Nonredundant and specialized functions of the zein prolamines control maize protein body formation and endosperm texture.
Abstract
Zeins, the maize (Zea mays) prolamin storage proteins, accumulate at very high levels in developing endosperm in endoplasmic reticulum membrane-bound protein bodies. Products of the multigene α-zein families and the single-gene γ-zein family are arranged in the central hydrophobic core and the cross-linked protein body periphery, respectively, but little is known of the specific roles of family members in protein body formation. Here, we used RNA interference suppression of different zein subclasses to abolish vitreous endosperm formation through a variety of effects on protein body density, size, and morphology. We showed that the 27-kilodalton (kD) γ-zein controls protein body initiation but is not involved in protein body filling. Conversely, other γ-zein family members function more in protein body expansion and not in protein body initiation. Reduction in both 19- and 22-kD α-zein subfamilies severely restricted protein body expansion but did not induce morphological abnormalities, which result from reduction of only the 22-kD α-zein class. Concomitant reduction of all zein classes resulted in severe reduction in protein body number but normal protein body size and morphology.
The zein storage proteins account for 70% of maize (Zea mays) endosperm protein but are devoid of essential amino acids Lys and Trp. As a result, the overall protein quality of maize grain is poor. Paradoxically, many studies have shown that high-level zein accumulation in endoplasmic reticulum (ER)-derived protein bodies normally plays a central role in vitreous endosperm formation, an essential characteristic underlying most maize functional grain properties. This relationship has been summarized in recent reviews (Holding and Larkins, 2006, 2009; Holding and Messing, 2013). Studies to understand this process more completely have been aimed at improving the protein quality of maize and were largely inspired by the opaque2 (o2) mutant. o2 has double the wild-type levels of Lys and Trp (Mertz et al., 1964) because it accumulates low levels of zein proteins and high levels of nonzein proteins that impart a better balance of essential amino acids. The poor agronomic properties associated with kernel opacity prevented implementation of o2, but subsequent breeding efforts led to the development of Quality Protein Maize (QPM) (Vasal et al., 1980; Geevers and Lake, 1992). QPM is a hard-kernel o2 variety that retains the high levels of Lys and Trp (Paez et al., 1969; Ortega and Bates, 1983). QPM has low levels of α-zeins, like o2, but the 27-kD γ-zein is increased 2- to 3-fold (Wallace et al., 1990; Geetha et al., 1991; Lopes and Larkins, 1991; Holding et al., 2011); the degree of endosperm vitreousness closely correlates with the level of 27-kD γ-zein protein (Lopes and Larkins, 1991), and this increase causes the observed elevation in protein body number in QPM (Lopes and Larkins, 1995; Moro et al., 1995) consistent with the proposed role of 27-kD γ-zein in protein body initiation. Some other opaque mutants are the result of aberrations in processes unrelated to protein bodies, such as amino acid biosynthesis, plastid development, and cytoskeletal function (Holding et al., 2010; Myers et al., 2011; Wang et al., 2012). This indicates that further work is needed to generate a more complete understanding of the factors that control endosperm maturation.
Although the highly duplicated nature of the α-zein gene families prevents the identification of loss-of-function mutants, several dominant opaque mutants have been characterized. These result from the accumulation of a defective zein that interferes with normal zein deposition and results in ER stress responses (Coleman et al., 1997; Kim et al., 2004, 2006). The very high-level accumulation of the hydrophobic α-zeins is made possible by their efficient packaging within a shell of cross-linked γ-zeins within ER membranes. The protein body ER membrane-specific Floury1 protein is a likely coordinating factor in the assembly process (Holding et al., 2007). Defective α-zeins or loss of specific γ-zeins cause improper α-zein packaging, varying degrees of protein body deformation, and abolition of vitreous endosperm. Although the α-zeins and the γ-zeins are deposited in two broad locations, there is little knowledge of the extent of functional redundancy within these classes. α-Zeins are encoded by three multimember 19-kD α (Z1A, Z1B, and Z1D) subfamilies and one multimember 22-kD (Z1C) subfamily (Song and Messing, 2003), whereas the 27-, 16-, and 50-kD γ-zeins and the 15-kD β-zein (a γ-zein family member; Woo et al., 2001) are encoded by single genes. Protein bodies first appear as small γ-zein accretions (Fig. 1, left), and later, α- and δ-zeins penetrate and appear as inclusions with the γ-zein backgrounds (Fig. 1, middle). Mature protein bodies of 1 to 2 μm have an even, round shape, with α-zeins confined within the core surrounded by a shell of γ- and β-zeins (Fig. 1, right). This model, developed from immunogold transmission electron microscopy (TEM) of chemically fixed endosperm samples, did not distinguish between the locations of 19- and 22-kD α-zeins (Lending and Larkins, 1989). However, antibodies specific to 22- and 19-kD α-zeins revealed that these proteins have distinct patterns of accumulation (Fig. 1, right). Whereas the 19-kD α-zein is found throughout the protein body core, the 22-kD α-zein is found only in a discrete ring at the interface between the 19-kD α-zein-rich core and the 27-kD γ-zein-rich peripheral region (Holding et al., 2007).
Figure 1.
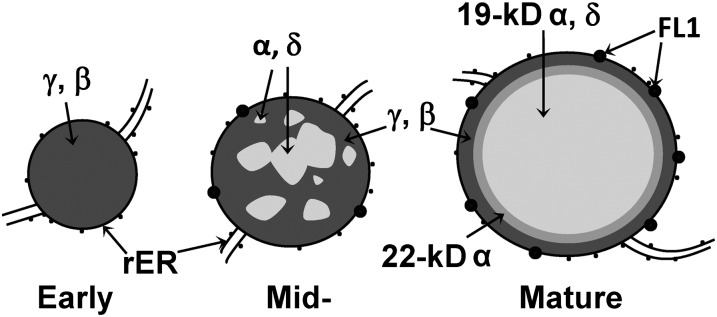
Current model for zein protein body architecture at early (left), mid- (middle), and mature stages.
In general, investigations of the temporal and spatial patterns of zein gene expression as well as an analysis of all classes of zein interactions reinforced the above model (Woo et al., 2001; Kim et al., 2002). It is thought that direct interactions between zeins are essential for their retention in the ER as properly formed protein bodies (Kim et al., 2002). Experiments using transgenic expression of both α- and γ-zeins in heterologous species shed some light on their interaction in protein body formation (Williamson et al., 1988; Bagga et al., 1995, 1997; Coleman et al., 1996). However, interpretation of such studies was limited because they did not take into account endogenous nonzein accessory factors such as Floury1 (Fig. 1), which are thought to be important determinants of protein body formation (Holding et al., 2007).
We now have the ability to routinely transform maize as well as an ability to selectively eliminate specific zein proteins, multiple products of gene subfamilies, or entire zein classes using RNA interference (RNAi). Previous use of RNAi to selectively eliminate specific γ-zein transcripts and α-zein subfamily transcripts have advanced our understanding of zein function and their inverse relationship to kernel protein quality (Segal et al., 2003; Huang et al., 2004, 2006; Wu et al., 2010; Wu and Messing, 2010, 2011), but questions concerning their distinct roles remain unanswered. This study took advantage of RNAi technology to address the extent of functional nonredundancy within the γ- and α-zein subclasses.
RESULTS
Zein RNAi Lines Abolish Vitreous Endosperm Formation through Discrete Reduction of Zein Transcripts and Proteins
We used three RNAi transgenes to target all α-zeins, a 27-kD γ-zein, and other γ-zein classes (16-kD γ-, 50-kD γ-, and 15-kD β-zeins). We also crossed these RNAi events to derive the pairwise and triplicate combinations that resulted in kernel opacity and the additive effects on zein depletion at the transcript and protein levels. The effects of each RNAi event on transcript level, protein level, and opacity are described in turn. For expression analysis of Z1A, Z1B, Z1C, and Z1D α-zein subfamilies, we used subfamily-specific primers previously described (Feng et al., 2009).
