Abstract
Autophagy is a cellular catabolic process that relies on the cooperation of autophagosomes and lysosomes. During starvation, the cell expands both compartments to enhance degradation processes. We found that starvation activates a transcriptional program that controls major steps of the autophagic pathway, including autophagosome formation, autophagosome-lysosome fusion, and substrate degradation. The transcription factor EB (TFEB), a master gene for lysosomal biogenesis, coordinated this program by driving expression of autophagy and lysosomal genes. Nuclear localization and activity of TFEB were regulated by serine phosphorylation mediated by the extracellular signal-regulated kinase 2, whose activity was tuned by the levels of extracellular nutrients. Thus, a mitogen-activated protein kinase-dependent mechanism regulates autophagy by controlling the biogenesis and partnership of two distinct cellular organelles.
Comment
The degradative pathway of macroautophagy has a critical role in many cellular processes, and recently important functions for autophagy in the liver have been demonstrated.1 Knowledge of the factors that regulate both basal levels of autophagy, and increases in function that occur with cellular stresses, is critical to understanding how defects in autophagic function lead to pathophysiological conditions. The majority of studies have focused primarily on a complex series of pathways that regulate formation of the autophagosome which is the double-membrane structure that sequesters cytosolic components and delivers them to the lysosome for degradation. Over 30 autophagy-related genes (ATGs) have been identified that control basal and inducible levels of autophagy through several distinct pathways.1 A physiological stimulus used to define these regulatory pathways is nutrient deprivation in cells or rodents. Nutrient insufficiency induces autophagy to degrade cellular components into substrates such as amino acids and free fatty acids necessary for the maintenance of energy homeostasis. With sufficient nutrients, amino acids and insulin activate the mammalian target of rapamycin (mTOR) to down regulate autophagy. The liver frequently serves as a model for the effects of starvation on autophagic function. Starvation-induced autophagy in mice decreases total hepatic protein content by 35% within 24 hours,2 indicating that autophagy is a potent degradative pathway requiring strict regulation.
Transcription factor EB (TFEB) was first cloned from a human B cell cDNA library by its ability to bind the adenoviral major late promoter. Sequence analysis demonstrated that TFEB has adjacent basic helix-loop-helix and leucine zipper domains,3 which places it in the micropthalmia-transcription factor E (MiT/TFE) subfamily along with the genes TFE3, TFEC and MITF.4 The function of TFEB remained unknown until Sardiello et al.5 identified by bioinformatics a consensus DNA sequence in the promoters of 96 lysosomal genes termed the CLEAR (Coordinated Lysosomal Expression and Regulation) motif. The CLEAR element overlapped with the DNA target site for the MiT/TFE family and expression studies demonstrated that TFEB specifically targeted the CLEAR motif to up regulate genes essential for lysosomal biogenesis and function.5
From these findings and knowledge of the existence of mTOR-independent regulation of autophagy genes with starvation,6 the present study by Ballabio and colleagues7 examined whether TFEB regulates autophagy. TFEB overexpression in several cell lines increased autophagosome formation and autophagic function whereas a knockdown inhibited autophagy.7 TFEB increased the expression of a number of autophagy genes containing a TFEB binding site, and a subsequent study from the same laboratory confirmed and extended the list of TFEB-regulated autophagy genes.8 In vivo TFEB overexpression also increased autophagosome number and levels of lysosomal and autophagic TFEB target genes in liver. In vitro nutrient deprivation led to TFEB dephosphorylation at Ser142 and translocation to the nucleus to increase gene transcription. The mitogen-activated protein kinase (MAPK) extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylated TFEB to maintain it in an inactive, cytosolic state. ERK1/2 is activated by growth factors making it logical that this kinase down regulates TFEB and autophagy in response to nutrients. Thus, the study demonstrates a central role for TFEB in controlling autophagosome formation in addition to lysosomal biogenesis to increase autophagy (Fig. 1).
Fig. 1.
