Reproductive organ development of the moss Physcomitrella patens is affected by two SHI/STY genes and the plant hormone auxin.
Abstract
In order to establish a reference for analysis of the function of auxin and the auxin biosynthesis regulators SHORT INTERNODE/STYLISH (SHI/STY) during Physcomitrella patens reproductive development, we have described male (antheridial) and female (archegonial) development in detail, including temporal and positional information of organ initiation. This has allowed us to define discrete stages of organ morphogenesis and to show that reproductive organ development in P. patens is highly organized and that organ phyllotaxis differs between vegetative and reproductive development. Using the PpSHI1 and PpSHI2 reporter and knockout lines, the auxin reporters GmGH3pro:GUS and PpPINApro:GFP-GUS, and the auxin-conjugating transgene PpSHI2pro:IAAL, we could show that the PpSHI genes, and by inference also auxin, play important roles for reproductive organ development in moss. The PpSHI genes are required for the apical opening of the reproductive organs, the final differentiation of the egg cell, and the progression of canal cells into a cell death program. The apical cells of the archegonium, the canal cells, and the egg cell are also sites of auxin responsiveness and are affected by reduced levels of active auxin, suggesting that auxin mediates PpSHI function in the reproductive organs.
In flowering plants, the gametophyte generation is reduced while the main plant body, including organs for reproduction, is formed by the diploid sporophyte generation. In Arabidopsis (Arabidopsis thaliana), the female gametophyte is restricted to only seven cells, including the egg and the binuclear central cell, and the male gametophyte is even more reduced, consisting of one vegetative cell and two generative cells (Berger and Twell, 2011). In contrast, mosses and other bryophytes, separated from the flowering plants by 450 million years (Rensing et al., 2008), are believed to represent extant remnants of the ancestral land plants in that they are gametophyte dominant with haploid reproductive organs (Wellman et al., 2003). In the model moss Physcomitrella patens, multiple male and female reproductive organs form at the tip of the gametophyte shoot in response to low temperature and short daylength (Hohe et al., 2002). Male ovoid antheridia produce slender biflagellated sperm cells, which access the egg cell of the pear-shaped female archegonia through an extended canal during fertilization (Kofuji et al., 2009).
It is not trivial to picture how the ancestral mode of sexual reproduction in land plants, mimicked by bryophytes, has evolved into that of flowering plants of today. However, based on their specialization in reproductive functions, traits homologous to those of flowering plant gametophytes could be expected to primarily exist in the reproductive organs of bryophytes. If so, remnants of common genetic and hormonal programs underlying the development of flowering plant gametophytes and bryophyte reproductive organs could be expected. Although the extent of recruitment of ancestral gametophytic programs into the sporophyte generation of flowering plants is not clear (Nishiyama et al., 2003; Bowman et al., 2007), examples have been reported (Menand et al., 2007; Saleh et al., 2011); hence, it is also possible that homologies in the initiation, morphogenesis, and function of reproductive organs of mosses and flowering plants may exist despite their occurrence in different generations.
Recently, Sakakibara et al. (2013) demonstrated that inactivation of moss class 2 KNOTTED LIKE HOMEOBOX (KNOX2) transcription factors induce the development of gametophyte leafy shoots from diploid embryos without meiosis, suggesting that PpKNOX2 acts to prevent the haploid-specific body plan from developing in the diploid phase. This result is intriguing, as it may suggest that loss of such a pathway potentially could have aided in the recruitment of haploid genetic tool kits into the diploid generation.
Candidate programs for reproductive development that potentially could be shared by flowering plants and mosses include those dependent on the plant hormone auxin, as it is ancient in its origin (Paponov et al., 2009) and plays important roles during the development of Arabidopsis gametophytes. Auxin is known to trigger physiological responses also in bryophytes (Johri and Desai, 1973; Ashton et al., 1979; Fujita et al., 2008), and homologs to seed plant genes involved in auxin synthesis, transport, perception, and signaling exist in P. patens (Rensing et al., 2008; Paponov et al., 2009; Eklund et al., 2010b; Prigge et al., 2010). Indications of the importance of auxin for gametophyte development in flowering plants include an auxin gradient, most likely established by the local activity of YUCCA (YUC) biosynthesis genes, important for cell type specification in the syncytial phase of the Arabidopsis female gametophyte (Pagnussat et al., 2009). Auxin also appears crucial for male gametophyte development, as mutations in genes encoding TRANSPORT INHIBITOR RESPONSE1 (TIR1)-related auxin receptors result in premature pollen maturation (Cecchetti et al., 2008). Furthermore, two endoplasmic reticulum-localized auxin transporters hypothesized to regulate intracellular auxin homeostasis were recently shown to be important for male gametophyte development (Dal Bosco et al., 2012; Ding et al., 2012).
While auxin transport facilitator genes exist in both bryophytes and flowering plants, it has been speculated that local biosynthesis may have a prominent role for the establishment of auxin maxima in the gametophyte generations, possibly reflecting the situation in early land plants (Fujita et al., 2008; Pagnussat et al., 2009; Eklund et al., 2010b). We have shown that auxin biosynthesis is regulated by SHORT INTERNODE/STYLISH (SHI/STY) family genes in both P. patens and Arabidopsis and that the Arabidopsis SHI/STY transcription factors directly bind to the promoters of YUC auxin biosynthesis genes and activate their expression (Sohlberg et al., 2006; Ståldal et al., 2008; Eklund et al., 2010a, 2010b).
Here, we have utilized PpSHI1 and PpSHI2 reporter and knockout lines (Eklund et al., 2010b), the auxin reporters GmGH3pro:GUS and PpPINApro:GFP-GUS, and the auxin-conjugating transgene PpSHI2pro:IAAL to study the role of auxin during reproductive development in P. patens. We have also added necessary resolution to previous descriptions of reproductive development in this moss (Kofuji et al., 2009). By utilizing this new reference information in our analysis of transgenics, we suggest that the PpSHI genes, and by inference also auxin, play important roles for reproductive organ development in moss. These findings are discussed in an evolutionary context with special reference to auxin-dependent regulation of the gametophyte generation and reproductive organs in flowering plants.
RESULTS
P. patens Reproductive Organ Formation Is Highly Organized But Differs in Phyllotactic Pattern from the Vegetative Shoot
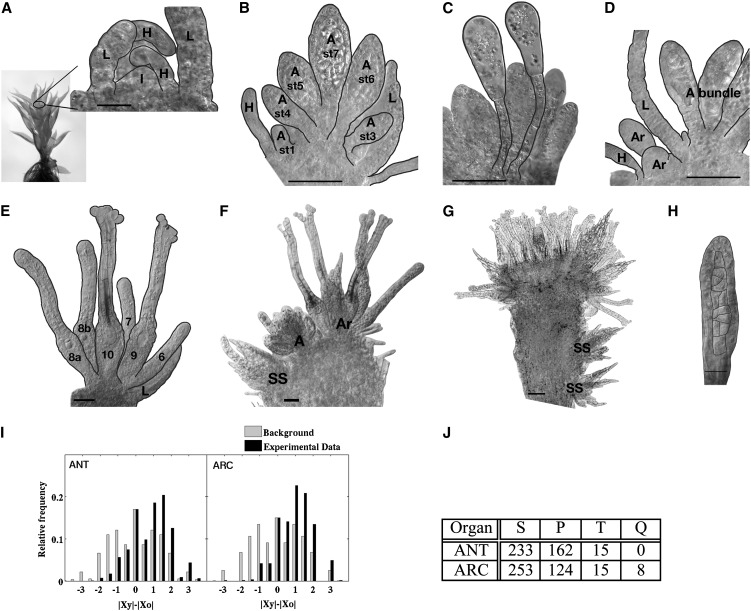
During vegetative shoot development, a single tetrahedral apical cell cleaves off daughter initials spirally, which gives rise to new leaf primordia (Harrison et al., 2009). Here, we show that the timing of the initiation and positioning of reproductive organ primorida during the first weeks after transition to an inductive environment (8 h of light/16 h of dark; 15°C) is highly reproducible in the conditions described in “Materials and Methods.” The first reproductive organ initial, giving rise to a male antheridium, forms in the very center of the primary shoot apex surrounded by young leaves and axillary hairs 6 or 7 d post induction (dpi; Fig. 1A). However, the primordium that follows is not formed in the center of the shoot but instead outside of the first antheridium, suggesting that the apical cell of the shoot already has produced a number of initials before entering into reproductive development or that new meristematic centers are initiated on the flanks of the first antheridium. This is true also for archegonia, where new initials form at the flanks of the previously initiated archegonium. As a consequence, the oldest organs are found in the middle of the bundle, while the youngest are detected in the bundle periphery (Fig. 1, B and E).
