Abstract
Sp1 is important for the transcription of many genes. Our previous studies have shown that Sp1 is degraded in normal cell, but it is preserved in cancer cells during mitosis and exists a priori in the daughter cells, ready to engage in gene transcription and thereby contributes to the proliferation and survival of cancer cells. The mechanism by which Sp1 is preserved in cancer cells during mitosis remains unknown. In this study, we observed that Sp1 strongly colocalized with cyclin-dependent kinase 1 (CDK1)/cyclin B1 during mitosis. Moreover, we showed that Sp1 is a novel mitotic substrate of CDK1/cyclin B1 and is phosphorylated by it at Thr 739 before the onset of mitosis. Phospho-Sp1 reduced its DNA-binding ability and facilitated the chromatin condensation process during mitosis. Mutation of Thr739 to alanine resulted in Sp1 remaining in the chromosomes, delayed cell-cycle progression, and eventually led to apoptosis. Screening of Sp1-associated proteins during mitosis by using liquid chromatography/mass spectrometry indicated the tethering of Sp1 to myosin/F-actin. Furthermore, phospho-Sp1 and myosin/F-actin appeared to exist as a congregated ring at the periphery of the chromosome. However, at the end of mitosis and the beginning of interphase, Sp1 was dephosphorylated by PP2A and returned to the chromatin. These results indicate that cancer cells use CDK1 and PP2A to regulate the movement of Sp1 in and out of the chromosomes during cell-cycle progression, which may benefit cancer-cell proliferation.
Keywords: Sp1, CDK1, PP2A, myosin, mitosis
Introduction
During interphase of the cell cycle, the transcription factor Sp1 has an important role in regulating the expression of genes involved in many cellular processes by binding to the promoter regions of its target genes. Sp1 binds specifically to the GC-rich promoter elements via 3 C2H2-type zinc finger regions at the C-terminus of the protein (Li and Davie, 2010) and regulates the transcriptional activity of target genes by using two major glutamine-rich transactivation domains localized at the N-terminus and the medial region (Li and Davie, 2010). Recent studies have shown that the DNA-binding affinity, transactivational activity, and protein stability of Sp1 may be regulated by posttranslational modifications such as glycosylation, ubiquitination, sumoylation, acetylation, and phosphorylation (Hung et al., 2006; Chuang et al., 2008; Wang et al., 2008; Tan and Khachigian, 2009; Li and Davie, 2010; Chuang and Hung, 2011). Among the posttranslational modifications of Sp1, phosphorylation is one of the most studied, especially with regard to its role during interphase of the cell cycle. For example, Sp1 serine or threonine residues are phosphorylated by different kinases, including DNA-dependent protein kinase, protein kinase A, and protein kinase C-ζ, and these phosphorylations cause an increase in the transcriptional activity of Sp1 by the enhancement of Sp1 binding to DNA (Chu and Ferro, 2005; Tan and Khachigian, 2009). On the other hand, some kinases can inhibit Sp1 function. During terminal liver differentiation, for example, casein kinase II modifies Thr579 on Sp1 and downregulates the DNA-binding ability of Sp1 (Chu and Ferro, 2005).
Previous studies have indicated that Sp1 accumulates in most types of cancer cells (Abdelrahim et al., 2004; Mertens-Talcott et al., 2007; Wang et al., 2008; Chuang et al., 2009; Li and Davie, 2010). Our recent study showing that Sp1 is modified by c-Jun NH2-terminal protein kinase 1 (JNK1) in mitosis, and phospho-Sp1 increases its protein stability, and is more present in cancer cells than that in primary noncancerous cells raised the possibility that the phosphorylation of Sp1 has an important role in the stability of Sp1 during mitosis (Chuang et al., 2008). Sp1 is known to be displaced from chromatin and remain stable in mitotic cancer cells, and then reserve Sp1 to daughter cells, thus apparently facilitating a quick start and the execution of cell growth (He and Davie, 2006; Chuang et al., 2008). However, the exact mechanism of how Sp1 modulates its DNA-binding activity to shuttle in and out of the chromosome during cell-cycle progression remains unknown.
Mitosis in vertebrates is triggered by cyclin-dependent kinase 1 (CDK1). In G2 stage, cyclin B1 is accumulated in cytoplasm, forms complex with CDK1, and then phosphorylated at Thr14 and Thr15, which causes the inactivation of cyclin B1 (Strebhardt, 2010). Until late G2 phase, CDK1 activation begins when it is dephosphorylated by phosphatase CDC25, and most of the cyclin B1/Cdk1 complexes are translocated rapidly from the cytoplasm into the nucleus while the nuclear envelope breaks down (Strebhardt, 2010). In this study, we discovered that CDK1 is a novel kinase to phosphorylate Sp1 at Thr739 during mitosis, and consequently, Sp1 DNA-binding ability is repressed. In addition, myosin/F-actin was found to interact with phospho-Sp1 and thereby results in its displacement from the chromatin, which is required for chromosome packaging during mitosis. Moreover, at the end of mitosis, PP2A was shown to dephosphorylate Sp1 to restore its DNA-binding activity. Finally, in N-methyl-N-nitrosourea (MNU)-induced mammary tumors, we found strong CDK1 and Sp1 accumulation. Therefore, the full phosphorylation of Sp1 is essential for cell-cycle progression of cancer cells.
Results
Phosphorylation of Sp1 in mitosis reduces its DNA-binding affinity
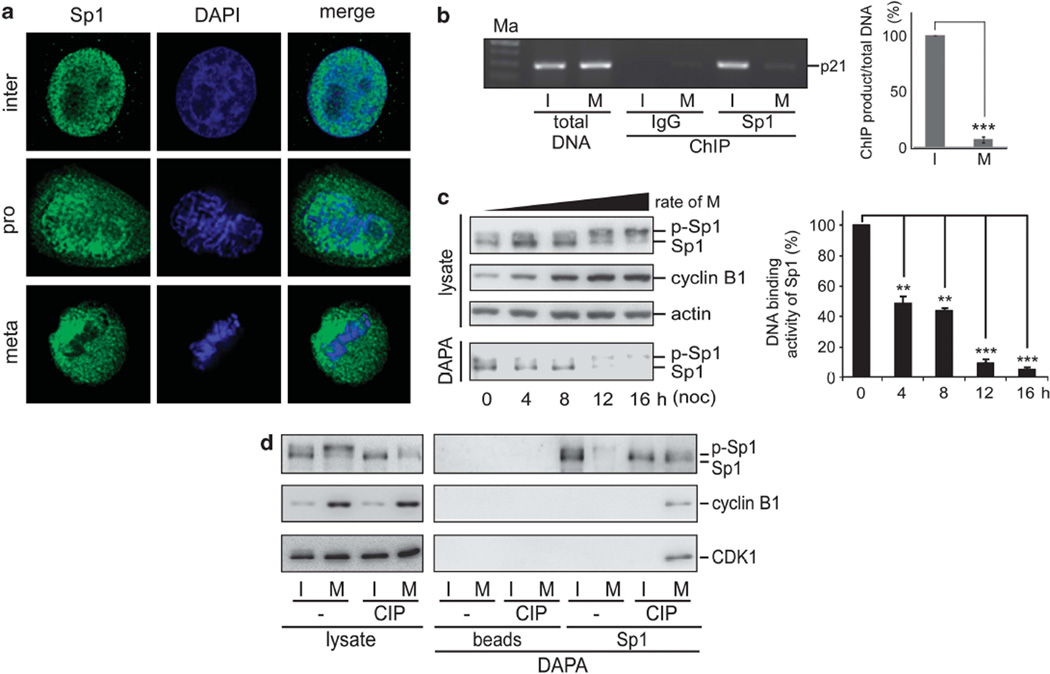
To further investigate the localization of Sp1 during different phases of the cell cycle, we investigated Sp1 distribution pattern in asynchronized cells by fluoromicroscopy. Sp1 was dissociated from the chromosomes during mitosis (Figure 1a). The results from chromatin immunoprecipitation (ChIP) assays confirmed that Sp1 significantly lost its DNA-binding activity during mitosis (Figure 1b). As the percentage of cells in mitosis increased during nocodazole treatment, the Sp1 signal shifted from the lower band to the upper one, as shown in western blotting (Figure 1c). Moreover, the DNA-binding ability of the shifted Sp1 was repressed (Figure 1c). After an alkaline phosphatase treatment, the upper band for the mitotic Sp1 shifted to the lower one, and Sp1 DNA-binding activity was rescued (Figure 1d). These data indicated that Sp1 was phosphorylated during mitosis and exhibited decreased Sp1 DNA-binding activity, but the question of which kinase(s) was responsible for phosphorylating Sp1 during mitosis remained unanswered.
Figure 1.
