with the Autism Diagnostic Interview-Revised. Journal of Autism & Developmental Disorders. 2003;33(4):427–433. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- Coombes SA, Corocos DM, Pavuluri MN, Vaillancourt DE. Maintaining force control despite changes in emotional context engages dorsomedial prefrontal and premotor cortex. Cerebral Cortex. 2011 doi: 10.1093/cercor/bhr141. (Epub June 17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D'Arcy M, deBerardinis R, Frackelton E, Kim C, Lantieri F, Muganga BM, Wang L, Takeda T, Rappaport EF, Grant SF, Berrettini W, Devoto M, Shaikh TH, Hakonarson H, White PS. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular Psychiatry. 2010;15(6):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Mead PR, Minshew NJ, Sweeny JA. Abnormal brain alteralization in high functioning autism. Journal of Autism and Developmental Disorders. 2003;33(5):539–543. doi: 10.1023/a:1025887713788. [DOI] [PubMed] [Google Scholar]
- Evans DW, Leckman JF, Carter A, Reznick JS, Henshaw D, King RA, Pauls D. Ritual, habit, and perfectionism: The prevalence and development of compulsive-like behavior in normal young children. Child Development. 1997;68(1):58–68. [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox-1 (A2BP1) controls neuronal excitation in the mamalian brain. Nature Genetics. 2011 doi: 10.1038/ng.841. (Epub May 29). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammock EA, Levitt P. Developmental expression mapping of a gene implicated in multiple neurodevelopmental disorders, a2bp1 (fox1) Develpomental Neuroscience. 2011;33(1):64–74. doi: 10.1159/000323732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Grant I, Matthews CG. Differences in neuropsychological test performance associated with age, education, and sex. In: Grant I, Adams KM, editors. Neuropsychological Assessment of Neuropsychiatric Disorders,1e. New York: Oxford; 1986. pp. 100–120. [Google Scholar]
- Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes & Development. 1994;8(13):1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature Genetics. 2003;34(1):27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO Journal. 2003;22(4):905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl TR, Shibata H, Vo T, Huynh DP, Pulst SM. Identification and expression of a mouse ortholog of A2BP1. Mammalian Genome. 2001;12(8):595–601. doi: 10.1007/s00335-001-2056-4. [DOI] [PubMed] [Google Scholar]
- Knights RM, Norwood JA. A Neuropsychological Test Battery for Children: Examiner's Manual. Ottawa: Robert M. Knights Psychological Consultants; 1980. [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nature Methods Journal. 2006;3(11):909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- Lam KS, Aman MG. The Repetitive Behavior Scale-Revised: independent validation in individuals with autism spectrum disorders. Journal of Autism and Developmental Disorders. 2007;37(5):855–866. doi: 10.1007/s10803-006-0213-z. [DOI] [PubMed] [Google Scholar]
- LeCouter A, Rutter M, Lord C, Rios P, Robertson S, Holdgrafer M, McLennan J. Autism Diagnostic Interview: A standardized investigator-based instrument. Journal of Autism And Davelopemtal Disorders. 1989;19:363–387. doi: 10.1007/BF02212936. [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes & Development. 2009;23(19):2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nature Reviews Neuroscience. 2007;8(11):819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore PC, Risi S. Autism Diagnositc Observation Schedule-WPS. Los Angeles: Western Psychological Services; 1999. [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, Cook E, Geschwind DH. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Research. 2005;33(2):714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Molecular and Cellular Biology. 1997;17(11):6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi MW, Kay M, D'Cruz AM, Guter S, Kapur K, Macmillan C, Stanford LD, Sweeney JA. Neurobehavioral abnormalities in first degree relatives of individuals with autism. Archives of General Psychiatry. 2010;67(8):830–840. doi: 10.1001/archgenpsychiatry.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Research. 2005;33(7):2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield RC. Assessment and Analysis of Handedness - Edinburgh Inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Rutter M, Bailey A, Lord C. Social Communication Questionnaire. Los Angeles: Western Psychological Services; 2003. [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Davis Wright NR, Dhodapkar RM, Dicola M, Dulullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O'Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Gunel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH, Jr, Geschwind D, Roeder K, Devlin B, State MW. Multiple recurrent de novo CNVs including deletions of the 7q11.23 Williams Syndrome region are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Human Molecular Genetics. 2000;9(9):1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- Sparrow SS, Chiccetti DV, Balla DA. Vineland Adaptive Behavior Scales. Second Edition. San Antonio: The Psychological Corporation; 2005. [Google Scholar]
- Spreen O, Gaddes WH. Developmental norms for 15 neuropsychological tests age 6 to 15. Cortex. 1969;5(2):170–191. doi: 10.1016/s0010-9452(69)80028-6. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Molecular and Cellular Biology. 2005;25(22):10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt DE, Mayka MA, Corcos DM. Intermittent visuomotor processing in the human cerebellum, parietal cortex, and premotor cortex. Journal of Neurophysiology. 2006;95(2):922–931. doi: 10.1152/jn.00718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voienagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011 doi: 10.1038/nature10110. (Epub May 26). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Abbreviated Scale of Intelligence. San Antonio: The Psychological Corporation; 1999. [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature Structural & Molecular Biology. 2009;16(2):130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes & Development. 2008;22(18):2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Molecular and Cellular Biology. 2007;27(3):830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]