Figure 1.
Wild-type reproductive organ initiation, organization, and positioning at the gametophore apex. A to H, DIC micrographs. Several leaves have been removed to reveal the reproductive organs. The focus plane is set approximately to the center of the longitudinal axis of the gametophore. A, First antheridium initial is formed. B, Antheridial bundle. C, Paraphyses. D, Archegonia initiated outside the antheridial bundle. E, Archegonial bundle. F, Secondary shoot with reproductive organs initiated to the left of the primary antheridial bundle. G, Gametophore apex, 10 weeks post initiation. H, Organ of mixed antheridial/archegonial identity. A, Antheridium; Ar, archegonium; H, hair; I, initial; L, leaf; SS, secondary shoot; st, stage (numbers define developmental stage; Fig. 2). Black lines mark cell or organ margins. Bars = 20 μm in A and H, 50 μm in B to F, and 100 μm in G. I and J, Statistical distributions of organ positions. I, Distribution showing the difference of distances from the center for the younger (|xy|) and the older (|xo|) organs in all pairs within the data set. This measure is positive, zero, or negative when the younger organ is farther from, at the same distance as, or closer to the center, respectively. The background distribution is generated by including all distance differences in the data set, regardless of the age of organs. For both antheridia and archegonia, there is a significant signal for positive values (P < 10−16). J, Part of the total number of organs appearing as singlets (S; one of the same stage), pairs (P; two at the same stage), triplets (T; three at the same stage), and quadruples (Q; four at the same stage) in bundles of antheridia (ANT) and archegonia (ARC) analyzed.
To confirm this pattern, we entered information about the age and position of individual organs for a large number of male and female bundles onto two-dimensional grids to allow for measurements and statistical analyses. Indeed, we found that a majority of younger (early stage) organs are present farther away from the center compared with older (late stage) organs (Fig. 1I). Most commonly, organs appear as singlets, but for 40% of the antheridia and 30% of the archegonia (Fig. 1J), organs of the same developmental stage appear as pairs, indicating that they are initiated at the same time during development.
Fourteen days after the first antheridium initial has formed (20 dpi), the primary antheridial bundle has, on average, 12 antheridia and one or two clavate paraphyses that are fully developed at 16 dpi. While paraphysis is a collective term describing a diverse set of sterile erect organs interspersed among reproductive organs in mosses and related plants, the paraphyses of P. patens and other Funariaceae species (Campbell, 1918; Parihar, 1966) have a very characteristic appearance, with a stalk, consisting of two to three elongated cells, on which a swollen apical cell with plenty of well-developed chloroplasts is situated (Fig. 1C). The first female archegonium is initiated just outside the antheridial bundle at 11 dpi, separated from the antheridia by at least one leaf (Fig. 1D), and additional archegonia are then initiated in a pattern similar to the primary antheridia, albeit at a slower pace. On average, only four archegonia can be found in the developing bundle 14 d after the first archegonium formed. Secondary shoots start to emerge in the axils of leaves at positions close to the apex from 28 dpi, often under the apical antheridial bundle at the opposite side of the primary archegonial bundle (Fig. 1F) or farther down the gametophore. After the development of only a few leaves, reproductive organ bundles form. At this time, new antheridia also form in the periphery of the primary archegonial bundle of the primary shoot. As a result, a mixture of leaves, antheridia, and archegonia is found in the main shoot apex a few weeks later (Fig. 1G). Furthermore, malformed archegonia as well as organs exhibiting characters of both antheridia and archegonia, a feature previously described also in several other mosses (Crum, 2001), are initiated at a low frequency in the archegonial bundles (compare Fig. 1H with stage 5 antheridium and archegonium in Fig. 2).
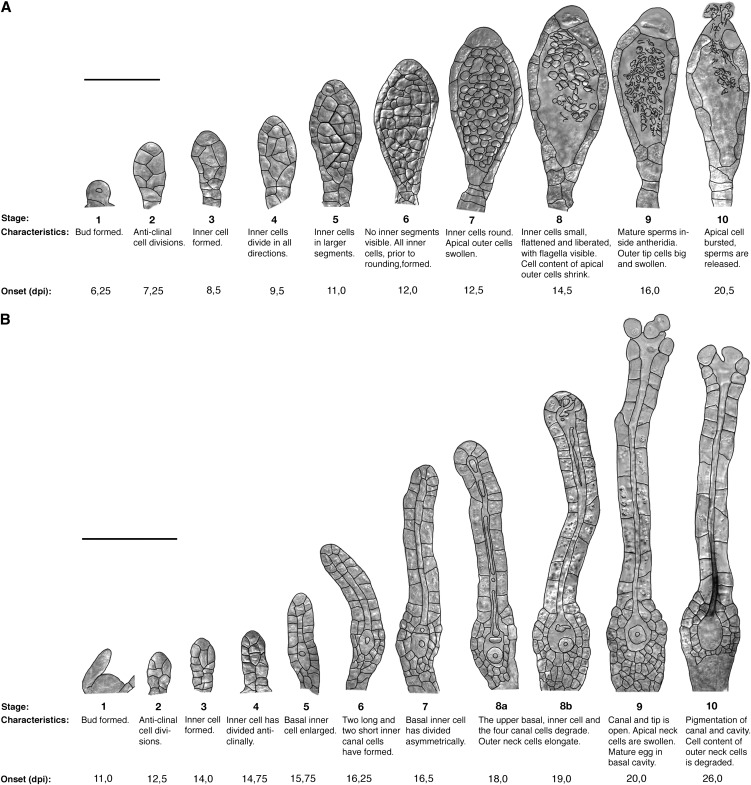
Figure 2.
Lateral view of stages defined for the development of P. patens reproductive organs, characteristics for each stage, and time to first organ formed. A, Male antheridia. B, Female archegonia. Black lines mark cell margins. Onset is the first day the specified stage was detected, estimated to the nearest 6 h. Bars = 50 μm in A and 100 μm in B.
In conclusion, the best time to study antheridia and archegonia is early during the reproductive phase, before the mixture of organs and organ identities occurs. For antheridia, 6 to 12 dpi is a good time, when no archegonia have yet been formed. Analysis of archegonia development should be performed during the formation of the first cluster (11–28 dpi), when proper archegonia, separated from antheridia by position and at least one leaf, can be readily observed. These are the stages that have been used to characterize the details of antheridia and archegonia development presented below.
Antheridial Morphogenesis Can Be Divided into 10 Discrete Stages
P. patens reproductive organ morphogenesis has previously only been described briefly (Kofuji et al., 2009). Here, we have divided the developmental program into well-defined and discrete spatial and temporal stages that are easily identifiable by light microscopy (Fig. 2; Supplemental Table S1). This division is a prerequisite for the detailed analysis of regulatory pathways controlling reproductive organ development in P. patens and has allowed us to conclude that reproductive organ morphogenesis in P. patens resembles that of mosses in general (Crum, 2001; Goffinet et al., 2009) and that of other Funariaceae species in particular (Campbell, 1918; Parihar, 1966; Lal and Bhandari, 1968). Figure 2A describes P. patens antheridium development. A bud-shaped initial cell (stage 1) and its daughter cells undergo a number of anticlinal divisions, forming a primordium with an increasing number of cells arranged in two files (stage 2). At stage 3, cells in the upper part divide periclinally, forming inner spermatogenous cell initials and an outer surrounding sterile cell layer. Inner cells divide in all directions (stage 4) and produce a few larger segments of cells (stage 5). Outer cells divide to keep up with the increasing size of the antheridium (stages 5 and 6). In the next stage (7), the inner cells acquire a round appearance. The outer cell layer initiates the production of a yellow pigment, and the apical-most cells start to swell and to confine the cell content to a small part of the cell. At stage 8, the inner cell mass has undergone the final cell division, giving rise to spermatids that now undergo spermatogenesis. At stage 9, slender, coiled, and biflagellated mature sperms have formed and the antheridium tip cells are round, big, and appear devoid of cell content. In the final stage (10), the swollen tip cells burst and the sperm cells are released, leaving the cavity empty. As a result, the antheridium collapses. Cells in the outer cell layer are now heavily pigmented (yellow-brownish) and appear empty. Under our growth conditions, the progress from the emergence of bud-shaped initials to sperm release takes 14 d, the first six stages lasting about 1 d each, while the last four stages, during which sperm development takes place, continues for 8 d (Fig. 2A; Supplemental Table S1).