Phosphorylation of Sp1 in mitosis mediates its DNA-binding affinity. (a) HeLa cells were grown on coverslips in DMEM, fixed using 4% paraformaldehyde, labeled with anti-Sp1 antibodies (green), and stained with DAPI for DNA(blue). The cells were examined using a confocal laser-scanning microscope. (b) Interphase (I) cells and mitotic (M) cells, which were treated with nocodazole for 16h were fixed by 1% formaldehyde for the ChIP assay with anti-Sp1 antibodies. DNA was then extracted from the sample for PCR with the sequences of p21WAF1/CIP1 promoter as the primers. After three independent experiments, the signal of Sp1 binding to the p21WAF1/CIP1 promoter was quantified. (c) HeLa cells were treated with nocodazole and collected after different time intervals (0, 4, 8, 12 and 16h), and then DNA affinity precipitation assay (DAPA) was performed with the Sp1 probe, containing Sp1-binding elements of the p21WAF1/CIP1 promoter. The lysates and the products of DAPA were analyzed with immunoblotting by using anti-Sp1, anti-cyclin B1 and anti-actin antibodies. After three independent experiments, DNA binding activity of Sp1 was quantified. (d) Interphase and mitotic cellular extracts were treated with or without alkaline phosphatase (CIP) for DAPA with the Sp1 probe and then immunoblotted with anti-Sp1, anti-cyclin B1 and anti-CDK1 antibodies. All of the P-values reaching statistical significance are marked on the graph (**P<0.01 and ***P<0.005).
CDK1/cyclin B1 phosphorylates Sp1 at Thr739 during mitosis
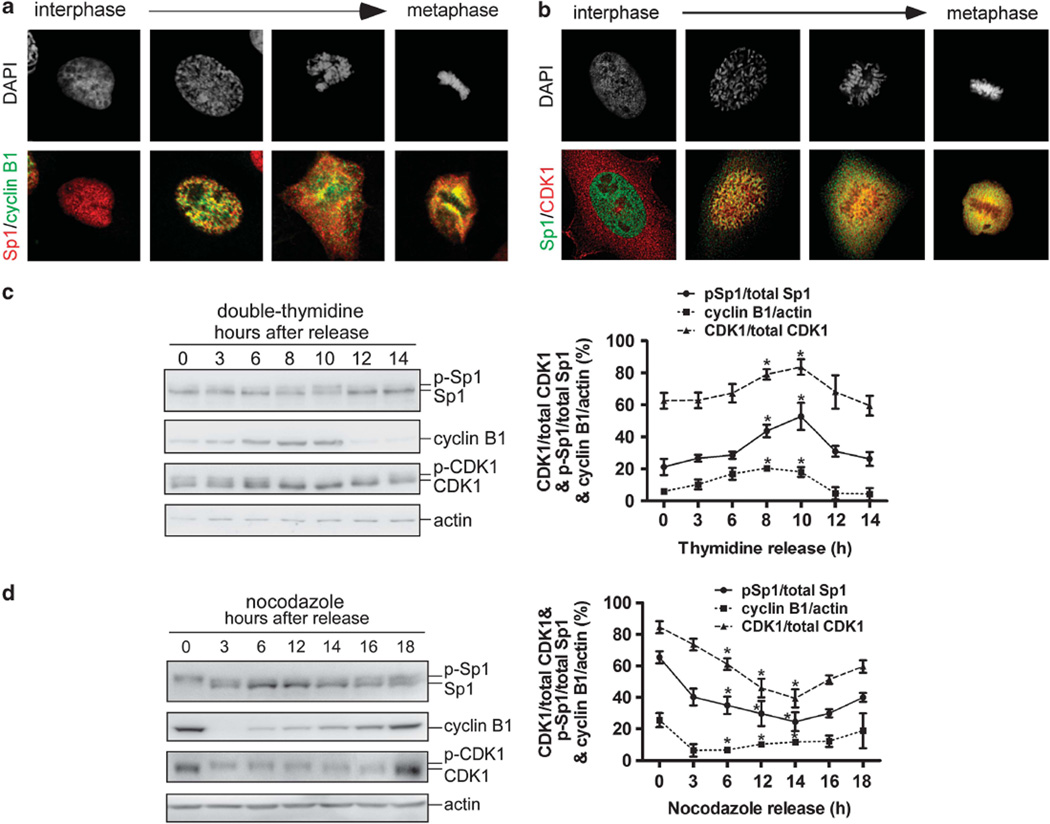
We observed that CDK1 and cyclin B1 were pulled down together with Sp1 after phosphatase treatment (Figure 1d), suggesting that CDK1/cyclin B1 might phosphorylate Sp1 during mitosis. We examined the localization of Sp1, cyclin B1, and CDK1 and found that Sp1 colocalized with cyclin B1 and CDK1 during mitosis (Figures 2a and b, Supplementary Figure S1 and Figure S4). In addition, we used thymidine (Figure 2c) and nocodazole (Figure 2d) to synchronize the cells in the G1/S phase and mitosis in order to observe the activation of CDK1 and the phosphorylation of Sp1 during cell-cycle progression, respectively. Approximately 10 h after removing thymidine, the levels of cyclin B1 and active CDK1 (hypophosphorylated CDK1) were increased along with an increase in Sp1 phosphorylation (Figure 2c). Furthermore, at the initiation of nocodazole release, the cells remained in early mitosis and showed high levels of cyclin B1, activation CDK1, and phospho-Sp1 (Figure 2d). Conversely, the levels of both activated CDK1 and phospho-Sp1 decreased as the cells entered interphase (Figure 2d). These results imply that CDK1 is responsible for phosphorylating Sp1 during mitosis.
Figure 2.
Sp1 and CDK1/cyclin B1 co-localize during mitosis, and there is a positive correlation between Sp1 phosphorylation and CDK1/cyclin B1 activation. (a, b) HeLa cells were grown on coverslips in DMEM, fixed using 4% paraformaldehyde, and double-labeled with anti-Sp1 (red) and anti-cyclin B1 (green) antibodies (a), or with anti-Sp1 (green) and anti-CDK1 (red) antibodies (b). DNA was stained with DAPI (blue). (c, d) The G1/S phase and mitotic cells were released into growth by removing thymidine (c) and nocodazole (d), respectively, and then harvested at different time points as indicated. Samples were immunoblotted with anti-Sp1, anti-cyclin B1, anti-CDK1 and anti-actin antibodies. The relative levels of phospho-Sp1, cyclin B1, and activated CDK1 were quantified from three independent experiments. The GraphPad Prism Software (San Diego, CA, USA) was used to determine the difference between two means. Data were expressed as mean±s.d. Two means were compared using Student’s t-test. *P<0.05 was considered as significantly different.
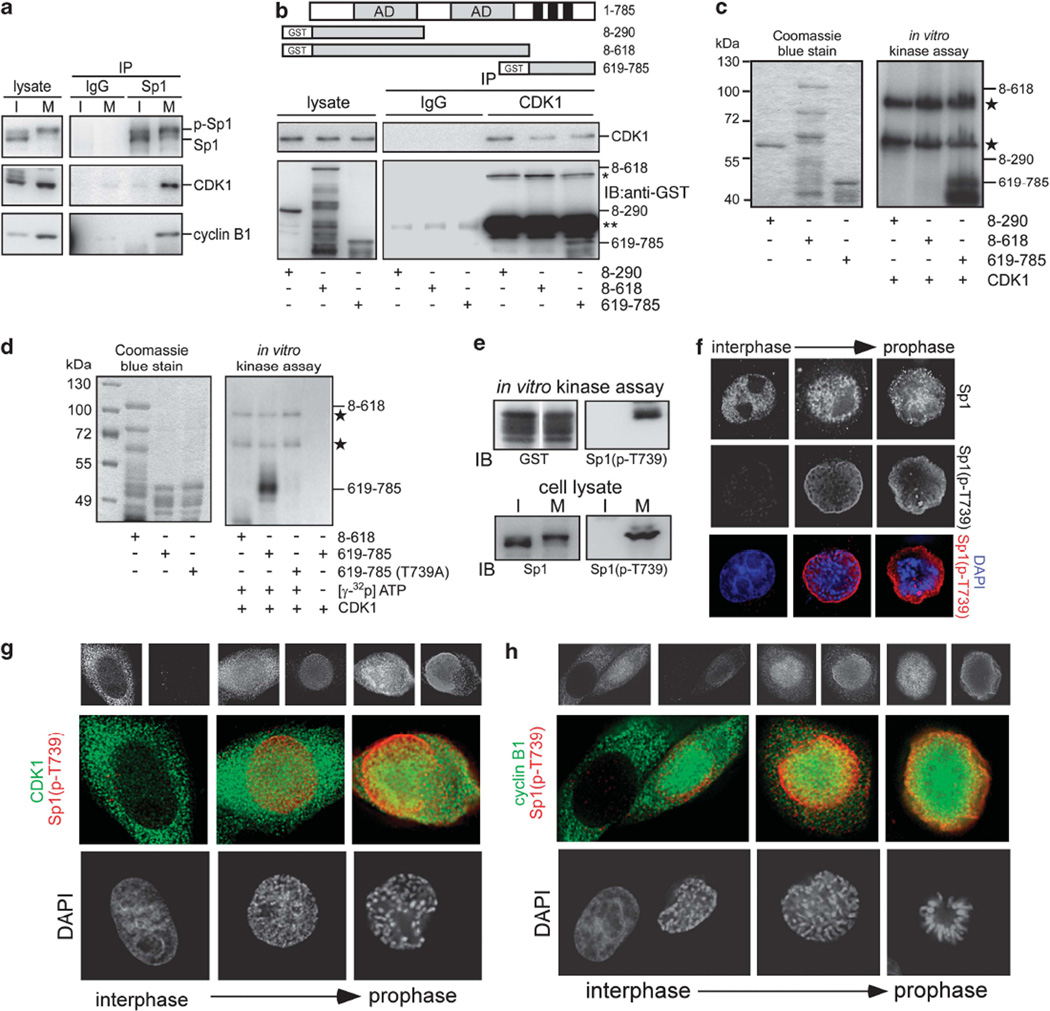
For further addressing the interaction between Sp1 and CDK1 in mitosis, an immunoprecipitation assay was performed (Figure 3a and Supplementary Figure S2). The results showed that Sp1 interacted with cyclin B1/CDK1 in mitosis but not in interphase. Three truncated Sp1 fragments were subsequently used for the CDK1 immunoprecipitation assay (Figure 3b). The data revealed that only the C-terminal DNA-binding domain of Sp1 could interact with CDK1. Furthermore, when we used the active CDK1/cyclin B1 to perform in vitro kinase assays, we found that CDK1 could phosphorylate the C-terminus of Sp1 at Thr739 (Figures 3c and d). Next, we used a phospho-peptide of Sp1, EGSG-TAT(p)PSALIT, as an antigen to generate an antibody that could recognize the phosphorylated Thr739 motif. The specificity of the antibody was verified by its recognition of CDK1-phosphorylated Sp1 and mitotic Sp1 (Figure 3e and Supplementary Figure S3). Immunofluorescence data showed that Sp1 was evenly distributed within the nucleus during interphase but no phospho-Sp1 signal was evident (Figure 3f). When the cells entered early prophase, CDK1 and cyclin B1 entered into nucleus, and the phospho-Sp1 signal increased in a parallel manner (Figures 3g and h, Supplementary Figure S5 and Figure S6). As the phospho-Sp1 signal became evident, it showed almost no colocalization with the DNA (Figure 3f, Supplementary Figure S5 and Figure S6). Moreover, the phospho-Sp1 signal continued to be detected but not colocalized with the DNA during the entire course of mitosis. In conclusion, these data clearly indicate that Sp1 is a substrate of CDK1/cyclin B1, and Thr739 phosphorylation of Sp1 might abolish its DNA binding activity during mitosis.
