The activity of class I TCP transcription factors is modulated by cellular redox agents both in vitro and in vivo, suggesting their role in developmental redox control in plants.
Abstract
TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR1 (TCP) transcription factors participate in plant developmental processes associated with cell proliferation and growth. Most members of class I, one of the two classes that compose the family, have a conserved cysteine at position 20 (Cys-20) of the TCP DNA-binding and dimerization domain. We show that Arabidopsis (Arabidopsis thaliana) class I proteins with Cys-20 are sensitive to redox conditions, since their DNA-binding activity is inhibited after incubation with the oxidants diamide, oxidized glutathione, or hydrogen peroxide or with nitric oxide-producing agents. Inhibition can be reversed by treatment with the reductants dithiothreitol or reduced glutathione or by incubation with the thioredoxin/thioredoxin reductase system. Mutation of Cys-20 in the class I protein TCP15 abolished its redox sensitivity. Under oxidizing conditions, covalently linked dimers were formed, suggesting that inactivation is associated with the formation of intermolecular disulfide bonds. Inhibition of class I TCP protein activity was also observed in vivo, in yeast (Saccharomyces cerevisiae) cells expressing TCP proteins and in plants after treatment with redox agents. This inhibition was correlated with modifications in the expression of the downstream CUC1 gene in plants. Modeling studies indicated that Cys-20 is located at the dimer interface near the DNA-binding surface. This places this residue in the correct orientation for intermolecular disulfide bond formation and explains the sensitivity of DNA binding to the oxidation of Cys-20. The redox properties of Cys-20 and the observed effects of cellular redox agents both in vitro and in vivo suggest that class I TCP protein action is under redox control in plants.
TCP transcription factors constitute a family of plant developmental regulators (Martín-Trillo and Cubas, 2010; Uberti Manassero et al., 2013). The name of the family stands for the three first characterized members: TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATING CELL FACTOR1 (Cubas et al., 1999). These factors bind DNA through the TCP domain, a conserved region of approximately 60 amino acids that also participates in protein dimerization. The TCP domain contains a basic N-terminal region involved in DNA recognition, followed by a helix-loop-helix (HLH) motif similar to the one present in basic helix-loop-helix (bHLH) transcription factors (Aggarwal et al., 2010; Martín-Trillo and Cubas, 2010). Target sites recognized by TCP proteins are different from those bound by bHLH proteins (Kosugi and Ohashi, 2002; Viola et al., 2012). This is probably due to the fact that the respective basic regions differ markedly in composition and structure. Based on sequence homology, two main classes of TCP domains can be described. Class II proteins affect leaf and petal development and also influence shoot branching, among other processes (Luo et al., 1996; Doebley et al., 1997; Nath et al., 2003; Palatnik et al., 2003; Aguilar-Martínez et al., 2007; Koyama et al., 2007; Nag et al., 2009). The role of class I proteins is less clear, but several studies indicated that they participate in developmental processes probably associated with the regulation of cell growth and proliferation. TCP20 has been linked to the regulation of the expression of the cyclin CYCB1;1 and ribosomal protein genes, thus establishing a link between cell growth and cell cycle control (Li et al., 2005). Accordingly, alteration of the function of TCP20 causes dramatic changes in plant development (Hervé et al., 2009). TCP15 also modulates the expression of cell cycle genes (Li et al., 2012) and is involved in leaf and inflorescence development and the regulation of auxin and cytokinin homeostasis (Kieffer et al., 2011; Steiner et al., 2012; Uberti Manassero et al., 2012). TCP16 and TCP11 participate in pollen development (Takeda et al., 2006; Viola et al., 2011). Other roles proposed for class I proteins include the coordination of mitochondrial biogenesis (Welchen and Gonzalez, 2006; Gonzalez et al., 2007; Giraud et al., 2010), the regulation of embryonic growth potential during germination (Tatematsu et al., 2008), and the regulation of the circadian clock. Particularly, TCP21 is known to interact with TOC1 and to bind to the CCA1 promoter, thus establishing a link between these core components of the clock (Pruneda-Paz et al., 2009).