Archegonial Development Also Follows a Pattern with Well-Defined Stages
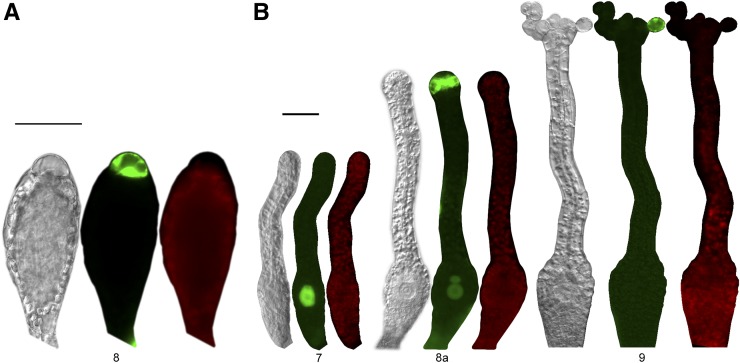
The first archegonium initiates as a dome-shaped protrusion (Fig. 2B, stage 1) flanked by axillary hairs and young leaves, and the first stages of development resemble those of early antheridia development. Cells divide primarily anticlinally, forming an archegonium primordium containing 10 to 14 cells (stage 2). However, whereas the primary inner cell formed during stage 3 divides in all directions in the antheridium, it divides strictly anticlinally in the archegonium (stages 4–6), resulting in five inner cells in a row. The basal-most inner cell, the egg cell precursor, enlarges (stage 5), while the four remaining inner cells form two elongated and slender basal cells and two shorter and wider apical canal cells (stage 6). In stage 7, the egg cell precursor divides asymmetrically, resulting in a large lower cell, later becoming the egg cell, and a small upper cell. The outer cells in the basal part of the developing archegonium divide both anticlinally and periclinally and produce two to three cell layers on the sides, and several cell layers below, of what is to become the egg cell cavity. The canal cell file of the apical elongated neck is surrounded by one layer of elongating outer cells, which has formed from anticlinal divisions. During stages 8a and 8b, the canal cells and the small upper basal cell undergo degradation. The protoplast retracts from the cell wall, which eventually degrades. Remnants of the two apical canal cells are still observed late at stage 8b, when a collapse of one or two of the archegonial tip cell inner walls facing the canal occurs. In parallel with the degradation of the upper basal cell at stage 8, the egg cell matures, becomes round, and by detachment from the surrounding cells is freed into the cavity that forms. At stage 9, the archegonium is fully developed and the apex has opened, exposing a canal down to the egg cavity. The process from dome formation to a mature stage 9 archegonium takes, on average, 9 d. The first archegonium that develops reaches stage 9 at 20 dpi, at the same time as the first antheridium bursts and releases the sperms (Fig. 2; Supplemental Table S1). Apical cells forming the opening of the canal have a distinct round, swollen appearance and little cell content. Chloroplasts are degraded in the neck starting at the apex, possibly marking the onset of cell death in the outer neck cells. In the last stage (10), a yellow-brownish pigment accumulates in the canal, and the outer neck cells completely lose their cell contents.
The PpSHI Genes Show Strict Spatial and Temporal Expression during Both Antheridial and Archegonial Development
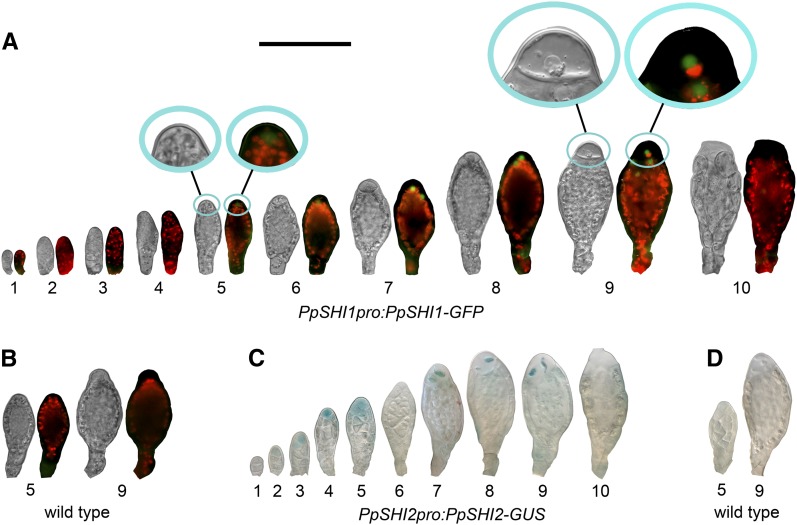
Using the PpSHI1pro:PpSHI1-GFP and PpSHI2pro:PpSHI2-GUS knockin reporter lines (Eklund et al., 2010b) as well as our new PpSHI1pro:PpSHI1-GUS knockin lines (see “Materials and Methods”), we could conclude that the two PpSHI genes show developmentally regulated and strongly overlapping expression during reproductive organ proliferation (Figs. 3 and 4; Supplemental Figs. S1–S3; Supplemental Table S2). As expected, the PpSHI1-GFP fusion protein showed a mostly nuclear localization. The PpSHI genes are active in the apical cells of the antheridia both during organ establishment (stages 2–5) and sperm maturation (stages 7–10), while there is a transient drop of expression at stage 6 (Fig. 3, A and C; Supplemental Fig. S1). Expression of the two genes in antheridia largely overlaps, although PpSHI1 expression shows a delayed onset and appears weaker, as indicated by the faint signals in the PpSHI1pro:PpSHI1-GFP line and the lack of consistent reporter activity in antheridia of the PpSHI1pro:PpSHI1-GUS line (data not shown). Although we could detect PpSHI2pro:PpSHI2-GUS activity in the early stages of antheridia formation (stages 2 and 3; Fig. 3C), reporter activity was not evident at the organ initiation site or at stage 1. However, in accordance with the expression of PpSHI genes during vegetative organ development, we found PpSHI2pro:PpSHI2-GUS, PpSHI1pro:PpSHI1-GUS, and PpSHI1pro:PpSHI1-GFP expression in axillary hairs surrounding the newly initiated reproductive organ primordium (Supplemental Fig. S4). This may suggest that PpSHI-related processes in the axillary hairs play a similar role during the initiation of both vegetative (Eklund et al., 2010b) and reproductive organs. No expression was detected in the mature clavate paraphyses in the antheridial bundles (Supplemental Fig. S5).
Figure 3.
PpSHI expression at different stages of antheridial development. A, PpSHI1pro:PpSHI1-GFP expression. B, Selected stages exemplifying the lack of a GFP signal in wild-type organs. C, PpSHI2pro:PpSHI2-GUS expression. D, Selected stages exemplifying the lack of a GUS signal in wild-type organs. For each developmental stage in A and B, both a light micrograph and an overlay image with GFP fluorescence (green) and chloroplast autofluorescence (red) are shown. Insets in A show magnified organ apices of selected stages. Bar = 100 μm.
Figure 4.
Expression of the PpSHI genes and the GH3 auxin-response reporter during different stages of archegonial development. A, PpSHI1pro:PpSHI1-GUS expression. B, PpSHI2pro:PpSHI2-GUS expression. C, GmGH3pro:GUS expression. D, Selected stages exemplifying the lack of a GUS signal in wild-type organs. Bar = 100 μm.