Figure 3.
CDK1/cyclin B1 interacts with Sp1 and phosphorylates Sp1 at Thr739 during mitosis. (a) Interphase and mitotic cellular extracts were used for the immunoprecipitation assay with anti-Sp1 antibodies in which IgG serves as a negative control and analyzed with anti-Sp1, anti-CDK1 and anti-cyclin B1 antibodies in immunoblotting. (b) Different truncated Sp1 proteins were synthesized and purified in E. coli and incubated with mitotic cell lysates for 2 h. Next, the samples were used for the immunoprecipitation assay with anti-CDK1 antibodies and then analyzed with immunoblotting using anti-GST and anti-CDK1 antibodies. The ‘*’ and ‘**’ are represented to heavy and light chains, respectively. (c, d) Four truncated Sp1 (8–290 a.a., 8–618 a.a., 619–785 a.a., and 619–785 a.a. (T739A)) proteins were combined with [γ-32P] ATP and activated CDK1/cyclin B1 for an in vitro kinase assay. Samples were then subjected to SDS–PAGE, gel drying, and X-ray exposure. Coomassie blue staining shows the protein level as the internal control. The two asterisks indicate recombinant CDK1 and cyclin B1 proteins that could be phosphorylated by CDK1/cyclin B1. (e) Phospho-Sp1 from in vitro kinase assay with activated CDK1/cyclin B1 and mitotic cell extracts were used for immunoblotting with anti-Sp1 phospho-T739, anti-GST, and anti-Sp1 antibodies. (f–h) Immunofluorescence was performed with anti-Sp1 (green) and anti-Sp1 phospho-T739 (red) antibodies (f), with anti-CDK1 (green) and anti-Sp1 phospho-T739 (red) antibodies (g), or with anti-cyclin B1 (green) and anti-Sp1 phospho-T739 (red) antibodies in asynchronous HeLa cells (h). DNA was stained with DAPI (blue).
Phosphorylation at Thr739 represses Sp1 DNA-binding affinity
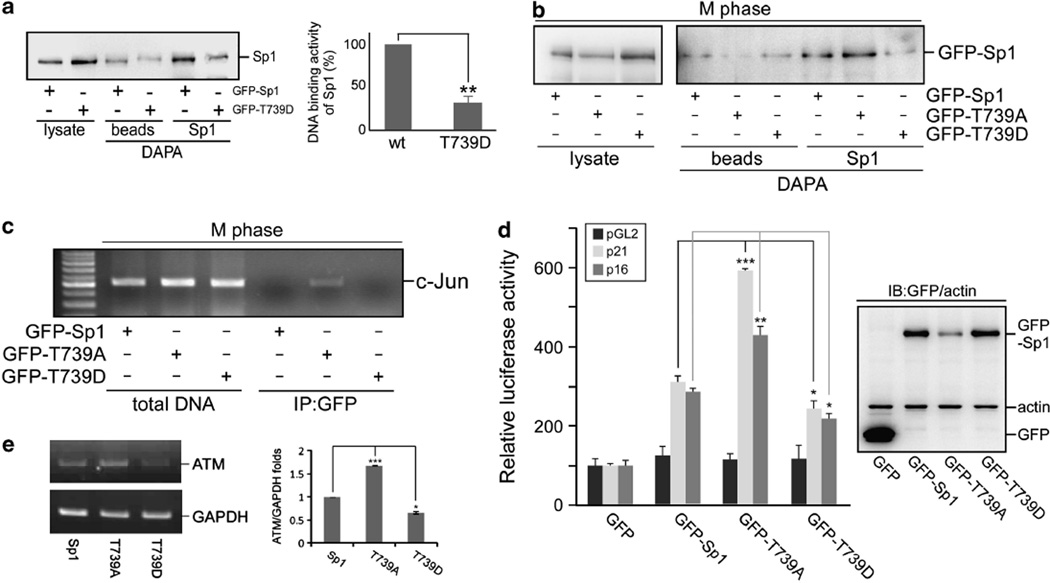
We next explored whether phosphorylation of Sp1 at Thr739 affects Sp1 DNA-binding activity. GFP-Sp1 and two Sp1 mutants, including GFP-Sp1 (T739A) and a mimic phosphorylated Sp1 construct with a Thr739 mutated to aspartic acid, GFP-Sp1 (T739D), were individually overexpressed in cells in order to study the DNA-binding ability of Sp1 in interphase (Figure 4a) or during mitosis (Figures 4b and c). The results indicated that GFP-Sp1 (T739D) possessed less DNA-binding activity in interphase relative to the wild-type Sp1. However, in mitosis, GFP-Sp1 (T739A) continued to bind to the Sp1-binding element and the c-Jun promoter region, but a weaker binding was observed for GFP-Sp1 and GFP-Sp1 (T739D). Finally, we used these constructs to study the importance of the Thr739 phospho-residue in Sp1 transcriptional activity during mitosis by measuring the promoter activity and the mRNA levels of Sp1 target genes such as p21WAF1/CIP1, p16INK4a and ataxia telangiectasia mutated (ATM) (Figures 4d and e and Supplementary Figure S8). GFP-Sp1 (T739A) strongly induced the promoter activity of p16INK4a and p21WAF1/CIP1 and the mRNA level of ATM during mitosis, but a weaker response was observed with GFP-Sp1 (T739D). To address the specificity of GFP-Sp1(T739D) in reducing DNA-binding activity, we also used other phosphorylation site in Sp1, Thr278, to address its DNA binding activity (Supplementary Figure S7). The results showed that no significant difference between GFP-Sp1 and GFP-Sp1(T278D) was found, implying Sp1 phosphorylated at Thr739 affected its DNA binding activity specifically. Thus, based on these results, CDK1 phosphorylates Sp1 at Thr739 as cells progress from the G2 phase into mitosis, and decreases Sp1 DNA-binding activity.
Figure 4.
Phosphorylation at Thr739 suppresses Sp1 DNA-binding activity. (a) Plasmids including pGFP-Sp1 and pGFP-Sp1 (T739D) were transfected individually into HeLa cells for 24h. The cell lysates were then used for the DAPA assay with the Sp1 probe, containing Sp1-binding elements of the p21WAF1/CIP1 promoter, and then analyzed with immunoblotting using anti-Sp1 antibodies. The relative levels of Sp1 were quantified from three independent experiments. (b, c) Plasmids including pGFP-Sp1, pGFP-Sp1 (T739A) and pGFP-Sp1 (T739D) were transfected into HeLa cells. After 24 h, the transfected cells were treated with nocodazole to synchronize the cells in mitosis. The mitotic cells were subsequently harvested and studied by the DAPA assay with the Sp1 probe (b) or by the ChIP assay with anti-GFP antibodies (c). In ChIP assay, the DNA samples were amplified by the PCR with the sequences of c-Jun promoter as the primers. (d) Empty vector, pGFP-Sp1, pGFP-Sp1 (T739A) and pGFP-Sp1 (T739D) were individually co-transfected with pGL2, pGL2-p21WAF1/CIP1 promoter or pGL2-p16INK4a promoter into HeLa cells for 24h, and the luciferase activity was then analyzed. In addition, these samples were analyzed by immunoblotting with anti-GFP and anti-actin antibodies. All of the experiments were done three times independently. (e) Cells were overexpressed with GFP-Sp1, GFP-Sp1 (T739A) or GFP-Sp1 (T739D), and were then synchronized in mitosis by nocodazole treatment. Total RNA was extracted from these cells and underwent RT–PCR using specific primers for ATM and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). From three independent experiments, the level of ATM mRNA was quantified. All of the P-values reaching statistical significance are marked on the graph (*P<0.05, **P < 0.01, and ***P<0.005).
