Abstract
Amyotrophic lateral sclerosis (ALS) research is undergoing an era of unprecedented discoveries with the identification of new genes as major genetic causes of this disease. These discoveries reinforce the genetic, clinical and pathological overlap between ALS and frontotemporal lobar degeneration (FTLD). Common causes of these diseases include mutations in the RNA/DNA-binding proteins, TDP-43 and FUS/TLS and most recently, hexanucleotide expansions in the C9orf72 gene, discoveries that highlight the overlapping pathogenic mechanisms that trigger ALS and FTLD. TDP-43 and FUS/TLS, both of which participate in several steps of RNA processing, are abnormally aggregated and mislocalized in ALS and FTLD, while the expansion in the C9orf72 pre-mRNA strongly suggests sequestration of one or more RNA binding proteins in pathologic RNA foci. Hence, ALS and FTLD converge in pathogenic pathways disrupting the regulation of RNA processing. This article is part of a Special Issue entitled RNA-Binding Proteins.
Keywords: Amyotrophic lateral sclerosis, Frontotemporal dementia, TDP-43, FUS/TLS, C9orf72, RNA processing
1. Introduction: ALS and FTLD are linked genetically, clinically and pathologically
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that primarily targets motor neurons resulting in progressive paralysis and death within a few years from onset. Just like Alzheimer's, Parkinson's and other neurodegenerative diseases, a proportion (∼10%) of ALS is dominantly inherited, while the remaining 90% (referred to as sporadic) do not have familial history. With the discovery of mutations in the gene encoding superoxide dismutase 1 (SOD1) as causative for 20% of inherited ALS (Rosen et al., 1993), and the generation of transgenic mice expressing ALS-causing SOD1 mutants (Bruijn et al., 1998; Gurney et al., 1994; Wong et al., 1995), the molecular era of deciphering disease mechanism was initiated. Indeed, most efforts to understand ALS pathogenesis over the last ∼19 years have focused on disease caused by SOD1 mutations. Collectively, the work of multiple groups worldwide has established that mutant SOD1 synthesized within motor neurons and their glial neighbors provokes non-cell autonomous disease from one or more toxic properties of mutant SOD1 (reviewed in Boillee et al., 2006; Ilieva et al., 2009; Pasinelli and Brown, 2006).
In 2006, a DNA/RNA-binding protein, called TAR DNA-binding protein (TDP-43) was found to be the major component of the cytoplasmic and ubiquitinated inclusions present in affected neurons of all sporadic ALS patients (Arai et al., 2006; Neumann et al., 2006) and in patients with frontotemporal lobar degeneration (FTLD), a neurodegenerative disorder characterized by behavioral and language disorders (Neary et al., 1998). This seminal discovery not only revolutionized the ALS field, but also significantly changed our perspective of neurodegeneration in general. Motor neuron disease and cognitive deficits of variable severity can be concomitant in patients or within families (Caselli et al., 1993; Lillo et al., 2012; Lomen-Hoerth et al., 2002; Neary et al., 2000; Ringholz et al., 2005) and the identification of a common pathological hallmark defined by TDP-43 and ubiquitin-positive, tau- and alpha-synuclein-negative cytoplasmic inclusions suggested that ALS and FTLD were part of a broad spectrum of neurodegeneration. In healthy neurons TDP-43 is localized mainly in the nucleus. In affected cells, however, ubiquitinated TDP-43 is mislocalized in the cytoplasm and is excluded from the nucleus, raising the possibility that loss of normal function of TDP-43 (Giordana et al., 2009; Neumann et al., 2006) and/or gain of toxic properties may contribute to ALS and FTLD pathogenesis. Supporting this view, TDP-43 pathology was recently shown to correlate with executive dysfunction in ALS patients (Brettschneider et al., 2012). Intriguingly, TDP-43 cytoplasmic accumulations and loss of nuclear localization in affected neurons are now reported in many neurodegenerative conditions besides ALS and FTLD, including a fraction of Alzheimer's, Parkinson's and Huntington's disease patients (reviewed in Lagier-Tourenne et al.). These observations implicate the functional disruption of TDP-43 in the pathogenesis and/or progression of many, if not all, of these conditions.
Evidence supporting a primary causative role for TDP-43 in ALS and FTLD was reported in 2008 with the discovery of mutations within this gene in ALS patients (Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008) and in rare FTLD patients (Benajiba et al., 2009; Borroni et al., 2009; Gitcho et al., 2009; Kovacs et al., 2009). Affected neurons of patients with TDP-43 mutations also present with cytoplasmic inclusions and nuclear loss (Van Deerlin et al., 2008), indicating that abnormal localization of TDP-43 represents the first mechanistic link between sporadic ALS and an inherited form caused by a known mutation.
The identification of TDP-43 as a major player in ALS and FTLD pathogenesis led to the discovery of ALS-causing mutations in a second gene, which encoded another DNA/RNA-binding protein called FUS/TLS for fused in sarcoma/translocated in liposarcoma (Kwiatkowski et al., 2009; Vance et al., 2009) in approximately 4% of familial ALS and in rare sporadic patients with no apparent familial history. Like TDP-43, FUS/TLS is mainly localized in the nuclei of unaffected neurons, but is partially cleared from those nuclei in neurons that contain cytoplasmic aggregations (Neumann et al., 2009; Tateishi et al., 2009; Vance et al., 2009). While most patients with FUS/TLS mutations develop a classical ALS phenotype without cognitive defect, occasionally, mutant FUS/TLS carriers develop either FTLD concurrently with motor neuron disease (Ticozzi et al., 2009) or FTLD in the absence of motor neuron deficits (Blair et al., 2009; Van Langenhove et al., 2010), providing further evidence that ALS and FTLD have clinical, pathological and genetic commonalities. Recent reports implicate TAF15 mutations in patients with ALS (Couthouis et al., 2011; Ticozzi et al., 2011). The fact that TAF-15 belongs to the same protein family as FUS/TLS (Bertolotti et al., 1999; Morohoshi et al., 1998), coupled with the observation that a fraction of RNA-binding proteins have exceptionally high aggregation propensities (Cushman et al., 2010), raise the likelihood that the list of misregulated RNA-binding proteins involved in ALS pathogenesis is still incomplete (Couthouis et al., 2011). TAF15 cytoplasmic accumulations were found in sporadic ALS patients (Couthouis et al., 2011), as well as patients with FTLD (Neumann et al., 2011), further supporting that the pathogenetic mechanisms of the two diseases are intertwined.