PpSHI2pro:PpSHI2-GUS is expressed in the apex also of young developing archegonia from at least stage 4 (Fig. 4B; Supplemental Figs. S2 and S3), which may suggest that the PpSHI genes play roles also in female reproductive organ morphogenesis. Apical expression of PpSHI1 in archegonial neck tip cells appears delayed compared with that of PpSHI2, just as in antheridia. However, we cannot exclude that PpSHI1 could be activated earlier, as the apical signals in the PpSHI1pro:PpSHI1-GFP line was very weak and only detected in less than 30% of the archegonia (Supplemental Fig. S3), and we could not find a consistent reporter activity in these cells in the PpSHI1pro:PpSHI1-GUS line. Both PpSHI genes are strongly expressed in the central cell file, consisting of the canal, upper basal, and egg cells. The onset of PpSHI2 expression again precedes that of PpSHI1, and after activation, both genes remain active in the canal cells and the upper basal cell until they degrade at stage 8 and in the egg cell throughout development. PpSHI1 differs from PpSHI2 by being expressed in the outer cells of the neck during stages 7 to 10, while PpSHI2 alone showed a low incidence of expression in the swollen cells lining the apical opening at stage 10.
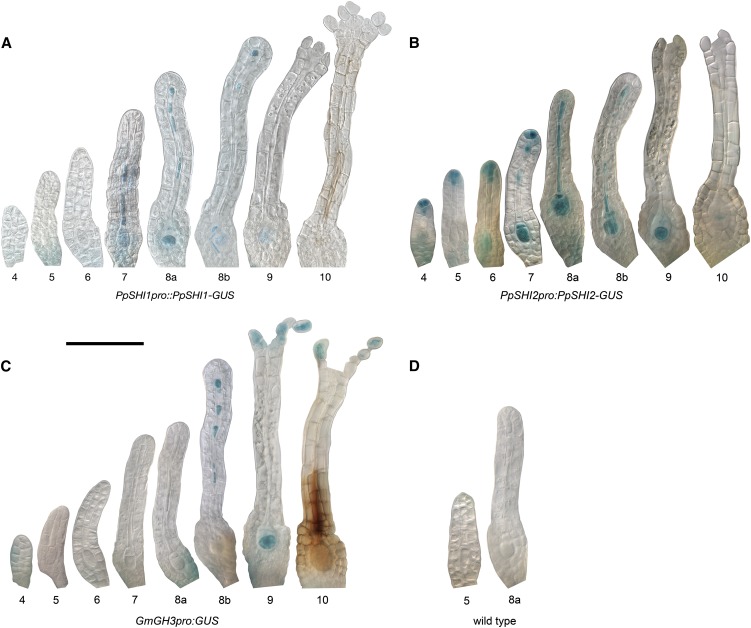
PpPINApro:GFP-GUS and GmGH3pro:GUS Expression Overlaps with the PpSHI Gene Activities during Archegonial Development, although GmGH3 Expression Is Delayed Compared with the PpPINA and PpSHI Reporters
Because the activity level of PpSHI genes affects auxin concentration in the vegetative shoot (Eklund et al., 2010b), we wanted to compare the PpSHI gene expression domains with those of other auxin-related genes. Therefore, we studied the activity of the auxin response reporter GmGH3pro:GUS (Bierfreund et al., 2003; Fujita et al., 2008) and of PpPINApro:GFP-GUS during reproductive organ development. The PIN-FORMED (PIN) family of auxin efflux carriers comprises four members in moss (Viaene et al., 2013), and based on their respective expression levels in tissues, referred to as “gametangia development” (Hruz et al., 2008), we selected the promoter of PpPINA for transcriptional studies in reproductive organs by creating a moss PpPINApro:GFP-GUS reporter line. It was suggested previously that the PpPINA protein localizes to the endoplasmic reticulum, where it may be involved in regulating cell-autonomous auxin homeostasis (Mravec et al., 2009); thus, we could expect the PpPINA gene to be active at auxin biosynthesis sites in moss. In developing antheridia, PpPINA promoter-driven GFP expression could be detected in the swollen apical cells at a short period during sperm maturation, lasting from late stage 8 to early stage 9 (Fig. 5A). PpPINA is also expressed in apical archegonial cells shortly before the reproductive organs open and remains active after opening at stage 9. In addition, we detect PpPINA activity in the maturating egg cell as well as in the upper basal cell (Fig. 5B; Supplemental Fig. S3; Supplemental Table S2). Although we could not detect any clear staining of GmGH3pro:GUS in antheridia or the paraphyses connected to them, expression was detected in axillary hairs, in the archegonial egg cell from stage 9, in the degrading canal cells, and in the swollen archegonial cells lining the apical opening at stage 10 (Fig. 4C; Supplemental Figs. S3, S4C, and S5C; Supplemental Table S2). These expression patterns overlap with that of PpSHI genes, but they are, especially for the auxin response reporter, initiated at later stages, suggesting that the PpSHI genes induce auxin biosynthesis at auxin transport and response sites during reproductive organ development.
Figure 5.
Stages of reproductive organ development exhibiting a GFP signal in the PpPINApro:GFP-GUS gene reporter line. A, Antheridia. B, Archegonia. For each developmental stage, a light micrograph, an image with GFP fluorescence (green), and an image with chloroplast autofluorescence (red) are shown. Bars = 50 μm.
Knockout of One of Two PpSHI Genes Results in Developmental Arrest in Both Male and Female Reproductive Organs
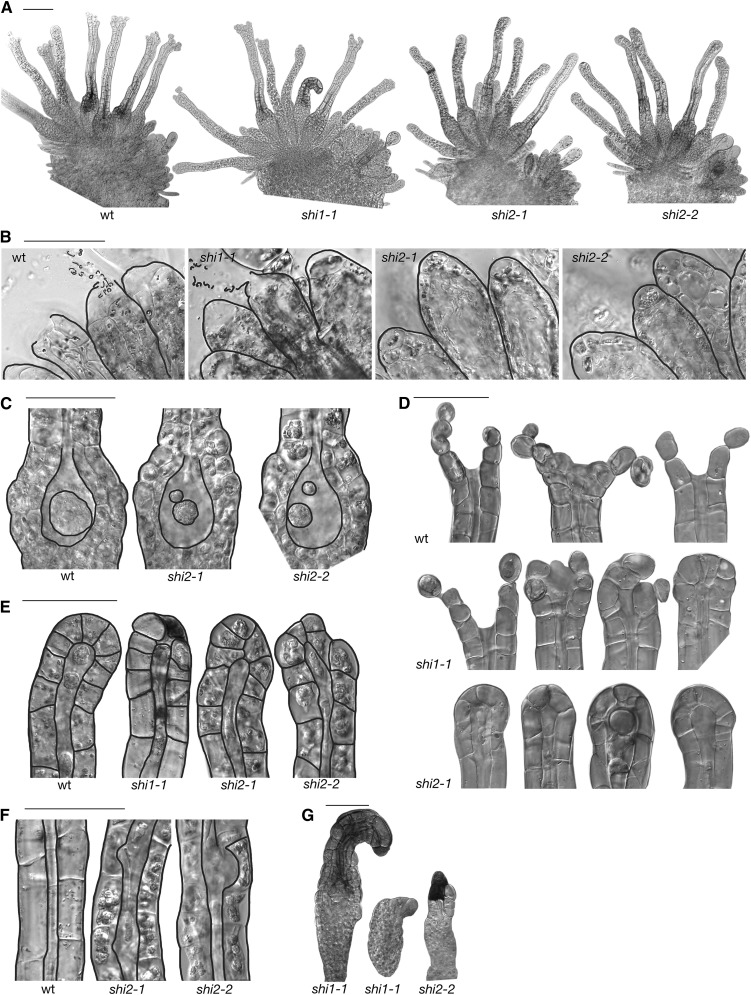
We next studied the reproductive development of three PpSHI knockout lines (Ppshi1-1, Ppshi2-1, and Ppshi2-2; Eklund et al., 2010b). In all lines, the initiation of antheridia and archegonia follows the same general pattern as in the wild type (Fig. 6A). During the first stages of development of antheridia, the Ppshi1 and Ppshi2 lines are indistinguishable from the wild type. However, Ppshi2-1 and Ppshi2-2 fail to open the antheridial apex (analyzed at 28 dpi [Supplemental Table S3], 39 dpi [Fig. 6A], and 67 dpi [data not shown]), correlating with a reduced swelling of the apical cell, and as a consequence, no sperms are released (Fig. 6B). In Ppshi1-1, the swelling of the apical cells is similar to that in the wild type, and most antheridia open, although a minor fraction fails (Supplemental Table S3).