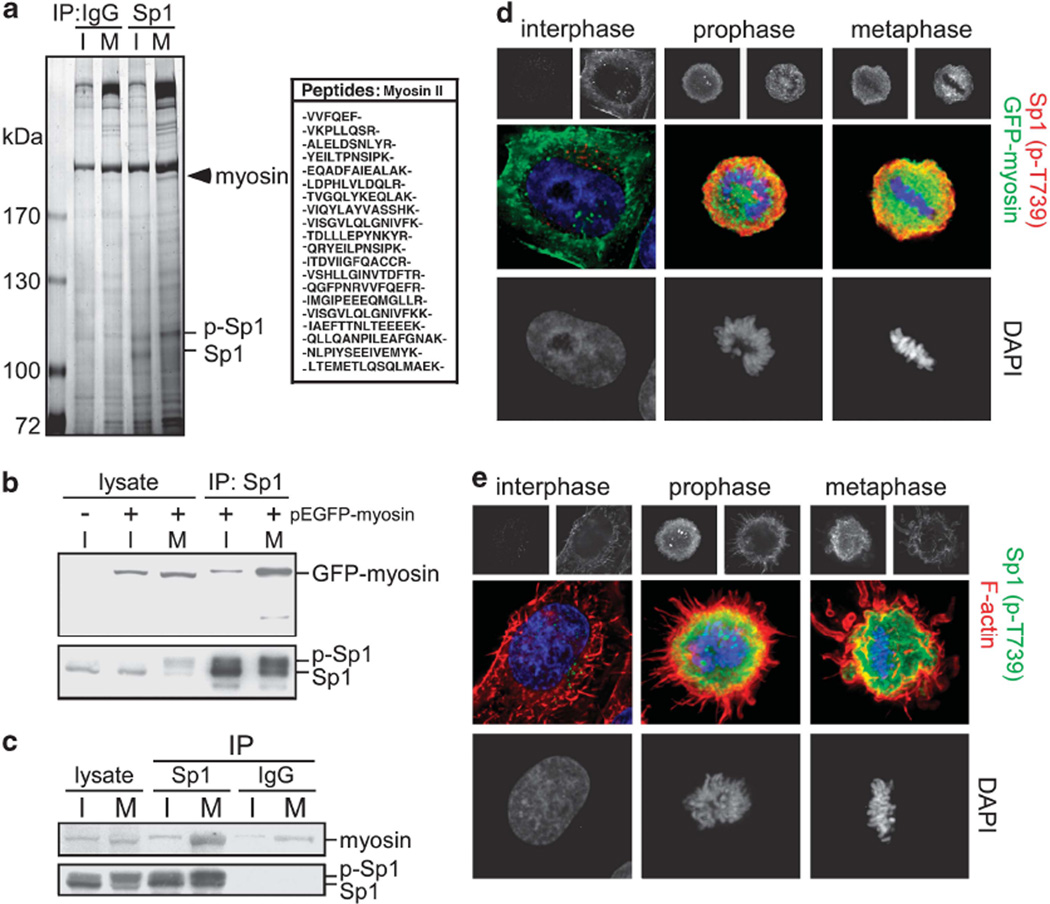
Mitotic phospho-Sp1 associates with myosin/F-actin
We observed phospho-Sp1 distribution during mitosis and found that Sp1 was not distributed evenly throughout the cell but remained at the border of the chromosomes (Supplementary Figure S5). We speculated that Sp1 may use the cytoskeleton to move in and out of chromosomes during cell-cycle progression. Thus, LTQ-OrbiTrap mass spectrometry was used to screen the Sp1-interacting proteins during mitosis. A number of Sp1-interacting proteins were identified and are shown in Supplementary Table 1. Ingenuity software analysis revealed a major Sp1-interacting group containing 23 cytoskeleton-related proteins such as actin, myosin and tropomyosin (Supplementary Figure S9). We also used liquid chromatography-mass spectrometry MS to confirm the interaction and found that myosin could interact with Sp1 during mitosis but not in interphase (Figure 5a). In addition, an immunoprecipitation assay using anti-Sp1 antibodies further confirmed that phospho-Sp1 could interact with myosin during the mitotic stage (Figures 5b and c). Next, we used fluoromicroscopy to observe the distribution of phospho-Sp1, Sp1, and myosin in asynchronized cells and found that GFP-myosin colocalized with phospho-Sp1 and Sp1 during mitosis (Figure 5d and Supplementary Figure S10). As myosin associates with F-actin, we also investigated the distribution of phospho-Sp1 and F-actin in asynchronized cells. The result showed that phospho-Sp1 co-localized with F-actin during mitotic stage (Figure 5e), and similar results were also observed that Sp1 and F-actin were co-localized at the border of cell in mitosis but not in interphase (Supplementary Figure S10). Together, these data suggest that Sp1 phosphorylation by CDK1 increases the interaction of Sp1 with myosin/F-actin to determine the distribution Sp1 during mitosis.
Figure 5.
Phospho-Sp1 interacts with myosin and F-actin during mitosis. (a) The cell extracts of interphase and mitosis were used for the immunoprecipitation assay with anti-Sp1 or IgG antibodies. The precipitated proteins were separated by gel and marked with silver staining for protein identification by MS/MS. (b) The pEGFP-myosin vector was transfected into HeLa cells for 24h and then cells were harvested for the immunoprecipitation assay with anti-Sp1 antibodies. Samples were analyzed with immunoblotting by using anti-GFP and anti-Sp1 antibodies. (c) Interphase and mitotic cellular extracts were used for the immunoprecipitation assay with anti-Sp1 or IgG antibodies. Samples were then immunoblotted with anti-myosin and anti-Sp1 antibodies. (d, e) Immunofluorescence was performed with anti-Sp1 phospho-T739 (red) antibodies and GFP signal in GFP-myosin expression cells (d) or with anti-Sp1 phospho-T739 (green) antibodies and Alexa Fluor 568-conjugated phalloidin that is a high-affinity probe for F-actin (red) in asynchronous HeLa cells (e). DNA was stained with DAPI (blue).
