Abstract
Up to 65% of deaths of young Asian elephants (Elephas maximus) between 3 mo and 15 yr of age in Europe and North America over the past 20 yr have been attributed to hemorrhagic disease associated with a novel DNA virus called elephant endotheliotropic herpesvirus (EEHV). To evaluate the potential role of EEHV in suspected cases of a similar lethal acute hemorrhagic disease occurring in southern India, we studied pathologic tissue samples collected from field necropsies. Nine cases among both orphaned camp and wild Asian elephants were identified by diagnostic PCR. These were subjected to detailed gene subtype DNA sequencing at multiple PCR loci, which revealed seven distinct strains of EEHV1A and one of EEHV1B. Two orphan calves that died within 3 days of one another at the same training camp had identical EEHV1A DNA sequences, indicating a common epidemiologic source. However, the high level of EEHV1 subtype genetic diversity found among the other Indian strains matches that among over 30 EEHV1 strains that have been evaluated from Europe and North America. These results argue against the previous suggestions that this is just a disease of captive elephants and that the EEHV1 virus has crossed recently from African elephant (Loxodonta africana) hosts to Asian elephants. Instead, both the virus and the disease are evidently widespread in Asia and, despite the disease severity, Asian elephants appear to be the ancient endogenous hosts of both EEHV1A and EEHV1B.
Keywords: Calves, EEHV1A, EEHV1B, Elephas maximus, orphans, proboscivirus, wild
INTRODUCTION
An acute hemorrhagic disease with 85% fatality rate was originally described in captive Asian elephants (Elephas maximus) in North American and European zoos (Ossent et al., 1990; Richman et al., 1999, 2000b; Fickel et al., 2003; Zong et al., 2007). Among 52 suspected cases since 1988, high levels of a novel herpes-virus called elephant endotheliotrophic herpesvirus (EEHV) have been confirmed by DNA PCR tests at the National Elephant Herpesvirus Laboratory (Washington, DC, USA) on blood or necropsy tissue from all 28 North American cases and 10 of 24 European cases (Zong et al., 2008; Garner et al., 2009; Latimer et al., 2011; Richman, unpubl. data). However, EEHV viruses have never been detected in numerous, random, unrelated elephant necropsy tissue samples or in blood from healthy asymptomatic elephants. The PCR test results show that EEHV disease occurs predominantly in Asian juveniles 1–8 yr old, and that there have been only 10 known survivors, all of which received timely and aggressive treatment with the human antiherpesvirus drugs famciclovir or ganciclovir (Schmitt et al., 2000; Brock et al., 2012). The EEHV disease is characterized by initial mild symptoms of facial edema and tongue cyanosis, which progresses rapidly to systemic viremia followed by hemorrhaging in all major internal organs (Richman et al., 2000a). Nuclear inclusion bodies and enveloped virions can be observed by light and electron microscopy, respectively, in microvascular endothelial cells, and high levels of viral DNA are found in white blood cells, serum, and necropsy tissue (Latimer et al., 2011; Zong, unpubl. data).
Most herpesviruses have become highly adapted to the natural hosts in which they have evolved and usually cause only mild localized disease in healthy animals. Therefore, the original description of a herpesvirus associated with rapid acute hemorrhagic disease in juvenile Asian elephants was unexpected (Richman et al., 1999). The idea that it might involve a cross-species transfer of a virus that was adapted to African elephants (Loxodonta africana) into naïve Asian elephants seemed to make sense of the unexpectedly severe pathology. Originally, two species of EEHV were described: EEHV1 was associated with 11 cases in captive Asian elephants, and EEHV2 was associated with the deaths of two young African elephants (Richman et al., 1999). The EEHV1 viruses found in those and all subsequent European and North American cases fall into two subgroups, referred to as EEHV1A and EEHV1B (Fickel et al., 2003; Richman, 2003), which are now known to represent two partially chimeric subspecies (Zong et al., 2008). EEHV1 and EEHV2 differ by an average of 25% at the nucleotide level across their genomes, whereas EEHV1A and EEHV1B differ by 15–55% at several small loci, but by no more than 1–2% over other large segments of their genomes (Richman, 2003; Ehlers et al., 2006; Zong et al., 2007; Richman, unpubl. data). Two more highly diverged species, EEHV3 and EEHV4 (35% different), were later identified from two lethal cases of hemorrhagic disease in Asian elephant calves in North America (Garner et al., 2009). Together with EEHV5 or EEHV6 (Latimer et al., 2011) and EEHV7 (Zong, unpubl. data), none of which have been associated with lethal disease, these species of elephant herpesviruses form a novel Proboscivirus genus that is most closely related to the roseoloviruses in the Betaherpesvirinae subfamily (Davison et al., 2010). The probosciviruses evidently diverged from all other mammalian herpesviruses over 100 million years ago when the ancestors of modern elephants diverged from all other placental mammals. Many of the captive elephant cases have been subjected to EEHV gene subtyping analysis, which has identified more than 30 strains of EEHV1A and six of EEHV1B (Zong et al., 2008; Zong, unpubl. data), indicating that this is primarily a sporadic and not an epidemic disease. Those studies have also shown that there has been no direct chain of transmission among cases at different elephant housing facilities or, in most cases, even among multiple afflicted progeny of a single breeding bull or cow elephant. Recent qualitative real-time PCR assays for EEHV1 have revealed that many asymptomatic Asian elephant herdmates of calves that died or survived viremic disease at the same North American housing facilities carry and periodically shed EEHV1A, EEHV1B, or EEHV5 in trunk secretions (Stanton et al., 2010; Atkins et al., in press; Stanton et al., in press).