α-Zein RNAi
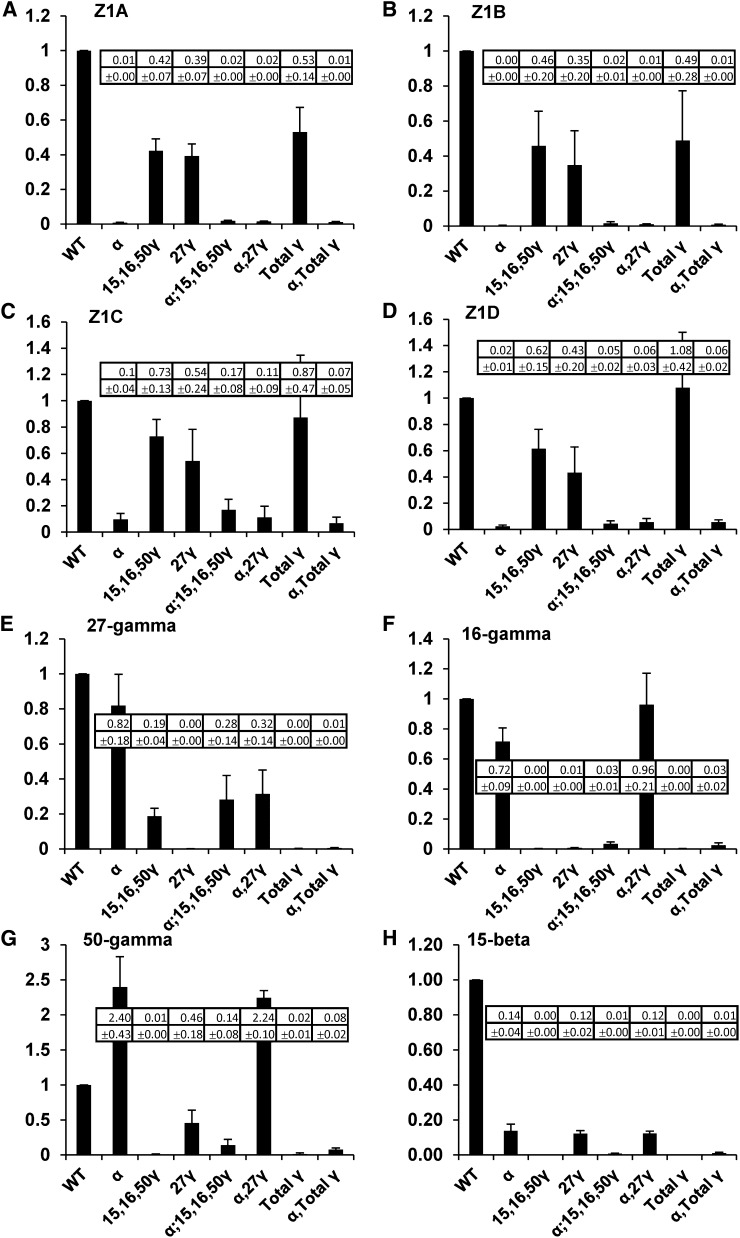
Our strategy for complete knockdown of 19- and 22-kD α-zeins involved a synthetic gene composed of approximately 220-bp conserved regions of a highly expressed member of each of the 19-kD α-zein subfamilies (Z1A, Z1B, and Z1D) and the 22-kD subfamily (Z1C; Fig. 2; Supplemental Fig. S2). Endosperm-specific expression of this RNAi cassette was driven by the 27-kD γ-zein promoter. To measure the effect of the RNAi transgene on endogenous α-zein genes, we used quantitative real-time reverse transcription (qRT)-PCR of endosperm RNA from kernels PCR genotyped using DNA extracted from their dissected embryos. The α-zein RNAi transgene reduced the level of detectable 19-kD subclass transcripts (Z1A and Z1B) to less than 1% of the level of the wild-type control (Fig. 3, A and B) and Z1D to approximately 2% to 5% of the wild-type control (Fig. 3D). The 22-kD subclass transcripts (Z1C) were reduced to approximately 10% of the wild-type level (Fig. 3C). These reductions were observed both in the single α-zein RNAi line and in combination with other RNAi constructs. Slight nonspecific reduction in expression of Z1A, Z1B, Z1C, and Z1D class transcripts also resulted from expression of both γ-zein RNAi constructs described below (Fig. 3, A–D). This may indicate that high-level expression of any transgene in the endosperm can reduce endogenous expression of α-zeins through competition for transcriptional machinery. Alternatively, improper protein body formation caused by γ-zein RNAi-mediated defects may cause negative feedback on expression of α-zeins.
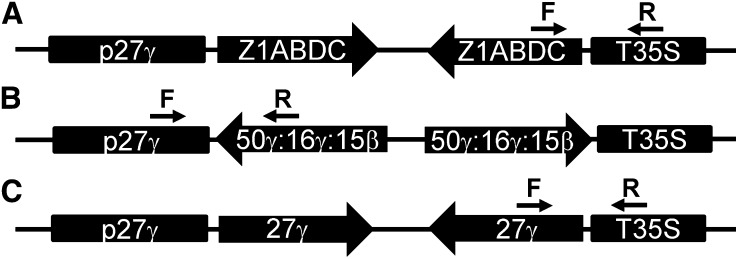
Figure 2.
Scheme of zein RNAi constructs with 27-kD γ-zein promoter (p27γ) and Cauliflower mosaic virus terminator (T35S).
Figure 3.
qRT-PCR expression analysis of α- and γ-zein genes in RNAi lines relative to wild-type nontransgenic (HiII). Each graph represents the measurement of a different zein gene in all the RNAi lines, which are shown on x axes. Values and sd (shown in insert) are the average of three biological replicate kernels and also incorporated three technical replicates of each measurement.
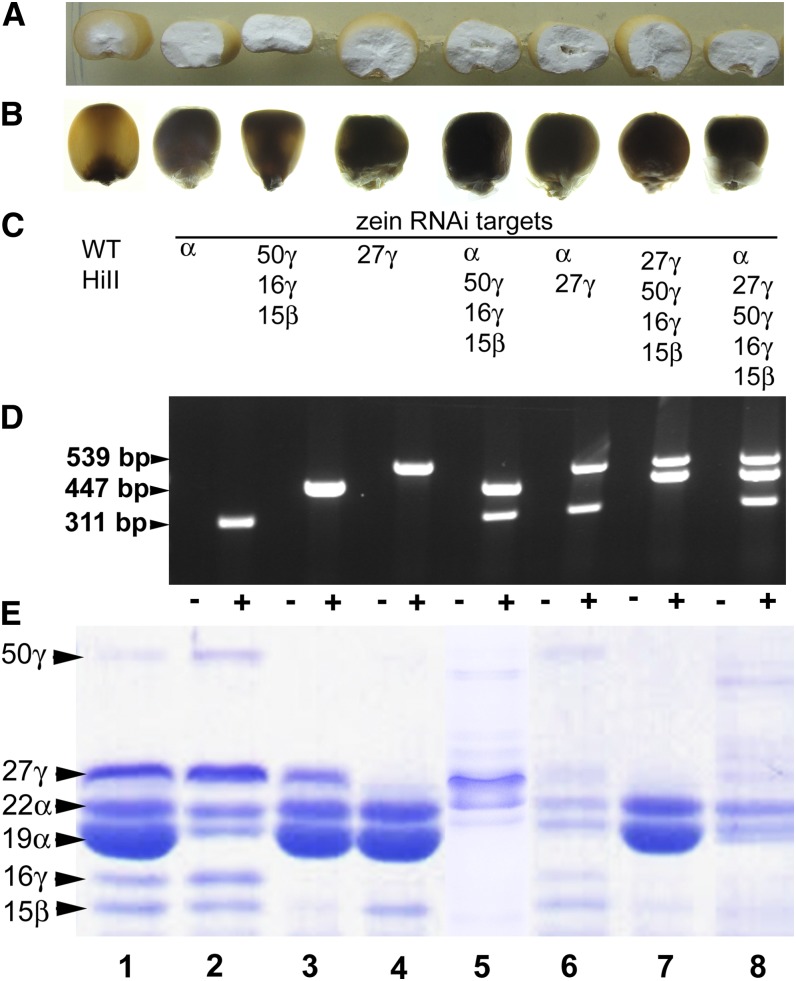
For the α-zein RNAi construct, we recovered multiple self-pollinated T1 ears from each of 11 events. Ten events had ears with a majority of completely opaque kernels and fewer normal, vitreous kernels. Seed fill was variable and often low in these ears, but we were able to make kernel counts and establish clear three-to-one opaque-to-vitreous segregation ratios in adequately filled ears from four events (Supplemental Table S1). HiII or B73 outcross ears from these events, where recovered, segregated approximately one to one, opaque to vitreous. These results indicate the presence of a single transgene locus and dominant action of the transgene. All opaque kernels showed dramatic reduction in total α-zeins in mature kernels (Supplemental Fig. S1). To isolate the specific effects of a transgene from nonspecific zein degradation as a result of improper protein body formation, we routinely analyzed zeins at 18 d after pollination (DAP) during midkernel filling (Fig. 4). This corresponds to the latest stage that developing kernels can be reliably fixed and infiltrated for TEM. At 18 DAP, endosperm from kernels containing the transgene accumulated only a small fraction of α-zein compared with the wild type (Fig. 4E). Curiously, α-zein RNAi resulted in a noticeable increase in the apparent abundance of 50-kD γ-zein (Fig. 4E, lanes 2 and 6; Supplemental Fig. S1; data not shown). This 50-kD γ-zein increase was also observed at the transcript level (Fig. 3G).