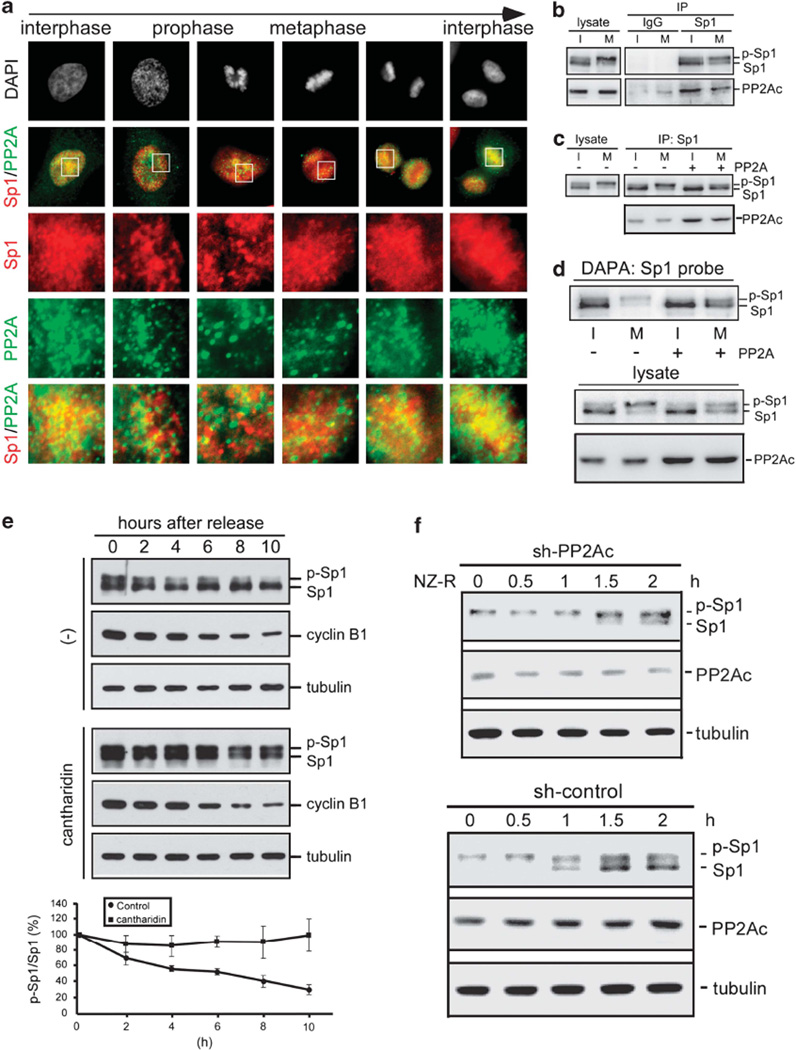
PP2A dephosphorylates Sp1 after mitosis
Sp1 is dephosphorylated after mitosis when cells enter interphase. However, the enzyme that dephosphorylates phospho-Sp1-Thr739 was still not known. PP2A reportedly dephosphorylates the Ser59 and Thr681 residues of Sp1 (Tan and Khachigian, 2009). We speculated that PP2A also dephosphorylates phospho-Sp1-Thr739 after mitosis. At first, we examined the distribution of Sp1 and PP2A during the cell cycle. Sp1 and PP2A colocalized during interphase, but the colocalization signal declined during mitosis (Figure 6a). The immunoprecipitation assay using anti-Sp1 antibodies confirmed that the interaction between Sp1 and PP2A was reduced as the cells entered mitosis (Figure 6b). An in vitro dephosphorylation assay by PP2A directly showed that mitotic phospho-Sp1 was the substrate of PP2A (Figure 6c). In addition, the DNA-binding activity of Sp1 was restored after phospho-Sp1 was incubated with PP2A (Figure 6d). In the next, we examined the effects of cantharidin, a PP2A inhibitor, on the level of Sp1 phosphorylation as the cells progressed from mitosis to interphase after nocodazole treatment (Figure 6e). HeLa cells without cantharidin treatment showed declining levels of phospho-Sp1. In contrast, the phospho-Sp1 levels were sustained after cantharidin treatment. Finally, we used shRNA of PP2A to knockdown PP2A, and then to study the phospho-Sp1 pattern after nocodazole releasement (Figure 6f). Data indicated that most of the up bands were shifted to low band after cell cycle entered into interphase again in the presence of PP2A. However, most of the up bands still remained in the up band in the absence of PP2A, indicating that PP2A can dephosphorylate phospho-Sp1 as cells enter into interphase again. In addition, previous studies reported that PP1 can interact with myosin, here we also used the PP1 to do in vitro phosphatase assay of phospho-Sp1 (Supplementary Figure S11). Data indicated that PP1 cannot dephosphorylate Sp1. These results thus indicated that Sp1 is phosphorylated by CDK1 during early mitosis and dephosphorylated by PP2A after mitosis, and Sp1 DNA-binding activity is regulated by CDK1 and PP2A in cell-cycle progression.
Figure 6.
PP2A dephosphorylates phospho-Sp1 at the end of mitosis. (a) HeLa cells were fixed with 1% paraformaldehyde for immunofluorescence double-labeling with anti-Sp1 antibodies (red) and anti-PP2A (green) antibodies. DNA was stained with DAPI (blue). (b) The cell extracts of interphase and mitosis were used for the immunoprecipitation assay with anti-Sp1 or IgG antibodies. Samples were then immunoblotted with anti-Sp1 and anti-PP2A antibodies. (c) Sp1 and phospho-Sp1 were purified by using anti-Sp1 antibodies from interphase and mitotic cell extracts, respectively. These products served as the substrate of PP2A in the in vitro dephosphorylation assay. Samples were then analyzed by immunoblotting with anti-Sp1 antibodies. The PP2A level was determined as an internal control. (d) Interphase and mitotic cellular extracts were treated with or without activated PP2A enzyme for DAPA assay with the Sp1-binding element probe and immunoblotted with anti-Sp1 antibodies. The level of Sp1 and PP2A were determined as the internal control. (e) The mitotic cells were treated with or without cantharidin and released into growth by removing nocodazole, and then harvested at different time points as indicated. Samples were analyzed by immunoblotting with anti-Sp1, anti-cyclin B1 and anti-tubulin antibodies in which tubulin served as an internal control. In addition, the phosphorylated level of Sp1 level was quantified from three independent experiments. (f) PP2A was knockdown by sh-PP2Ac (up panel) or sh-control (low panel) in HeLa cells, and then cells were synchronized in mitosis by nocodazole treatment. Samples collected at various time periods after nocodazole releasement (NZ-R) were analyzed by immunoblotting using anti-Sp1, anti-PP2Ac and anti-tubulin antibodies.
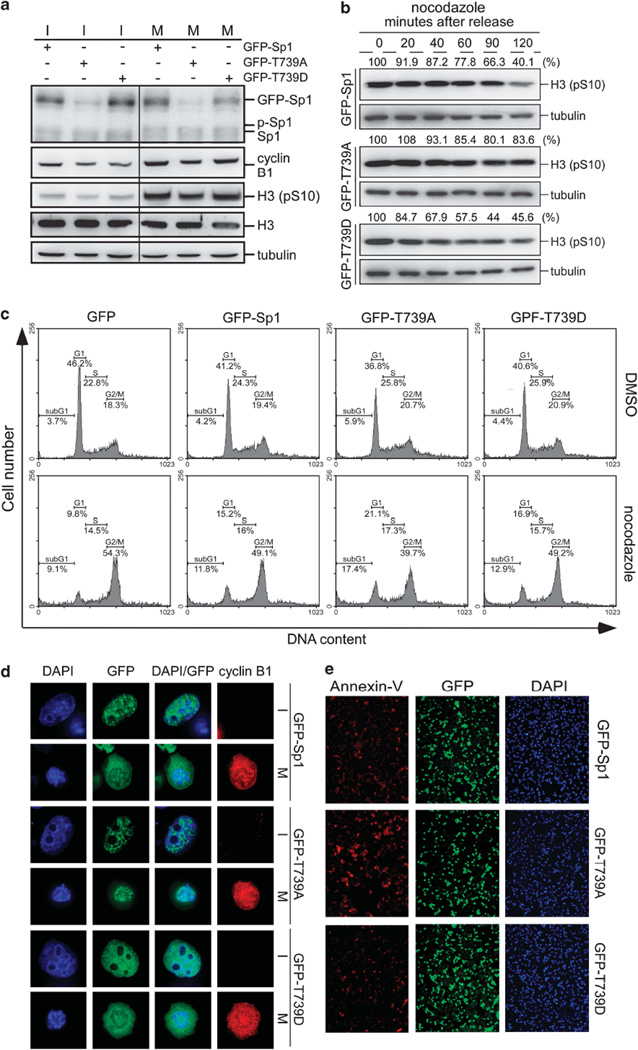
Underphosphorylation of Sp1 interferes with cell-cycle progression into mitosis and is lethal to cancer cells during mitosis
As a transcription factor, Sp1 regulates a number of target genes by binding to their promoters during interphase. However, it is unclear why Sp1 needs to leave the chromosome as the cell cycle enters mitosis and what happens if Sp1 is unable to undergo phosphorylation and continues to bind DNA during mitosis. Various Sp1 constructs, including GFP-Sp1, GFP-Sp1 (T739D), and GFP-Sp1 (T739A) were individually transfected into the cells, and these cells were treated with nocodazole for synchronization of mitosis. We then studied the interference of various Sp1 constructs on mitotic progression by assessing the cyclin B1 and phospho-histone H3-Ser10 levels. The results indicated that the levels of cyclin B1 and phospho-histone H3-Ser10 were decreased by GFP-Sp1 (T739A) expression, but not by GFP-Sp1 or GSP-Sp1 (T739D) expression (Figure 7a). After removing nocodazole to release the transfected cells into growth, GFP-Sp1- or GFP-Sp1 (T739D)-expressing cells exited mitosis quickly, as evidenced by the rapidly decreasing level of histone H3 phosphorylation at Ser10, whereas GFP-Sp1 (T739A)-expressing cells exited mitosis slowly (Figure 7b). Furthermore, the flow cytometry data revealed that cells with expressed wild-type Sp1 and expressed Sp1 (T739D) mutant had more mitotic fraction under nocodazole treatment (Figure 7c). However, when the cells were expressed with the unphosphorylated Sp1 (T739A) mutant, 21.1% of cells remained at the G1 fraction under nocodazole treatment, and more sub-G1 (17.4%) populations were observed (Figure 7c). These results indicated that the expression of unphosphorylated Sp1 (T739A) interferes with cell-cycle progression into mitosis. Fluorescent microscopy of cells synchronized in mitosis by nocodazole treatment for 24 h revealed that most of the GFP-Sp1 (T739A) mutant protein remained inside the chromatin, whereas GFP-Sp1 and GFP-Sp1 (T739D) did not (Figure 7d). The annexin-V signal indicated that GFP-Sp1 (T739A) induced more apoptosis than GFP-Sp1 (T739D) and GFP-Sp1 (Figure 7e) when cells were treated with nocodazole, suggesting that the repression of DNA packaging by unphosphorylated Sp1 results in apoptosis. Thus, in cancer cells, the phosphorylated form of Sp1 during mitosis is required for chromatin condensation, chromosome packaging and cell-cycle progression.
Figure 7.