We and others originally speculated that EEHV disease in Asian zoo elephants might be associated with opportunities for direct or indirect contact with and transmission from African zoo elephants (Richman et al., 1999, 2000b). However, some cases occurred at facilities that had never cohoused African elephants and there have been a number of anecdotal suggestions of a similar disease occurring in range countries, including India, Thailand, Cambodia, Myanmar, and Nepal, with one preliminary published report of an EEHV1A-positive case in Cambodia (Reid et al., 2006). Current evidence indicates that only EEHV2, EEHV3, EEHV6, and EEHV7 (but not EEHV1) are found in healthy asymptomatic African elephants (Zong, unpubl data), whereas only EEHV1A, EEHV1B, and EEHV5 have been found in healthy asymptomatic Asian zoo elephants (Schaftenaar et al., 2010; Stanton et al., 2010; Hardman et al., 2012; Atkins et al., in press). Therefore, we wished 1) to confirm the presence of EEHV disease in the wild in an Asian range country, 2) to address the question of which EEHV species might be causing disease there, and 3) to reassess the origins of pathogenic EEHV1.
MATERIALS AND METHODS
Tissue and DNA recovery from field necropsies
Among the 15 South India cases examined, two were captive-born, privately owned elephants; six were wild-born juveniles being reared as orphans at working or training camps or (in one case) at the Arigna Anna Zoological Park, Chennai; and seven were from free-ranging wild herds. The locations of the wild cases were distributed relatively uniformly across the entire South Western Ghats, including both sides of the Palakhad Gap, which separates two major subpopulations of elephants in southern India, the largest remaining wild population of E. maximus in Asia. In most cases, liver or heart necropsy tissue was collected and preserved in alcohol or directly frozen for long-term storage at −45 C. In one positive case, only a viremic blood sample was available without any necropsy tissue and in another only lymph-node tissue was available. DNA was extracted from stored frozen necropsy tissue samples after mincing with the use of standard procedures (Latimer et al., 2011).
PCR amplification
PCR amplification and gel purification procedures and most of the PCR primers used for the U38/POL, U60/TERex3, U71/gM, U77/HEL, and U51/vGPCR loci were described by Stanton et al. (2010, (in press) or Latimer et al. (2011). Primers used for the U48/gH-TK locus encompassing the N-terminal part of U48/glyH and the C-terminal end of U48.5 (TK) were as follows:
| L1 | LGH7981 5′-CT[A/G]CATT[T/G][A/ C]CCAAAGTATGGAAGTA-3′ |
| L2 | LGH7982 5′-C[G/A]T[C/T]TATATCATCAAA[A/G]AC[C/T]TCACA-3′ |
| R2 | LGH7984 5′-CAGCCTTCAAGCGGCATACACTG-3′ |
| R1 | LGH7985 5′-GGTAGGTTCACCTACATGGAACTTC-3′ |
First round R1/L1=1,080 base pairs (bp), second round A L1/R2=920 bp, second round B L2/R1=1,040 bp, third round A R2/ L2=860 bp.
Gene subtype DNA sequence analysis
All DNA samples were initially subjected to PCR amplification with first- or second-round primers for both PAN-POL and EEHV1-specific DNA polymerase (U38/ POL). DNA samples from the nine cases that consistently gave correctly sized PCR products were further characterized by additional first-, second-, or third-round seminested PCR analysis at up to five other previously well-characterized EEHV1 gene loci. We consider that the other six potential cases are still inconclusive, possibly because the necropsy tissue DNA available was degraded and of low quality, or because those calves instead died from non-EEHV related causes.
Correctly sized PCR products were purified after agarose gel electrophoresis with a Qiagen Gel Extraction kit (Qiagen, Valencia, California, USA) and subjected to direct-cycle sequencing on both strands with the ABI PRISM DigDye Terminator v3.1 cycle sequencing kit and analyzed on an ABI 310 DNA Sequencer (Life Technologies Inc., Carlsbad, California, USA) or at Macrogen Inc. (Rockville, Maryland, USA). All DNA sequence editing, analysis, and manipulation was performed with the use of Assembly-LIGN, Clustal-W, and distance-based phylogenetic trees under a neighbor-joining tree program with Tamura Nei logic as implemented in MacVector v. 7 (Symantec Corp., Mountain View, California, USA). Similar trees prepared after alignment in MUSCLE and tree building by maximum likelihood in MEGA5 had identical topography (not shown). Graphic alignments comparing the patterns of sequence polymorphisms were prepared in Geneious, where deviations from consensus are shown in green (A), blue (C), black (G), or red (T).
The Indian EEHV1 PCR DNA sequence results described here are deposited in Gen-Bank under accessions JX011032-JX011038 (U38/POL), JX011039-JX011046 (U48/gH-TK), JX011047-JX011055 (U51/vGPCR1), JX011056-JX011062 (U60/TERex3), JX011063-JX011071(U71/gM), and JX011072-JX011079 (U77/HEL). The comparative data used in the phylogenetic trees and graphic alignments from prototype North American EEHV cases EEH-V1A(NAP18, NAP11), EEHV1B(NAP14, NAP19), EEHV2(NAP12), and EEHV6(NAP35) come from GenBank accessions HM568528, HM568515, HM568541, HM568556, HM568564, and JN983113 (U71-gM), HM568525, HM568513, HM568538, HM568552, and JN983120 (vGPCR1), HM568529, HM568515, HM568542, HM568557, HM568564, and JN983126 (HEL) plus HM568525, HM568513, HM568538, HM568552, HM568561, and JN983119 (gH-TK), as well as JN633913 (NAP33, vGPCR1), JN633921 (NAP45, vGPCR1), and JQ300058 (NAP42-D, gH-TK). After manual sequence read editing, the assembly, Clustal-W or MUSCLE alignments and distance-based neighbor-joining or maximum-likelihood phylogenetic tree dendrograms were generated as implemented in MacVector v. 7.0 or Geneious v. 5.4.6.