Figure 4.
Kernel phenotypes of zein RNAi lines. A, Half kernels showing vitreous or opaque endosperm. B, Kernels on a light box showing vitreousness or opacity. C, RNAi transgenes present in A, B, D, and E. D, Embryo genomic PCR showing genotype of kernels shown in E, which were used for TEM analysis. PCRs for double and triple construct lines were conducted separately and combined for agarose gel electrophoresis. Minus symbol refers to nontransgenic HiII control kernel, and plus symbol refers to the transgenic kernel. Size for PCR product in base pair is indicated. E, SDS-PAGE of purified zeins from developing endosperms of genotypes shown in D.
16-kD γ-Zein, 50-kD γ-Zein, and 15-kD β-Zein RNAi
γ-Zein family members 27-kD, 50-kD, and 16-kD γ-zeins and the 15-kD β-zein share a common location in the protein body periphery and have been assumed to have redundant function. To address the question of functional redundancy, we made transgenic lines containing two different RNAi constructs. The first of these used a synthetic gene composed of 212-bp segments of the 50-kD and 16-kD γ-zeins and the 15-kD β-zein (Fig. 2; Supplemental Fig. S2). This construct, whether alone or in combination with other RNAi constructs, reduced the abundance of all of the three targeted γ-zein proteins to very low levels, usually less than 1% of the wild-type level (Fig. 3, F–H). This construct also reduced to level of the 27-kD γ-zein to approximately 20% to 30% of the wild-type level (Fig. 3E), probably due to the similarity of the 16-kD and 27-kD γ-zeins, and such cross repression was previously described (Wu and Messing, 2010).
We recovered self-pollinated T1 ears, often from several plants per event, from seven events. Six events had ears that apparently segregated three to one, semiopaque to vitreous. However, because the semiopaque phenotype on the light box was subtle and kernel numbers were often low, it was not possible to obtain rigorous segregation ratios. HiII or B73 outcross ears from these events, where recovered, segregated approximately one to one, semiopaque to vitreous, but for the above reason, segregating kernel counts were not possible. These results suggest the presence of a single transgene locus and dominant action of the transgene. One of the seven events produced ears with 100% opaque kernels, whether self pollinated or outcrossed to HiII or B73, indicating the presence of more than one transgenic locus or possibly insertion of the transgene into a gene causing opacity. The dominant phenotype driven by this transgene is described as semiopaque because the kernels showed partial light transmittance (Fig. 4B). Although scarce in normal kernels, the 50-kD γ-zein was rendered undetectable in developing and mature kernels of this line. The 16-kD γ-zein ranged from reduced (Fig. 4E, lane 5) to undetectable (Fig. 4E, lane 3) in kernels containing this transgene. Though still detectable, the 15-kD β-zein was substantially reduced in this line. Through sequence similarity to the 16-kD γ-zein and consistent with the reduced transcript level, the 27-kD γ-zein was usually slightly reduced by this transgene, consistent with its reduced transcript level.
27-kD γ-Zein RNAi
We sought to specifically target 27-kD γ-zein knockdown using an RNAi hairpin composed of the entire 27-kD γ-zein open reading frame (ORF; Fig. 2). At the transcript level, this transgene resulted in 1% or less of the wild-type transcript level of both 27-kD and 16-kD γ-zeins (Fig. 3, E and F) and partial suppression (46% of the wild type) of the 50-kD γ-zein (Fig. 3G) and 15-kD β-zein (12% of the wild type; Fig. 3H). At the protein level, this transgene resulted in total suppression of the 27-kD γ-zein as well as substantial suppression of 16-kD γ-zeins (Fig. 4E, lane 4). When combined with the α-zein RNAi, the suppression of the 27-kD γ-zein and the 16-kD γ-zein were less pronounced both at the RNA level (Fig. 3, E and F) and the protein level (Fig. 4E, lane 6). The reduced 50-kD γ-zein transcript correlated with reduced 50-kD γ-zein protein below wild-type levels (Fig. 4E, lane 4), but the protein was present at normal levels in mature kernels (not shown). Interestingly, when combined with the α-zein RNAi, as described above, the suppressive effect of this transgene on the 50-kD γ-zein was outweighed because both the transcript (Fig. 3G) and the protein (Fig. 4E, lane 6) are substantially increased. Despite the fact that this transgene substantially reduced 15-kD β-zein transcript, it did not markedly reduce the level of 15-kD β−zein protein (Fig. 4E, lanes 4 and 6).
We recovered self-pollinated T1 ears, often several ears per event, from four events. Two of these events had ears that apparently segregated three to one, opaque to vitreous. Although this transgene resulted in more complete opacity than 50-kD, 16-kD, and 15-kD RNAi transgenes, light box opacity was not as clear as the α-zein RNAi. This ambiguity, as well as low kernel numbers, again prevented us from obtaining segregation ratios. HiII or B73 outcross ears from these events, where recovered, segregated approximately one to one, opaque to vitreous. The other two events produced self or outcross ears, with all opaque kernels indicating the presence of more than one transgenic locus.
RNAi Effects on Protein Body Density, Size, and Morphology as Well as Zein Distribution Suggest Nonredundant Roles of Zeins in Protein Body Formation
Figures 5, 6, and 7 show low-magnification TEM analysis of protein body density (where density refers to number of protein bodies per unit area, not buoyant density), high-magnification TEM analysis of protein body size and morphology, and high-magnification TEM immunogold analysis of zein distribution, respectively. To account for natural variation in protein body size and density, these analyses were standardized by sampling the fourth subaleurone starchy endosperm cell layer. In our growth conditions, at 18 DAP, the fourth subaleurone cell layer had densely clustered protein bodies in wild-type endosperm, whereas the number and size of protein bodies increased in a gradient in the first through third cell layers. In immunogold TEM, control labeling without primary antibody showed no nonspecific secondary antibody labeling. α-Zein antiserum showed very low labeling outside protein bodies, and the γ-zein antibody showed no labeling outside protein bodies.