The strongest genetic link between ALS and FTLD was reported in late 2011 with the identification of hexanucleotide repeat expansions in the first intron of the C9orf72 gene as causative for ALS, FTLD or concomitant ALS-FTLD disease (DeJesus-Hernandez et al., 2011; Gijselinck et al., 2011; Renton et al., 2011). Today, expansions in the C9orf72 represent the most frequent genetic cause of ALS and patients carrying the mutation harbor characteristic pathologic hallmarks. In addition to TDP-43 pathology, patients with C9orf72 expansion display ubiquitin and p62 positive, TDP-43 negative cytoplasmic and intranuclear inclusions in various CNS regions including the cerebellum and hippocampus (Al-Sarraj et al., 2011; DeJesus-Hernandez et al., 2011; Murray et al., 2011; Troakes et al., 2011).
Not all ALS-causing genes, however, overlap with FTLD phenotypes. The recently discovered mutations in an X-linked gene, UBQLN2, encoding ubiquilin 2 is responsible for ∼1% of familial ALS (Daoud and Rouleau, 2011; Deng et al., 2011). And while some patients carrying UBQLN2 mutations present with concomitant dementia, no reported UBQLN2-positive cases showed a pure FTLD phenotype in the absence of motor system involvement (Deng et al., 2011; Millecamps et al., 2011b). Another recently identified ALS-causing gene is the OPTN gene encoding optineurin or optic neuropathy inducing protein, whose mutations were recently identified in Japanese familial ALS (Iida et al., 2011; Maruyama et al., 2010). OPTN mutations in Caucasian populations were reported to be either extremely rare (Del Bo et al., 2011; Sugihara et al., 2011; Tumer et al., 2012; van Blitterswijk et al., 2011) or completely absent (Millecamps et al., 2011a) and to date no association between OPTN mutations and FTLD phenotype has been found (Rollinson et al., 2012). The latter seems also to be true for SOD1, whose mutations seem to be strictly associated with ALS disease phenotype, in contrast to TDP-43, FUS/TLS and C9orf72.
2. Misregulated RNA processing is a convergent causative basis for ALS and FTLD
The mechanisms by which TDP-43 and FUS/TLS trigger neurodegeneration are at the earliest stages of investigation and it is at present unresolved as to whether neurodegeneration is due to a loss of function, a gain of toxic property, or a combination of the two arising from their sequestration into nuclear or cytoplasmic aggregates. If we consider the loss of function hypothesis, then the next question is: what are the physiological roles of TDP-43 and FUS/TLS whose interruption occurs in affected cells? While these are not fully elucidated and there is no evidence that TDP-43 and FUS/TLS act together, they are both structurally related to the family of heterogeneous ribonucleoproteins (hnRNP) and have been involved in multiple levels of RNA processing including transcription, splicing, transport and translation. Such multifunctional proteins could have roles in coupling transcription with splicing and other RNA processes (Andersson et al., 2008; Bertolotti et al., 1999; Buratti and Baralle, 2008; Hirose and Manley, 2000; Kameoka et al., 2004; Law et al., 2006; Tan and Manley, 2009). It is therefore easy to imagine the devastating consequences of their functional disruption for cellular homeostasis.
The alternative hypothesis proposes that a gain of toxic property of TDP-43 and FUS/TLS cytoplasmic inclusions drives disease. Several of the neurotoxic mechanisms that have been proposed to result from the aggregates of different misfolded protein involved in ALS and other neurodegenerative disorders (Ilieva et al., 2009), may be triggered by TDP-43 or FUS/TLS inclusions. The observation that ALS-linked mutations increase the stability of TDP-43 protein (Guo et al., 2011; Johnson et al., 2009; Ling et al., 2010) supports the view that protein aggregation may initiate the pathogenic cascade in ALS. More-over, TDP-43 and FUS/TLS are involved in the formation of stress granules, cytoplasmic foci containing RNA in complex with RNA binding proteins that appear transiently under cellular stress (Andersson et al., 2008; Bosco et al., 2011; Colombrita et al., 2009; Dewey et al., 2011; Gal et al., 2011; Ito et al., 2011; Liu-Yesucevitz et al., 2010; McDonald et al., 2011; Meyerowitz et al., 2011; Moisse et al., 2009; Wang et al., 2008a). Hence it is plausible that the physiologic TDP-43- or FUS/TLS-containing stress granules may transform into pathogenic inclusions during neurodegeneration. The observation that stress granule proteins partition in the TDP-43 and FUS/TLS pathologic inclusions of ALS patients supports this view (Dormann et al., 2011; Fujita et al., 2008; Liu-Yesucevitz et al., 2010). Therefore, sequestration of specific cellular RNAs with-in cytoplasmic TDP-43 and FUS/TLS inclusions may deplete the cell of essential RNA components, contributing to pathogenesis. The latter, if true, could explain the observation that TDP-43 and FUS/TLS binding to RNA is linked to their cytotoxicity independently of their propensity to aggregate (Elden et al., 2010; Sun et al., 2011; Voigt et al., 2010).
Just like TDP-43 and FUS/TLS, the pathogenic mechanism of the recently discovered hexanucleotide expansions in the C9orf72 gene is not understood, but there are two possible pathways emerging from early observations in these patients. The first one is a mechanism linked to haploinsufficiency of C9orf72 supported by the 50% reduction of C9orf72 transcript levels observed in patients with expansions (DeJesus-Hernandez et al., 2011; Gijselinck et al., 2011). As there is currently no functional or structural information on the C9orf72 protein, there is no evidence on the consequences of a C9orf72 haploinsufficiency. The second possibility is that the expanded RNA forms pathogenic foci (DeJesus-Hernandez et al., 2011) that trap one or more RNA binding protein(s). This mechanism of RNA toxicity resulting in the depletion and loss of function of specific RNA binding protein(s) with affinity for the expanded RNA has been established in other neurological diseases, especially myotonic dystrophy types 1 and 2 (Fugier et al., 2011; Kanadia et al., 2003; Mankodi et al., 2000, 2003; Margolis et al., 2006; Miller et al., 2000; Ranum and Cooper, 2006), or Fragile X-associated tremor ataxia syndrome (FXTAS) (Hagerman and Hagerman, 2004; Iwahashi et al., 2006; Jin et al., 2007; Sellier et al., 2011; Tassone et al., 2004; Willemsen et al., 2003). If a similar mechanism is demonstrated in ALS, it will further emphasize the crucial role of RNA misregulation in ALS pathogenesis. In addition, the role of RNA metabolism in ALS is further underscored by diseasecausing mutations in angiogenin (Greenway et al., 2006; Li and Hu, 2011; Wu et al., 2007) and senataxin (Chen et al., 2004), two proteins involved in RNA processing, as well as by the recognition of intermediate length polyglutamine expansions in ataxin-2, another RNA binding protein, as a risk factor for ALS (Elden et al., 2010).