Sp1 phosphorylation during mitosis is required for chromatin condensation and cell-cycle progression. (a) Plasmids including pGFP-Sp1, pGFP-Sp1 (T739A) and pGFP-Sp1 (T739D) were individually transfected into HeLa cells for 24h. These cells were then treated with or without the nocodazole for another 16 h and collected for immunoblotting with anti-Sp1, anti-cyclin B1, anti-histone H3 phospho-S10, anti-histone H3 and anti-tubulin antibodies. (b) Plasmids including pGFP-Sp1, pGFP-Sp1 (T739A) and pGFP-Sp1 (T739D), were individually transfected into HeLa cells, and cells were arrested in mitosis by nocodazole. After removal of nocodazole, an equal number of cells was released into growth and then harvested at different time points as indicated. Samples were analyzed by immunoblotting with anti-histone H3 and phospho-S10 antibodies, and the level of phospho-histone H3 was also quantified as a percentage of relative optical density. (c, d) Empty vector, pGFP-Sp1, pGFP-Sp1 (T739A) and pGFP-Sp1 (T739D) were individually transfected into HeLa cells for 24 h. These cells were then treated with or without the nocodazole for another 16h and were fixed for flow cytometry assay by PI staining (c) or for immunofluorescence assay with anti-GFP (green) and anti-cyclin B1 (red) antibodies (d). DNA was stained with DAPI (blue); Cyclin B1 was used as mitotic phase marker. (e) Plasmids including pGFP-Sp1, pGFP-Sp1 (T739A), and pGFP-Sp1 (T739D) were individually transfected into HeLa cells, and cells were arrested in mitosis by nocodazole. The cells were then fixed in order to detect the signal of annexin-V by using immunofluorescence assay with Alexa 568-labeled annexin V. Green fluorescence signal derived from various GFP-Sp1 proteins, and DAPI staining shows the DNA content.
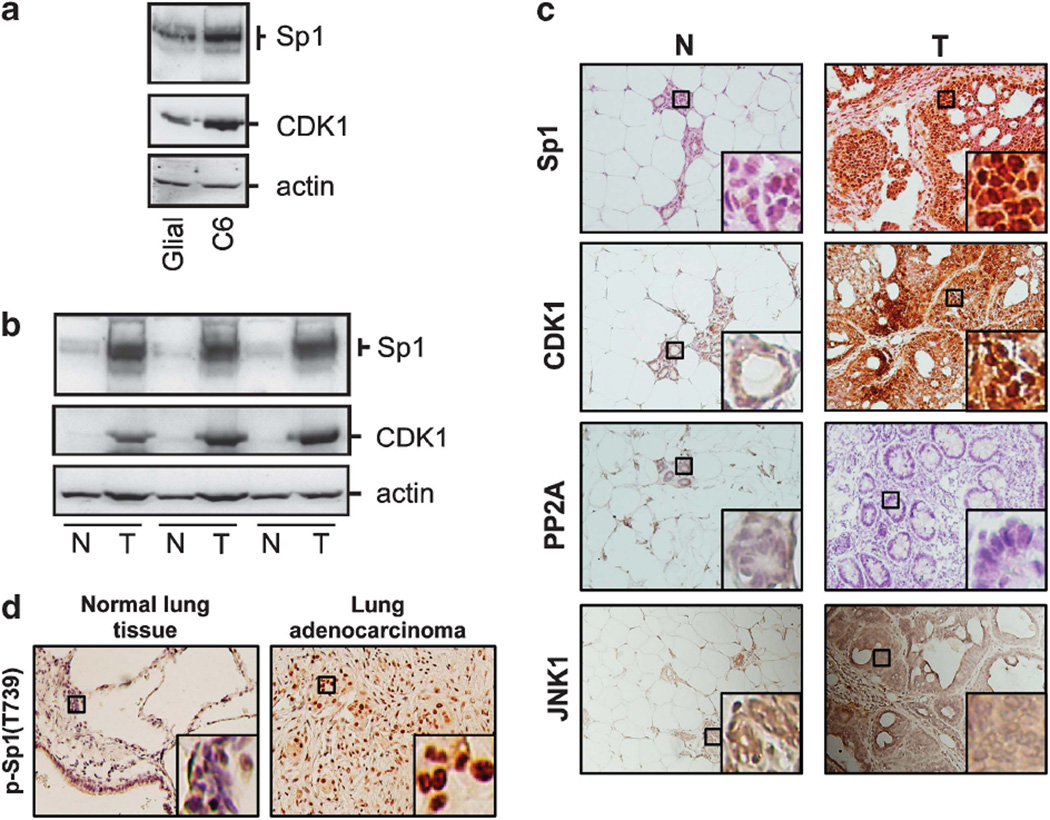
Sp1 and CDK1 accumulate in MNU-induced mammalian tumor tissue and cancer cells
Our present studies indicated that CDK1 affects Sp1 function during mitosis. However, both our studies and even the previous ones have shown that Sp1 accumulates in certain human cancers such as cervical cancer, colon cancer, thyroid cancer, gastric cancer and breast cancer (Chiefari et al., 2002; Wang et al., 2003; Abdelrahim et al., 2004; Hosoi et al., 2004; Mertens-Talcott et al., 2007; Chuang et al., 2008). To further address whether the levels of Sp1 and CDK1 are parallel elevation during tumorigenesis that CDK1 can regulate Sp1 DNA-binding activity in cell-cycle progression, the levels of Sp1 and CDK1 were compared between primary glial cells and glioma C6 cells (Figure 8a) and between the normal mammary tissue and MNU-induced mammary adenocarcinoma in rats (Figures 8b and c). The results showed a positive correlation for increased Sp1, CDK1 and JNK1 levels and an inverse correlation for decreased PP2A level in cancer cells and tumor tissue. In addition, by using anti-p-Sp1(T739) antibodies, we also determined the phospho-Sp1(T739) level in clinical lung cancer specimens (Figure 8d). Data revealed that the phospho-Sp1(T739) signal was increased in lung cancer tissue compared with normal lung tissue. This implies that Sp1 phosphorylation modulated by CDK1 and PP2A to modulate Sp1 DNA-binding activity and protein level in cell-cycle progression is important for tumorigenesis.
Figure 8.
The levels of Sp1 and CDK1 are accumulated in glioma cells and MNU-induced tumor. (a) Rat glioma C6 cell line and primary glial cells from postnatal day 0 to day 1 rat pups were harvested and studied by immunoblotting with anti-Sp1 and anti-CDK1 antibodies. The actin was used as an equal loading control. In addition, rats intraperitoneally injected with 50 mg/kg MNU were killed after 8 weeks, and the normal and tumor tissues were then harvested. (b, c) Sp1, CDK1, PP2A and JNK1 level were determined in the normal (N) and tumor (T) mammalian tissues by immunoblotting (b) or by immunohistochemistry (c) using anti-Sp1, anti-CDK1, anti-PP2A and anti-JNK1 antibodies, respectively. The actin is as an internal control. (d) The p-Sp1(T739) level was determined by immunohistochemistry using anti-p-Sp1(T739) antibodies in the human clinical normal and lung adenocarcinoma tissue samples. Sections were counterstained with hematoxylin to determine the tissue and tumor type.
Discussion
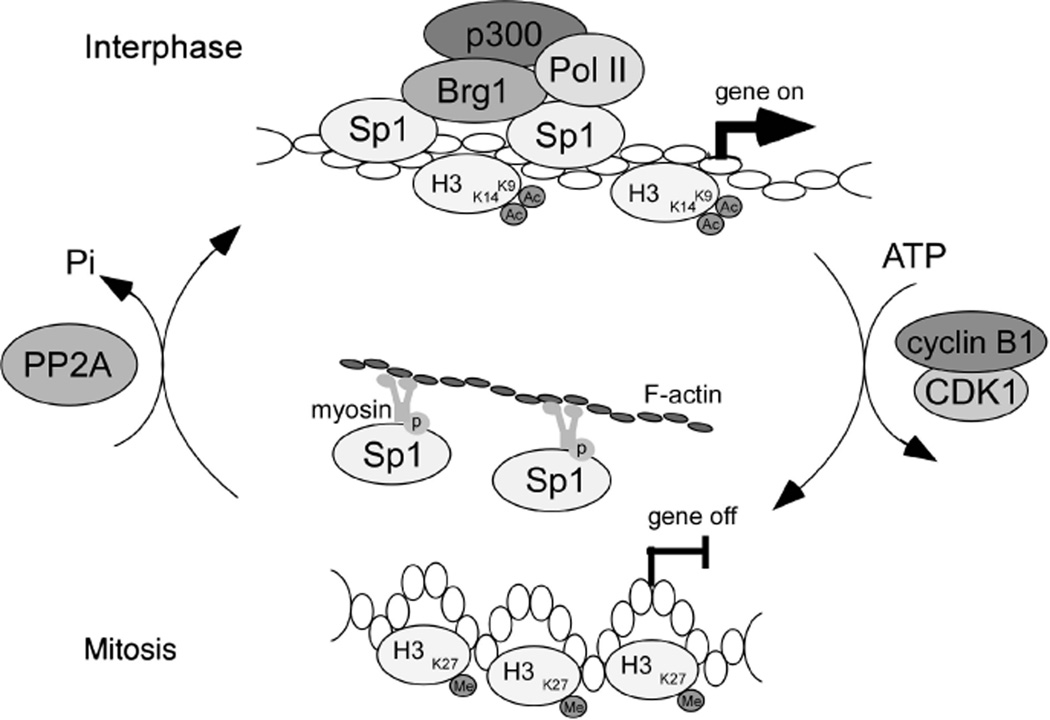
Sp1, the first transcription factor to be cloned, has major physiological roles in normal cells and in cancer biology (Safe and Abdelrahim, 2005). In this study, we clarified how cancer cells use posttranslational modifications to modulate the DNA-binding activity of Sp1 during cell-cycle progression (Figure 9). Our data indicate that the cyclin B1/CDK1 complex phosphorylates Sp1 at Thr739 during mitosis. The phosphorylation represses the DNA-binding activity of Sp1 to disrupt the basal transcription complex (Figure 4 and Supplementary Figure S12), which leads to the formation of chromatin condensation during mitosis. During mitosis, myosin/ F-actin is critical for displacing phospho-Sp1 from the chromatin, but at the end of mitosis, PP2A dephosphorylates Sp1 to restore its DNA-binding activity.