Figure 6.
Reproductive organ defects of the Ppshi1-1, Ppshi2-1, and Ppshi2-2 single knockouts. A, Overview of the reproductive organs at the gametophore apex, 39 dpi. B, Mature antheridia at stage 10, 28 dpi. C, Basal part of archegonia at stage 9, 28 dpi. D, Archegonial apices at stage 10, 39 dpi. E, Archegonial apices at stage 8. F, Part of the archegonial neck showing the central canal at stage 8b. G, Malformed archegonia. All images are DIC micrographs. wt, Wild type. Bars = 100 μm in A and 50 μm in B to G.
Similarly, no deviations from the wild type could be detected during early archegonial development (stages 1–7). However, at stage 8, degradation of the upper basal cell in the archegonium fails in the Ppshi2 lines, resulting in an extra cell in the egg cavity (Fig. 6C; Supplemental Table S3). In addition, the development of the egg cell appears arrested, and the apical opening that normally occurs at the transition to stage 9 fails, leaving the apex closed throughout development (Fig. 6D; Supplemental Table S3). We have analyzed the apices at 28 dpi (Supplemental Table S3), 39 dpi (Fig. 6A), and 67 dpi (data not shown) without detecting any fully open archegonia in Ppshi2-1 or Ppshi2-2, whereas the relative abundance of stage 8b archegonia is steadily increased with time. In Ppshi1-1, opening of the apex does occur in several of the archegonia, but the surrounding neck cells do not swell to the same extent as in the wild type. In 3%, 30%, 59%, and 55% of the mature wild-type, Ppshi1-1, Ppshi2-1, and Ppshi2-2 archegonia, respectively, we see malformation of the canal, such as widening at positions where cross walls of the canal cells were previously situated, as well as malformed tips (Fig. 6, E and F). In addition, we observe a low frequency of severely malformed archegonia in both Ppshi1 and Ppshi2 (Fig. 6G) at 28 dpi. Degradation of chloroplasts in the wild-type neck cells starts from the organ apex, while it is initiated farther down in the two Ppshi2 lines, leaving the apical part green for a longer time (data not shown).
In summary, both Ppshi1 and Ppshi2 have defects in the formation of the archegonia canal, egg cell maturation, degradation of the upper basal cell, and opening of the apical tips of both archegonia and antheridia. These defects correlate well with the expression domains of the two genes and also in the observed delay and weaker signal strength of PpSHI1 in the apical cells, resulting in a more severe opening defect in Ppshi2 mutant lines. These data suggest that when one of the PpSHI genes is deleted, archegonial development is completely or partially arrested at late stage 8.
Inactivation of Auxin at PpSHI Expression Sites Mimics the PpSHI Knockouts
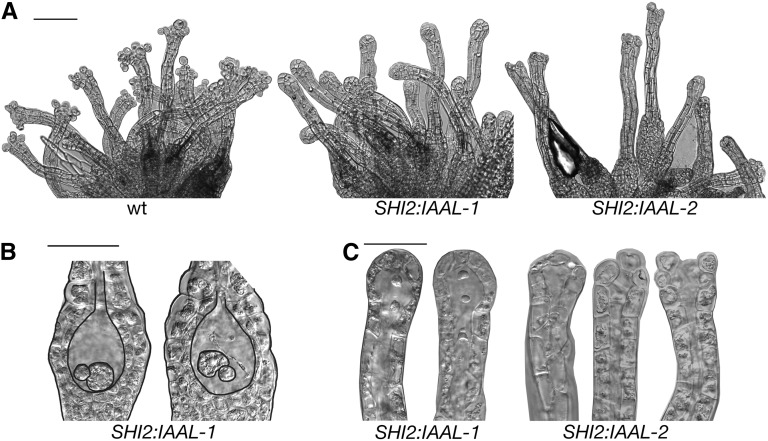
To test if the arrest in Ppshi archegonia development could be a result of reduced auxin levels in PpSHI expression domains, we produced moss lines expressing the Pseudomonas syringae subspecies savastanoi indole-3-acetic acid-lysine synthetase (iaaL) gene (Glass and Kosuge, 1986; Romano et al., 1991), encoding an auxin-conjugating enzyme converting free auxin to inactive Lys conjugates, from the PpSHI2 promoter. Two out of five verified transgenic lines showed phenotypic deviations from the wild type, and the severity of the deviations correlated with the expression level of PpSHI2pro:IAAL, analyzed by real-time quantitative PCR (Fig. 7; Supplemental Table S3; Supplemental Fig. S6). PpSHI2pro:IAAL-1 archegonia fail to degrade the upper basal cell at stage 8, and remnants of the canal cells can also be observed late in development. The egg cell is small and appears arrested in development (Fig. 7B). Additionally, opening of the apices fails, leaving the archegonia closed. Apices were analyzed both 32 and 72 dpi (Supplemental Table S3; Fig. 7, A and C) without detecting any fully open archegonia. The effect on archegonia development in the PpSHI2pro:IAAL-2 line is more subtle, and we only detected defects in opening of the apex. A fraction of the archegonia fail to open, and many open only partially (Fig. 7, A and C). In summary, PpSHI2pro:IAAL-1 archegonia phenocopy those of Ppshi2 and PpSHI2pro:IAAL-2 archegonia resemble those of Ppshi1, suggesting that the observed defects in the PpSHI knockout reproductive organs are caused by a reduction in free auxin levels and that high levels of active auxin in the PpSHI expression domains are required for the differentiation of egg, ventral, apical, and canal cells.
Figure 7.
Archegonial defects of the PpSHI2pro:IAAL lines. A, Overview of archegonia, 72 dpi. B, Basal part of archegonia, 32 dpi. C, Archegonial apices, 32 dpi. All images are DIC micrographs. wt, Wild type. Bars = 100 μm in A and 50 μm in B and C.
DISCUSSION
Here, we report the role of PpSHI genes and auxin during reproductive organ development. As this relies heavily on a reference framework for the detailed analysis of reproductive development in P. patens, we also report temporal and positional information of organ initiation and the division of antheridial and archegonial development into discrete numbered stages. Such reference information is crucial for unraveling the mechanisms underlying reproductive development, as it allows standardized descriptions of expression patterns and facilitates the identification of subtle deviations from normal development.
The Formation of P. patens Primary Male and Female Reproductive Organs Is Highly Ordered, and Their Maturation Is Synchronized
Monoecious mosses can be classified based on the positioning of reproductive organ clusters on the leafy shoot and the occurrence of male and female organs in the same or in separate such clusters (Crum, 2001; Goffinet et al., 2009). Examination of a P. patens leafy shoot kept in inductive conditions for 10 weeks (Fig. 1G) offers a view where male and female organs are found interspersed in the same clusters (synoicy), positioned both in the apex of the main shoot (acrocarpy) and on reduced lateral branches. However, by following the progress from the very onset of reproductive development, we here show that the road toward this rather complex situation is highly ordered. It passes transient stages where male and female organs are found in distinct and clearly separated clusters at the apex of the main shoot, while the mixing of organ types within the same clusters and reproductive development in secondary branches follows only later.
Reproductive development in P. patens starts with the initiation of male antheridia, while the onset of female archegonia formation is evident only several days later. However, our data suggest that the first female reproductive organs, although induced later, mature almost at the same time as the first male organs. This complies with the general rule of monoecious mosses (Stark, 2002) and indicates that the timing of male and female reproductive organ maturation poses no barrier to self-fertilization in P. patens.