3. RNA processing alterations in ALS patients
Early studies have analyzed RNA expression profiles in affected postmortem tissues of sporadic ALS patients. While there is a large variation in the results reported, probably reflecting the heterogeneity in genetic background, disease stage and tissue preservation, neuroinflammatory pathways were uniformly seen activated in patients' spinal cords (Malaspina et al., 2001) and motor cortices (Wang et al., 2006) and celldeath-associated genes are significantly upregulated in isolated motor neurons (Jiang et al., 2005). Importantly, splicing alterations were also reported in ALS patients (Lin et al., 1998; Rabin et al., 2010; Xiao et al., 2011), some of which may be directly related to TDP-43 misregulation (Xiao et al., 2011). Additionally, errors in adenosine (A) to inosine (I) RNA editing have been described in ALS patients (Kawahara et al., 2004), potentially resulting from loss of the RNA editing enzyme ADAR2 (adenosine deaminase acting on RNA2) in neurons with TDP-43 cytoplasmic inclusions (Aizawa et al., 2010).
Altogether, the above observations reinforce the notion that abnormal RNA-processing contributes to ALS pathogenesis, albeit the role of TDP-43 or FUS/TLS in these modifications has not been accurately examined. It is now crucial to determine the normal functions of TDP-43 and FUS/TLS and to identify the set of alterations in RNA processing that define a TDP-43- and FUS/TLS-dependent disease signature.
4. Identification of TDP-43 RNA targets highlights its multifunctional role in RNA processing
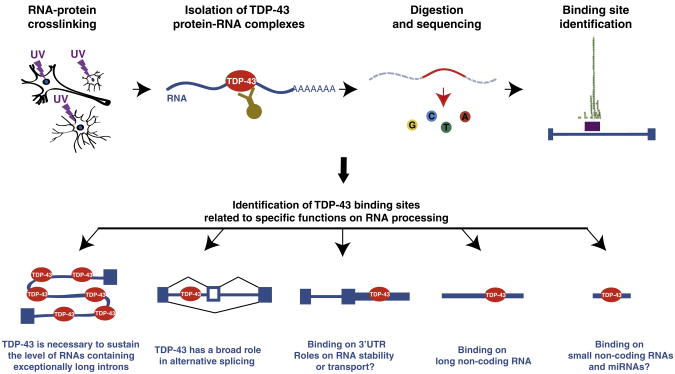
Recent studies using revolutionary DNA sequencing technologies have provided initial insights on the normal functions of TDP-43 within the central nervous system. Until recently, only candidate approaches could be used to identify the RNA targets for specific RNA-binding proteins or aberrant RNA splice isoforms related to diseases. Advances in DNA sequencing technology have provided powerful tools for exploring gene regulation in remarkable detail. Indeed, using cross-linking, immunoprecipitation and high-throughput sequencing or CLIP-seq (Ule et al., 2003; Licatalosi et al., 2008; Yeo et al., 2009) or an alternative technique that omits the crosslinking step (RNA-immunoprecipitation or RIP-seq), TDP-43 RNA targets within the normal central nervous system were comprehensively defined (Polymenidou et al., 2011; Sephton et al., 2011; Tollervey et al., 2011). More than 6000 TDP-43 RNA targets have defined an RNA–protein interaction map, demonstrating a broad role of TDP-43 in RNA processing. While binding of TDP-43 on transcripts corresponding to approximately 30% of the mouse genome may seem like an overwhelming number, most RNA binding proteins, whose RNA targets have been previously comprehensively identified, show similar prolific binding patterns. For example, the RNA binding protein Nova was shown to bind 3754 genes in the mouse brain (Licatalosi et al., 2008), Fox2 binds 1876 genes in human embryonic stem cells (Yeo et al., 2009), the miRNA-binding protein Argonaute associates with 3093 or 829 genes in Caenorhabditis elegans (Zisoulis et al., 2010) or human cells (Chi et al., 2009), respectively, and the human RNA-binding proteins Pumilio 2 (PUM2) and Quaking (QKI) have 3000 and 2500 RNA targets, respectively (Hafner et al., 2010). In all these cases, including TDP-43 and FUS/TLS (Hoell et al., 2011), the multiple binding partners of these proteins reflect the multifunctional nature of their physiological roles in regulating RNA metabolism.
Consistent with in vitro studies (Buratti et al., 2001), in vivo TDP-43 binding sites are enriched in sequences containing stretches of GU repeats (Polymenidou et al., 2011; Sephton et al., 2011; Tollervey et al., 2011). The majority of the TDP-43 binding sites lie within intronic regions more than 2 kb away from the nearest exon–intron boundary. This finding is surprising for a protein that previously was best characterized for its role on splicing regulation through binding just 10 bases upstream of an alternatively spliced exon on the pre-mRNA of CFTR, the gene mutated in cystic fibrosis (Buratti et al., 2001). Although most TDP-43 binding sites are found within introns, it is noteworthy that more than a thousand RNAs are bound in their 3′ untranslated regions (3′UTR), consistent with multiple roles of TDP-43 on nuclear RNA processing as well as within the cytoplasm. Indeed, binding onto 3′UTRs has been linked with a role in RNA stability or transport for local translation, processes that are crucial for neuronal integrity (Dahm et al., 2007; Kindler et al., 2005). Importantly, TDP-43 — as well as FUS/TLS — is found in RNA granules translocating to dendritic spines upon different neuronal stimuli (Belly et al., 2005; Elvira et al., 2006; Fujii et al., 2005; Kanai et al., 2004; Wang et al., 2008b). In addition, loss of TDP-43 reduces dendritic branching as well as synaptic formation in Drosophila neurons (Feiguin et al., 2009; Lu et al., 2009). Collectively, these results suggest that TDP-43 could play a role in the modulation of neuronal plasticity by altering mRNA transport and local translation through binding onto the 3′UTR of specific mRNAs in neurons. Interestingly, TDP-43 binding sites were identified on the 3′UTR of several genes involved in ALS pathogenesis including FUS/TLS, the light chain of neurofilament (NFL) and the glutamate transporter EAAT2 (Polymenidou et al., 2011).
