Abstract
The catalytic mechanism by which the hairpin ribozyme accelerates cleavage or ligation of the phosphodiester backbone of RNA has been incompletely understood. There is experimental evidence for an important role for an adenine (A38) and a guanine (G8), and it has been proposed that these act in general acid-base catalysis. In this work we show that a large reduction in cleavage rate on substitution of A38 by purine (A38P) can be reversed by replacement of the 5′-oxygen atom at the scissile phosphate by sulfur (5′-PS), which is a much better leaving group. This is consistent with A38 acting as the general acid in the unmodified ribozyme. The rate of cleavage of the 5′-PS substrate by the A38P ribozyme increases with pH log-linearly, indicative of a requirement for a deprotonated base with a relatively high pKa. On substitution of G8 by diaminopurine, the 5′-PS substrate cleavage rate at first increases with pH and then remains at a plateau, exhibiting an apparent pKa consistent with this nucleotide acting in general base catalysis. Alternative explanations for the pH dependence of hairpin ribozyme reactivity are discussed, from which we conclude that general acid-base catalysis by A38 and G8 is the simplest and most probable explanation consistent with all the experimental data.
Keywords: RNA catalysis, catalytic mechanism, 5′-phosphorothiolate substitution
Introduction
RNA-mediated catalysis is important in biologically-significant RNA processing events and in the condensation of amino acids in the ribosome 1. Contemporary ribozymes also offer a glimpse of how phosphoryl transfer reactions might have been catalyzed in an early ‘RNA world’, postulated to have played a key role in the early development of life on the planet 2. Yet the chemical mechanisms of even the relatively simple ribozymes presently known to exist are incompletely understood, and sometimes the subject of considerable debate.
The nucleolytic ribozymes are a group of five species that carry out a site-specific cleavage or ligation of the phosphodiester backbone of RNA, in order to process replication intermediates, transcripts or to control gene expression. Despite sharing little similarity in sequence or structure, these ribozymes each bring about cleavage by nucleophilic attack of an O2′ on the adjacent phosphorus atom (Figure 1A).
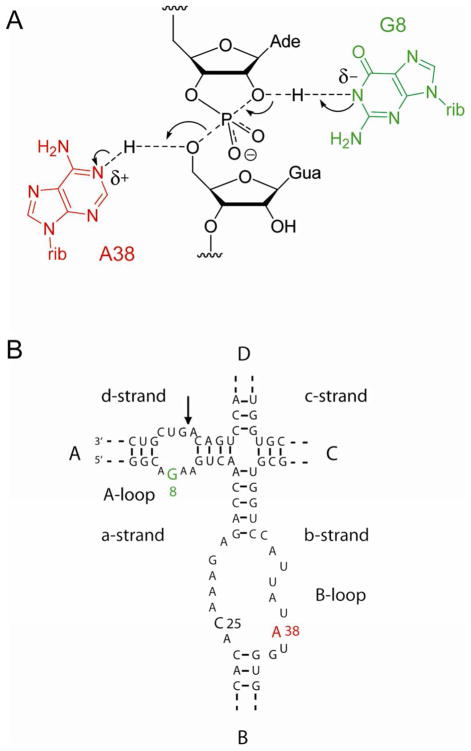
Figure 1.
The hairpin ribozyme and a proposed reaction mechanism.
A. Proposed mechanism of cleavage and ligation based on general acid-base catalysis. This depicts the transition state; the arrows show the flow of electrons for cleavage.
B. The sequence of the hairpin ribozyme used here. The ribozyme comprises four strands labelled a through d.
In principle the reaction could be accelerated by transition state stabilization (e.g. by hydrogen bonding, or by electrostatic stabilization of the formally-dianionic transition state), general acid-base catalysis (enhancing hydroxyl nucleophilicity by deprotonation, and stabilization of the oxyanion leaving group by protonation) or by conformational facilitation of the trajectory into an in-line transition state, and it is quite possible that multiple processes contribute to the overall rate enhancement.
In this paper we address the catalytic mechanism of the hairpin ribozyme, derived from the (−) strand of the tobacco ringspot virus satellite RNA 5, 6. The secondary structure of the ribozyme-substrate complex 7 is based upon a four-way helical junction (Figure 1B) 8–10. Adjacent arms (A and B) contain internal loops that undergo intimate interaction to generate the active conformation of the ribozyme (11–13). Within these loops, two nucleotides have been identified as particularly critical to catalytic activity, G8 in loop A on the opposite strand to the scissile phosphate 11–13, and A38 in loop B 14. We have previously noted 15 that the arrangement of these components is very similar to that in the Varkud satellite ribozyme, where there is good evidence that the corresponding guanine (G638) acts as general base and the adenine (A756) as general acid in the cleavage reaction 16. Crystal structures of the hairpin ribozyme both in the four-way junction form 17 and the minimal hinged version (lacking helices C and D) 18, 19 show that G8 and A38 are close to the scissile phosphate, and in the structure of a transition state analog 20 A38 and G8 are hydrogen bonded to O5′ and O2′ respectively, thus positioned in a manner that is consistent with their proposed roles in general acid-base catalysis (Figure 1A).