RESULTS
The deaths of up to 20 young Asian elephant calves within south India since 1997 have been suspected of being caused by an acute disseminated viral hemorrhagic disease first described in captive zoo and circus elephants (Richman et al., 1999). Field necropsies were carried out on 15 suspected cases of sudden hemorrhagic deaths in juveniles or subadults that came to the attention of the Kerala Forest Department in southern India between 2007 and 2011. Most revealed edema and tongue cyanosis as well as typical hemorrhagic features in the heart and liver consistent with those described previously in young captive Asian elephants that had died suddenly of EEHV disease (Richman et al., 2000). Representative examples of gross pathologic features are provided (Fig. 1).
Figure 1.
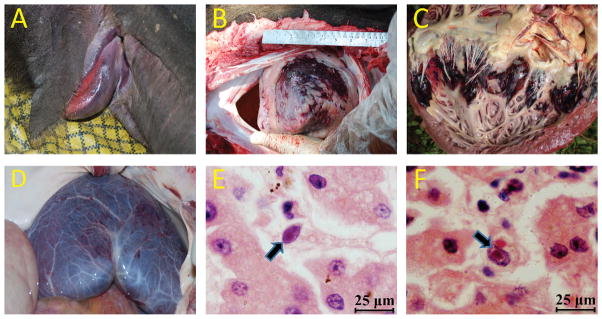
Pathologic changes of elephant endotheliotropic herpesvirus (EEHV) –associated disease found during field necropsy. (A) Asian elephant (Elephas maximus) with cyanosis of the tongue attributed to EEHV disease; (B) epicardial surface of the heart (apex view) showing severe extensive hemorrhage; (C) ventricular endocardial surface of the heart showing multifocal areas of ecchymotic hemorrhage; (D) serosal membrane surface of the liver showing diffusely scattered petechial hemorrhage; (E,F) photomicrograph of two capillary endothelial cells containing typical basophilic intranuclear viral inclusion bodies from necropsy liver tissue. H&E stain, bar=25 μm.
The US National Elephant Herpesvirus Laboratory at the National Zoological Park in Washington, DC, and the Johns Hopkins University in Baltimore, Maryland, working with the Kerala Forest Department, have established a diagnostic EEHV laboratory in Wayanad Wildlife Sanctuary in Kerala. Initial PCR analysis carried out there with the use of both PAN POL and specific EEHV1 POL primers on DNA samples from stored frozen tissues identified nine of the 15 suspected cases tested that that were sufficiently well preserved and abundant to yield high levels of PCR products that were positive for EEHV1 after first- or second-round amplification (Fig. 2). Features and sources of these nine positive cases are listed in Table 1. From our previous semiquantitative analyses and plasmid DNA reconstruction experiments with limiting dilution steps with conventional PCR (Latimer et al., 2011) or real-time PCR analysis (Stanton et al., 2010), we determined that strong first-round positive bands with PAN-POL EEHV primers from PBMC DNA represent levels of viral DNA ranging from 1 million to 75 million copies or genome equivalents per milliliter. Blood and necropsy DNA samples from healthy and known latently infected elephants not undergoing primary or reactivated infections have always been negative, even after third-round conventional PCR. Our best estimates of the detection limits for first-, second-, and third-round PCR as used here are 375,000, 15,000, and 600 genome equivalents per milliliter of blood. For whole-blood DNA with strong first-round PCR products this represents 1–75 million genome equivalents per 5 million PBMCs. Although we did not have blood samples from the India cases, the necropsy tissue DNA samples used here averaged the same 50–75 μg/mL as for the quantitated US PBMC DNA samples. Therefore, when combined with the evidence for viral inclusion bodies within a small subset of the vascular endothelial cells (assume 1%) in sufficiently well-preserved Indian case necropsy tissue, we can estimate the total amount of viral DNA per infected endothelial cell of 20–1,500 viral genome equivalents per cell, essentially the same as in the captive elephant cases. This value is compatible with active acute lytic viral replication, not latent-state infection.
Figure 2.
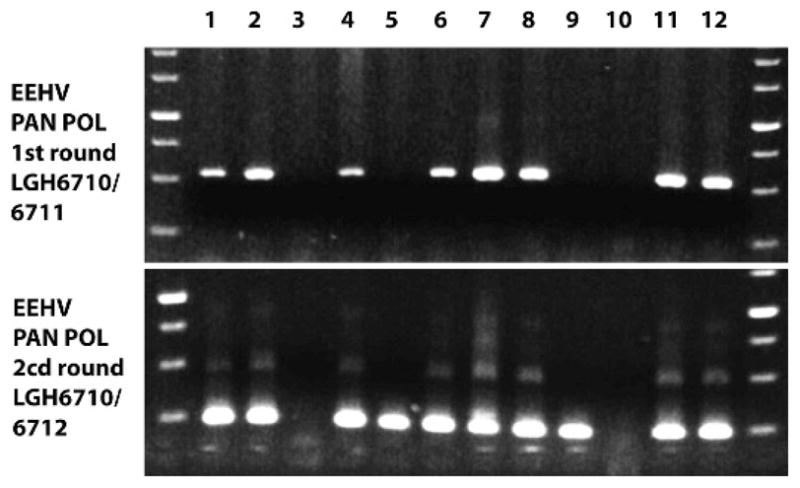
Representative agarose gel–ethidium bromide PCR band results obtained after first-round (520 base pairs [bp]) and second-round seminested (250 bp) amplification with elephant endotheliotropic herpesvirus PAN-POL primers from 12 samples of necropsy tissue DNA from Asian elephants (Elephas maximus). Lanes 1, 2, and 12, different tissues from case IP07; 3, case IP10 (negative); 4–8 multiple tissue samples from case IP06; 9, case IP01; 10, case IP04 (negative); 11, case IP11.