5. TDP-43 regulation of its RNA targets converge in long synaptic transcripts and non-coding RNAs
Amidst the plethora of TDP-43 binding sites, it is crucial to determine the functional events that matter the most both physiologically and pathologically. To identify the contribution of TDP-43 in maintaining the levels and splicing patterns of RNAs, antisense oligonucleotide (ASO) silencing was used to deplete TDP-43 within the normal central nervous system of adult mice (Polymenidou et al., 2011). TDP-43 mRNA was degraded via endogenous RNase H digestion, which specifically recognizes ASO-pre-mRNA (DNA/RNA) hybrids. These tools enabled definition of RNA-processing alterations resulting from loss of function of TDP-43, using a genome-wide approach with high-throughput sequencing (known as RNAseq) (Parkhomchuk et al., 2009). Following ASO-mediated depletion of TDP-43, levels of 601 mRNAs were changed and 965 splicing events were altered (Polymenidou et al., 2011).
In order to pinpoint changes in expression that resulted from direct interactions of TDP-43 protein with its target mRNAs, the RNA-binding sites for TDP-43 (obtained by CLIP-seq) were compared with the degree of expression change upon TDP-43 knockdown (RNA-seq). While very few TDP-43 binding sites were identified among genes whose mRNAs were upregulated after suppression of TDP-43, a striking enrichment of multiple binding sites was observed within the genes that were downregulated upon depletion of TDP-43 (Polymenidou et al., 2011). In fact, the 100 most downregulated genes upon TDP-43 reduction contained an average of ∼37 TDP-43 binding sites per pre-mRNA and 12 genes contained more than 100 binding sites.
Another striking property of the most downregulated genes upon TDP-43 reduction was that they contained exceptionally long introns; on average, they were six-fold larger than the introns from unaffected or upregulated genes. Notably, pre-mRNAs with exceptionally long introns are most prominently represented in brain-enriched transcripts (Ameur et al., 2011; Polymenidou et al., 2011). Strikingly, several of these RNAs encode proteins with crucial roles in synaptic activity/function and have previously been implicated in neurological diseases including Parkin (Park2), Neurexin 1 and 3 (Nrxn1, Nrxn3) and Neuroligin 1 (Nlgn1). Among this TDP-43 target group, Neurexin 3 pre-mRNA was independently found downregulated in autopsy tissues from patients with TDP-43 pathology (Tollervey et al., 2011). It is important to emphasize that not all neuronal genes are affected by TDP-43 loss. For example, while Neuroligin 1, a long-intron-containing gene is strongly affected, other members of the Neuroligin family that contain substantially smaller introns (Neuroligin 2 and 3) are neither significantly bound by TDP-43, nor altered in levels after TDP-43 depletion.
These results thus identify a novel conserved role for TDP-43 in regulating a subset of very long intron-containing brainenriched genes and suggest for the first time a mechanism underlying the selective neuronal vulnerability from the interruption of TDP-43 function (Polymenidou et al., 2011). Nonetheless, the molecular events underlying this regulation have not been established. One possibility is that TDP-43 binding within long introns protects transcripts from undergoing nonsense-mediated mRNA decay by facilitating splicing events and preventing the introduction of premature termination codons. Indeed, TDP-43 may be involved in compacting the structure of RNA through multiple binding sites within long introns to facilitate their splicing. Another possibility is that TDP-43 affects RNA polymerase elongation, similar to what has been shown for another splicing regulator, SC35 (Lin et al., 2008). Future studies are needed to elucidate these mechanisms.
TDP-43 also interacts with and regulates the levels of long non-coding RNAs such as MALAT1 (Polymenidou et al., 2011; Tollervey et al., 2011) and NEAT1, which was markedly increased in expression in FTLD brains with TDP-43 pathology (Tollervey et al., 2011). The binding behavior of TDP- 43 on small non-coding RNAs (sncRNAs) and mature micro-RNAs (miRNAs), however, remains unexplored. Nevertheless, association of TDP-43 with proteins involved in the miRNA biogenesis and maturation such as Drosha (Gregory et al., 2004), argonaute 2 and DDX17 (Freibaum et al., 2010; Ling et al., 2010) suggest that TDP-43 may participate in these pathways. Importantly, alterations in a set of miRNAs were reported in cultured cells following TDP-43 knockdown (Buratti et al., 2010). These include the miRNAs, let-7b and miR-663 expression, whose levels are down- and upregulated, respectively upon TDP-43 reduction and which were shown to bind TDP-43. Fig. 1 summarizes the main conclusions of the unbiased screens for TDP-43 binding sites in the nervous system.
Fig. 1.
Roles of TDP-43 on RNA processing via binding on different RNA species, as identified by unbiased screens utilizing high-throughput technologies (Polymenidou et al., 2011; Sephton et al., 2011; Tollervey et al., 2011). While TDP-43 was shown to bind to the miRNAs let-7b and miR-663, thereby regulating their expression (Buratti et al., 2010), the complete list of mature miRNA and small non-coding RNA that TDP-43 associates with remains to be established.
It is noteworthy that while TDP-43 binds more that 6000 RNAs, only a small fraction of these change in splicing or expression when TDP-43 is depleted. So, what may be the purpose of TDP-43 binding on all these RNAs that seem to remain unaffected upon its loss? RNA binding may alter translation of some targets, as suggested for TDP-43 by a recent study (Fiesel et al., 2011), or facilitate the transport of others, as described above. Moreover, RNA-binding proteins often function in complexes and some redundancy may occur to ensure the most vital RNA-processing events. In agreement with this view, TDP-43 binding on UG elements in the vicinity of splice junctions was independently found to lack regulatory power in many instances (Passoni et al., 2011). Finally, given the reported role of TDP-43 in stress responses, one could speculate that in some instances, binding of TDP-43 to RNA is “inert” under physiological conditions and only becomes functional under cellular stress.