However, the role of nucleobase-mediated general acid-base catalysis in the hairpin ribozyme, and particularly the role of G8, has been controversial. This originally stems from the pH dependence of the cleavage and ligation reactions. In contrast to the VS ribozyme (that exhibits a bell-shape dependence of reaction rate on pH consistent with a role for two nucleobases of relatively low and high pKa values 15), the rates of the hairpin ribozyme reactions increase with pH to approximately neutrality, and then remain at a plateau at higher pH values 11, 13, 21. This appears to suggest that the reactions depend on the ionization of a single group. Coupled with the results of exogenous base rescue experiments, this led to the suggestion that A38 acts not as a general acid, but instead by the electrostatic stabilization of the transition state 13, 22. However, when G8 is substituted by a nucleobase of lower pKa the cleavage reaction rate falls with pH to generate a bell-shaped dependence 11, 23, 24, and it is clear that these modified hairpin ribozymes, just like the VS ribozyme, require two groups to be in the appropriate ionisation state for activity.
5′-Phosphorothiolate (5′-PS) substitution of the scissile phosphate provides a powerful means to examine the proposed role of nucleobases in general acid-base catalysis, and has previously illuminated the mechanisms of the hepatitis delta virus 25 and VS ribozymes 16. Sulfur is a far better leaving group than oxygen, and does not require protonation by a general acid. If substitution of a particular nucleobase leads to loss of catalytic activity because it normally (i.e. in the unmodified case) protonates the leaving group as a general acid, then activity should be restored by the 5′-PS substitution. By contrast, if the nucleobase accelerates the reaction by acting as a general base or by other mechanisms such as transition state stabilization, the restorative effect of the 5′-PS substitution should be much smaller. In addition, if the general acid no longer carries out its catalytic function in the presence of the 5′-PS group, the general base can then be studied in isolation. We critically examined the role of A38 and G8 in the hairpin ribozyme by this approach. The results are consistent with their proposed function in general acid-base catalysis.
Experimental Section
Construction of a hairpin ribozyme with a substrate strand containing an ApsG dinucleotide with a 5′-phosphorothiolate linkage
The chemical synthesis of A[2′-O-(o-nitrobenzyl)]-psG is described in the Supporting Information and Scheme S1. A substrate strand containing this dinucleotide was prepared by ligation to the 5′- and 3′ oligonucleotides. The hairpin ribozyme was constructed by hybridization of four oligonucleotides as shown in Figure S1. All methods and sequences are fully described in the Supporting Information.
Ribozyme kinetics
Cleavage of the radioactively 5′-32P labelled substrate strand was studied under single-turnover conditions as described in Wilson et al. 16. The 2-hydroxyl of the adenosine was deprotected using UV irradiation for 15 min under neutral conditions. Following equilibration to 25°C, reactions were started by adding MgCl2. Standard reaction conditions for the hairpin ribozyme cleavage were 10 mM MgCl2 at 25°C using various buffers at 25 mM concentration depending on pH and 50 mM NaCl (pH 5–5.5 acetate, pH 5.5–6.75 MES, pH 6.75–8 HEPES, pH 8–9 TAPS). Products of the cleavage reaction were analyzed via denaturing PAGE and quantified by phosphorimaging.
Analysis of ribozyme kinetics and analysis of pH dependence of reaction rate
Substrate cleavage was studied under single-turnover conditions as fully described in the Supporting Information. The pH dependence of observed cleavage rates was fitted to either a single-ionization model incorporating a pH-independent contribution observable at low pH (k0),
| [2] |
or a double-ionization model 26 :
| [3] |
where kcleave is the intrinsic rate of the cleavage reaction and pKA1 and pKA2 are the acid dissociation constants of two titrating functional groups.
Results
The construction of the hairpin ribozyme
In this study we have focussed on the natural form of the hairpin ribozyme, based on a four-way junction 8 (Figure 1B). This was constructed by the hybridization of four synthetic oligonucleotides, including the radioactively [5′-32P]-labelled substrate strand d in which the scissile phosphate was either unmodified (5′-PO) or incorporated a 5′-phosphorothiolate linkage 16, 25, 27. Complete sequences for each strand are given in the Supporting Information. For synthetic convenience the substrate strand was extended 3′ to the cleavage site beyond the six nucleotides used in previous studies 21, 23. We therefore shortened strand a in order to retain just three base pairs with the 3′ product of cleavage, that is known to result in rapid dissociation of the 3′ product 28 and thus to eliminate the reverse reaction (i.e. ligation). This new construct exhibited a cleavage rate that was indistinguishable from that observed for a hairpin ribozyme species with a 5′-extended a-strand 23. The 5′-PS substrate strand included o-nitrobenzyl protection of the 2′-hydroxyl of the adenosine to prevent premature activation of the nucleophile; its synthesis is described in the Supporting Information.
The activity of a hairpin ribozyme with a purine at position 38 is restored by 5′-phosphorothiolate substitution in the substrate
Substrate strand cleavage was studied under single-turnover conditions immediately following deprotection of the 2′-hydroxyl nucleophile using ultraviolet irradiation. Products of ribozyme cleavage were separated by gel electrophoresis, and quantified by phosphorimaging. The unmodified substrate (i.e. 5′-PO) strand exhibited no detectable cleavage after 20 min incubation under standard conditions (25 mM HEPES (pH 7.5), 50 mM NaCl, 10 mM MgCl2 at 25°C) in the absence of the ribozyme (Figure 2A).