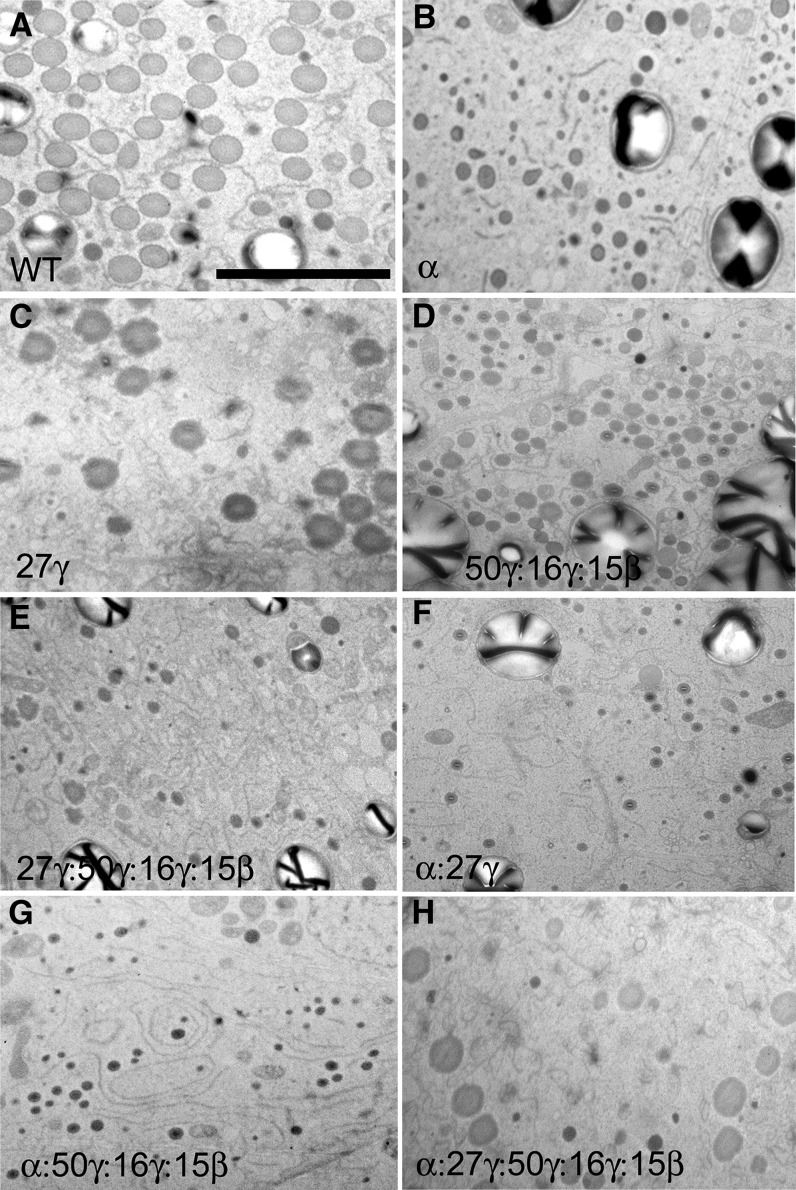
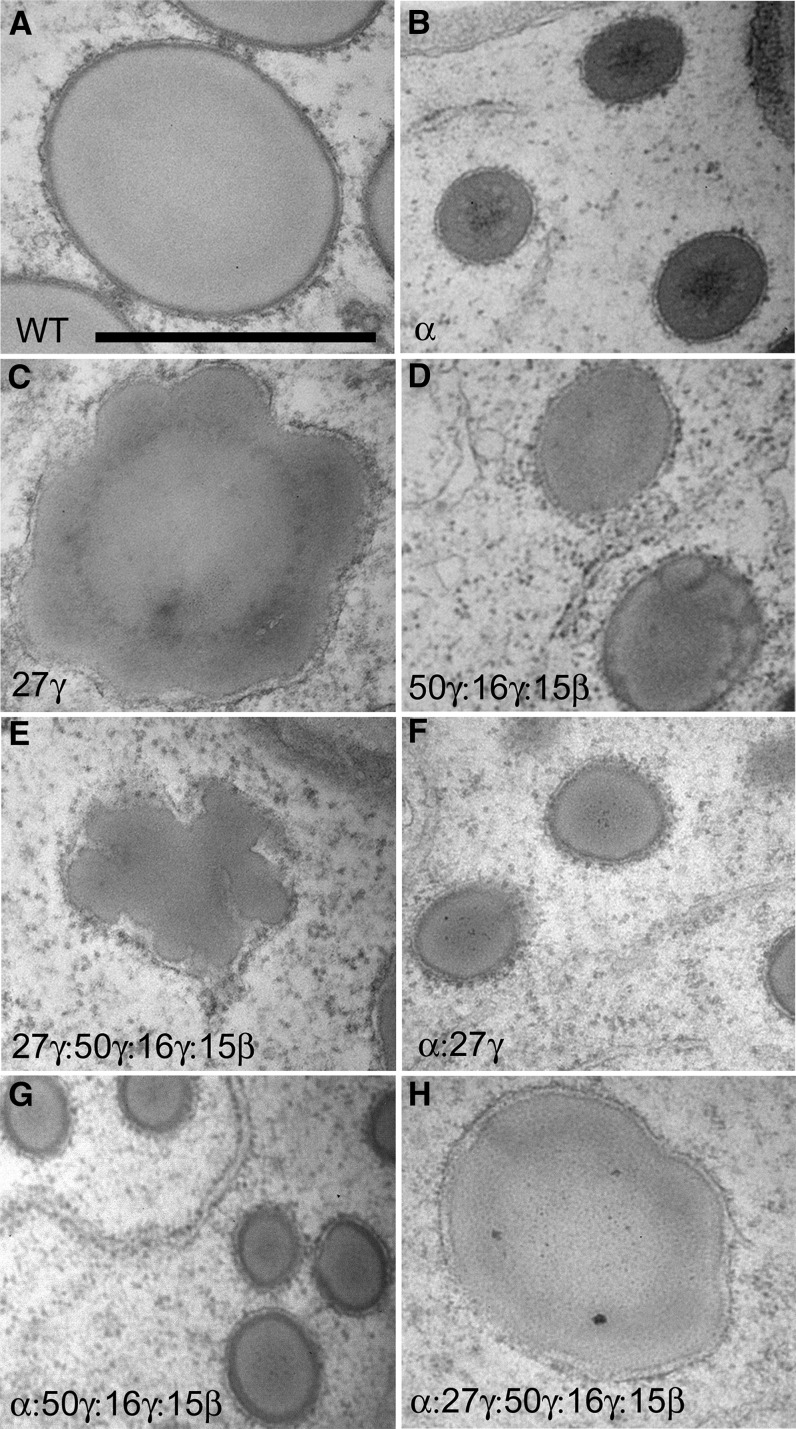
Figure 5.
TEM analysis showing protein body density in fourth subaleurone starchy layer of 18-DAP endosperm in zein RNAi lines and their crosses. Bar = 5 μm (A; refers to all panels). RNAi transgenes present are shown in bottom left of each section.
Figure 6.
TEM analysis showing protein size and morphology in fourth subaleurone starchy layer of 18-DAP endosperm in zein RNAi lines and their crosses. Bar = 1 μm (A; refers to all panels). RNAi transgenes present are shown in bottom left of each section.