A largely unexplored aspect of TDP-43 (and FUS/TLS) biology is their role in transcription regulation through their DNA-binding properties. Indeed, TDP-43 was shown to bind TG-repeat-rich DNA, both single stranded (Acharya et al., 2006; Buratti and Baralle, 2001) and double-stranded (Kuo et al., 2009). Originally identified as a transcriptional repressor that binds to transactive response (TAR) DNA of the human immunodeficiency virus type 1 (HIV-1) (Ou et al., 1995), TDP-43 was also reported to bind the promoter of the mouse SP-10 gene that is required for spermatogenesis (Abhyankar et al., 2007; Acharya et al., 2006; Lalmansingh et al., 2011). In both instances, TDP-43 represses transcription by binding these DNA regulatory elements, but little is known about the mechanisms of this transcriptional repression (reviewed in Buratti and Baralle, 2008). While the above observations strongly suggest critical roles of TDP-43 in transcription regulation of a few gene targets, a comprehensive protein/DNA interaction map still remains to be defined.
Furthermore, it will be crucial to identify which of the TDP-43 regulated events are altered in ALS patients. Whole tissue transcriptome and/or splicing profiling may not accurately reveal the most relevant alterations but rather reflect modifications of the cellular composition in tissues with neuronal death and glial activation. While laser capture microdissection of motor neurons (Rabin et al., 2010) is a substantial technical improvement towards this goal, the reported heterogeneity of affected cells in the spinal cords of ALS patients (Bodansky et al., 2010; Giordana et al., 2009) may mask some of the changes occurring in the most severely affected neurons. Single cell transcriptome analysis (Tang et al., 2011) or pre-selection of cells that are at the same disease stage might shed light to this outstanding question. Another promising approach is the use of relatively homogeneous populations of neurons differentiated from induced pluripotent stem (iPS) cells generated from somatic cells isolated from ALS patients (Dimos et al., 2008; Han et al., 2011).
Similar high-throughput sequencing approaches to those followed for TDP-43 are underway to identify FUS/TLS RNA targets and RNA processing changes upon FUS/TLS depletion. Since the mutation or cytoplasmic aggregation of either FUS/TLS or TDP-43 cause similar disease phenotypes in humans, and both proteins are involved in the same RNA-processing steps (Lagier-Tourenne et al., 2010), a plausible hypothesis is that the RNA-targets affected by both proteins may be the most relevant for disease. Deciphering the function of both TDP-43 and FUS/TLS will determine if these two RNA-binding proteins act through common pathways to trigger and/or exacerbate the progression of neuronal death.
6. Autoregulatory mechanisms sustain the levels of TDP-43 protein
The multifunctional role and physiological importance of TDP-43 is evidenced by early embryonic lethality in mice with homozygous disruption in the Tardbp gene (Kraemer et al., 2010; Sephton et al., 2010; Wu et al., 2010) and by autoregulatory mechanisms that have evolved to maintain its levels (Ayala et al., 2011; Igaz et al., 2011; Polymenidou et al., 2011; Xu et al., 2010) to avoid toxicity caused by elevated TDP-43 expression (Shan et al., 2010; Tsai et al., 2010; Wils et al., 2010; Xu et al., 2010). In fact, TDP-43 was found highly expressed in the developing nervous system, with lower levels in adult rodents that are sustained with age (Huang et al., 2010), confirming the vital role of this protein for the homeostasis of the adult nervous system.
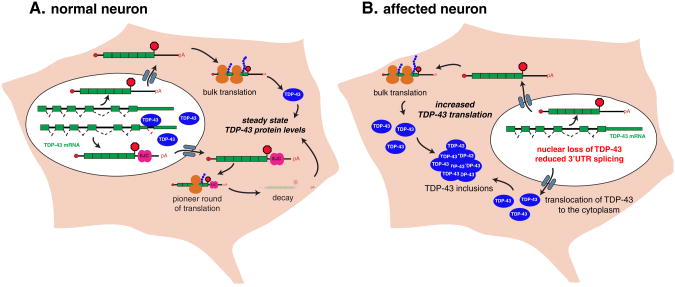
Evidence for tight regulationofTDP-43 levels came from cellular and animal models where over-expression of exogenous TDP-43 induced striking reduction of endogenous TDP-43 RNA and protein levels (Ayala et al., 2011; Igaz et al., 2011; Polymenidou et al., 2011; Xu et al., 2010). The genome-wide identification ofTDP-43 binding sites revealed that TDP-43 protein binds withinan intron residing in the 3′UTR ofits own premRNA (Ayala et al., 2011; Polymenidou et al., 2011; Sephton et al., 2011). Binding of TDP-43 causes the removal of this intron, evidenced by the dramatic increase of this 3′UTR-spliced TDP-43 isoform, upon overexpression of TDP-43 in cultured cells (Polymenidou et al., 2011). Locations of introns are demarcated by the presence of exon-junction complexes (EJCs) deposited on the mature mRNA proximal to the exon–exon junction. The pioneer round of translation normally ejects EJCs upstream of the stop codon, the presence of an unperturbed EJCelicits the nonsense mediated decay (NMD) pathway which causes degradation of the TDP-43 message (Ayala et al., 2011; Polymenidou et al., 2011). Interestingly, many other RNA binding proteins are auto-regulated via the NMD pathway (Dredge et al., 2005; Lareau et al., 2007; Ni et al., 2007; Saltzman et al., 2008; Sureau et al., 2001; Wollerton et al., 2004). The NMD pathway, however, is not the only possible autoregulatory mechanism shown for TDP-43 (Lee et al., 2012). Other mechanistic contributors to TDP-43 autoregulation include the promotion by TDP-43 overexpression in cultured cells of RNA-instability in an exosome-dependent fashion, suggesting a complementary pathway for sustaining its levels (Ayala et al., 2011). Additional mechanisms, such as miRNA-mediated regulation may also contribute to the maintenance of TDP-43 levels. Indeed, conserved binding sites for the miRNA miR-143 were identified in TDP-43 molecules of multiple vertebrates (Trakooljul et al., 2009). In the future, it will be important to clarify the level of contribution of each pathway to the autoregulation of TDP-43 in the mouse and human nervous system.
Whatever the underlying molecular mechanism(s), the tight autoregulation of TDP-43 provides an attractive model for a feed-forward mechanism driving disease progression in ALS patients. In cells with cytoplasmic accumulations of TDP-43 and consequent nuclear clearance of the protein, the production of stable TDP-43 mRNA will be expected to increase, which may further promote the growth of the cytoplasmic aggregates, as more and more TDP-43 protein gets incorporated into the growing aggregates (Fig. 2). The observed increased levels of TDP-43 mRNA in motor neurons from ALS patients (Rabin et al., 2010) supports this view.