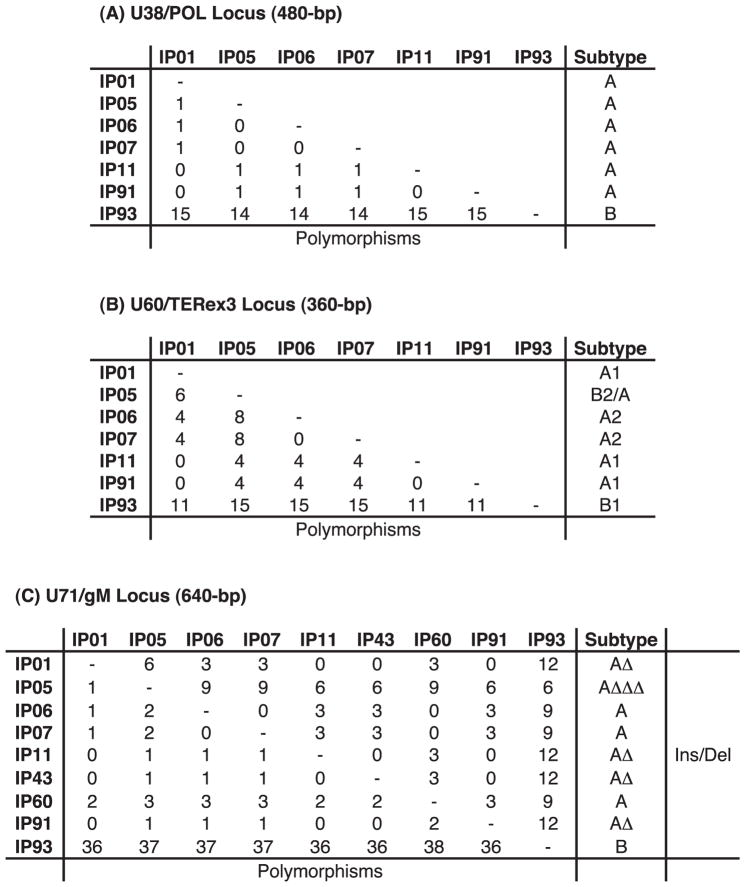
Table 1.
Summary of features and PCR results for nine cases of elephant endotheliotropic herpesvirus (EEHV) disease in Asian elephants (Elephas maximus) from India.
| Case number | Place | PCR loci subtypes or polymorphisma
|
Key features or commentsb | |||||
|---|---|---|---|---|---|---|---|---|
| vGPCR | U71/gM | POL | TER | TK/gH | HEL | |||
| IP01 | Muthanga | A2 | Δ++, 289C | A, 960C | A1 | A1 | 603T,636A, 774G, 825T, 930G | “Sunni”, M, 12 yo, 2005, captive born |
| IP05 | Thekkady | E | ΔΔΔ,289C,562T | — | B2/A | F | 764T | Wild, M, 5 yo, 2007 |
| IP06c | Kodanad-1 | E | +++, 289T | A | A2,211T | C3 | 603T,636A | “Nirangen”, M, 2 yo, 2007, rescued wild born |
| IP07c | Kodanad-2 | E | +++, 289T | A | A2,211T | C3 | 603T,636A | “Aswathi”, F, 2 yo, 2007, rescued wild born |
| IP11 | Pathiri-1 | E/A | Δ++, 289C | A, 960C | A1 | E | 36A,774G,825T,930G | Wild, F, 6 yo, 2008 |
| IP43 | Vandalur | D1 | Δ++, 289C | — | — | C1 | 603T,636A,774G,825T | “Chellama”, F, 8 yo, 2009, rescued wild born |
| IP60 | Thrissur-1 | D2/A | +++, 88T,442C | — | — | D** | 774G,825T,930G | Wild, F, 8 yo, 2010 |
| IP91 | Thirunelli | A2 | Δ++, 289C | A, 960C | D* | 858A | Wild, M, 18 m, 2011 | |
| IP93 | Thrissur-2 | B1 | B | B | B1 | — | — | M, 7 yo, 2011, captive-born (Andaman Island) |
Either overall gene subtype assignments (A, B, C1, C3, D*, D**, E/A, etc.) or the positions of minor strain polymorphism variability are indicated for each of the six PCR loci analyzed. For the U71/gM and HEL PCR loci, all data=A subtype if B is not stated. Polymorphisms are defined by changes from the consensus nucleotide at numbered standard nucleotide positions within each PCR locus. Bold type indicates unique polymorphisms found only in India. — = not done; +/Δ = presence/absence of 3× inserted/deleted codon triplets.
Word in quotations is given name of elephant; M = male; F = female; yo = years old; year = year sample taken.
These two cases had identical DNA sequences at all PCR loci tested, including in multiple tissue samples examined from each. Two DNA preparations from each were also found to be identical for cases IP05, IP11, IP43, and IP60.
Detailed gene subtype DNA sequencing analysis at multiple PCR loci was performed yielding unambiguous DNA sequence data for four to six tested PCR loci in each case (Table 1). The first three PCR loci used, U38/POL, U60/TER, and U76-U77/HEL represent well conserved core genes that serve primarily to distinguish unambiguously between the EEHV1A and EEHV1B subspecies, which differ at these loci by 15/480, 11/360 and 20/640 common nucleotides, respectively (about 3% each), but otherwise display relatively few additional characteristic polymorphisms. The primers at these loci and U71-gM also have the potential to identify DNA from most other species of EEHV in the absence of EEHV1, but only EEHV1 was detected.