Figure 9.
Schematic diagram illustrates CDK1 and PP2A regulating the phosphorylation of Sp1 to recycle Sp1 back to the daughter cells of cancer. This mechanism is very important for Sp1 in order to reuse Sp1 more quickly and more economically to increase Sp1 level to help cell growth and proliferation.
Previous studies have shown that Sp1 activation might augment the potential of tumor cells by inducing overexpression of downstream genes such as vascular endothelial growth factor, urokinase plasminogen activator and epithelial growth factor receptor (Abdelrahim et al., 2004; Benasciutti et al., 2004; Hosoi et al., 2004; Lou et al., 2005; Safe and Abdelrahim, 2005). The role of Sp1 as an essential transcription factor for a number of genes in regulating cell growth, angiogenesis, and survival has been shown in pancreatic, gastric and colorectal cancers (Han and Kudlow, 1997; Wang et al., 2003; Abdelrahim et al., 2004; Hosoi et al., 2004). As many posttranslational modifications were reported to modulate Sp1 activity (Chu and Ferro, 2005; Tan and Khachigian, 2009; Li and Davie, 2010), the rigorous control of posttranslational Sp1 modification may determine its endogenous role under various conditions. Various Sp1 residues could be phosphorylated by different kinases under various physiological conditions in order to modulate its transcriptional activity, DNA-binding activity, or protein stability (Chu and Ferro, 2005; Chuang et al., 2008; Tan and Khachigian, 2009). However, the functional roles of phosphorylated Thr739 depend on the target genes. This might be due to the different repertoire of binding partners at specific target gene promoters under different physiological conditions in cells (Chu and Ferro, 2005). In the present study, we showed that phosphorylation of Sp1 at Thr739 represses its DNA-binding activity during mitosis and Sp1 phosphorylation is essential for the progression of cell cycle. The data presented here support the notion that Sp1 phosphorylation during mitosis is important for cell-cycle progression. First, mutation of Thr739 to Ala caused accumulation of Sp1 in the chromosome during mitosis and additionally resulted in delayed mitotic progression and cell death (Figure 7). Second, we previously reported that Sp1 overexpressed by adeno-Sp1 virus infection is produced at such high level that could not be phosphorylated completely by endogenous kinases during mitosis, resulting in cell apoptosis (Chuang et al., 2009). Finally, this study indicates that Sp1 must leave chromosome for chromosome package in mitosis. To this end, Sp1 is degraded during mitosis in primary cells (Chuang et al., 2008). In cancer cells, Sp1 is stabilized and out of chromosome by phosphorylation modification at Thr739 by CDK1 in mitosis. Therefore, appropriate Sp1 phosphorylation modulated by CDK1/ PP2A might be important for Sp1 accumulation and facilitate cell-cycle progression.
According to previous studies and the present study, phosphorylation of Sp1 at a critical residue (Thr739) by ERK1/2, JNK1 and CDK1 may be important for the normal progression of various physiological functions in cancer cells (Milanini-Mongiat et al., 2002; Bonello and Khachigian, 2004; Chuang et al., 2008). A previous study indicated that the Thr739 residue of Sp1 is phosphorylated by ERK1/2 during interphase and that phospho-Sp1-Thr739 has an important role in the regulation of Sp1 transcriptional activity in interphase (Milanini-Mongiat et al., 2002; Bonello and Khachigian, 2004). In addition to phosphorylation of Sp1 in interphase, the Thr739 residue of Sp1 is also phosphorylated by JNK1 during mitosis, which affects Sp1 protein stability (Chuang et al., 2008). JNK1 phosphorylates Sp1 at Thr739 and Thr278 in mitosis to shield Sp1 from ubiquitin-dependent degradation. Sp1 phosphorylation at Thr739 also prevents the interaction between Sp1 and the ubiquitin E3-ligase (Wang et al., 2011). In addition, we also examined the relative contribution of JNK1 and CDK1 to Sp1phosphorylation. We found that CDK1 is more important than JNK1 in phosphorylating Sp1 (Supplementary Figure S13).
Our results indicating that CDK1/cyclin B1 is the major kinase that phosphorylates Sp1 suggests that CDK1 has an important role in mitosis by phosphor-ylating Sp1 at Thr739, resulting in Sp1 lacking DNA affinity. Although our results indicate that Thr739 phosphorylation is important for the regulation of Sp1 DNA-binding activity, previous studies have shown that phosphorylation of Sp1 zinc fingers is also important for the regulation of Sp1 DNA-binding activity (Armstrong et al., 1997). It is possible that different mechanisms exist to regulate Sp1 DNA-binding activity under different physiological conditions. In this study, we focused on Sp1 loss of DNA-binding activity during mitosis, whereas the previous studies focused on interphase. In addition, we indicated that Sp1 (T739D) expressed inside cells shows a significant loss of DNA-binding activity in mitosis (Figure 4) but the exogenous Sp1 (T739D) from Escherichia coli does not have altered DNA-binding activity (data not shown). This implies that other factors inside of cells cooperate with phospho-Sp1 to further repress Sp1 DNA-binding activity during mitosis. One possibility for these factors is phosphorylation-dependent peptidyl-prolyl cis–trans isomerases such as Pin1 and FKBP12, or phosphorylation-dependent kinases such as glycogen synthase kinase (GSK) 3. The PPIase-substrate interaction is regulated by phosphorylated modification of the substrate, with the PPIase catalyzing a conformational change to regulate the function of the substrate, including the regulation of protein stability, transcriptional activity, protein–protein interactions and phosphorylation status (Lu and Zhou, 2007; Guo et al., 2010; Saxena et al., 2010). The other possibility is that the phosphorylation of Sp1 at Thr739 leads to further phosphorylation by other kinase(s) to block its DNA-binding activity. Previous studies have indicated that heat shock transcription factor 1 (HSF-1) activity is suppressed by serial phosphorylation, and ERK phosphorylates HSF-1 at Ser307, which then allows GSK 3β to phosphorylate Ser 303 to negatively regulate both the DNA-binding and transcriptional activities of HSF-1 (He et al., 1998; Xavier et al., 2000). In addition, a recent study showed that GSK 3β inhibits the DNA binding and transcriptional activity through the Ser369-Ser373-Ser377 phosphorylation of the Runx 2 transcription factor, which mediates endothelial cell migration and invasion during tumor angiogenesis (Kugimiya et al., 2007). However, the mechanism by which the DNA-binding activity of Sp1 is affected by phosphorylation at Thr739 needs to be studied further.
Materials and methods
Cell culture and transfection
Human cervical adenocarcinoma HeLa cells and glioma C6 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum, 100µg/ml streptomycin sulfate, and 100 U/ml penicillin G sodium at 37°C and 5% CO2. Transfection of HeLa cells with expression vectors was done using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s directions.
DNA affinity precipitation assay
The Sp1-binding oligonucleotide 5′-CCCGCCTCCTTGAGGCGGGCCCGGGCGGGGCGG-3′, localized from −82 to −50 bp within the promoter of p21WAF1/CIP1, was biotinylated at 5′ termini and then annealed with their complementary strands. The assay was performed by incubating 1 µg of biotin-labeled probe with 300 µg of cell extract in 500 ml of binding buffer that contained 60 mM KCl, 12mM HEPES, pH 7.9, 4mM Tris-HCl, 5% glycerol, 0.5mM EDTA, 1 mM dithiothreitol. After it was incubated for 1 h at 4°C, DNA–protein complexes were then incubated with 30 µl of streptavidin-agarose (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 4°C. DNA-protein complexes were then washed three times in the binding buffer.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed according to a method described previously (Boulon et al., 2002; Chang et al., 2005; Hung et al., 2006). Formaldehyde-fixed DNA–protein complex were immunoprecipitated with 5µg of normal rabbit IgG, anti-GFP, or anti-Sp1 antibodies. Immunoprecipitated DNA was analyzed by PCR. The primer sequences for promoter of p21WAF1/CIP1 in PCR analyses were as follows: (5′-ACCAACGC AGGCGAGGGACT-3′ and 5′-CCGGCTCCACAAGGAAC TGA-3′). The primer sequences for promoter of c-Jun in PCR analyses were as follows: (5′- TAGACAGTCAAACCCCAA GA-3′ and 5′- CTAGCTCTGGGCAGTTAGAG-3′).