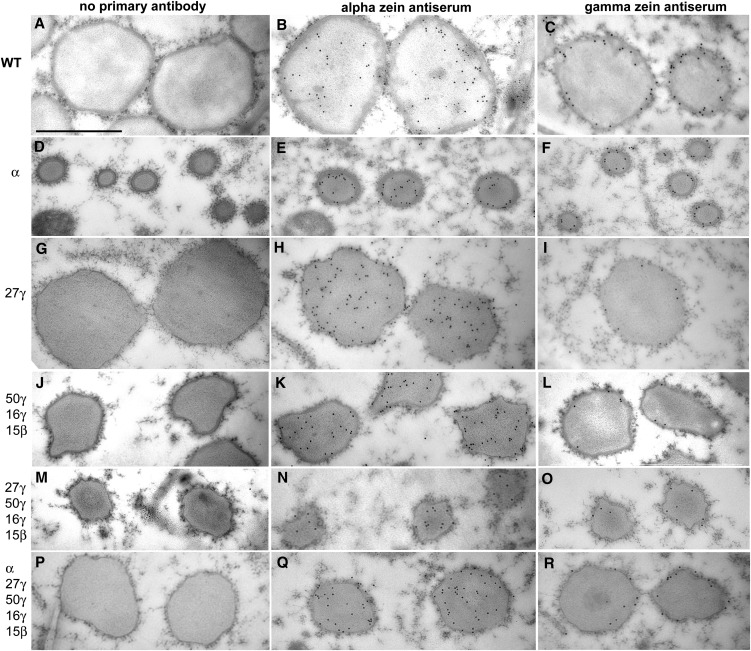
Figure 7.
Immunogold TEM analysis showing α- and γ-zein distribution in protein bodies of fourth subaleurone cell layer of 18-DAP endosperm in zein RNAi lines and their crosses. Bar = 1 μm (A; refers to all panels). RNAi transgenes present are shown in bottom left of each section. α-Zein antibody was raised to total α-zeins, and γ-zein antibody was raised to 27-kD γ-zein.
α-Zein RNAi
Dramatic reduction of total α-zeins resulted in a similar effect to the o2 mutant. α-Zein filling was reduced, which resulted in protein bodies approximately one-quarter to one-third of their normal diameter (Figs. 5B, 6B, and 7, D–F). Unlike o2 however, no reduction in protein body density was observed, indicating that α-zeins are not required for protein body initiation (Fig. 5B; Table I). Normally, α-zeins are located in the center of the protein body and are largely encapsulated by and excluded from an outer peripheral ring of γ-zeins (Fig. 7, B and C). α-Zein and γ-zein distribution appeared normal in α-zein RNAi protein bodies (Fig. 7, E and F), and residual α-zeins were densely packaged within the smaller protein bodies. Notably, unlike selective repression of the 22-kD α-zein, where the normal ratio of 22- to 19-kD α-zeins is disturbed (Segal et al., 2003), we did not observe any abnormal protein body shape, such as lobing, aggregation, or reticulation.
Table I. Protein body number per 100 μm2 of fourth subaleurone starchy endosperm cell layer.
Protein body counts were made by sampling 100-μm2 regions, which excluded nuclei and cell walls, within 18-DAP fourth subaleurone endosperm cells. Average is derived from counts of four TEM sections (two independent kernels and two independent embedments of each kernel).
| Genotype | Protein Body Number |
|---|---|
| per 100 μm2 ± sd | |
| Wild type (HiII) | 50.0 ± 3.2 |
| α-Zein RNAi | 51.5 ± 5.0 |
| 27-kD γ-zein RNAi | 20.4 ± 3.4b |
| 16- and 50-kD γ-zein and 15-kD β-zein RNAi | 46.0 ± 3.8 |
| 27-, 16-, and 50-kD γ-zein and 15-kD β-zein RNAi | 12.0 ± 2.1b |
| α-Zein and 27-kD γ-zein RNAi | 25.5 ± 3.4b |
| α-Zein, 16- and 50-kD γ-zein, and 15-kD β-zein RNAi | 36.3 ± 6.6a |
| α-Zein, 27-, 16-, and 50-kD γ-zein, and 15-kD β-zein RNAi | 15.0 ± 1.8b |
Significantly different from the wild type at the 0.01 level. bSignificantly different from the wild type at the 0.001 level using a two-tailed Student’s t test.
27-kD γ-Zein RNAi
Elimination of the 27-kD γ-zein and the severe reduction of the 16-kD γ-zein caused by this RNAi transgene resulted in substantial reduction in protein body number per unit area (Fig. 5C; Table I), consistent with the role of the 27-kD γ-zein in protein body initiation, although this does not rule out the 16-kD γ-zein as also playing a role in initiation. Interestingly, protein body size was normal in this line (Figs. 5C, 6C, and 7, G–I), suggesting that the 27-kD γ−zein is not required for the bulk of protein body filling. Minor distortions in protein body shape were usually observed (Figs. 6C and 7, G and H) that may result from α-zein not being properly encapsulated by γ-zein and is supported by an increased amount of α-zein deposited in the protein body periphery and membrane (Fig. 7H). The dark-staining protein body periphery was visibly thinner (Figs. 6C and 7, G–I), and γ-zein was less abundant, as suggested by a noticeable reduction in the number of gold particles (Fig. 7I).
16-kD γ-Zein, 50-kD γ-Zein, and 15-kD β-Zein RNAi
Severe reduction of these three γ-zein family proteins, while leaving the 27-kD γ-zein only slightly reduced, had very different effects. First, protein body number was normal (Fig. 5D; Table I), suggesting the other γ-zein family members are not required for protein body initiation. However, protein body size was reduced by approximately 50%, suggesting that some or all of these zeins have roles in protein body expansion. Although these smaller protein bodies showed minor shape distortions (Fig. 7, J–L), the dark-staining protein body periphery was of normal appearance (Fig. 7, J–L). Furthermore, α-zein did not appear ectopically in the protein body periphery, suggesting the 27-kD γ-zein plays the dominant role in packaging α-zeins and consistent with it being by far the most abundant of the γ-zein family members.
Total γ-Zein Family RNAi
Combining the 27-kD γ-zein RNAi to the 16-kD γ-zein, 50-kD γ-zein, and 15-kD β-zein RNAi resulted in very low levels of all the γ-zein family proteins (Fig. 3E). Effects of this were additive in terms of severely reduced protein body density (Fig. 5E), reduced protein body size, and moderate to severe protein body distortion or reticulation (Figs. 6E and 7, M–O).
α-Zein RNAi Combined with Individual γ-Zein RNAi Events
Combination of the 27-kD γ-zein RNAi or the 16-kD γ-zein, 50-kD γ-zein, and 15-kD β-zein RNAi to the α-zein RNAi resulted in protein body size reductions similar to the α-zein RNAi alone (Fig. 5, F and G). Although protein bodies in 16-kD γ-zein, 50-kD γ-zein, and 15-kD β-zein RNAi × α-zein RNAi had a slightly reduced density (Table I; Fig. 5G), the 27-kD γ-zein × α-zein RNAi combination resulted in a similar severe loss of protein body density to that of 27-kD γ-zein RNAi alone (Table I; Fig. 5F). This supports the suggestion that the 27-kD γ-zein has the major role in protein body initiation.
α-Zein RNAi Combined with Total γ-Zein RNAi Events
To achieve low levels of all the major α- and γ-zeins, we combined all three RNAi constructs. This resulted in the lowest density of protein bodies (Table I; Fig. 5H). However, protein body size ranged from only slightly reduced to normal, compared with the wild type, when all classes of zeins were reduced (Figs. 5H, 6H, and 7, P–R). α-Zein and γ-zein distribution and density in these sparse protein bodies were relatively normal (Fig. 7, Q–R).
DISCUSSION
The ability to use RNAi to dominantly reduce or eliminate either single gene or whole gene family products, coupled with routine transformation of maize, has enhanced our ability to decipher the structural functions of zein storage protein classes. Because of the high level of α-zein gene duplication, recessive knockout mutations for functional analysis are not recoverable, but we now have the technology to selectively or collectively eliminate α-zein subfamilies. This study sought to address unresolved questions about the differential roles of zeins in protein body formation. First, are the colocalized γ- and β-zeins functionally redundant or do they have specific roles? Second, what is the effect of total α-zein knockdown and does the 22-kD α-zein function to constrain 19-kD α-zeins in the protein body center? The data also provided new insight into role of transcript levels in determining zein protein abundance.