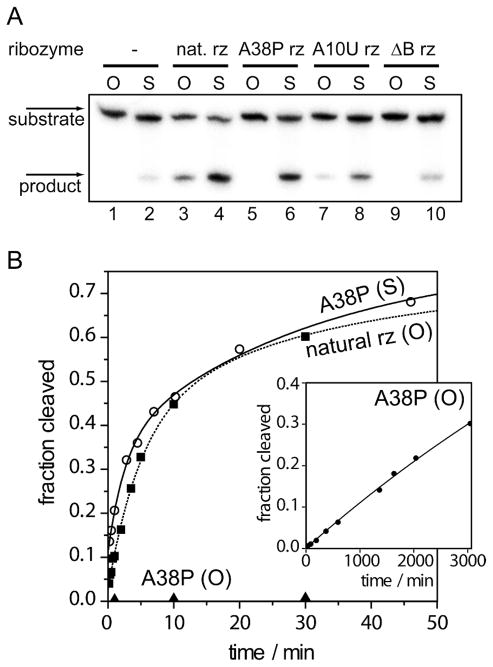
Figure 2.
Cleavage of the 5′-PO or 5′-PS substrate by some hairpin ribozyme variants.
The substrate strand was [5′-32P]-labelled, and reaction products were separated by electrophoresis in 20 % polyacrylamide and subjected to phosphorimaging.
A. Gel electrophoresis. The 5′-PO (tracks 1, 3, 5, 7 and 9) and 5′-PS (track 2, 4, 6, 8 and 10) substrates were incubated with no ribozyme (track 1, 2), the natural hairpin ribozyme (track 3, 4), A38P (track 5, 6), A10U (7, 8) or ΔB ribozyme where loop B was removed by complementation (track 9, 10) for 20 min. under standard conditions.
B. Reaction progress of the cleavage reaction under standard conditions, fitted to double exponential functions. Open circles: 5′-PS + A38P ribozyme, filled squares: 5′-PO + natural ribozyme, filled triangles and filled circles (inset): 5′-PO + A38P ribozyme.
Only a very small fraction of the corresponding 5′-PS substrate was cleaved under these conditions. Hybridization with strands b–d to form the hairpin ribozyme led to significant cleavage of the 5′-PO substrate. The 5′-PS substrate strand is cleaved faster, so that a greater extent of cleavage following the 20 min incubation was observed; this might be anticipated if the general acid protonating the leaving group is significantly deprotonated at pH 7.5 (see below). Graphs of reaction progress are shown in Figure 2B, and measured rates tabulated in Table 1.
Table 1.
Rates of cleavage for different hairpin ribozymes measured under single-turnover conditions in 25 mM HEPES (pH 7.5), 50 mM NaCl, 10 mM MgCl2 at 25°C incorporating an unmodified substrate (5′-PO) or a 5′-phosphorothiolate linked substrate (5′-PS). Each rate constant is the average of ≥ 3 independent measurements.
| ribozyme | 5′-PO, kobs/min−1 | 5′-PS, kobs/min−1 | 5′-PS kobs/5′-PO kobs |
| natural ribozyme | 0.51 ± 0.08 | 5.8 ± 0.8 | 11 |
| none | u.d.* | 0.004 ± 0.003 | -- |
| ΔB ribozyme | u.d.* | 0.020 ± 0.002 | -- |
| G8DAP | 0.011 ± 0.002 | 2.0 ± 0.2 | 180 |
| G8I | 0.02 ± 0.02 | 0.17 ± 0.03 | 8 |
| A10U | 0.0147 ± 0.0002 | 0.17 ± 0.003 | 12 |
| C25U | 0.014 ± 0.003 | 0.025 ± 0.002 | 2 |
| A38P | 0.00015 ± 0.00003 | 0.5 ± 0.1 | 3,600 |
| G8DAP/A38P | u.d* | 0.14 ± 0.04 | -- |
u.d. denotes a rate of < 10−5 min−1, i.e. essentially undetectable activity after > 40 h incubation.
Ribozymes with substitutions detrimental to activity each cleave the 5′-PS substrate more rapidly than the 5′-PO substrate (Figure 2A, Table 1). However, the ribozyme in which A38 has been substituted by purine (A38P) stands out. It is the substitution most deleterious to cleavage of the 5′-PO substrate, yet it shows the greatest extent of restoration of activity by the 5′-PS substitution (3,600-fold). The almost complete restoration of cleavage activity for the A38P ribozyme in the presence of the 5′-PS linkage is consistent with the assignment of A38 as the putative general acid in the cleavage reaction catalyzed by the hairpin ribozyme.
The pH-dependence of activity of the A38P ribozyme is consistent with general acid-base catalysis
The data presented above are consistent with A38 acting as a general acid to protonate the 5′-oxyanion leaving group in cleavage, but it is necessary to examine activity as a function of pH for this to be substantiated and to examine the role of the putative general base. The cleavage and ligation rates of the hairpin ribozyme are pH dependent with identical shapes. They increase with pH until neutrality is reached and remain constant over the measurable pH range thereafter 13, 21. The proposed general acid-base catalysis mechanism for hairpin ribozyme cleavage requires a protonated A38 and a deprotonated G8 as active components (Figure 1). Thus the observed rate of cleavage (kobs) will be given by:
| [1] |
where fA and fB are the fractions of A38 and G8 in their protonated and deprotonated states respectively (which can be calculated for any pH from their assumed pKa values), and kcat is the rate of cleavage by ribozyme with acid and base in their required forms. Following the approach of Bevilacqua 26, in Figure 3A we simulate the expected pH profile of the reaction for pKa values of 6 and 10.5, corresponding to A38 and G8 respectively 13, 21, 29.