Fig. 2.
Model for a feed forward autoregulatory mechanism driving disease progression in TDP-43 proteinopathies. A. In normal neurons, steady state of TDP-43 protein levels is tightly regulated by binding of TDP-43 onto the 3′UTR of its own mRNA. High levels of nuclear TDP-43 protein increase the splicing of an alternative intron in the 3′UTR with the deposition of an exon-junction complexes (EJC) located 3′ of the stop codon. After the pioneer round of translation, the stop codon is recognized as a premature termination codon and the transcript is targeted for degradation via a nonsense-mediated RNA decay (NMD) mechanism. This figure depicts one of several possible autoregulatory mechanisms for TDP-43 and alternative mechanisms, such as exosome-mediated decay or miRNA-mediated regulation have also been proposed. B. In affected neurons with TDP-43 cytoplasmic inclusions and loss of nuclear TDP-43, the production of stable TDP-43 mRNA is expected to increase the levels of TDP-43 proteins that may be trapped to further promote the growth of cytoplasmic aggregates.
7. Perspectives and open questions
While we are witnessing a time of remarkable progress in our understanding of the complex roles of TDP-43, FUS/TLS and other RNA binding proteins in the homeostasis and degeneration of the nervous system, there is undeniably a long list of key, outstanding questions that must be addressed in the near future. Included here are what are the FUS/TLS-dependent RNA-processing events and what is the overlap between the TDP-43 and FUS/TLS regulated events? Which, if any, are the RNA binding proteins trapped in the RNA-foci of patients with C9orf72 expansions and what are the downstream misregulated RNA-processing events? Pertinently, is there convergence of the three seemingly distinct pathways driving disease, i.e. TDP-43, FUS/TLS and C9orf72 expansions? Are familial ALS cases caused by SOD1, optineurin or ubiquilin 2 mutations triggered by similar mechanisms, or do multiple pathogenic mechanisms lead to the same disease phenotype? Are there more repeat expansions causing ALS and FTLD? Why are TDP-43 and FUS so commonly aggregated in neurodegeneration? Is this due to their normal roles in the cellular stress response pathway? What defines the disease phenotype in ALS and FTLD? Is there truly sporadic disease, and what causes it? Or are there multiple genetic and/or environmental factors triggering sporadic ALS?
As long and challenging as the above list may seem, the next few years of ALS and FTLD research will be even more exciting than the past has been and new discoveries will empower therapeutic interventions for these devastating conditions.
Acknowledgments
M.P. was the recipient of a long-term fellowship from the international Human Frontier Science Program Organization. C.L-T. was the recipient of the Milton-Safenowitz post-doctoral fellowship from the Amyotrophic Lateral Sclerosis Association and is supported by a development grant from the Muscular Dystrophy Association. The authors receive support from the National Institute of Neurological Disorders and Stroke, K99NS075216 to M.P. and R37NS27036 to D.W.C, as well as from the National Institutes of Health (HG004659 and GM084317 to GWY), the California Institute for Regenerative Medicine (RB1-01413 to G.W.Y) and the Stem Cell Program at the University of California, San Diego (G.W.Y). G.W.Y. is an Alfred P. Sloan Research Fellow (2011–2012).
Contributor Information
Don W. Cleveland, Email: dcleveland@ucsd.edu.
Gene W. Yeo, Email: geneyeo@ucsd.edu.
References
- Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J Biol Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- Acharya KK, Govind CK, Shore AN, Stoler MH, Reddi PP. cis-requirement for the maintenance of round spermatid-specific transcription. Dev Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Sawada J, Hideyama T, Yamashita T, Katayama T, Hasebe N, Kimura T, Yahara O, Kwak S. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010;120:75–84. doi: 10.1007/s00401-010-0678-x. [DOI] [PubMed] [Google Scholar]
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive taunegative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendano-Vazquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, Baralle FE. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011 doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, Legallic S, Salachas F, Hannequin D, Decousus M, Lacomblez L, Guedj E, Golfier V, Camu W, Dubois B, Campion D, Meininger V, Brice A. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Bell B, Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, Blumbergs PC, Vucic S, Kiernan MC, Nicholson GA. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry. 2009 doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- Bodansky A, Kim JM, Tempest L, Velagapudi A, Libby R, Ravits J. TDP-43 and ubiquitinated cytoplasmic aggregates in sporadic ALS are low frequency and widely distributed in the lower motor neuron columns independent of disease spread. Amyotroph Lateral Scler. 2010;11:321–327. doi: 10.3109/17482961003602363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, Padovani A. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat. 2009;30:E974–E983. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr, Sapp P, McKenna-Yasek D, Brown RH, Jr, Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2011;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J, Libon DJ, Toledo JB, Xie SX, McCluskey L, Elman L, Geser F, Lee VM, Grossman M, Trojanowski JQ. Microglial activation and TDP-43 pathology correlate with executive dysfunction in amyotrophic lateral sclerosis. Acta Neuropathol. 2012 doi: 10.1007/s00401-011-0932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, De Conti L, Stuani C, Romano M, Baralle M, Baralle F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- Caselli RJ, Windebank AJ, Petersen RC, Komori T, Parisi JE, Okazaki H, Kokmen E, Iverson R, Dinapoli RP, Graff-Radford NR, et al. Rapidly progressive aphasic dementia and motor neuron disease. Ann Neurol. 1993;33:200–207. doi: 10.1002/ana.410330210. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, Dejesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, Epstein J, Chiang A, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Solski JA, Williams KL, Mojsilovic-Petrovic J, Ingre C, Boylan K, Graff-Radford NR, Dickson DW, Clay-Falcone D, Elman L, McCluskey L, Greene R, Kalb RG, Lee VM, Trojanowski JQ, Ludolph A, Robberecht W, Andersen PM, Nicholson GA, Blair IP, King OD, Bonini NM, Van Deerlin V, Rademakers R, Mourelatos Z, Gitler AD. Feature article: from the cover: a yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm R, Kiebler M, Macchi P. RNA localisation in the nervous system. Semin Cell Dev Biol. 2007;18:216–223. doi: 10.1016/j.semcdb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Daoud H, Rouleau GA. A role for ubiquilin 2 mutations in neurodegeneration. Nat Rev Neurol. 2011;7:599–600. doi: 10.1038/nrneurol.2011.163. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bo R, Tiloca C, Pensato V, Corrado L, Ratti A, Ticozzi N, Corti S, Castellotti B, Mazzini L, Soraru G, Cereda C, D'Alfonso S, Gellera C, Comi GP, Silani V. Novel optineurin mutations in patients with familial and sporadic amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82:1239–1243. doi: 10.1136/jnnp.2011.242313. [DOI] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P, III, Good SK, Johnson BA, Herz J, Yu G. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–1108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, Neumann M, Haass C. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2011;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomic. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, Weber SS, Supper J, Zell A, Kahle PJ. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugier C, Klein AF, Hammer C, Vassilopoulos S, Ivarsson Y, Toussaint A, Tosch V, Vignaud A, Ferry A, Messaddeq N, Kokunai Y, Tsuburaya R, de la Grange P, Dembele D, Francois V, Precigout G, Boulade-Ladame C, Hummel MC, de Munain AL, Sergeant N, Laquerriere A, Thibault C, Deryckere F, Auboeuf D, Garcia L, Zimmermann P, Udd B, Schoser B, Takahashi MP, Nishino I, Bassez G, Laporte J, Furling D, Charlet-Berguerand N. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Fujita K, Ito H, Nakano S, Kinoshita Y, Wate R, Kusaka H. Immunohistochemical identification of messenger RNA-related proteins in basophilic inclusions of adult-onset atypical motor neuron disease. Acta Neuropathol. 2008;116:439–445. doi: 10.1007/s00401-008-0415-x. [DOI] [PubMed] [Google Scholar]
- Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32(2323):e27–e40. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Baumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2011;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Giordana MT, Piccinini M, Grifoni S, De Marco G, Vercellino M, Magistrello M, Pellerino A, Buccinna B, Lupino E, Rinaudo MT. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol. 2009 doi: 10.1111/j.1750-3639.2009.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, III, Bigio EH, Caselli R, Baker M, Al-Lozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Bigio EH, Mishra M, Johnson N, Weintraub S, Mesulam M, Rademakers R, Chakraverty S, Cruchaga C, Morris JC, Goate AM, Cairns NJ. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009;118:633–645. doi: 10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr, Hardiman O. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, Liu J, Xu M, Yang Y, Wang C, Zhang D, Bigio EH, Mesulam M, Shen Y, Xu Q, Fushimi K, Wu JY. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SS, Williams LA, Eggan KC. Constructing and deconstructing stem cell models of neurological disease. Neuron. 2011;70:626–644. doi: 10.1016/j.neuron.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, Hafner M, Borkhardt A, Sander C, Tuschl T. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xia PY, Zhou H. Sustained expression of TDP-43 and FUS in motor neurons in rodent's lifetime. Int J Biol Sci. 2010;6:396–406. doi: 10.7150/ijbs.6.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, Lee VM. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2011 doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Hosono N, Sano M, Kamei T, Oshima S, Tokuda T, Kubo M, Nakamura Y, Ikegawa S. Optineurin mutations in Japanese amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011 doi: 10.1136/jnnp.2010.234963. [DOI] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Seki M, Tsunoda Y, Uchiyama H, Suzuki N. Nuclear transport impairment of amyotrophic lateral sclerosislinked mutations in FUS/TLS. Ann Neurol. 2011;69:152–162. doi: 10.1002/ana.22246. [DOI] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, Niwa J, Tanaka F, Doyu M, Yoshida M, Hashizume Y, Sobue G. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kameoka S, Duque P, Konarska MM. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GG, Murrell JR, Horvath S, Haraszti L, Majtenyi K, Molnar MJ, Budka H, Ghetti B, Spina S. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov Disord. 2009;24:1843–1847. doi: 10.1002/mds.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer BC, Schuck T, Wheeler JM, Robinson LC, Trojanowski JQ, Lee VM, Schellenberg GD. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010 doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco JD, LeClerc AD, Tamrazian E, Van den Berg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis PN, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance M, Yan J, Ticozza N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009 doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmansingh AS, Urekar CJ, Reddi PP. TDP-43 is a transcriptional repressor: the testis-specific mouse acrv1 gene is a TDP-43 target in vivo. J Biol Chem. 2011;286:10970–10982. doi: 10.1074/jbc.M110.166587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- Lee EB, Lee VM, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hu GF. Angiogenin-mediated rRNA transcription in cancer and neurodegeneration. Int J Biochem Mol Biol. 2011;1:26–35. [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo P, Savage S, Mioshi E, Kiernan MC, Hodges JR. Amyotrophic lateral sclerosis and frontotemporal dementia: a behavioural and cognitive continuum. Amyotroph Lateral Scler. 2012;13:102–109. doi: 10.3109/17482968.2011.639376. [DOI] [PubMed] [Google Scholar]
- Lin CL, Bristol LA, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein JD. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- Lu Y, Ferris J, Gao FB. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspina A, Kaushik N, de Belleroche J. Differential expression of 14 genes in amyotrophic lateral sclerosis spinal cord detected using gridded cDNA arrays. J Neurochem. 2001;77:132–145. doi: 10.1046/j.1471-4159.2001.t01-1-00231.x. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- Meyerowitz J, Parker SJ, Vella LJ, Ng D, Price KA, Liddell JR, Caragounis A, Li QX, Masters CL, Nonaka T, Hasegawa M, Bogoyevitch MA, Kanninen KM, Crouch PJ, White AR. C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol Neurodegener. 2011;6:57. doi: 10.1186/1750-1326-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Boillee S, Chabrol E, Camu W, Cazeneuve C, Salachas F, Pradat PF, Danel-Brunaud V, Vandenberghe N, Corcia P, Le Forestier N, Lacomblez L, Bruneteau G, Seilhean D, Brice A, Feingold J, Meininger V, LeGuern E. Screening of OPTN in French familial amyotrophic lateral sclerosis. Neurobiol Aging. 2011a;32(557):e11–e13. doi: 10.1016/j.neurobiolaging.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Corcia P, Cazeneuve C, Boillee S, Seilhean D, Danel-Brunaud V, Vandenberghe N, Pradat PF, Le Forestier N, Lacomblez L, Bruneteau G, Camu W, Brice A, Meininger V, Leguern E, Salachas F. Mutations in UBQLN2 are rare in French amyotrophic lateral sclerosis. Neurobiol Aging. 2011b doi: 10.1016/j.neurobiolaging.