Targeted Zein Transcript Reduction Causes Major Zein Protein Reduction, while Minor Nonspecific Transcript Reductions Do Not Substantially Affect Protein Level
The chimeric α-zein RNAi construct achieved the targeted reduction of Z1A, Z1B, Z1C, and Z1D transcripts. Z1C (22-kD) class transcripts were reduced to approximately 10% of wild-type levels, and the protein was not reduced to quite the same extent as the 19-kD class (Fig. 4E, lane 2). In the case of the 19-kD α-zein classes, we detected less than 1% of the relative wild-type level of transcript. Although dramatically reduced, reduction of the 19-kD α-zein protein does not appear to be as profound as the transcript. Moreover, we observed a nonspecific transcript reduction of all four classes of α-zein caused by the γ-zein RNAi construct. Despite transcript reductions of around 50%, these nonspecific effects were not seen at the protein level, suggesting that transcript level is in excess and zein abundance is also likely governed by available amino acids and translational machinery. Another nonspecific transcript reduction was that of the 15-kD β-zein, which was caused by both the α-zein RNAi and the 27-kD γ-zein RNAi constructs. Although the basis for these substantial transcript reductions is unknown, they do not result in noticeably lower-than-wild-type levels of 15-kD β-zein protein, again suggesting that this protein is not primarily limited by transcript level.
It was observed that the 27-kD γ-zein RNAi also suppressed the 16-kD γ-zein at the RNA and protein level. However, when combined with the α-zein RNAi, while the 27-kD γ-zein remains suppressed, the 16-kD γ-zein suppression is largely lifted at both the RNA and protein level (Figs. 3F and 4E, lane 6). A similar effect was observed with the 50-kD γ-zein, whose transcript and protein was increased in both the γ-zein RNAi construct lines when combined with the α-zein RNAi. Together, these may suggest that with reduced α-zein synthesis, nonspecific effects of other RNAi events may be less pronounced because of increased availability of translational resources.
Separable Roles of γ-Zeins in Protein Body Initiation and Filling
Because γ-zein family member genes are generally not duplicated, it is perhaps surprising that recessive mutants have not been recovered for these, although a double null mutant for the 10- and 18-kD δ-zein was recovered (Wu et al., 2009), which had no observable effects on protein body morphology (Wu and Messing, 2010). With careful design, it is possible to knock down all γ-zein family members individually, but sequence conservation, such as between the 27- and 16-kD γ-zeins, as well as the high cost and time input for working with maize transgenic plants place restrictions on the experimental design. For example, Wu and Messing (2010) chose to simultaneously target the 27- and 16-kD γ-zeins with a single construct, suppress the 15-kD β-zein with another construct, and not target the 50-kD γ-zein because of its low abundance on SDS-PAGE gels. However, using our protocol, we find the 50-kD γ-zein abundance approaching that of the 16-kD γ- and 15-kD β-zeins, and by maturity, it is usually highly abundant (Supplemental Fig. S1). During zein purification, nonzeins are separated by precipitation with 70% (v/v) ethanol (Wallace et al., 1990). When a lower ethanol percentage is used at this step (45%), we have found that, although there is more contamination with nonzeins, the 50-kD γ-zein band is even more intense (not shown). We therefore targeted the 50-kD γ-zein with RNAi.
The 27-kD/16-kD γ-zein and 15-kD β-zein RNAi events were previously reported to cause only very minor morphological changes to protein bodies (Wu and Messing, 2010). Although an incidence of very small protein bodies was shown in both cases, the majority of protein bodies were of normal size, although standardization of which subaleurone endosperm cell layer was used for TEM imaging was not indicated (Wu and Messing, 2010). Our 27-kD γ-zein RNAi construct was similar because it resulted in complete elimination of 27-kD γ-zein and almost complete elimination of the 16-kD γ-zein. While we also showed that protein body size is normal in developing kernels of this RNAi mutant, later-stage protein body filling is compromised because most protein bodies had a distorted, undulating surface. Our study also incorporated quantitative counts of protein body density. Because protein body density in the developing endosperm was reduced by more than 50%, this suggests that the 27-kD and/or 16-kD γ-zeins are required for protein body initiation. Our 16-kD γ-, 50-kD γ-, 15-kD β-zein constructs completely eliminated the 16-kD γ-zein as well as the 50-kD γ-zein and strongly reduced the 15-kD β-zein. Significantly, this construct did not reduce protein body density, thus indicating that the16-kD γ-zein does not function in protein body initiation. This construct did result in a significant reduction in protein body size. Because this work and the previous study (Wu and Messing, 2010) showed that RNAi of 27-kD γ-, 16-kD γ-, or 15-kD β-zeins (but not the 50-kD γ-zein) do not markedly reduce protein body size, a significant role for the 50-kD γ-zein in α-zein-driven protein body expansion is supported. Interestingly, we consistently observed increased 50-kD γ-zein (transcript and protein) in α-zein RNAi lines, suggesting that the 50-kD γ-zein has a closer association to the central α-zein core than other γ-zeins and can partially compensate the loss of α-zeins. Combination of the γ-zein family RNAi constructs resulted in addition of the distortion and the low-density phenotypes. The combined RNAi in the previous study (Wu and Messing, 2010; 27-kD γ-, 16-kD γ-, and 15-kD β-zeins) did not target the 50-kD γ-zein and again did not reduce protein body size, which further supports a major role for the 50-kD γ-zein in protein body filling. Taken together, our results suggest nonredundant initiation and filling roles of γ-zeins.
Reduction of Both α-Zein Classes Affects Protein Body Size but Not Number or Morphology
At the outset of this work, previous efforts to use RNAi to eliminate α-zeins had been aimed solely at creating a dominant, nonpleiotropic low-zein line for Lys improvement. The first targeted only the 22-kD α-zein (Segal et al., 2003), and the second was private-sector work that targeted both the 19- and 22-kD α-zein but did not address the morphological effects on protein bodies (Huang et al., 2006). We required such a total α-zein RNAi line both as a nonpleiotropic, dominant opaque line for testing the activity of overexpressed o2 (QPM) modifier genes as well as to try to dissect possible nonredundant function between the 19- and 22-kD α-zeins. A total α-zein RNAi line that incorporated a dominant GFP marker was subsequently published, but morphological protein body data were not reported (Wu and Messing, 2011).
Protein bodies in o2 endosperm are small (Geetha et al., 1991), presumably because of reduced accumulation of both 22-kD and 19-kD α-zeins, the former resulting from reduced transcriptional activation and the latter as an indirect effect (Schmidt et al., 1990). Protein body density in cells is also reduced (Geetha et al., 1991), consistent with the fact that the mutation also indirectly reduces 27-kD γ-zein accumulation. However, o2 exhibits no misshapen protein body morphologies, as is seen in dominant negative α-zein mutants (Coleman et al., 1997; Gillikin et al., 1997; Kim et al., 2004). The 22-kD α-zein-specific RNAi event, which has substantial residual 19-kD α-zein, also has abnormally lobed and distorted protein bodies whose size is not significantly reduced over the wild type (Wu and Messing, 2010). This leads to the suggestion that the bulk of protein body expansion is driven by the 19-kD α-zein, which is consistent with its location throughout the protein body core within a ring of the 22-kD α-zein (Holding et al., 2007). This also suggests that the 22-kD α-zein may have a structural role in packaging the 19-kD α-zeins and may explain why protein body lobing and distortion result from specific reduction of the 22-kD α-zein. In support of this hypothesis, when we reduced both α-zein classes equivalently with RNAi, protein body morphology and zein distribution were normal and only a reduced size resulted.
Our results show that reduced protein body size results from reduced total α-zein as well as reduction in some but not all γ-zein classes. Reduced protein body initiation results specifically from the reduced 27-kD γ-zein and not from reductions in other γ-zein family members. However, when all the RNAi events were combined, all zeins were reduced roughly in proportion. Surprisingly, this resulted in protein bodies of relatively normal size and shape but at very low density. This indicates that correct stoichiometric ratio of the different zeins is essential for vitreous endosperm formation through correctly formed, concentrically organized protein bodies (Fig. 1).