Figure 3.
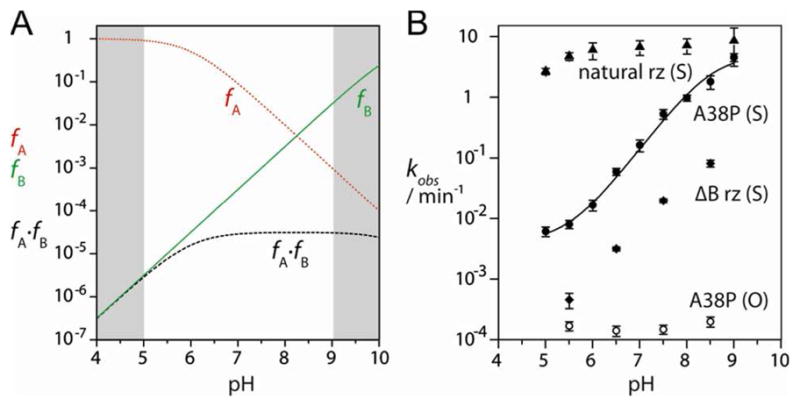
The pH dependence of cleavage rates for the 5′-PO and 5′-PS substrate with various ribozyme constructs.
A. Simulation of the reaction rate as a function of pH for a general acid of pKa = 6 and a general base of pKa = 10.5. The fractions of the protonated acid (fA – red dotted line), unprotonated base (fB- green solid line) and fA × fB (dashed line) are plotted as a function of pH. In all simulations, the shaded regions lie outside the experimentally accessible range of pH; note that the reduction in fA × fB at high pH falls outside the observable region. If general acid catalysis is not required, the pH dependence should simply follow fB.
B. Experimental cleavage rates for the 5′-PO and 5′-PS substrate with various ribozyme constructs are plotted on a logarithmic scale as a function of pH. Filled triangles, 5′-PS substrate + natural-sequence ribozyme; filled circles, 5′-PS substrate + A38P ribozyme; filled diamonds, 5′-PS substrate + ΔB ribozyme (lacking loop B); open circles, 5′-PO substrate + A38P ribozyme. The data for the 5′-PS substrate + A38P ribozyme have been fitted to a single ionization model with additional pH-independent component as discussed in the text, yielding an apparent pKa of 8.5± 0.2.
The rise in rate at low pH is due to the deprotonation of the guanine (i.e. the rise in fB, which occurs over the whole range of the titration), while the formation of the plateau is due to the deprotonation of the adenine (i.e the fall in fA) offsetting the rise in fB. Thus the pKa measured is that of the adenine. If the 5′-PS substitution obviates the requirement for leaving group protonation, then a general acid is no longer required for catalysis and the pH profile of activity would be expected to reflect only the extent of deprotonation of the general base (i.e. fB, not the product fA × fB). Thus the rate of cleavage of the 5′-PS substrate would be expected to increase in a log-linear manner over the observable pH range, diverging from the pH profile for cleavage of the 5′-PO substrate at the pKa of the general acid. Experimentally the observed activity is almost constant over the range of pH investigated (Figure 3B). Thus either the cleavage of the 5′-PS substrate is pH independent, and thus not subject to general base catalysis by G8, or the cleavage reaction is not rate-limiting under these conditions.
In order to resolve this ambiguity we investigated the pH dependence of activity of the A38P ribozyme (Figure 3B), where the rates of cleavage are lower. For the unmodified 5′ PO substrate the cleavage rate remains constant at approximately kobs = 10−4 min−1 over the measureable pH range. However, for the 5′-PS substrate, the cleavage rate rises monotonically with increasing pH and is approximately log-linear over the range of pH 6 to 8. However, the deviation from linearity below pH 6 is not consistent with general base catalysis alone. This deviation could be due to the presence of an additional pH-independent reaction pathway that contributes significantly to catalysis at low pH. For example, this might involve removal of a proton from the 2′-OH nucleophile by the pro-R non-bridging oxygen of the scissile phosphate as proposed on the basis of computational modeling of the reaction pathway 30. Alternatively protonation of a nucleobase close to the cleavage site such as A10 might contribute to catalysis 31. This could stabilize the transition state and result in the rate enhancement observed at low pH. Fitting the data to a single-ionization model incorporating a low-pH channel to catalysis yields an apparent pKa of 8.5 ± 0.2 for the general base. Thus, allowing for an additional contribution to catalysis observable at low pH, the data for the A38P ribozyme are consistent with a general acid-base cleavage mechanism with the nucleobase at position 38 acting as the general acid and a species with high pKa acting as the general base.
Nucleotide substitutions of G8 in the hairpin ribozyme influence the pH profile for cleavage of the 5′-PS substituted substrate strand
The log-linear increase in cleavage rate with pH for the 5′-PS substrate catalyzed by the A38P ribozyme is consistent with general base catalysis by G8, but to test this conclusion we have substituted it by a different nucleotide with a markedly lower pKa. This should result in a predictable change in the pH-activity profile (Figure 4A).
Figure 4.