Organ Phyllotaxis Differs between P. patens Vegetative and Reproductive Development
During vegetative shoot development, organs are initiated from a single apical cell that gives rise to daughter organ initials in a spiral pattern, with the youngest organ initial closest to the center (Harrison et al., 2009). Here, we show that organ initiation in P. patens switches to a different pattern following the reproductive phase transition, with the oldest organ in the center and the youngest farthest out from the center. This pattern suggests either the existence of a number of preformed initials or the initiation of new meristematic centers at or near the base of new reproductive organs. It has previously been suggested that the first archegonium in moss species forming archegonia bundles is derived from the apical cell, while the following archegonia may originate from branch initials of segments below (Goffinet et al., 2009).
At this stage, we can only speculate about the reasons for the major differences between leaf and reproductive organ positioning. Before the initiation of leaf primordia, an axillary hair forms at the shoot apex (Harrison et al., 2009). As these organs are the predominant sites of auxin responses and the activity of auxin biosynthesis regulators (Eklund et al., 2010b), we have previously suggested that axillary hairs may be important providers of auxin to the organ initials and organ primordia (Eklund et al., 2010b). Axillary hairs surround the initiation site of reproductive organs as well. As no new axillary hairs are formed in the reproductive organ bundles, organ initiation may rely on signals from the already existing axillary hairs, thus directing organ initiation from cells close to the surrounding hairs and peripheral to the central reproductive organ. However, further studies are required in order to test this hypothesis.
Instead of axillary hairs, paraphyses are formed in direct connection to the male reproductive organs (Fig. 1C). They have been proposed to participate in pathogen protection, vegetative propagation, reproductive organ dehiscence, and sperm dispersal (Reese, 1955; Schofield, 1981; Glime, 2007). As we could not connect them to PpSHI activity, auxin responses, or antheridia initiation (Supplemental Fig. S5), their role may be confined to later stages in antheridia development or function.
SHI/STY Family Members Are Essential in Reproductive Organ Development in Both P. patens and Arabidopsis
Arabidopsis SHI/STY family members are active at the earliest time point of leaf, lateral root, and female reproductive organ primordia initiation (Kuusk et al., 2002, 2006; Eklund et al., 2011). In addition, they are required for the morphogenesis of both vegetative and reproductive organs. SHI/STY members in barley (Hordeum vulgare) have also been shown to be important for gynoecium morphogenesis (Yuo et al., 2012), suggesting that their function in female reproductive organ development may be conserved among angiosperms. We have shown that Arabidopsis SHI/STY family members act as DNA-binding transcriptional activators that control the expression of YUC genes (Sohlberg et al., 2006; Eklund et al., 2010a). They can also activate genes encoding enzymes regulating cell expansion as well as a number of transcription factors (Ståldal et al., 2012). Our recent work has also demonstrated that the P. patens SHI/STY homologs are required for auxin biosynthesis and vegetative development, including proposed auxin-dependent traits such as chloronema-to-caulonema ratio and shoot internode elongation (Eklund et al., 2010b). Here, we demonstrate that, like the Arabidopsis homologs, PpSHI genes may be essential for the morphogenesis of reproductive organs. We detected malformations of the Ppshi knockout archegonia at a higher frequency compared with wild-type archegonia (data not shown). The expression of PpSHI genes during the early stages of reproductive primordia development also suggests that the PpSHI genes, like the Arabidopsis SHI/STY genes, could be essential for organ outgrowth or patterning. However, considering the high similarity in protein sequence, as well as the strong overlap in expression patterns, the single knockouts, in this respect, could be seen as hypomorphic, indicating that both genes need to be knocked out in order to give a phenotypic effect on organ outgrowth. We have so far failed to recover double mutant lines, suggesting that double mutants are defective in cell division or differentiation; therefore, it has not yet been possible to test this hypothesis with mutant analyses. In addition, studies of PpSHI overexpressor lines (Eklund et al., 2010b) have not been informative, as the strong PpSHI overexpressor lines are too distorted to produce reproductive organs, and the weak lines show no developmental defects.
This suggests that the ancestral function of SHI/STY proteins has been conserved during the evolution of land plants. However, since the Gransden 2004 laboratory strain used to produce the transgenic lines fails to produce enough sporophytes for analysis, we still lack knowledge concerning the putative role of these transcription factors in the moss sporophyte, and we cannot distinguish between a possible recruitment of these genes from the early land plant sporophyte to the dominating sporophyte generation of angiosperms or from the early land plant gametophyte to the angiosperm sporophyte.
In addition to being essential for organ morphogenesis and differentiation, the PpSHI genes also appear to be involved in specifying cells destined to die. The PpSHI genes and the auxin response reporter are active in the archegonial canal cells, the upper basal cell, and apical cells, all of which appear to go through programmed cell death, suggesting that auxin biosynthesis in these cells directly or indirectly induces a terminal differentiation leading to death. Interestingly, when PpSHI genes are ectopically expressed in leaf cells, these cells appear to enter a death pathway (Eklund et al., 2010b). It has previously been suggested that auxin may be inducing genetic processes, eventually leading to a final differentiation including programmed cell death in angiosperms. For example, Crawford and Yanofsky (2011) provide data suggesting that the auxin response genes ARF6 and ARF8 indirectly induce programmed cell death of the transmitting tract of Arabidopsis gynoecia by up-regulating the expression of the HALF FILLED gene.
Auxin Function during Egg Cell Differentiation May Be Conserved
Our data also indicate that auxin is important during female gamete differentiation not only in the angiosperm gametophyte, as described previously (Pagnussat et al., 2009), but also in mosses. We show that auxin responses inducing the GmGH3 promoter activity is cell and stage specific in moss archegonia and that these responses peak slightly later than the expression of the two PpSHI genes, previously shown to affect auxin biosynthesis rate, and the auxin transport facilitator and/or cell autonomous auxin homeostasis gene PpPINA. Intriguingly, although auxin responsiveness of the unfertilized egg cell of Marchantia polymorpha has so far not been detected (Ishizaki et al., 2012), auxin biosynthesis, homeostasis, and responsiveness appear important for the final differentiation of the egg and upper basal cells in P. patens. It has been hypothesized that auxin biosynthesis driven by YUC genes in the micropylar end of the Arabidopsis gametophyte, followed by diffusion of auxin, creates an auxin gradient, detected by DR5::GFP activity, over the eight gametophyte nuclei along the gametophyte (Pagnussat et al., 2009). The different auxin levels appear to induce distinct differentiation processes, leading to different fates of the seven cells formed by the eight nuclei. This also includes the egg cell, which is positioned in the intermediate- to high-auxin end of the gradient. When the activity of a subset of auxin response genes (ARFs) was down-regulated in the embryo sac by artificial microRNA transgenes, the three cells at the micropylar end, including the egg cell, had identity defects (Pagnussat et al., 2009). Our data thus suggest that the role of auxin during egg cell differentiation in angiosperms was derived from ancestral land plants. This is an important new finding that links developmentally regulated processes between bryophytes and flowering plants.
MATERIALS AND METHODS
Plant Material and Culture Conditions
The Physcomitrella patens subspecies patens strain Gransden 2004, here called the wild type, is the background of Ppshi1-1, Ppshi2-1, Ppshi2-2, PpSHI2pro:PpSHI2-GUS, PpSHI1pro:PpSHI1-GUS, PpSHI1pro:PpSHI1-GFP, GmGH3pro:GUS (Bierfreund et al., 2003; Eklund et al., 2010b), PpPINApro:GFP-GUS, and PpSHI2pro:IAAL. To obtain fresh homogenous starting material, protonemal tissue was subcultivated two to three times with a 7- to 10-d interval on cellophane-covered plates (90 mm in diameter) containing BCD medium (Thelander et al., 2007) supplemented with 5 mm ammonium tartrate and 0.8% agar and grown at 25°C under constant white light from fluorescent tubes (FL40SS W/37; Toshiba) at 30 μmol m−2 s−1 in a Sanyo MLR-350 light chamber with side irradiation. For reproductive organ experiments, small pieces of protonemal tissue were shaped into 2-mm balls and inoculated on solid BCD medium in 25-mm-deep petri dishes (90 mm in diameter), six balls per plate. After growth under the above conditions for 5 weeks, several gametophores, each with approximately 20 leaves, had formed on each colony, and the plates were transferred to gametangia-inducing conditions, 15°C and 8 h of light (30 μmol m−2 s−1) per day.