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisse K, Volkening K, Leystra-Lantz C, Welch I, Hill T, Strong MJ. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1249:202–211. doi: 10.1016/j.brainres.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- Murray ME, Dejesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB, Rademakers R, Dickson DW. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, Boone K, Miller BL, Cummings J, Benson DF. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Mann DM. Cognitive change in motor neurone disease/amyotrophic lateral sclerosis (MND/ALS) J Neurol Sci. 2000;180:15–20. doi: 10.1016/s0022-510x(00)00425-1. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzschmar HA, Munoz DG, Kusaka H, Yokota O, Ang LC, Bilbao J, Rademakers R, Haass C, Mackenzie IR. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Passoni M, De Conti L, Baralle M, Buratti E. UG repeats/TDP-43 interactions near 5′ splice sites exert unpredictable effects on splicing modulation. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011 doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin SJ, Kim JM, Baughn M, Libby RT, Kim YJ, Fan Y, La Spada A, Stone B, Ravits J. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet. 2010;19:313–328. doi: 10.1093/hmg/ddp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringholz GM, Appel SH, Bradshaw M, Cooke NA, Mosnik DM, Schulz PE. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65:586–590. doi: 10.1212/01.wnl.0000172911.39167.b6. [DOI] [PubMed] [Google Scholar]
- Rollinson S, Bennion J, Toulson G, Halliwell N, Usher S, Snowden J, Richardson A, Neary D, Mann D, Pickering-Brown SM. Analysis of optineurin in frontotemporal lobar degeneration. Neurobiol Aging. 2012;33(425):e1–e2. doi: 10.1016/j.neurobiolaging.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2011;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Good SK, Atkin S, Dewey CM, Mayer P, III, Herz J, Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han YH, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011 doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci U S A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K, Maruyama H, Kamada M, Morino H, Kawakami H. Screening for OPTN mutations in amyotrophic lateral sclerosis in a mainly Caucasian population. Neurobiol Aging. 2011;32(1923):e9–e10. doi: 10.1016/j.neurobiolaging.2011.03.024. [DOI] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Lao K, Surani MA. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8:S6–S11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Garcia-Arocena D, Khandjian EW, Greco CM, Hagerman PJ. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J Med Genet. 2004;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi T, Hokonohara T, Yamasaki R, Miura S, Kikuchi H, Iwaki A, Tashiro H, Furuya H, Nagara Y, Ohyagi Y, Nukina N, Iwaki T, Fukumaki Y, Kira JI. Multiple system degeneration with basophilic inclusions in Japanese ALS patients with FUS mutation. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0621-1. [DOI] [PubMed] [Google Scholar]
- Ticozzi N, Silani V, LeClerc AL, Keagle P, Gellera C, Ratti A, Taroni F, Kwiatkowski TJ, Jr, McKenna-Yasek DM, Sapp PC, Brown RH, Jr, Landers JE. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology. 2009;73:1180–1185. doi: 10.1212/WNL.0b013e3181bbff05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V, Bosco DA, Shaw CE, Brown RH, Jr, Landers JE. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakooljul N, Hicks JA, Liu HC. Identification of target genes and pathways associated with chicken microRNA miR-143. Anim Genet. 2009;41:357–364. doi: 10.1111/j.1365-2052.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- Troakes C, Maekawa S, Wijesekera L, Rogelj B, Siklos L, Bell C, Smith B, Newhouse S, Vance C, Johnson L, Hortobagyi T, Shatunov A, Al-Chalabi A, Leigh N, Shaw CE, King A, Al-Sarraj S. An MND/ALS phenotype associated with C9orf72 repeat expansion: Abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathology. 2011 doi: 10.1111/j.1440-1789.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- Tsai KJ, Yang CH, Fang YH, Cho KH, Chien WL, Wang WT, Wu TW, Lin CP, Fu WM, Shen CK. Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med. 2010;207:1661–1673. doi: 10.1084/jem.20092164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumer Z, Bertelsen B, Gredal O, Magyari M, Nielsen KC, Lucamp, Gronskov K, Brondum-Nielsen K. A novel heterozygous nonsense mutation of the OPTN gene segregating in a Danish family with ALS. Neurobiol Aging. 2012;33:e1–e5. doi: 10.1016/j.neurobiolaging.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, van Vught PW, van Es MA, Schelhaas HJ, van der Kooi AJ, de Visser M, Veldink JH, van den Berg LH. Novel optineurin mutations in sporadic amyo-trophic lateral sclerosis patients. Neurobiol Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, Steinbart E, McCluskey L, Grossman M, Neumann M, Wu IL, Yang WS, Kalb R, Galasko DR, Montine TJ, Trojanowski JQ, Lee VM, Schellenberg GD, Yu CE. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, Van den Broeck M, Mattheijssens M, Peeters K, De Deyn PP, Cruts M, Van Broeckhoven C. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. doi: 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy DM, Wright P, Ganeslingam J, Williams KL, Tripathi VB, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo J, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009 doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Simmons Z, Liu W, Boyer PJ, Connor JR. Differential expression of genes in amyotrophic lateral sclerosis revealed by profiling the post mortem cortex. Amyotroph Lateral Scler. 2006;7:201–210. doi: 10.1080/17482960600947689. [DOI] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008a;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Shen CK. TDP-43: an emerging new player in neurodegenerative diseases. Trends Mol Med. 2008b;14:479–485. doi: 10.1016/j.molmed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, Hagerman PJ, Mientjes EJ, Oostra BA. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Groote CC, Van Broeckhoven C, Kumar-Singh S. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsensemediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, Sisodia SS, Cleveland DW, Price DL. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, Xin W, Sims K, Hu GF. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol. 2007;62:609–617. doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- Xiao S, Sanelli T, Dib S, Sheps D, Findlater J, Bilbao J, Keith J, Zinman L, Rogaeva E, Robertson J. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol Cell Neurosci. 2011;47:167–180. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Xu YF, Gendron TF, Zhang YJ, Lin WL, D'Alton S, Sheng H, Casey MC, Tong J, Knight J, Yu X, Rademakers R, Boylan K, Hutton M, McGowan E, Dickson DW, Lewis J, Petrucelli L. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]