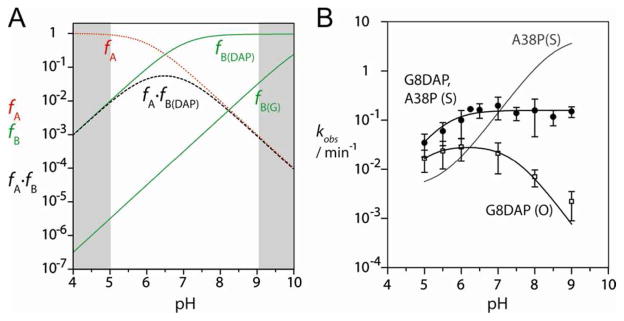
The pH dependence of ribozyme cleavage rates for the 5′-PO and 5′-PS substrate where G8 has been substituted by diaminopurine.
A. Simulation of reaction rate as a function of pH for an acid of pKa = 6 and a base of pKa = 7 (corresponding to DAP). The fractions of the protonated acid (fA – red dotted line), unprotonated base (fB- green solid line) and fA × fB (dashed line) are plotted as a function of pH. Note that the pH dependence should now be bell-shaped within the observable range. In addition, the fraction of unprotonated base (fB) for a pKa = 10.5 is also plotted to simulate ribozyme with an unmodified G8 (solid line).
B. Experimental cleavage rates (plotted on a logarithmic scale) for the 5′-PO and 5′-PS substrates + G8DAP hairpin ribozyme. Filled circles, 5′-PS substrate + A38P, G8DAP ribozyme (data fitted to a single ionization model with k0 = 0, yielding an apparent pKa of 5.5 ± 0.2); open squares, 5′-PO substrate + G8DAP ribozyme (data fitted to a double ionization model, pKa1 = 5.1 ± 0.1, pKa2 = 7.3 ± 0.1). The fit to the data for the 5′PS substrate + A38P ribozyme has been plotted for comparison.
For this purpose we investigated the activity of a modified hairpin ribozyme with 2,6-diaminopurine (pKa = 5.1) at position 8 (G8DAP). This substitution leads to a bell-shaped pH profile with a centre near pH = 6 for the cleavage of the 5′-PO substrate (Figure 4B), because the saturation of the deprotonation of the base is now within the observable pH range. A similar pH dependence has previously been found for the minimal form of the hairpin ribozyme with the same G8DAP substitution 11. Fitting the curve to a double-ionization model gives two apparent pKa values of 5.1 ± 0.1 and 7.3 ± 0.1, possibly corresponding to A38 and G8DAP respectively.
In order to determine the effect of a G8DAP substitution on the pH profile for cleavage of the 5′-PS substituted substrate, we investigated the activity of a ribozyme with both A38P and G8DAP substitutions. This ribozyme has no detectable activity towards the 5-PO substrate, while the pH profile for the cleavage of the 5′-PS substrate rises up to pH = 6.5 and remains constant at higher pH (Figure 4B). The apparent pKa value of 5.5 ± 0.2 is consistent with the deprotonation of 2,6-DAP. The shift in apparent pKa on substitution of G8 with a nucleobases of lower pKa supports the proposition that the nucleobase at position 8 is acting as a general base in hairpin ribozyme catalysis.
Activity of hairpin ribozyme lacking loop B on substrate carrying a 5′-PS substitution
If the main function of loop B is to deliver A38 to act as the general acid for the cleavage reaction, it might be anticipated that ribozyme with loop A alone could retain some level of activity when the substrate carries the 5′-PS substitution. We therefore investigated a hairpin ribozyme variant in which only loop A is present by inclusion of an alternative b strand that complements loop B while preserving the four-way junction (termed ΔB ribozyme). This ΔB ribozyme exhibits negligible cleavage activity on the 5′-PO substrate (Table 1), confirming that loop B (which includes A38) is absolutely required for catalysis. By contrast, the presence of ΔB ribozyme produces a 5-fold greater rate of cleavage of the 5′-PS substrate compared to the 5′-PS substrate alone (Table 1). However, the rate is significantly lower than that observed for the natural and A38P ribozymes. The reaction rate exhibits log-linear dependence on pH, almost parallel to the A38P hairpin ribozyme but with lower absolute rates (Figure 3B). This is consistent with G8 acting as general base in both cases.
Discussion
5′-Phosphorothiolate substitution of the scissile phosphate was used to address the functions of A38 and G8 in hairpin ribozyme catalysis, and in particular their proposed roles as general acid and general base respectively. 5′-PS substitution obviates the need for protonation of the leaving group, rendering the action of the general acid unnecessary. The effects of modifications to the ribozyme that decrease the activity of the general acid are potentially suppressed by the 5′-PS substitution whereas other modifications should affect 5-PO and 5′-PS substrates to a similar extent. Thus the identity of the general acid can be established and the function of the general base can be studied in isolation. The effects of nucleobase substitutions to the hairpin ribozyme and the pH dependence of their rates are consistent with guanine at position 8 acting as general base and adenine at position 38 acting as general acid to provide a significant contribution to catalysis in the hairpin ribozyme.
The activity of an impaired ribozyme containing a purine nucleobase at position 38, which generates negligible cleavage of a 5′-PO substrate, was restored to the level of the natural ribozyme by 5′-PS substitution at the cleavage site. All other substitutions were suppressed by the 5′-PS modification to a much smaller extent (Table 1). This is consistent with A38 acting as a general acid in protonating the leaving group. If the contribution of A38 to catalysis is solely or indeed substantially through electrostatic stabilization of the transition state, as has been proposed 14, an effect of this magnitude would not be expected.