Phenotypic Analysis
For phenotypic analysis, tissue was harvested at time points after induction as indicated in “Results.” Gametophores positioned at similar distances from the colony center were selected to ensure the analysis of gametophores of similar age and size. Using a dissecting microscope (Leica MZ16), on agar plates to prevent gametophores from drying, leaves close to apex were detached. The entire upper parts of the leafless gametophores were mounted on objective glasses, in 30% glycerol, under coverslips. Reproductive organs were observed and photographed using a Leica DMI4000B microscope with differential interference contrast (DIC; Nomarski) optics, a Leica DFC360FX camera, and the LAS AF (Leica Microsystems) software. Adobe Photoshop CS3 was used to merge two to three photographs taken at 63× or 100× magnification to allow visualization of entire late-stage archegonia, to remove background, and to mark borders and cells for better visualization in some figures. For analysis of the timing of bundle and organ formation, as well as for the progression of antheridial and archegonial stages, eight to 20 gametophores were harvested starting 4 d after transfer to gametangia-inducing conditions, every day for the first 3 weeks and thereafter twice per week for another 3 weeks. Gametophores were thereafter harvested at additional time points as indicated in “Results.” For all individual gametophores harvested at each time point, the number of antheridia and archegonia in each stage, their position at the apex, and the number of paraphyses were registered. This was performed for two independent biological experiments. Time to onset of the stages of reproductive organ development was calculated using a Microsoft Excel trend line using the data points surrounding the approximate maximum stage count increase (the first three to five stage count measurements). The average for the two independent experiments was used to calculate the final results.
GUS Staining and Fluorescence Microscopy
For histochemical staining of reproductive organs, gametophores stripped of leaves were incubated in GUS staining solution (Mattsson et al., 2003) for 10 to 12 h at room temperature followed by (1) water for 10 to 60 min, (2) ethanol:acetic acid mix (6:1) for 1 to 3 h, (3) 99.9% ethanol twice for 1 min each, and (4) 70% ethanol before analysis using a Zeiss Axioplan microscope with a Leica DFC295 camera and LAS core imaging software. For fluorescence microscopy, stripped gametophores were analyzed using a Leica DMI4000B inverted microscope (HXC PL fluotar 40× objective) with a Leica DFC360FX camera and LAS AF software. DIC (Nomarski) optics was used to capture light micrographs, an L5 fluorescein isothiocyanate/GFP band-pass filter for GFP fluorescence, and a rhodamine/red fluorescent protein long-pass filter for chloroplast autofluorescence.
PpSHI1pro:PpSHI1-GUS, PpPINApro:GFP-GUS, and PpSHI2pro:IAAL Lines
A PpSHI1 genomic fragment terminating immediately prior to the stop codon was amplified with primers 165BamHI_II_F and PpSHI153 (Supplemental Table S4) and inserted into the BamHI site of the vector pPpGUS (Eklund et al., 2010b). A NotI/XbaI fragment in the resulting product was exchanged for a genomic fragment amplified with primers 5′PpUTR_NotI and 3′PpUTR_XbaI (Supplemental Table S4) and covering the 3′ untranslated region of PpSHI1 to produce plasmid pPpSHI1-iGUS. The targeting construct (Supplemental Fig. S6A) was released by XbaI digestion prior to transformation. To create the PpPINApro:GFP-GUS lines, a 2-kb fragment containing the promoter sequence of PpPINA was cloned into the Gateway vector pDONRP4P1r using the primer pair pPpPINAF and pPpPINAR extended with the attP1 and attP4 sequences (Supplemental Table S4). This vector, together with pEN-L1-F-L2 (GFP) and pEN-R2-S-L3 (GUS), was recombined through two MultiSite Gateway intermediary vectors into a final destination vector. This vector further contains integration sites for the Pp108 locus and the neomycin resistance gene nptII (driven by the 35S promoter). The vector was linearized with PmeI prior to transformation. To construct the PpSHI2pro:IAAL lines, a 2-kb PpSHI2 promoter fragment was PCR amplified using primers SHI2_F_Hind3 and SHI2_R_KpnI and inserted in front of IAAH by cutting the fragment and vector pG2NBL-IAAH-NOST (Blilou et al., 2005) with HindIII and KpnI. The resulting PpSHI2pro:IAAH sequence was released with KpnI and SapI, blunted, and inserted into vector pCMAK1 (http://moss.nibb.ac.jp/), which had been cut with EcoRI and NdeI, deleting the modified 35S promoter cassette, and blunted. Finally, the IAAH sequence was released from pCMAK1 by HindIII/SacI cleavage and exchanged for IAAL (Romano et al., 1991), which had been PCR amplified, using primers IAAL_pCMAK_F2 and IAAL_pCMAK_R1, from the TOPO vector pCR2.1-IAAL and cut with the same restriction enzymes, creating PpSHI2pro:IAAL. The construct was NotI linearized before transformation. Following transformation into P. patens protoplasts (Schaefer et al., 1991), transformants were selected on 50 μg mL−1 G418 (G9516; Sigma; PpSHI1pro:PpSHI1-GUS and PpPINApro:GFP-GUS) or 50 μg mL−1 zeocine (Invitrogen; PpSHI2pro:IAAH). PCR genotyping of stable transformants was performed with the primers shown in Supplemental Figure S6 and Supplemental Table S4.
Quantitative Real-Time PCR
RNA extraction, complementary DNA synthesis, and quantitative real-time PCR were performed essentially as described previously (Sohlberg et al., 2006). Protonemal moss tissue was used for RNA extraction, 1 μg of total RNA was used for complementary DNA synthesis, and PCR was carried out on a Bio-Rad iCycler MyiQ real-time machine. The data represent averages of three biological replicates, each quantified in three technical replicates. The samples were normalized using PpACTIN as an internal control. The gene-specific primers are listed in Supplemental Table S4; PpACTIN primers have been published (Prigge et al., 2010).
Quantifying the Positions of Organs within the Bundles
To quantify the positions of the reproductive organs, shoots were harvested 10 to 15 dpi and 21 to 30 dpi for studies of antheridia and archegonia, respectively, and prepared for microscopy as above. Using DIC optics, 63 antheridial and 77 archegonial primary bundles were screened, from a lateral view, moving the focus plane manually through the bundle. The positions within the bundles of the basal part of all organs, including information on their stages, were plotted on a square grid to the nearest 45° in relation to surrounding organs, generating a two-dimensional geometric description for the organ positions within a bundle. The center of the bundles was defined as the position of the oldest (latest stage) organ (or centroid if more than one) on the grid. We calculated the distance between any organ and this center using Euclidian distances on the grid and did a pairwise comparison of distances for organs of different stages within a single bundle. A distribution of the difference of such distances between younger (|xy|) and older (|xo|) organs was compared with a background distribution representing all distance differences in the data set, regardless of the age of organs. P values were calculated from a binomial expansion as the probability of having frequencies of data in a specific bin at least as extreme as in the experimental data, assuming that the probabilities for each interval are governed by a background distribution. We repeated the analysis with different distance measures as well as different definitions of the bundle center, which gave consistent results.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Apical expression of PpSHI1 and PpSHI2 during antheridial development.
Supplemental Figure S2. Expression of the PpSHI1pro:PpSHI1-GFP reporter during archegonial development.
Supplemental Figure S3. Expression of PpSHI1, PpSHI2, GmGH3 and PpPINA during archegonial development.
Supplemental Figure S4. PpSHI and GmGH3 are expressed in axillary hairs surrounding the reproductive organ initial in the apex.
Supplemental Figure S5. PpSHI and GmGH3 are not expressed in mature clavate paraphyses.
Supplemental Figure S6. Generation of PpSHI1pro:PpSHI1-GUS, PpPINApro:GFP-GUS and PpSHI2pro:IAAL lines using gene targeting by homologous recombination.
Supplemental Table S1. Initiation times for developmental stages of antheridia and archegonia.
Supplemental Table S2. Number of reproductive organs analysed for each reporter line during different stages of development.
Supplemental Table S3. Percentage of total number of organs analysed exhibiting the stated phenotypic character, for wt, PpSHI knock-out mutants and PpSHI2pro:IAAL.