Immunofluorescence and confocal microscopy
The cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) in PBS according to a method described previously (Chuang et al., 2008). Immunostaining was conducted with primary antibodies such as anti-Sp1, anti-PP2Ac (Upstate and Abcam, Cambridge, MA, USA), anti-CDK1, anti-cyclin B1, anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or anti-Sp1 phospho-Thr739 antibodies made by Kelowna International Scientific Inc. (Taipei, Taiwan). The cells were then treated with Alexa Fluor 488-conjugated goat anti-mouse or rabbit immunoglobulin G (IgG) and Alexa Fluor 568-conjugated goat anti-mouse or rabbit IgG polyclonal antibodies (Invitrogen). The F-actin staining was done with Alexa Fluor 568-conjugated Phallotoxin (Invitrogen). Finally, the cells were mounted in 90% glycerol containing 4’-6-diamidino-2-phenylindole (DAPI) (Invitrogen), and examined using a confocal laser-scanning microscope (FluoView FV 1000; Olympus, Center Valley, PA, USA) or immunofluorescence microscope (Delta Vision Personal DV; Applied Precision, Issaquah, WA, USA). The images were analyzed with softWoRx software (Applied Precision).
In vitro CDK1/cyclin B1 kinase assay
For the in vitro phosphorylation analysis, the different GST-Sp1 fragments and the point mutants of Sp1 were purified from E. coli BL21 (DE3). These different Sp1 proteins and active CDK1/cyclin B1 (New England Biolabs, Ipswitch, MA, USA) were used to examine Sp1 phosphorylation in vitro. Each reaction (20µl) contained 1 µg of purified Sp1, 50 ng of active CDK1/cyclin B1, 2mCi of [γ-32p] ATP (Amersham Biosciences, Piscataway, NJ, USA), 1 mM ATP (Sigma-Aldrich), and 2µl of 10 × kinase buffer containing 500 mM HEPES, pH 7.4, 100 mM MgCl2, 10 mM EGTA, and 10 mM dithiothreitol. The phosphorylation reactions were incubated at 30°C for 15 min. After incubation, one-half of the reaction mixture was added to 10 µl of 2 × electrophoresis sample buffer, which was then heated to 95 °C for 5 min. Proteins in the mixtures were immediately separated using SDS-polyacrylamide gel electrophoresis (PAGE).
In vitro phosphatase assay
The whole mitotic cell lysate or purified (γ-32p labeled) Sp1 by anti-Sp1 antibody were incubated with PP2A (Promega, Madison, WI, USA) in dephosphorylation buffer (50 mM Tris-HCl, pH 7.5, 20mM MgCl2, 1 mM dithiothreitol, 0.5mM EDTA) for 10 min at 30 °C. The sample was then analyzed by SDS-PAGE and was detected by immunoblotting with anti-Sp1 or autoradiography.
Fluorescence-activated cell sorting (FACS) analysis
The cells were washed with ice-cold PBS and fixed in cold absolute ethanol at 4 °C for 4 h. The cells were then washed twice with PBS. After treating with 10 µg/ml RNase A (Qiagen, Valencia, CA, USA) at 37 °C for 30 min, cells were stained with 50 µg/ml propidium iodide (Sigma-Aldrich) at room temperature for 30 min. Finally, cells were analyzed using a flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA, USA).
The materials and methods of western blots, immunoprecipitation, reverse transcription–PCR, in vitro calf intestinal alkaline phosphatase (CIP) assay, immunohistochemistry, experimental animals and cultured rat primary glial cells are in Supplementary Information.
Supplementary Material
Acknowledgements
This work was supported by the National Cheng Kung University project of the Program for Promoting Academic Excellence and Developing World Class Research Centers, together with grants NSC 97-2320-B-006-016-MY3 and NSC 97-2311-B-006-002-MY3 obtained from the National Science Council, Taiwan.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Abdelrahim M, Smith R, 3rd, Burghardt R, Safe S. Role of Sp proteins in regulation of vascular endothelial growth factor expression and proliferation of pancreatic cancer cells. Cancer Res. 2004;64:6740–6749. doi: 10.1158/0008-5472.CAN-04-0713. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Barry DA, Leggett RW, Mueller CR. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J Biol Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- Benasciutti E, Pages G, Kenzior O, Folk W, Blasi F, Crippa MP. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood. 2004;104:256–262. doi: 10.1182/blood-2003-08-2661. [DOI] [PubMed] [Google Scholar]
- Bonello MR, Khachigian LM. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J Biol Chem. 2004;279:2377–2382. doi: 10.1074/jbc.M308254200. [DOI] [PubMed] [Google Scholar]
- Boulon S, Dantonel JC, Binet V, Vie A, Blanchard JM, Hipskind RA, et al. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol Cell Biol. 2002;22:7769–7779. doi: 10.1128/MCB.22.22.7769-7779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LK, Chung JY, Hong YR, Ichimura T, Nakao M, Liu ST. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res. 2005;33:6528–6539. doi: 10.1093/nar/gki956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiefari E, Brunetti A, Arturi F, Bidart JM, Russo D, Schlumberger M, et al. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer. 2002;2:35. doi: 10.1186/1471-2407-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Ferro TJ. Sp1: regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Chuang JY, Hung JJ. Overexpression of HDAC1 induces cellular senescence by Sp1/PP2A/pRb pathway. Biochem Biophys Res Commun. 2011;407:587–592. doi: 10.1016/j.bbrc.2011.03.068. [DOI] [PubMed] [Google Scholar]
- Chuang JY, Wang YT, Yeh SH, Liu YW, Chang WC, Hung JJ. Phosphorylation by c-Jun NH2-terminal Kinase 1 Regulates the Stability of Transcription Factor Sp1 during Mitosis. Mol Biol Cell. 2008;19:1139–1151. doi: 10.1091/mbc.E07-09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JY, Wu CH, Lai MD, Chang WC, Hung JJ. Overexpression of Sp1 leads to p53-dependent apoptosis in cancer cells. Int J Cancer. 2009;125:2066–2076. doi: 10.1002/ijc.24563. [DOI] [PubMed] [Google Scholar]
- Guo T, Cornea RL, Huke S, Camors E, Yang Y, Picht E, et al. Kinetics of FKBP12.6 binding to ryanodine receptors in permeabilized cardiac myocytes and effects on Ca sparks. Circ Res. 2010;106:1743–1752. doi: 10.1161/CIRCRESAHA.110.219816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Meng YH, Mivechi NF. Glycogen synthase kinase 3beta and extracellular signal-regulated kinase inactivate heat shock transcription factor 1 by facilitating the disappearance of transcriptionally active granules after heat shock. Mol Cell Biol. 1998;18:6624–6633. doi: 10.1128/mcb.18.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Davie JR. Sp1 and Sp3 foci distribution throughout mitosis. J Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- Hosoi Y, Watanabe T, Nakagawa K, Matsumoto Y, Enomoto A, Morita A, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol Cell Biol. 2006;26:1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugimiya F, Kawaguchi H, Ohba S, Kawamura N, Hirata M, Chikuda H, et al. GSK-3beta controls osteogenesis through regulating Runx2 activity. PLoS One. 2007;2:e837. doi: 10.1371/journal.pone.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–283. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Lou Z, O’Reilly S, Liang H, Maher VM, Sleight SD, McCormick JJ. Down-regulation of overexpressed sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 2005;65:1007–1017. [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- Milanini-Mongiat J, Pouyssegur J, Pages G. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J Biol Chem. 2002;277:20631–20639. doi: 10.1074/jbc.M201753200. [DOI] [PubMed] [Google Scholar]
- Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Saxena UH, Owens L, Graham JR, Cooper GM, Hansen U. Prolyl isomerase Pin1 regulates transcription factor LSF (TFCP2) by facilitating dephosphorylation at two serine-proline motifs. J Biol Chem. 2010;285:31139–31147. doi: 10.1074/jbc.M109.078808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- Tan NY, Khachigian LM. Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol. 2009;29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- Wang YT, Chuang JY, Shen MR, Yang WB, Chang WC, Hung JJ. Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process. J Mol Biol. 2008;380:869–885. doi: 10.1016/j.jmb.2008.05.043. [DOI] [PubMed] [Google Scholar]
- Wang YT, Yang WB, Chang WC, Hung JJ. Interplay of posttranslational modifications in Sp1 mediates Sp1 stability during cell cycle progression. J Mol Biol. 2011;414:1–14. doi: 10.1016/j.jmb.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Xavier IJ, Mercier PA, McLoughlin CM, Ali A, Woodgett JR, Ovsenek N. Glycogen synthase kinase 3beta negatively regulates both DNA-binding and transcriptional activities of heat shock factor 1. J Biol Chem. 2000;275:29147–29152. doi: 10.1074/jbc.M002169200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.