Developmental processes in plants show much more plasticity than those in animals. This has been assigned to the sessile nature of the plant lifestyle. As a consequence, plant developmental processes are usually influenced by changes in environmental conditions. It is becoming increasingly evident that many processes that link developmental responses to environmental changes operate through the modification of redox conditions within plant cells (Potters et al., 2007; Meyer, 2008). In addition, modifications in redox conditions are known to occur during cell proliferation (Pellny et al., 2009; Diaz-Vivancos et al., 2010), and the influence of redox agents on several aspects of plant development has been well documented (May et al., 1998; Vernoux et al., 2000; Foreman et al., 2003; Xing et al., 2005; Cairns et al., 2006; Carol and Dolan, 2006; Pasternak et al., 2008). Redox agents act as signaling molecules to modify the expression of certain genes, and they also react with proteins and other cellular components, modifying their properties and activating signal transduction pathways (Pei et al., 2000; Tavakoli et al., 2001; Noctor et al., 2002; Gupta and Luan, 2003; Buchanan and Balmer, 2005; Fey et al., 2005; Foyer and Noctor, 2005; Hicks et al., 2007; Terrile et al., 2012). A paradigmatic case of this behavior is the direct modification of a transcription factor that produces changes in its DNA-binding properties and, as a consequence, in the expression of its target genes.
There are several examples of transcription factors whose activity is modified by redox agents in plants (Tron et al., 2002; Heine et al., 2004; Comelli and Gonzalez, 2007; Serpa et al., 2007; Shaikhali et al., 2008). The best studied case is perhaps the NPR1-TGA system (Després et al., 2003; Mou et al., 2003; Lindermayr et al., 2010). NPR1 is a coactivator that is converted to its reduced state through a salicylic acid-dependent pathway. This leads to the translocation of NPR1 to the nucleus and interaction with the transcription factor TGA1 that is also modulated by redox conditions and induces the expression of pathogenesis-related genes. Considering the importance of redox changes in plant function, it is conceivable that many other proteins are subjected to modifications by redox agents. In this work, after the identification of a conserved Cys in the HLH region of class I TCP transcription factors, we demonstrate that several redox agents reversibly modulate the capacity of these factors to interact with DNA and to modulate transcription. The results suggest that redox changes operate in vivo to influence the activity of these factors within plant cells.
RESULTS
The DNA-Binding Activity of Class I TCP Proteins Is Modulated by Redox Conditions
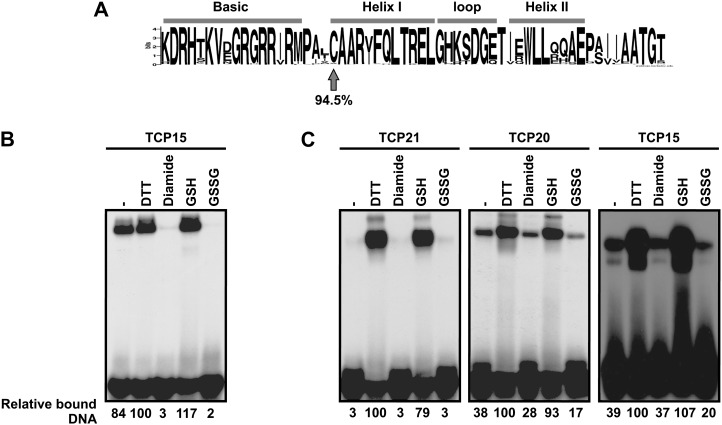
Comparison of class I TCP domain sequences shows the presence of a conserved Cys at position 20 (Fig. 1A). This Cys is located at the beginning of helix I of the putative HLH domain. Considering that Cys residues are involved in redox modulation of the activity of different proteins, we tested the effect of redox treatments on the DNA-binding activity of three different class I TCP proteins from Arabidopsis (Arabidopsis thaliana): TCP15, TCP20, and TCP21. These proteins contain only one Cys, Cys-72 in TCP15, Cys-98 in TCP20, and Cys-51 in TCP21, all located at position 20 of the TCP domain. For this purpose, full-length TCP15 (amino acids 1–325) and TCP21 (amino acids 1–239) as well as a shorter form (amino acids 1–157, comprising the entire TCP domain plus 78 and 22 residues located N and C terminal to it, respectively) of TCP20 were expressed in recombinant form fused to the maltose-binding protein (MBP) in Escherichia coli and purified. Incubation of recombinant TCP15 with the oxidant diamide produced a pronounced decrease in its DNA-binding activity (Fig. 1B). A similar effect was observed after treatment with oxidized glutathione (GSSG), indicating that the protein is sensitive to redox conditions. Incubation in the presence of the reducing agents dithiothreitol (DTT) or reduced glutathione (GSH), on the other hand, did not produce changes in DNA-binding activity (Fig. 1B), probably reflecting the fact that the purified protein is in its reduced state. We then dialyzed TCP15, TCP20, and TCP21 overnight in buffer lacking reducing agents and tested the DNA-binding activity of these proteins after incubation with redox agents. In this case, treatments with reducing agents produced an increase in DNA binding, while oxidants were ineffective (Fig. 1C). This most likely indicates that the proteins were oxidized during dialysis. The results obtained suggest that redox interconversions modulate the DNA-binding capacity of the class I TCP proteins tested.