The positive samples were also analyzed with EEHV1-specific primer sets for three strain variable PCR gene loci, encompassing parts of the U71/myristylated tegument protein plus U72/glycoprotein M region (U71-gM), the G-protein coupled receptor (U51/vGPCR1), and an overlapping glycoprotein-H (U48/gH) plus thymidine kinase (U48.5/TK) region (gH-TK). These latter three loci are much further diverged than the others and each clusters into multiple unlinked subtypes. The results obtained were compared to those from similar extensive analyses of EEHV genomes carried out previously on most known European and North American hemorrhagic disease cases (Fig. 3). For U71-gM, the A and B subtypes differ at the DNA level by an average of 39/750 nucleotides (5%), plus some deletions or insertions, and there is a rare third intermediate subtype (C). In the case of the vGPCR1 protein, there are five major EEHV1 subtype clusters known (A, B, C, D, and E) plus several additional chimeric forms E/A, D2/A, or other distinctive variants (A1, A2, B1, B2, B3, D1, and D2) encompassing a hot spot with up to nine adjacent variable amino acids, including deletions and insertions. Within the hypervariable gH coding segment of the gH-TK locus, all EEHV1B strains differ from the EEHV1As by 35% at the amino-acid level, with the EEHV1A strains themselves splitting further into five more subtype clusters (A, C, D, E, and F) that differ by 7–15% at the amino acid level.
Figure 3.
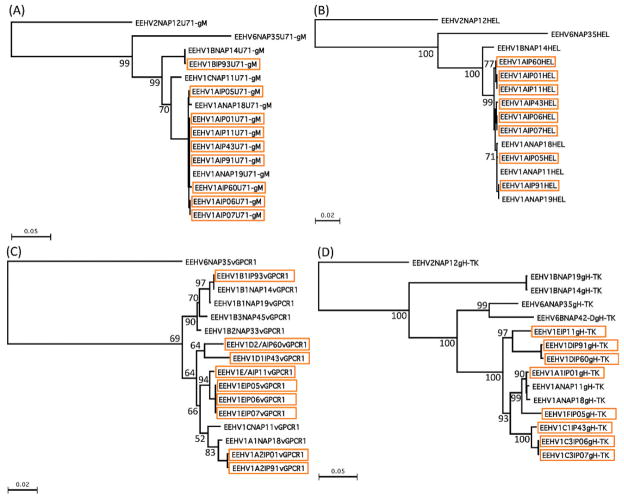
Phylogenetic distance–based dendrograms showing patterns of DNA level divergence and subtyping among Indian elephant endotheliotropic herpesvirus (EEHV1) cases (boxed in orange) in comparison with prototype examples of EEHV1A, EEHV1B, EEHV6, and EEHV2 at the four most variable PCR sequence loci: (A) U71/gM, (B) U77/HEL, (C) U51/vGPCR1, and (D) U48/gH-TK. Bootstrap values are indicated at branch points.
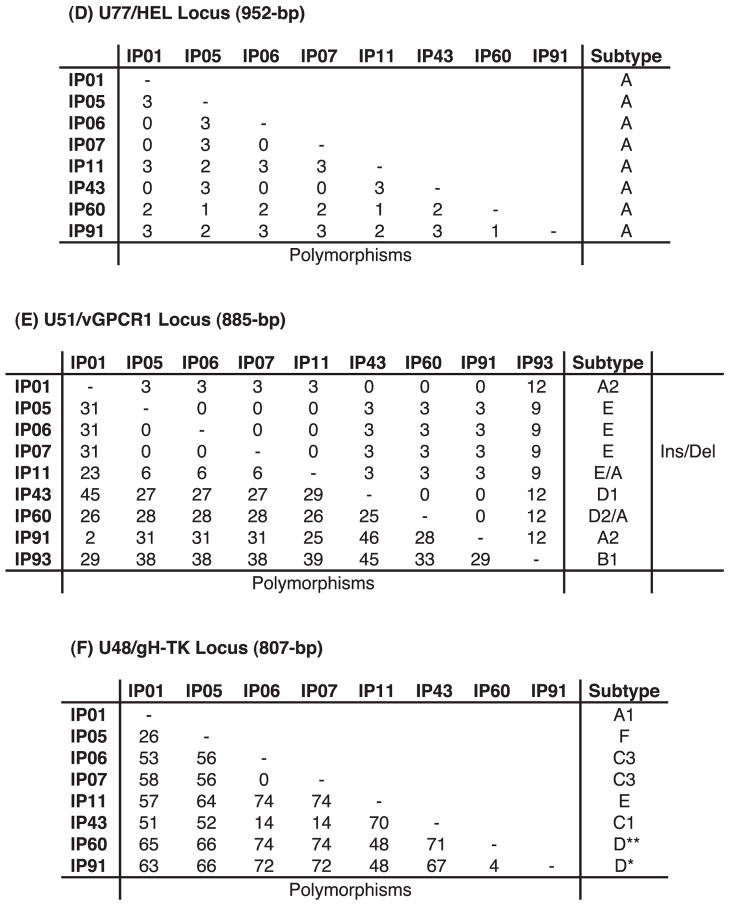
Overall, the DNA sequencing results revealed that eight of the nine positive Indian cases involved EEHV1A strains, whereas one was an EEHV1B strain. Furthermore, all but two were readily distinguishable as independent unrelated strains (Table 1). The exceptions were from two orphan calves at Kodanad Camp that died within 3 days of one another in December 2007. These had identical DNA sequences at all six PCR loci tested (totaling over 4,000 bp), including over 1,500 bp tested from tissue of each of five organs sampled for calf 1 and in both the heart and liver for calf 2. Several other instances of two afflicted calves at the same location being infected almost simultaneously with identical EEHV1 strains have also been observed in Europe and North America. Remarkably, among the two most hypervariable PCR loci tested, the Indian cases included four of the five known major vGPCR1 subtypes and five of the six known major glycoprotein-H subtypes, thus encompassing almost all of the overall genetic range of EEHV1 variants observed to occur among the 31 Asian elephant cases analyzed in a similar way from Europe and North America. Phylogenetic comparisons of the viruses from all nine Indian cases with selected prototype EEHV1A, EEHV1B, EEHV2, and EEHV6 cases at the U71-gM, vGPCR1, HEL, and gH-TK loci are presented in Figure 3, and graphic alignment presentations of the nucleotide polymorphism patterns for the same four variable loci are presented in Figure 4. Although a small number of novel polymorphisms were detected, all nine Indian examples were closely related to known subtypes at each locus examined. Comparisons of the overall distribution of EEHV1 subtypes for the vGPCR1 and gH-TK loci between Indian, North American, and European cases are illustrated in Figure 5. The actual numbers of nucleotide polymorphisms and deletion or insertion differences found among the individual Indian EEHV1 strains at all six PCR loci are presented in Figure 6.