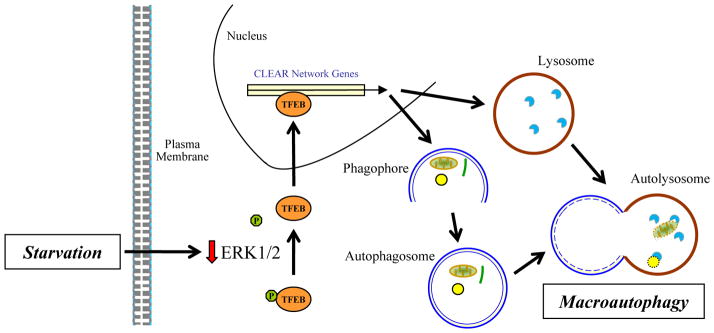
Regulation of autophagy by TFEB. In response to the loss of extracellular nutrients by starvation, there is decreased intracellular ERK1/2 activity. The reduction in ERK1/2 leads to dephosphorylation of cytosolic TFEB at Ser142. Dephosphorylation at this site allows nuclear translocation of TFEB where it increases transcription for a number of CLEAR network genes that encode for critical autophagic and lysosomal proteins. The increase in these proteins promotes autophagosome formation and lysosomal biogenesis to increase levels of autophagy.
A weakness of the study is its reliance on ectopically expressed TFEB and failure to examine endogenous TFEB protein trafficking. The effects of mTOR signaling on TFEB were also not examined. Another recent study indicates that mTOR up regulates TFEB.9 Although TFEB phosphorylation regulated nuclear localization, Ser142 was not involved.9 Thus, other mTOR-regulated phosphorylation sites may influence TFEB cellular localization. The divergent findings between the two studies may be secondary to differences in what constituted a nutritionally deprived cell culture medium.
The findings from this study elevate the importance of the lysosome in autophagy from a passive dumping site for autophagosomal contents to an actively regulated component of the autophagic process. Coordinate up regulation of both lysosomes and autophagosomes might prevent the problem of generating too many cargo-filled autophagosomes that overwhelm the degradative capacity of lysosomes. A mismatch between the numbers of autophagosomes and lysosomes could have dire consequences for the cell. The study emphasizes the need to focus more on whether defects in autophagy are secondary to lysosomal problems and possibly TFEB. Steatosis inhibits autophagic function in hepatocytes,10 and this decrease in autophagy has been attributed to both defects in autophagosome/lysosome fusion,11 and decreased expression of ATGs.12 It is possible that defects in TFEB regulation contribute to a multifactorial impairment in autophagic function in fatty liver disease.
The study by Settembre et al.7 also delineates another critical function for MAPK signaling. Studies in nonhepatic cells have shown that the MAPK c-Jun N-terminal kinase (JNK) up regulates autophagy through phosphorylation of Bcl-2 family members,13 although the existence of this pathway in hepatocytes which lack Bcl-2 remains unproven. ERK1/2 and JNK, which are frequently activated in tandem by cellular stresses, may counterbalance each other’s effect on autophagy. That ERK1/2 down regulates autophagy contradicts the concept of ERK1/2 signaling as cytoprotective since autophagy generally promotes survival. Interestingly, although oxidant stress is considered a major inducer of autophagy, hepatocyte oxidant stress associated with ERK1/2 activation failed to increase levels of autophagy.14 The effects of JNK and ERK1/2 on autophagic function specifically in hepatocytes need to be examined.
The study does not provide direct evidence that endogenous TFEB regulates hepatocyte autophagy in vivo, however this is likely given the strong evidence of TFEB function and TFEB’s high expression in liver.15 However, hepatocyte knockout/knockdown studies of TFEB need to be performed. Whether TFEB mediates increases in autophagy to stimuli other than starvation also needs to be examined. Recently a chemical stimulator of autophagy has been shown to be an effective treatment for murine α1-antitrypsin deficiency.16 A number of other hepatic diseases including nonalcoholic and alcoholic fatty liver disease, viral hepatitis and liver cancer may benefit from autophagy-directed therapies.1 By establishing a central role for TFEB in the regulation of autophagy, this study identifies this protein as a potential therapeutic target.
References
- 1.Czaja MJ. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895–1908. doi: 10.1053/j.gastro.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 3.Carr CS, Sharp PA. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 5.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 6.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 9.Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–4616. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- 16.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant α1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]