Figure 1.
The DNA-binding activity of class I TCP proteins is modulated by redox agents. A, Sequence logo constructed with the TCP domain sequences of 218 class I TCP proteins from different species. Position 20 is indicated with the arrow. B, TCP15 (50 ng) purified with buffer lacking 2-ME was incubated in the presence of 25 mm DTT, 10 mm diamide, 25 mm GSH, or 25 mm GSSG, as indicated. The control (–) was incubated under similar conditions without redox agents. After incubation, the DNA-binding activity of TCP15 was analyzed by EMSA. C, A similar experiment as in B using TCP21, TCP20, and TCP15 that had been previously dialyzed in the absence of redox agents as described in “Materials and Methods.” In B and C, the amount of bound DNA, relative to the sample incubated with DTT, is shown below the EMSA images. Experiments were repeated at least three times with similar results.
Cys-20 of the TCP Domain Is Responsible for the Redox Sensitivity of Class I Proteins
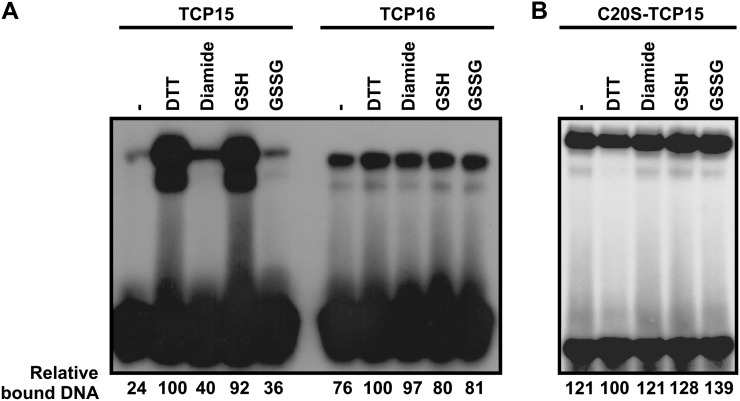
The three class I proteins tested contain only one Cys, located at position 20 of the TCP domain. It is logical to assume, then, that this Cys may be the target of redox modifications that affect DNA binding. To reinforce this, we tested the effect of redox conditions on the DNA-binding activity of TCP16, a class I protein that contains Val instead of Cys at position 20 of the TCP domain. TCP16 shows lower DNA-binding activity than TCP15 but is not affected by redox conditions (Fig. 2A). We also mutagenized Cys-20 to Ser in TCP15 and analyzed the effect of this mutation on the DNA-binding activity and the redox properties of the protein. Inclusion of Ser-20 did not affect the DNA-binding activity of TCP15 (data not shown), but the protein became insensitive to modifications of redox conditions (Fig. 2B). We conclude that Cys-20 is not required for DNA binding but is responsible for the redox sensitivity of class I TCP proteins.
Figure 2.
Cys-20 of the TCP domain is required for redox sensitivity. A, The redox sensitivity of TCP16, which contains Val instead of Cys at position 20 of the TCP domain, was analyzed as described in Figure 1. B, A similar experiment as in A with a TCP15 mutant protein that contains Ser instead of Cys-20. The amount of bound DNA, relative to the sample incubated with DTT, is shown below the EMSA images. Experiments were repeated at least three times with similar results.