This conclusion is in accord with previous evidence, reviewed in Wilson and Lilley 29. A ribozyme with N1-deazaadenine at position 38 has no measureable activity although it folds into an apparently unperturbed active structure 32. Replacement of A38 with isoguanine, which has the N6 exocyclic amine of adenine but a pKa similar to guanine, gives a ribozyme with negligible activity at neutral pH. But at high pH its cleavage activity is as great as the native ribozyme 14, consistent with general acid-base catalysis by two nucleobases of high pKa. Replacement of A38 with 8-azaadenine, which has a lower pKa, resulted in a corresponding decrease in the apparent pKa of ribozyme cleavage 33, and nucleotide analogue interference mapping revealed a strong interference at position 38 from adenosine analogues of low pKa 31, consistent with ionization of N1 influencing ribozyme activity. Finally, A38 has an elevated ground state microscopic pKa of 5.46 in a precatalytic conformation of the hairpin ribozyme, as measured by Raman crystallography 34, which correlates well with the macroscopic apparent pKa of hairpin ribozyme activity. We consider that these results, together with our new data, demonstrate that A38 protonates the 5′-O leaving group in the cleavage reaction of the hairpin ribozyme.
The cleavage rate of the 5′-PS substrate by the A38P hairpin ribozyme exhibits the log-linear dependence on pH expected if the nucleophile is activated by a general base of high pKa. In contrast, the activity of the natural hairpin ribozyme towards the 5′-PS substrate is almost constant over the pH range investigated. In this case either the cleavage of the 5′-PS substrate is pH independent, and thus either not subject to general base catalysis by G8, or the cleavage reaction is not rate-limiting under these conditions. A pH-independent activation of the nucleophile is not consistent with the data for the natural ribozyme (see below) but cannot be excluded for cleavage of the 5′-PS substrate. However we favour the possibility that a conformational change is limiting the activity of the ribozyme. We have previously shown that the rate of oscillation by the four-way junction between parallel and antiparallel states is ~100 min−1 in the standard buffer used here 21, 35. As the loop-loop interaction necessary for ribozyme activity occurs with the junction in the antiparallel state, the dynamics of the junction sets an upper limit on the activity of the ribozyme that is only one order of magnitude greater than the rates observed for cleavage of the 5′-PS substrate by the natural ribozyme. The extensive remodelling of both loops A and B to form the active ribozyme, revealed by comparison of the NMR structures of the isolated loops 36, 37 and the crystal structure of the ribozyme 17, presumably occurs subsequently to initial contact and may further limit the rate of formation of the active ribozyme 38. The observation that the natural ribozyme and a ribozyme with a G8DAP substitution exhibit almost identical maximum rates of cleavage of the 5-PS substrate yet very different rates of cleavage of the 5′-PO substrate (Table 1) also suggests that conformational changes may limit the activity of these ribozymes.
The importance of remodelling loop A to obtain an active structure is emphasized by our data for the ΔB ribozyme (Table 1). These results indicate that cleavage of the 5′-PS substrate can proceed in the absence of loop B, and that in this situation a species of high pKa is activating the nucleophile. But the lower rate compared to the A38P ribozyme shows that the interaction between loop A and loop B is important in generating the catalytically competent conformation. This is seen also in the data for the A10U and C25U substitutions, which weaken the loop-loop interaction through disruption of the ribose zipper and a critical G+1:C25 base pair respectively (11, 15, 36). The limited suppression of these defects by the 5′-PS substrate is consistent with an impairment in loop A remodelling.
The pH dependence of cleavage of the 5′-PS substrate by the A38P hairpin ribozyme is consistent with the action of a general base of high pKa. We have previously argued that the substitution of G8 with nucleobases of lower pKa provides direct support for G8 fulfilling this role 29. The bell-shaped pH dependence and apparent pKa values observed on substitution of G8 with diaminopurine, 2-aminopurine 11, imidazole 23 or 8-azaguanine 24 are consistent with the nucleobase at position 8 acting in concert with A38 in general acid-base catalysis. Similarly, rescue of an abasic lesion at position 8 by exogenous cytosine or 2-aminopyridine 13 is consistent with the hypothesis that the exogenous base is acting as a general base during cleavage. Furthermore, the structure of the cleaved ribozyme has G8 positioned to act as a general acid in the ligation reaction, with N1 donating a hydrogen bond to the O2′ leaving group 20. If it so acts, then by the principle of microscopic reversibility it should act as a general base during cleavage. To test if G8 does fulfil this role we investigated the cleavage of the 5′-PS substrate by a ribozyme with 2,6-diaminopurine at position 8, retaining the A38P substitution. The pH dependence of activity reflects the deprotonation of a base with a much lower apparent pKa, consistent with that of 2,6-diaminopurine. Since the pH profile is expected to correspond to the protonation state of the general base in this experiment, the lower apparent pKa supports the proposition that the nucleobase at position 8 is directly involved as general base in the catalytic mechanism of hairpin ribozyme cleavage.
It is interesting to note that the apparent pKa values of nucleobases at position 8 measured in the context of the A38P and 5′-PS substitutions (8.5 for guanine and 5.5 for 2,6-diaminopurine) are substantially lower than those inferred from the pH profiles for ribozymes with A38 and a natural substrate (10.6 for guanine 29, 7.3 for 2,6-diaminopurine). This might reflect limits to activity that result from conformational change. Alternatively the loss of hydrogen bonding potential due to the A38P and 5′-PS substitutions may result in a change in the local environment of the nucleobase at position 8.