Figure 4.
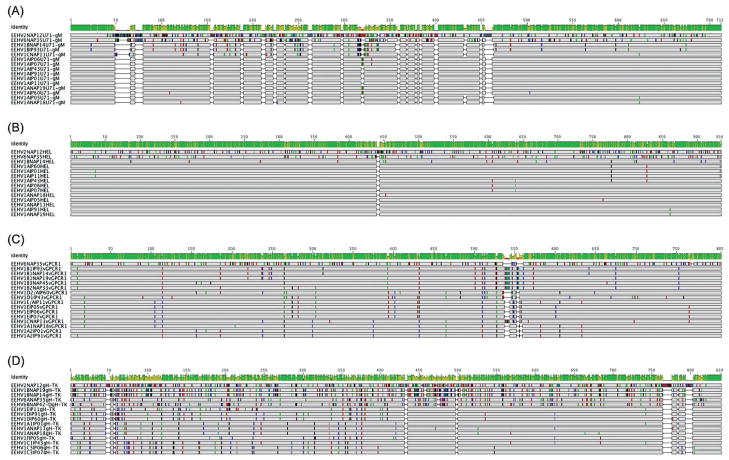
Graphic presentation generated from Geneious of the patterns of DNA sequence polymorphisms (including gaps) across the four most variable PCR loci: (A) U71-gM, (B) U77/HEL, (C) U51/vGPCR, and (D) U48/gH-TK for all Indian cases (IP#) compared to aligned prototype examples of EEHV1A, EEHV1B, EEHV6, and EEHV2.
Figure 5.
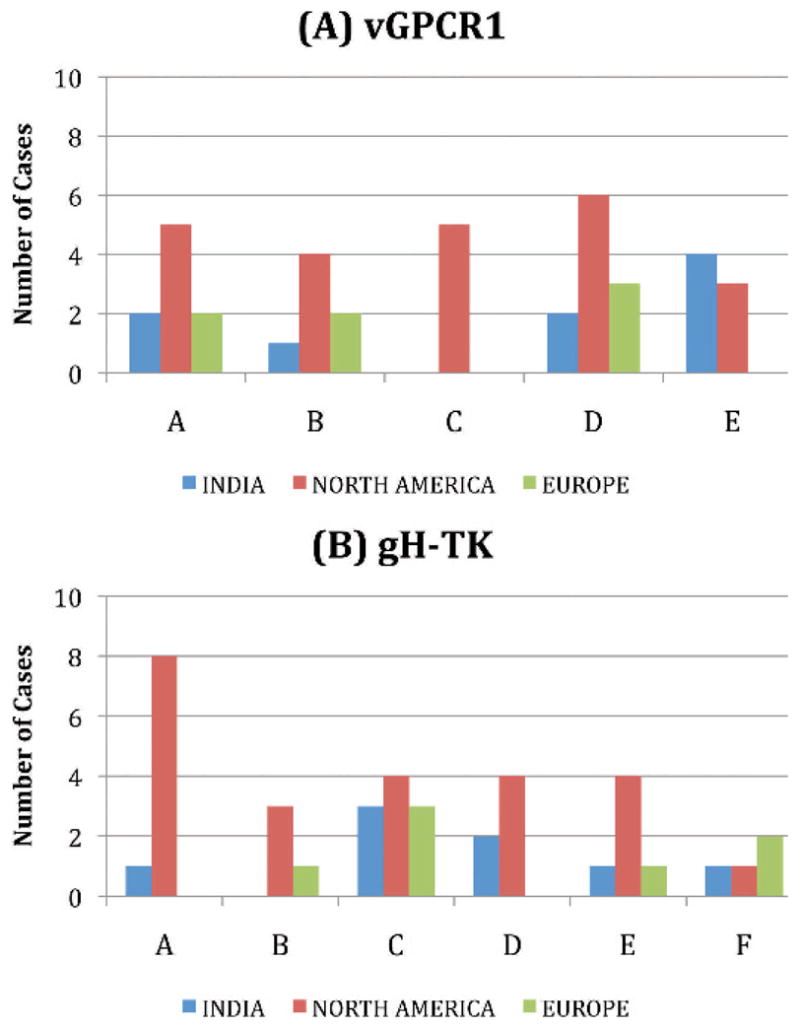
Comparison of the major elephant endotheliotropic herpesvirus (EEHV1) gene subtype distribution patterns among Indian, North American, and European hemorrhagic disease cases at the two most variable PCR loci examined: (A) U51/vGPCR1 and (B) U48/gH-TK.
Figure 6.
Direct side-by-side nucleotide level differences observed among nine Indian elephant endotheliotropic herpesvirus (EEHV1) cases at all six PCR loci: (A) U38/POL, (B) U60/TERex3, (C) U71/ gM, (D) U77/HEL, (E) U51/vGPCR1, and (F) U48/gH-TK. Nucleotide insertion or deletion (Ins/Del) differences found at the U71/gM and U51/vGPCR1 loci are also indicated.