Class I TCP Proteins Form Intermolecular Disulfide Bonds under Oxidation Conditions
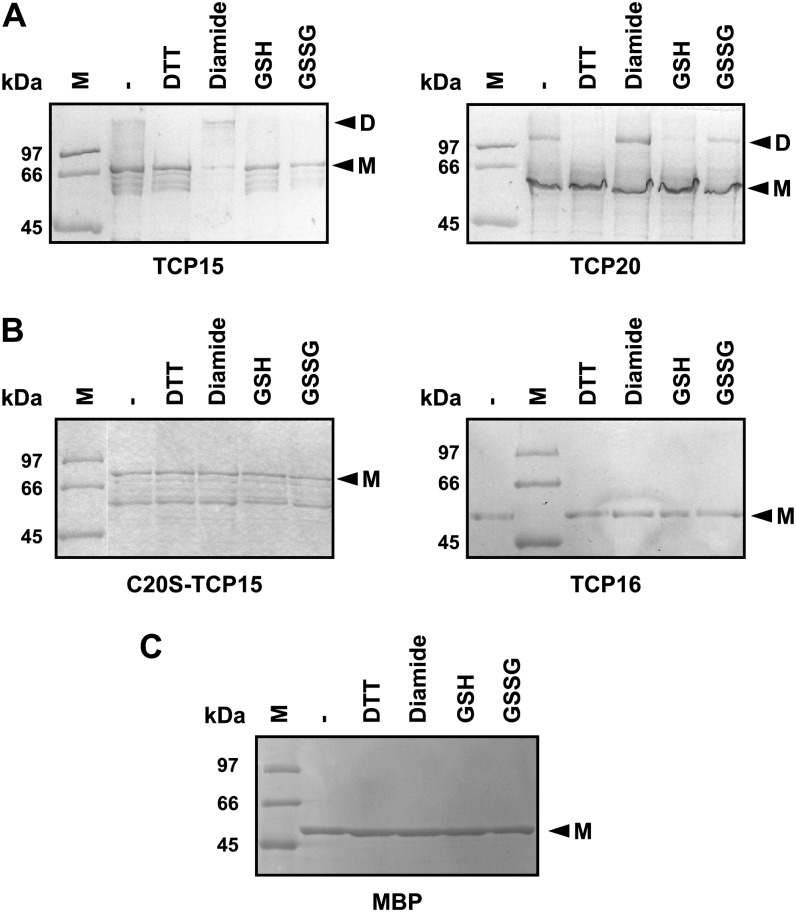
The fact that the proteins analyzed contain only one Cys suggests that if oxidation leads to the formation of disulfide bonds, these must form between Cys residues located in different polypeptides. To analyze this, we tested the migration of oxidized and reduced forms of TCP proteins in nonreducing SDS-PAGE. As observed in Figure 3A, species with the mobility of dimers were observed after treatment with diamide, and the amounts of these species were significantly reduced after treatment with DTT or GSH. This is indicative of the formation of intermolecular disulfide bonds. Since incubation with oxidizing agents produces a decrease in binding capacity, it can be postulated that the formation of intermolecular disulfide bonds affects the interaction of class I proteins with DNA. TCP16 and C20S-TCP15, on the other hand, migrated as monomers under all conditions tested (Fig. 3B), indicating that Cys-20 is responsible for the formation of intermolecular covalent bonds under oxidation conditions. Particularly, TCP15 and its mutant showed several bands that migrate faster than the monomer when analyzed in either reducing (data not shown) or nonreducing SDS-PAGE (Fig. 3, A and B). We speculate that these are truncated products of the recombinant proteins rather than impurities that copurify with them. This was confirmed by western-blot analysis of reduced and oxidized MBP-TCP15 fusions with anti-MBP antibodies (Supplemental Fig. S1). Since fusions to MBP were used in these experiments, we also tested the effect of redox conditions on the migration of MBP alone. In agreement with the fact that MBP does not contain Cys residues, no changes in protein mobility were observed after treatments with reducing or oxidizing agents in this case (Fig. 3C).
Figure 3.