We further note that the natural ribozyme exhibits significantly faster cleavage of the 5′-PS substrate than the 5′-PO substrate, even at pH = 5, where A38 is expected to be almost fully protonated. This implies a corresponding enhancement in kcat. This may be due to a greater intrinsic reactivity because sulfur is a better leaving group, although a similar enhancement in activity at low pH was not observed for the HDV 25 or VS ribozymes 16. Alternatively, the increase in rate may be due to a greater fraction of ribozyme molecules having an active conformation, a possibility suggested by the crystal structure of an inactive hairpin ribozyme with a 2′-O-methyl substitution of the nucleophile 17. Despite G8 being in proximity to the nucleophile and a near-optimal in-line orientation of nucleophile and leaving group, an unmodified ribozyme having the same conformation as the crystal structure would not be reactive since A38 lies some distance from the leaving group. Yet in the context of the 5′-PS substitution, where protonation of the leaving group by A38 is not required, such a structure should be active and may account for the enhanced rate.
General acid-base catalysis by A38 and G8 is fully consistent with the data, but can other mechanisms provide an alternative explanation? The equivalent pH dependence observed for cleavage and ligation reactions 13, 21 significantly constrains the possible mechanisms. Equivalent pH profiles are readily explained by the ionization of two groups (Figure 5).
Figure 5.
General acid-base catalysis yields equivalent pH dependence for cleavage and ligation. The roles of general acid and base will be exchanged between cleavage and ligation reactions. However, the rates of reaction are proportional to fA × fB, which have the same shape for both forward and reverse reactions, although the magnitudes are very different.
A. Simulation of reaction rate as a function of pH for an acid of pKa = 6 and a base of pKa = 10.5, corresponding to the proposed mechanism for cleavage. The upper panel plots the fractions of the protonated acid (fA) and unprotonated base (fB) in red dotted and green solid lines respectively, with fA × fB shown in the lower panel.
B. Simulation of the reaction rate as a function of pH for an acid with pKa = 10.5 and a base with pKa = 6 corresponding to the proposed mechanism for ligation, following the presentation in part A. Note that the pH dependence of fB and the product fA × fB are experimentally indistinguishable for the ligation reaction, in contrast to the cleavage reaction shown in A.
The curves of the product fA × fB as a function of pH have an identical shape for the cleavage and ligation reactions, and while the magnitude of fA × fB is much higher for ligation in the case illustrated, this will be offset by a lower kcat due to both nucleobases being in a less reactive, uncharged state 26. In contrast, ionization of a single group that participates in proton transfer would yield inverted pH profiles for cleavage and ligation. In principle, the pH dependence of the ligation reaction of the hairpin ribozyme is consistent with a single group with pKa ~ 6, such as A38, acting as a general base in a step-wise mechanism where formation of the bond between the 5′-O and P is rate limiting. In this case the rate of reaction would be proportional to the magnitude of fB (Figure 5B). But by the principle of microscopic reversibility the cleavage reaction rate would be proportional to fA for the same group (Figure 5A), and thus an inverted profile would be expected in this case; this is not consistent with observation. However ionization of a single group is consistent with equivalent profiles for cleavage and ligation if the group is involved in electrostatic transition state stabilization or indirectly influences activity, for example by affecting a rate-limiting conformational change.
Accepting that A38 protonates the leaving group in the cleavage reaction, equivalent pH dependence of cleavage and ligation reactions is only consistent either with A38 acting together with a second ionizable group, or with the reaction not being limited by chemistry. Yet in the latter scenario, the state of protonation of A38 should not affect the pH dependence of the reaction, whereas there is good evidence that titration of A38 does determine the pH dependence of the reaction, as discussed above. We conclude that A38 acting in concert with a second ionizable group is the only mechanism that is consistent with the data.
The identity of the functional group activating the nucleophile in the cleavage reaction has been the subject of considerable investigation. A shuttle mechanism in which A38 removes a proton from the nucleophile and donates it to the leaving group has been proposed 39, but this mechanism is necessarily step-wise and would be expected to yield inverted pH profiles for cleavage and ligation. Both the pro-R and pro-S non-bridging oxygens have been considered 30, 39, 40. But the non-bridging oxygens are fully deprotonated over the observable pH range. Thus the dependence of cleavage activity on pH would be expected to parallel the fractional protonation of the acid (fA in Figure 5A), which is opposite to what is observed. However removal of a proton from the nucleophile by a non-bridging oxygen could in principle explain the data for the cleavage of the 5′-PS substrate by the natural ribozyme, but not those for the A38P ribozyme. It may also explain the deviation from linearity below pH 6 observed in the cleavage of the 5′-PS substrate by the A38P ribozyme.
All the available data support a high pKa for the second ionisable group. Of the nucleobases with a high pKa only G8 lies close to the O2′ in the crystal structure. The hairpin ribozyme functions efficiently in the absence of divalent cations so a magnesium ion-bound hydroxide ion is not necessary for activity 3, 41–43. Although divalent cations may contribute to catalysis under some conditions they cannot account for the low pKa observed with 2,6-diaminpurine at position 8. Bevilacqua 26 has argued that water lacks the necessary catalytic power but it remains a possibility that for the natural ribozyme, where the pKa of the general base is outside of the observable range, a water molecule activated by a specific hydrogen bonding network removes a proton from the nucleophile instead of G8. This role for water is plausible so long as the upper pKa is not observable, but is difficult to reconcile with the lowered pKa for the general base observed for ribozymes with substitutions at position 8 (such as G8DAP) that exhibit a bell-shaped pH profile. It seems probable that these ribozymes use a species other than water to activate the nucleophile, and in each case the observed pKa for the general base is consistent with the nucleobase substituted for G8 taking this role. In the absence of evidence to the contrary we strongly favour the simplest and most consistent hypothesis that the pH dependence of hairpin ribozyme activity arises from general acid-base catalysis by A38 and G8.