MATERIALS AND METHODS
Constructs
The α-zein RNAi cassette harbored a synthesized gene (GenScript) containing 220-bp conserved regions derived from the GenBank accession numbers EU116446, EU116472, EU116497, and EU116477 for the 19- and 22-kD subfamilies. The synthetic gene was assembled into the hairpin element in the base vector pUC18-RNAi (gift from Heriberto Cerutti [University of Nebraska-Lincoln]), a plasmid that carries the second intron of the Arabidopsis (Arabidopsis thaliana) small nuclear ribonuclear protein D1 (locus At4g02840). The intron is delineated by a set of compatible cohesive end restrictions sites that facilitate assembly of inverted repeats. The hairpin structure was subsequently subcloned between the 27-kD γ-zein promoter and T35S terminator of transcription within the binary vector pPZP212 (Hajdukiewicz et al., 1994).
For genotyping plants, a primer in the Z1A, Z1B, Z1D, and Z1C gene (5′-GCAGCAAGGTTTCCTAGCAC) and T35S poly A sequence (5′-ATGCTCAACACATGAGCGAA) were used to give a 311-bp band.
For the 50-kD γ-, 16-kD γ-, and 15-kD β−zein RNAi, a synthetic gene was made and inserted with the 27-kD γ-zein promoter in pPZP212 as described above. For the synthetic gene, 212-bp regions were selected from GenBank accessions AF371263 (50-kD), DQ400403 (16-kD), and NM_001112534 (15-kD). For genotyping plants, a 27-kD γ-zein promoter primer (5′-GATGAACCATCGACGTGCTA) was used with a primer within the synthetic RNAi gene (5′-CCATGGGTGGACTCTACCAG) to give a 447-bp band.
For the 27-kD γ-zein RNAi, the entire ORF was amplified (forward primer 5′-ATGAGGGTGTTGCTCGTTGCC and reverse primer 5′-TCAGTGGGGGACACCGCC) for insertion into the RNAi hairpin structure. For genotyping primers in the 27-kD γ-zein, ORF (5′-ACTGATGCCTCAGGAACTCG) and T35S poly A sequence (5′-ATGCTCAACACATGAGCGAA) were used to give a 539-bp band.
For the pZ1C:27γ construct, the 27-kD ORF was inserted with the α-zein Z1C promoter in pPZP212. For genotyping, a primer with the Z1C promoter (5′-CGAAATCGAGTAGATGCCATA) was used with a 27-kD γ-zein ORF primer (5′-GGGCGGTTGAGTAGGGTAGT) to give a 486-bp band. Constructs were transformed into Agrobacterium tumefaciens and into immature HiII maize (Zea mays) embryos and regenerated using an established procedure (Sattarzadeh et al., 2010).
Genotyping, Zein Protein Analysis, and Fixation for Microscopy
Given the highly readily discernible effects of reducing individual zein species at the protein level, we used protein level as the criterion for selecting lines for TEM analysis. For RNAi lines with dominant phenotypes, opaque T2 kernels were planted. RNAi and the pZ1C:27γ plants were selected for the presence of the transgene using leaf genomic PCR using DNA extracted according to the urea method (Holding et al., 2008). For the 18-DAP stage, kernels were removed, without damaging the base of the kernels, using a razor blade and treated as follows. Kernels were placed embryo-side down on a clean glass plate and sliced in half longitudinally through the embryo. The embryo halves were placed immediately into DNA extraction buffer on ice, one-half of the endosperm was frozen in liquid nitrogen for protein analysis, and the other half kernel was dissected for fixation. Keeping the pericarp intact, the half kernel was placed cut-side down and a 1- to 2-mm longitudinal central section was taken and placed into 5 mL fixative (2% Suc, 0.1m sodium cacodylate, pH 7.4, and 5% glutaraldehyde). Samples were fixed at 4°C for at least 1 week while genotyping and SDS-PAGE of zeins were conducted to identify transgenic kernels for further processing. Embryo DNA was extracted using microfuge-scale urea DNA extraction and blue plastic pestles. Endosperm zeins were extracted as previously described (Wallace et al., 1990).
Zein Gene Expression Analysis
We extracted endosperm RNA, DNase treated, made complementary DNA, and measured the relative levels of all α- and γ-zein subclasses using qRT-PCR as described previously (Holding et al., 2011) with the following modifications. The control gene used in this case was tubulin. RNA was extracted from individual halves of developing endosperms from frozen kernels derived from plants already genotyped as described in the previous section. The other half was used for zein extractions to confirm identity (not shown), and each kernel was genotyped using the dissected embryo as described in the previous section (not shown but illustrated in kernels used for TEM and protein analyses; Fig. 4). In these analyses, we compared RNA from three kernels positive for the transgene(s) from the same ear and, thus, the same event to exemplify the effect of each transgene. While expression levels could vary between different events of each RNAi construct, the purpose here was to exemplify the effects of each transgene on the expression of all zeins. However, for the wild-type control RNA, each of the three kernels was taken from separate ears to be sure to exclude ear-to-ear variation that might otherwise bias the relative expression levels of all RNAi construct lines. We averaged the three technical replicates first, and then the final average is between the biological replicate kernels. Zein primers are listed in Supplemental Table S2.
TEM
Selected kernel segments were further dissected following fixation to remove the apical and basal kernel portions, leaving two 1-mm midkernel pieces with pericarp still attached for orientation. Samples were postfixed by 1% osmium tetroxide. Samples were then dehydrated in a graded series of ethanol (30%, 50%, 70%, 95%, and 100%), followed by 100% acetone. Dehydrated samples were infiltrated in Epon 812 resin (Electron Microscopy Sciences) using a resin:acetone (1:2 and 2:1) and 100% resin series before polymerization at 60°C overnight. Ninety-nanometer sections were cut using a Diatome diamond knife and a LKB Ultrotome III microtome and attached to carbon- and Formvar-coated grids (Electron Microscopy Sciences). Sections were stained with 4% lead citrate and 3% uranyl acetate and air dried before being viewed on a Hitachi 7500 TEM.
Immunogold TEM
Selected kernel segments were further dissected and dehydrated as for regular TEM, except that the osmium tetroxide post fixation and acetone infiltration were omitted, and the samples were infiltrated in LR White resin (Electron Microscopy Sciences) before sectioning as for TEM. For immunogold labeling, grids were hybridized in 50-μL drops on Parafilm strips as follows. The grids were incubated for 30 min in blocking solution (pH 8.2) containing 0.2% bovine serum albumin and 0.06% Tween 20 in 20 mm Tris-HCl and 500 mm NaCl. Rabbit primary antisera were added (α-zein at 1:2,000 and γ-zein at 1:100) and incubated overnight at 4°C before three 10-min washes in 100 μL blocking solution. Secondary antibody (goat anti-rabbit, 15 nm, gold-conjugated; Electron Microscopy Sciences) was added at 1:20 dilution for 1 h at room temperature. The grids were washed three times and rinsed three times in deionized water and air dried. Grids were poststained in 4% lead citrate and 3% uranyl acetate and air dried again before being viewed on a Hitachi 7500 TEM.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU116446, EU116472, EU116497, and EU116477 for the 19- and 22-kD alpha zein genes represented in the chimeric alpha zein RNAi gene. For the gamma zein genes represented in the chimeric RNAi gene, accession numbers are AF371263 (50-kD), DQ400403 (16-kD), and NM_001112534 (15-kD).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Purified zeins from transgenic mature kernels.
Supplemental Figure S2. Design of synthetic zein RNAi transgenes.
Supplemental Table S1. Segregation ratios of opaque versus vitreous kernels from T1 ears of selected α-zein RNAi events.
Supplemental Table S2. Primers for qRT-PCR of zein expression.