DISCUSSION
We conclude that sporadic EEHV1-associated hemorrhagic disease is distributed across the entire southern India elephant population and that it displays gross and microscopic pathologic features that are indistinguishable from those found in the highly lethal disease documented previously in captive Asian elephants in Europe and North America. Most importantly, the very wide range of genetic diversity observed among these nine Indian EEHV1 cases is not compatible with an earlier model suggesting that this disease resulted from cross-species transmission of EEHV1 from African to Asian elephants. The latter interpretation was largely based on the apparent finding of EEHV1 viral DNA in archival paraffin block specimens of skin nodules from several African elephant calves that were imported from Zimbabwe to Florida in the mid-1980s. However, we have not been able to reproduce that result upon reexamination of the same archival skin nodule specimens and no other examples of EEHV1 DNA have been detected in necropsy samples from African elephants. Instead, multiple examples of EEHV2, EEHV3, EEHV6, and EEHV7 have all been found in lung and skin nodules from eight asymptomatic adult African elephants from South Africa, Kenya, and the United States (Zong, unpubl.; V. Pearson, pers. comm.). Similarly, only EEHV2 or EEHV6 have been detected in blood or necropsy samples from just three known rare cases of hemorrhagic or viremic disease in African elephants in North America. We interpret that the earlier result was most likely a contamination error and that both EEHV1A and EEHV1B are natural endogenous viruses that coevolved with Asian elephant hosts rather than in African elephants. Relatively recent introduction (even several thousand years ago) from just one or a few exogenous sources would have resulted in a very limited number of strains and a very restricted level of genetic variation. Current evidence suggests that EEHV4 and EEHV5 (Latimer et al., 2011) may also be endogenous to Asian elephants, but whether they are distributed much less abundantly or are just less pathogenic than EEHV1A and EEHV1B is not known. Only one death of a zoo Asian elephant calf with hemorrhagic disease caused by EEHV3 has been documented (Garner et al., 2009), but otherwise EEHV3 has been found on multiple occasions in benign lung and skin nodules from African elephants, and cross-species infections thus appear to be rare.
Herpesvirus genomes evolve relatively slowly and are generally highly stable, yet individual human cytomegaloviruses (HCMV), for example, display considerable hypervariability in certain genes that may represent relics of ancient genomic divergence that has subsequently become scrambled through extensive recombination. The vGPCR1 and glycoprotein-H subtype hypervariability in EEHV1 appears to indicate a similar ancient species population-drift effect. We had anticipated that the multiple subtypes of EEHV1 originated, and might still be distributed nonuniformly, in Asian elephant subpopulations, especially considering that the Nilgiri Eastern Ghat E. maximus population itself displays relatively narrow nuclear and mitochondrial DNA haplotype diversity compared to that of other subpopulations (Vidya et al., 2005). However, this was not the case; instead most known subtypes were present, even within this small number of cases from southern India, representing almost the entire known genetic diversity of EEHV1. This result implies that EEHV1 is likely to be an ancient infection carried by, transmitted between, and widespread among all Asian elephant populations.
Evidently, both the A and B chimeric variants of EEHV1, but seemingly not the other EEHV2 to EEHV7 species of probosciviruses, commonly produce severe hemorrhagic pathology upon primary infection within all present-day Asian elephant populations, whether born and reared in the wild or under human care. Since the late 1980s, acute primary EEHV1 viremic disease has afflicted more than 20% of Asian elephant calves born in North America, with an observed fatality rate of >80%. Why a virus that would normally be expected to be well adapted to and benign in its natural hosts displays such fulminant pathology remains to be explained, and the possibility that some other agent or cofactor contributes to the disease cannot be ruled out. The incidence of disease in the wild is unknown, but if it is similar to that in Europe and North America, the effects on future reproductive success and survival of this endangered species could be great. Additional studies on the prevalence of infections by the EEHV1, EEHV4, and EEHV5 viruses and of EEHV-associated hemorrhagic disease, especially in free-ranging wild elephants and in all Asian range countries, are needed.
Acknowledgments
Financial support for these studies, for the setup and operation of the diagnostic laboratory in Wayanad, Kerala, and for travel of team members from NEHL and Johns Hopkins University to India came from the International Elephant Foundation, the Smithsonian Institution, and the Kerala Forest Department. Studies at Johns Hopkins University were supported in part by National Institutes of Health research grant R01 AI24576 to G.S.H. Field research by A.Z. was supported in part by the US Fish and Wildlife Services Asian Elephant Conservation Fund. We thank T. V. Anilkumar for the inclusion body photomicrographs and R. Thirumurugan of the Arigna Anna Zoological park in Chennai for providing tissue samples.