Supplementary Material
Acknowledgments
We thank Scott McPhee for oligoribonucleotide synthesis, the Deutsche Forschungsgemeinschaft (SKS research fellowship), and Cancer Research UK (DMJL lab) and NIH (Grant: R01AI081987 to JAP) for financial support.
Footnotes
Supporting Information Available
The chemical synthesis of A[2′-O-(o-nitrobenzyl)]-psG is described in the Supporting Information and Scheme S1. The ligation of the substrate and construction of the hairpin ribozyme are fully described in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lilley DMJ, Eckstein F, editors. Ribozymes and RNA Catalysis. Royal Soc. Chemistry; Cambridge: 2008. pp. 1–318. [Google Scholar]
- 2.Lilley DMJ, Sutherland J, editors. The chemical origins of life and its early evolution. Vol. 366. Royal Society Publishing; London: 2011. pp. 2851–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. Chem & Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 4.Roth A, Nahvi A, Lee M, Jona I, Breaker RR. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buzayan JM, Gerlach WL, Bruening G. Nature. 1986;323:349–353. [Google Scholar]
- 6.Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 8.Murchie AIH, Thomson JB, Walter F, Lilley DMJ. Molec Cell. 1998;1:873–881. doi: 10.1016/s1097-2765(00)80086-6. [DOI] [PubMed] [Google Scholar]
- 9.Walter F, Murchie AIH, Lilley DMJ. Biochemistry. 1998;37:17629–17636. doi: 10.1021/bi9821115. [DOI] [PubMed] [Google Scholar]
- 10.Walter NG, Burke JM, Millar DP. Nature Struct Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- 11.Pinard R, Hampel KJ, Heckman JE, Lambert D, Chan PA, Major F, Burke JM. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson TJ, Zhao ZY, Maxwell K, Kontogiannis L, Lilley DMJ. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmin YI, Da Costa CP, Fedor MJ. J Molec Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 14.Kuzmin YI, Da Costa CP, Cottrell JW, Fedor MJ. J Molec Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Wilson TJ, McLeod AC, Lilley DMJ. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson TJ, Li NS, Lu J, Frederiksen JK, Piccirilli JA, Lilley DMJ. Proc Natl Acad Sci USA. 2010;107:11751–11756. doi: 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rupert PB, Ferré-D’Amaré AR. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 18.Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacElrevey C, Salter JD, Krucinska J, Wedekind JE. RNA. 2008;14:1600–1616. doi: 10.1261/rna.1055308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupert PB, Massey AP, Sigurdsson ST, Ferré-D’Amaré AR. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 21.Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DMJ, Ha T. Nature Struct Molec Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 22.Lebruska LL, Kuzmine I, Fedor MJ. Chem Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- 23.Wilson TJ, Ouellet J, Zhao ZY, Harusawa S, Araki L, Kurihara T, Lilley DMJ. RNA. 2006;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Cottrell JW, Scott LG, Fedor MJ. Nature Chem Biol. 2009;5:351–357. doi: 10.1038/nchembio.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das SR, Piccirilli JA. Nature Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 26.Bevilacqua PC. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 27.Li NS, Frederiksen JK, Koo SC, Lu J, Wilson TJ, Lilley DMJ, Piccirilli JA. Nucleic Acids Res. 2011;39:e31. doi: 10.1093/nar/gkq1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fedor MJ. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 29.Wilson TJ, Lilley DMJ. RNA. 2011;17:213–221. doi: 10.1261/rna.2473711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mlynsky V, Banas P, Walter NG, Sponer J, Otyepka M. J Phys Chem B. 2011;115:13911–13924. doi: 10.1021/jp206963g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suydam IT, Levandoski SD, Strobel SA. Biochemistry. 2010;49:3723–3732. doi: 10.1021/bi100234v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spitale RC, Volpini R, Heller MG, Krucinska J, Cristalli G, Wedekind JE. J Amer Chem Soc. 2009;131:6093–6095. doi: 10.1021/ja900450h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cottrell JW, Scott LG, Fedor MJ. J Biol Chem. 2011;286:17658–17664. doi: 10.1074/jbc.M111.234906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE. J Amer Chem Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DMJ, Ha T. Proc Natl Acad Sci USA. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai ZP, Tinoco I. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 37.Butcher SE, Allain FH, Feigon J. Nature Struct Biol. 1999;6:212–216. doi: 10.1038/6651. [DOI] [PubMed] [Google Scholar]
- 38.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Proc Natl Acad Sci U S A. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ditzler MA, Sponer J, Walter NG. RNA. 2009;15:560–575. doi: 10.1261/rna.1416709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nam K, Gao J, York DM. J Amer Chem Soc. 2008;130:4680–4691. doi: 10.1021/ja0759141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hampel A, Cowan JA. Chem & Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- 42.Nesbitt S, Hegg LA, Fedor MJ. Chem & Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 43.Young KJ, Gill F, Grasby JA. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.