Supplementary Material
Acknowledgments
We thank Brian Larkins and Brenda Hunter for supplying the zein antisera.
Glossary
- ER
endoplasmic reticulum
- QPM
Quality Protein Maize
- TEM
transmission electron microscopy
- RNAi
RNA interference
- qRT
quantitative real-time reverse transcription
- DAP
d after pollination
- ORF
open reading frame
References
- Bagga S, Adams H, Kemp JD, Sengupta-Gopalan C. (1995) Accumulation of 15-kilodalton zein in novel protein bodies in transgenic tobacco. Plant Physiol 107: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Adams HP, Rodriguez FD, Kemp JD, Sengupta-Gopalan C. (1997) Coexpression of the maize δ-zein and β-zein genes results in stable accumulation of Δ-zein in endoplasmic reticulum-derived protein bodies formed by β-zein. Plant Cell 9: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Clore AM, Ranch JP, Higgins R, Lopes MA, Larkins BA. (1997) Expression of a mutant α-zein creates the floury2 phenotype in transgenic maize. Proc Natl Acad Sci USA 94: 7094–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Herman EM, Takasaki K, Larkins BA. (1996) The maize γ-zein sequesters α-zein and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. Plant Cell 8: 2335–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha KB, Lending CR, Lopes MA, Wallace JC, Larkins BA. (1991) opaque-2 modifiers Increase γ-zein synthesis and alter its spatial distribution in maize endosperm. Plant Cell 3: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geevers HO, Lake JK (1992) Development of modified opaque-2 maize in South Africa. In ET Mertz, ed, Quality Protein Maize. American Society of Cereal Chemists, St. Paul, pp 49–78 [Google Scholar]
- Gillikin JW, Zhang F, Coleman CE, Bass HW, Larkins BA, Boston RS. (1997) A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol 114: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng LN, Zhu J, Wang G, Tang YP, Chen HJ, Jin WB, Wang F, Mei B, Xu ZK, Song RT. (2009) Expressional profiling study revealed unique expressional patterns and dramatic expressional divergence of maize α-zein super gene family. Plant Mol Biol 69: 649–659 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Holding DR, Hunter BG, Chung T, Gibbon BC, Ford CF, Bharti AK, Messing J, Hamaker BR, Larkins BA. (2008) Genetic analysis of opaque2 modifier loci in quality protein maize. Theor Appl Genet 117: 157–170 [DOI] [PubMed] [Google Scholar]
- Holding DR, Hunter BG, Klingler JP, Wu S, Guo X, Gibbon BC, Wu R, Schulze JM, Jung R, Larkins BA. (2011) Characterization of opaque2 modifier QTLs and candidate genes in recombinant inbred lines derived from the K0326Y quality protein maize inbred. Theor Appl Genet 122: 783–794 [DOI] [PubMed] [Google Scholar]
- Holding DR, Larkins BA. (2006) The development and importance of zein protein bodies in maize endosperm. Maydica 51: 243–254 [Google Scholar]
- Holding DR, Larkins BA (2009) Zein storage proteins. In BA Larkins, AL Kriz, eds, Molecular Genetic Approaches to Maize Improvement. Springer Verlag, Berlin, pp 269–268 [Google Scholar]
- Holding DR, Meeley RB, Hazebroek J, Selinger D, Jung R, Larkins BA. (2010) Identification and characterization of the maize arogenate dehydrogenase gene family. J Exp Bot 61: 3663–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding DR, Messing J (2013) Evolution, structure, and function of prolamin storage proteins. In P Becraft, ed, Seed Genomics. John Wiley & Sons, New York, pp 139–158 [Google Scholar]
- Holding DR, Otegui MS, Li BL, Meeley RB, Dam T, Hunter BG, Jung R, Larkins BA. (2007) The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Frizzi A, Florida CA, Kruger DE, Luethy MH. (2006) High lysine and high tryptophan transgenic maize resulting from the reduction of both 19- and 22-kD α-zeins. Plant Mol Biol 61: 525–535 [DOI] [PubMed] [Google Scholar]
- Huang SS, Adams WR, Zhou Q, Malloy KP, Voyles DA, Anthony J, Kriz AL, Luethy MH. (2004) Improving nutritional quality of maize proteins by expressing sense and antisense zein genes. J Agric Food Chem 52: 1958–1964 [DOI] [PubMed] [Google Scholar]
- Kim CS, Gibbon BC, Gillikin JW, Larkins BA, Boston RS, Jung R. (2006) The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response. Plant J 48: 440–451 [DOI] [PubMed] [Google Scholar]
- Kim CS, Hunter BG, Kraft J, Boston RS, Yans S, Jung R, Larkins BA. (2004) A defective signal peptide in a 19-kD α-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol 134: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Woo Ym YM, Clore AM, Burnett RJ, Carneiro NP, Larkins BA. (2002) Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. Plant Cell 14: 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending CR, Larkins BA. (1989) Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA. (1991) γ-Zein content is related to endosperm modification in Quality Protein Maize. Crop Sci 31: 1655–1662 [Google Scholar]
- Lopes MA, Larkins BA. (1995) Genetic-analysis of opaque2 modifier gene activity in maize endosperm. Theor Appl Genet 91: 274–281 [DOI] [PubMed] [Google Scholar]
- Mertz ET, Bates LS, Nelson OE. (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145: 279–280 [DOI] [PubMed] [Google Scholar]
- Moro GL, Lopes MA, Habben JE, Hamaker BR, Larkins BA. (1995) Phenotypic effects of opaque2 modifier genes in normal maize endosperm. Cereal Chem 72: 94–99 [Google Scholar]
- Myers AM, James MG, Lin QH, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW. (2011) Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega EI, Bates LS. (1983) Biochemical and agronomic studies of two modified hard-endosperm opaque-2 maize (Zea-mays-L) populations. Cereal Chem. 60: 107–111 [Google Scholar]
- Paez AV, Helm JL, Zuber MS. (1969) Lysine content of opaque-2 maize kernels having different phenotypes. Crop Sci 9: 251–253 [Google Scholar]
- Sattarzadeh A, Fuller J, Moguel S, Wostrikoff K, Sato S, Covshoff S, Clemente T, Hanson M, Stern DB. (2010) Transgenic maize lines with cell-type specific expression of fluorescent proteins in plastids. Plant Biotechnol J 8: 112–125 [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. (1990) Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci USA 87: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Song RT, Messing J. (2003) A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RT, Messing J. (2003) Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci USA 100: 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasal SK, Villegas E, Bjarnason M, Gela W, Goertz P (1980) Genetic modifiers and breeding strategies in developing hard endosperm opaque2 materials. In WG Pollmer, RH Phillips, eds, Quality Traits of Maize for Grain and Silage Use. Martinus Nijhoff, London, pp 37–73 [Google Scholar]
- Wallace JC, Lopes MA, Paiva E, Larkins BA. (1990) New methods for extraction and quantitation of zeins reveal a high content of γ-zein in modified opaque-2 maize. Plant Physiol 92: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Wang F, Wang G, Wang F, Zhang XW, Zhong MY, Zhang J, Lin DB, Tang YP, Xu ZK, et al. (2012) Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24: 3447–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JD, Galili G, Larkins BA, Gelvin SB. (1988) The synthesis of a 19 kilodalton zein protein in transgenic petunia plants. Plant Physiol 88: 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Hu DWN, Larkins BA, Jung R. (2001) Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Holding DR, Messing J. (2010) γ-Zeins are essential for endosperm modification in quality protein maize. Proc Natl Acad Sci USA 107: 12810–12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Messing J. (2010) RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol 153: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YR, Goettel W, Messing J. (2009) Non-Mendelian regulation and allelic variation of methionine-rich Δ-zein genes in maize. Theor Appl Genet 119: 721–731 [DOI] [PubMed] [Google Scholar]
- Wu YR, Messing J. (2011) Novel genetic selection system for quantitative trait loci of quality protein maize. Genetics 188: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.