LITERATURE CITED
- Atkins L, Zong J, Tan J, Mejia A, Heaggans SY, Nofs S, Stanton JJ, Flanagan JP, Howard L, Latimer E, et al. EEHV-5, A newly recognized elephant herpesvirus associated with clinical and subclinical infections in captive Asian elephants (Elephas maximus) J Zoo Wildl Med. doi: 10.1638/1042-7260-44.1.136. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock AP, Isaza R, Hunter RP, Richman LK, Montali RJ, Schmitt DL, Koch DE, Lindsay WA. Estimates of the pharmacokinetics of famciclovir and its active metabolite penciclovir in young Asian elephants (Elephas maximus) Am J Vet Res. 2012;73:1996–2000. doi: 10.2460/ajvr.73.12.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Dural G, Marschall M, Schregel V, Goltz M, Hentschke J. Endotheliotropic elephant herpesvirus, the first betaherpesvirus with a thymidine kinase gene. J Gen Virol. 2006;87:2781–2789. doi: 10.1099/vir.0.81977-0. [DOI] [PubMed] [Google Scholar]
- Fickel J, Lieckfeldt D, Richman LK, Streich WJ, Hildebrandt TB, Pitra C. Comparison of glycoprotein B (gB) variants of the elephant endotheliotropic herpesvirus (EEHV) isolated from Asian elephants (Elephas maximus) Vet Microb. 2003;91:11–21. doi: 10.1016/s0378-1135(02)00264-x. [DOI] [PubMed] [Google Scholar]
- Garner MM, Helmick K, Ochsenreiter J, Richman LK, Latimer E, Wise AG, Maes RK, Kiupel M, Nordhausen RW, Zong JC, et al. Clinicopathologic features of fatal disease attributed to new variants of endotheliotropic herpesviruses in two Asian elephants (Elephas maximus) Vet Pathol. 2009;46:97–104. doi: 10.1354/vp.46-1-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman K, Dastjerdi A, Gurrala R, Routh A, Banks M, Steinbach F, Bouts T. Detection of elephant endotheliotropic herpesvirus type 1 in asymptomatic elephants using TaqMan real-time PCR. Vet Rec. 2012;170:205. doi: 10.1136/vr.100270. [DOI] [PubMed] [Google Scholar]
- Latimer E, Zong J, Heaggans S, Richman L, Hayward G. Detection and evaluation of novel herpesviruses in routine and pathological samples from Asian and African elephants: Identification of two new probosciviruses (EEHV5 and EEHV6) and two new gammaherpesviruses (EGHV3B and EGHV5) Vet Microbiol. 2011;147:28–41. doi: 10.1016/j.vetmic.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossent P, Guscetti F, Metzler A, Lang E, Rübel A, Hauser B. Acute and fatal herpesvirus infection in a young Asian elephant (Elephas maximus) Vet Pathol. 1990;27:131–133. doi: 10.1177/030098589002700212. [DOI] [PubMed] [Google Scholar]
- Reid C, Hildebrandt T, Marx N, Hunt M, Thy N, Reynes J, Schaftenaar W, Fickel J. Endotheliotropic elephant herpes virus (EEHV) infection. The first PCR-confirmed fatal case in Asia. Vet Q. 2006;28:61–64. doi: 10.1080/01652176.2006.9695209. [DOI] [PubMed] [Google Scholar]
- Richman L, Montali R, Cambre R, Schmitt D, Hardy D, Hildbrandt T, Bengis R, Hamzeh F, Shahkolahi A, Hayward G. Clinical and pathological findings of a newly recognized disease of elephants caused by endotheliotropic herpesviruses. J Wildl Dis. 2000a;36:1–12. doi: 10.7589/0090-3558-36.1.1. [DOI] [PubMed] [Google Scholar]
- Richman L, Montali RJ, Hayward GS. Review of a newly recognized disease of elephants caused by endotheliotrophic herpesviruses. Zoo Biol. 2000b;19:383–392. doi: 10.7589/0090-3558-36.1.1. [DOI] [PubMed] [Google Scholar]
- Richman LK. PhD Thesis. The Johns Hopkins University; Baltimore, Maryland: 2003. Pathological and molecular aspects of fatal endotheliotropic herpesviruses of elephants; p. 374. [Google Scholar]
- Richman LK, Montali RJ, Garber RL, Kennedy MA, Lehnhardt J, Hildebrandt T, Schmitt D, Hardy D, Alcendor DJ, Hayward GS. Novel endotheliotropic herpesviruses fatal for Asian and African elephants. Science. 1999;283:1171–1176. doi: 10.1126/science.283.5405.1171. [DOI] [PubMed] [Google Scholar]
- Schaftenaar W, Reid C, Martina B, Fickel J, Osterhaus AD. Nonfatal clinical presentation of elephant endotheliotropic herpes virus discovered in a group of captive Asian elephants (Elephas maximus) J Zoo Wildl Med. 2010;41:626–632. doi: 10.1638/2009-0217.1. [DOI] [PubMed] [Google Scholar]
- Schmitt D, Hardy D, Montali R, Richman L, Lindsay W, Isaza R, West G. Use of famciclovir for the treatment of endotheliotrophic herpesvirus infections in Asian elephants (Elephas maximus) J Zoo Wildl Med. 2000;31:518–522. doi: 10.1638/1042-7260(2000)031[0518:UOFFTT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Stanton JJ, Zong JC, Latimer E, Tan J, Herron A, Hayward GS, Ling PD. Detection of pathogenic elephant endotheliotropic herpesvirus in routine trunk washes from healthy adult Asian elephants (Elephas maximus) by use of a real-time quantitative polymerase chain reaction assay. Am J Vet Res. 2010;71:925–933. doi: 10.2460/ajvr.71.8.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton JJ, Zong J, Eng C, Howard L, Flanagan J, Stevens S, Schmitt D, Wiedner E, Graham D, Junge R, et al. Kinetics of viral loads and genotypic analysis of elephant endotheliotropic herpesvirus-1 infection in captive Asian elephants (Elephas maximus) J Zoo Wildl Med. doi: 10.1638/1042-7260-44.1.42. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidya TN, Fernando P, Melnick DJ, Sukumar R. Population differentiation within and among Asian elephant (Elephas maximus) populations in southern India. Heredity (Edinb) 2005;94:71–80. doi: 10.1038/sj.hdy.6800568. [DOI] [PubMed] [Google Scholar]
- Zong J-C, Latimer E, Heaggans S, Richman L, Hayward G. Pathogenesis and molecular epidemiology of fatal elephant endotheliotropic disease associated with the expanding Probosci-virus genus of the Betaherpesvirinae. Proceedings of the International Elephant Conservation and Research Symposium; Orlando, Florida. 2–3 November; Azle, Texas: International Elephant Foundation; 2007. pp. 23–35. [Google Scholar]
- Zong J-C, Latimer E, Heaggans SY, Richman LK, Hayward GS. Viral gene subtyping of eighteen North American cases of EEHV hemorrhagic disease. Proceedings of the International Elephant Conservation and Research Symposium; Pattaya, Thailand. Azle, Texas: International Elephant Foundation; 2008. p. 63. [Google Scholar]