Oxidation of TCP proteins that contain Cys-20 leads to the formation of intermolecular bonds. A and B, TCP proteins (1 µg of MBP-TCP fusion) that either contain (A) or lack (B) Cys-20 within the TCP domain were incubated with redox agents as described in Figure 1. C, Purified MBP was subjected to a similar treatment. After treatment, the molecular mass of the proteins was analyzed in a nonreducing SDS-PAGE. Arrowheads point to species with the mobility of monomers (M) or dimers (D). MW, Molecular mass markers. Experiments were repeated at least three times with similar results.
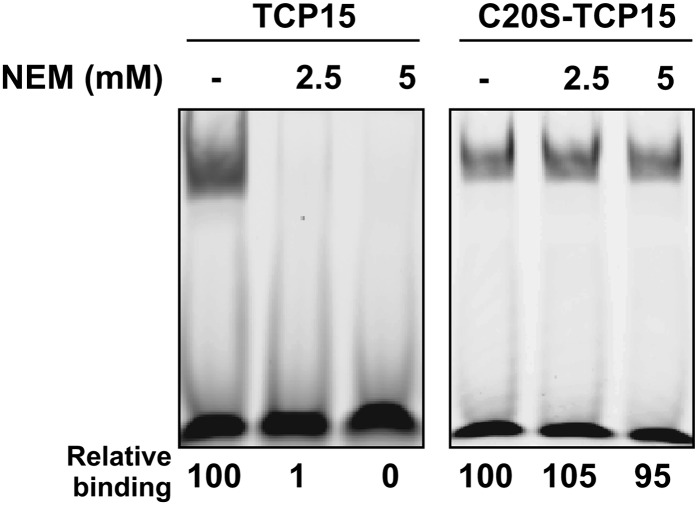
A somewhat different behavior was observed for TCP15 in the presence of GSSG, since covalent dimer formation was not observed under this condition (Fig. 3A), in spite of the fact that GSSG treatment inhibited DNA binding (Fig. 1B). One possibility is that TCP15 forms a mixed disulfide with glutathione under these conditions and that this modification affects the interaction of the protein with DNA. In agreement with this, modification of TCP15 with the Cys alkylating agent N-ethylmaleimide (NEM) produced a complete loss of DNA-binding activity (Fig. 4), suggesting that the presence of a bulky group at Cys-20 precludes binding to DNA.
Figure 4.
TCP15 is inactivated by modification of Cys-20. TCP15 and C20S-TCP15 were incubated with 2.5 or 5 mm NEM in the dark during 20 min at 25°C. After treatment, the DNA-binding activity of the proteins was analyzed by EMSA. The amount of bound DNA, relative to the untreated sample (–), is shown below the EMSA images. The experiment was repeated at least three times with similar results.
To ascertain the oligomerization state of the TCP proteins in solution, we subjected purified MBP-TCP20 (active, nondialyzed) to gel filtration on a calibrated column. The results (Supplemental Fig. S2) indicated that the protein forms oligomers of high Mr that probably correspond to hexamers. Similar results (the presence of oligomers with higher Mr than dimers) were obtained with TCP15 (data not shown). This is somewhat unexpected, since it is assumed that TCP proteins form dimers, and Aggarwal et al. (2010) have observed that the class II TCP domain of TCP4 behaves as a dimer in solution when fused to MBP. We do not know the reason for this different behavior, but it is interesting that the proteins we have used contain other protein segments in addition to the TCP domain. One possibility is that these segments contribute to the formation of higher order structures. The formation of higher order oligomers has also been observed for the bHLH protein MyoD in several studies (Starovasnik et al., 1992; Fairman et al., 1993; Wendt and Thomas, 1997).