Supplemental Table S4. List of oligonucleotides used.
Supplementary Material
Acknowledgments
We thank Isabel Valsecchi and Chen Song for technical assistance, Jian Xu for the pG2NBL-IAAH-NOST plasmid, Lars Ostergaard for the pCR2.1-IAAL plasmid, Mitsuyasu Hasebe for the pCMAK1 plasmid, and Magnus Eklund, Tom Martin, and two anonymous reviewers for valuable comments on the manuscript.
Glossary
- dpi
days post induction
- DIC
differential interference contrast
References
- Ashton NW, Grimsley NH, Cove DJ. (1979) Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144: 427–435 [DOI] [PubMed] [Google Scholar]
- Berger F, Twell D. (2011) Germline specification and function in plants. Annu Rev Plant Biol 62: 461–484 [DOI] [PubMed] [Google Scholar]
- Bierfreund NM, Reski R, Decker EL. (2003) Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep 21: 1143–1152 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K. (2007) Green genes: comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Campbell DH (1918) The Structure and Development of Mosses and Ferns (Archegoniatae). MacMillan, New York, pp 195–203 [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford BC, Yanofsky MF. (2011) HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138: 2999–3009 [DOI] [PubMed] [Google Scholar]
- Crum HA (2001) Structural Diversity of Bryophytes. University of Michigan Herbarium, Ann Arbor [Google Scholar]
- Dal Bosco C, Dovzhenko A, Liu X, Woerner N, Rensch T, Eismann M, Eimer S, Hegermann J, Paponov IA, Ruperti B, et al. (2012) The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J 71: 860–870 [DOI] [PubMed] [Google Scholar]
- Ding Z, Wang B, Moreno I, Dupláková N, Simon S, Carraro N, Reemmer J, Pěnčík A, Chen X, Tejos R, et al. (2012) ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun 3: 941. [DOI] [PubMed] [Google Scholar]
- Eklund DM, Cierlik I, Ståldal V, Claes AR, Vestman D, Chandler J, Sundberg E. (2011) Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol 157: 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ståldal V, Valsecchi I, Cierlik I, Eriksson C, Hiratsu K, Ohme-Takagi M, Sundström JF, Thelander M, Ezcurra I, et al (2010b) The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22: 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Thelander M, Landberg K, Ståldal V, Nilsson A, Johansson M, Valsecchi I, Pederson ERA, Kowalczyk M, Ljung K, et al (2010a) Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens. Development 137: 1275–1284 [DOI] [PubMed] [Google Scholar]
- Fujita T, Sakaguchi H, Hiwatashi Y, Wagstaff SJI, Ito M, Deguchi H, Sato T, Hasebe M. (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10: 176–186 [DOI] [PubMed] [Google Scholar]
- Glass NL, Kosuge T. (1986) Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi. J Bacteriol 166: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glime JM (2007) Bryophyte Ecology, Vol. 1. Physiological Ecology. Michigan Technological University (Houghton, MI), Botanical Society of America (St. Louis, MO), and International Association of Bryologists. www.bryoecol.mtu.edu (August 16, 2011)
- Goffinet B, Buck WR, Shaw AJ (2009) Morphology, anatomy and classification of the Bryophyta. In B Goffinet, AJ Shaw, eds, Bryophyte Biology, Ed 2. Cambridge University Press, Cambridge, UK, pp 55–138 [Google Scholar]
- Harrison CJ, Roeder AHK, Meyerowitz EM, Langdale JA. (2009) Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr Biol 19: 461–471 [DOI] [PubMed] [Google Scholar]
- Hohe A, Rensing SA, Mildner M, Lang D, Reski R. (2002) Day length and temperature strongly influence sexual reproduction and expression of a novel MADS-box gene in the moss Physcomitrella patens. Plant Biol 4: 595–602 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Nonomura M, Kato H, Yamato KT, Kohchi T. (2012) Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J Plant Res 125: 643–651 [DOI] [PubMed] [Google Scholar]
- Johri MM, Desai S. (1973) Auxin regulation of caulonema formation in moss protonema. Nat New Biol 245: 223–224 [DOI] [PubMed] [Google Scholar]
- Kofuji R, Yoshimura T, Inoue H, Sakakibara K, Hiwatashi Y, Kurata T, Aoyama T, Ueda K, Hasebe M (2009) Gametangia development in the moss Physcomitrella patens. Annu Plant Rev 36: 167–181 [Google Scholar]
- Kuusk S, Sohlberg JJ, Long JA, Fridborg I, Sundberg E. (2002) STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129: 4707–4717 [DOI] [PubMed] [Google Scholar]
- Kuusk S, Sohlberg JJ, Eklund DM, Sundberg E. (2006) Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J 47: 99–111 [DOI] [PubMed] [Google Scholar]
- Lal M, Bhandari NN. (1968) The development of sex organs and sporophyte in Physcomitrium cyathicarpum Mitt. Bryologist 71: 11–20 [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T. (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. (2007) An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Krecek P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin-I T, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K, et al (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc Natl Acad Sci USA 100: 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. (2009) Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, Palme K. (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parihar NS (1966) Bryophyta, Ed 5. Central Book Depot, Allahabad, India, pp 215–223 [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M. (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20: 1907–1912 [DOI] [PubMed] [Google Scholar]
- Reese WD. (1955) Regeneration of some moss paraphyses. Bryologist 58: 239–241 [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Romano CP, Hein MB, Klee HJ. (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev 5: 438–446 [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Ando S, Yip HK, Tamada Y, Hiwatashi Y, Murata T, Deguchi H, Hasebe M, Bowman JL. (2013) KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 339: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Saleh O, Issman N, Seumel GI, Stav R, Samach A, Reski R, Frank W, Arazi T. (2011) MicroRNA534a control of BLADE-ON-PETIOLE 1 and 2 mediates juvenile-to-adult gametophyte transition in Physcomitrella patens. Plant J 65: 661–674 [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd JP, Knight CD, Cove DJ. (1991) Stable transformation of the moss Physcomitrella patens. Mol Gen Genet 226: 418–424 [DOI] [PubMed] [Google Scholar]
- Schofield WB. (1981) Ecological significance of morphological characters in the moss gametophyte. Bryologist 84: 149–165 [Google Scholar]
- Sohlberg JJ, Myrenås M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E. (2006) STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J 47: 112–123 [DOI] [PubMed] [Google Scholar]
- Ståldal V, Cierlik I, Chen S, Landberg K, Baylis T, Myrenås M, Sundström JF, Eklund DM, Ljung K, Sundberg E. (2012) The Arabidopsis thaliana transcriptional activator STYLISH1 regulates genes affecting stamen development, cell expansion and timing of flowering. Plant Mol Biol 78: 545–559 [DOI] [PubMed] [Google Scholar]
- Ståldal V, Sohlberg JJ, Eklund DM, Ljung K, Sundberg E. (2008) Auxin can act independently of CRC, LUG, SEU, SPT and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytol 180: 798–808 [DOI] [PubMed] [Google Scholar]
- Stark LR. (2002) Phenology and its repercussions on the reproductive ecology of mosses. Bryologist 105: 204–218 [Google Scholar]
- Thelander M, Nilsson A, Olsson T, Johansson M, Girod PA, Schaefer DG, Zryd JP, Ronne H. (2007) The moss genes PpSKI1 and PpSKI2 encode nuclear SnRK1 interacting proteins with homologues in vascular plants. Plant Mol Biol 64: 559–573 [DOI] [PubMed] [Google Scholar]
- Viaene T, Delwiche CF, Rensing SA, Friml J. (2013) Origin and evolution of PIN auxin transporters in the green lineage. Trends Plant Sci 18: 5–10 [DOI] [PubMed] [Google Scholar]
- Wellman CH, Osterloff PL, Mohiuddin U. (2003) Fragments of the earliest land plants. Nature 425: 282–285 [DOI] [PubMed] [Google Scholar]
- Yuo T, Yamashita Y, Kanamori H, Matsumoto T, Lundqvist U, Sato K, Ichii M, Jobling SA, Taketa S. (2012) A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. J Exp Bot 63: 5223–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.