Binding to DNA Protects TCP15 from Inactivation by Redox Agents
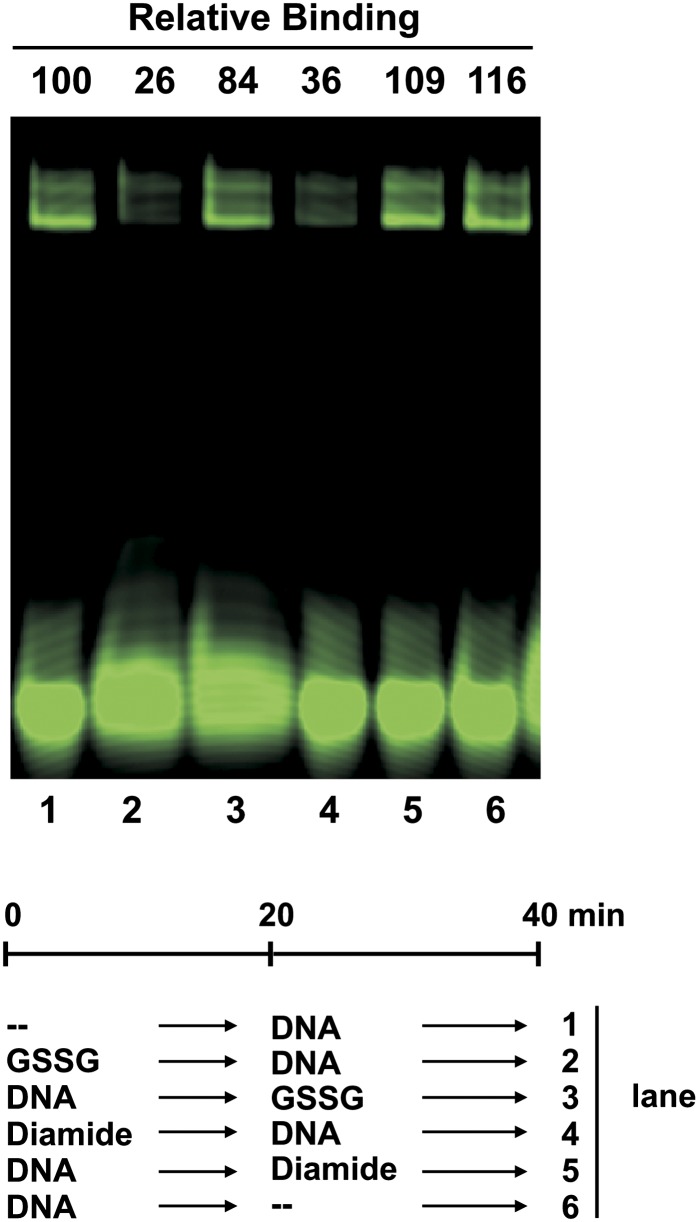
Our results suggest that class I TCP proteins must be in the reduced state to be able to interact with DNA. Since Cys-20 is located at the beginning of helix I, near the basic region that presumably establishes contacts with DNA, it can be envisaged that binding to DNA may protect TCP proteins from redox agents. To analyze this, we performed treatments with diamide and GSSG before and after allowing TCP15 to interact with DNA and then analyzed the amount of protein-DNA complex formed. In these experiments, a double-stranded oligonucleotide labeled with the fluorescent dye 6-carboxyfluorescein (6-FAM) was used. This allowed the use of higher amounts of DNA, thus increasing the proportion of TCP15 that is present as a protein-DNA complex before treatment. As observed in Figure 5, the amount of bound DNA was decreased to about 30% relative to controls when TCP15 was treated with oxidants before the addition of DNA, but these treatments were ineffective when TCP15 was previously allowed to form a complex with DNA. The observed protection from oxidation when TCP15 is bound to DNA indicates that Cys-20 becomes inaccessible to oxidants in the TCP15-DNA complex, perhaps due to its close proximity to the DNA-binding surface of the protein.
Figure 5.
DNA protects TCP15 from inactivation by oxidants. TCP15 (770 ng of MBP-TCP15 fusion; 0.5 μm) purified with buffer lacking 2-ME was incubated in the presence of 10 mm diamide or 25 mm GSSG, as indicated, before or after the addition of DNA (100 ng; 0.25 μm double-stranded oligonucleotide labeled with 6-FAM). Controls (–) were incubated under similar conditions without redox agents. After incubation, the amount of TCP15-DNA complex present in each reaction was analyzed by EMSA. The amount of bound DNA relative to control lane 1 is specified above each lane and was calculated from signal intensities obtained with a Typhoon (GE Healthcare) scanner. The experiment was repeated at least three times with similar results.
Cellular Redox Agents Modulate the DNA-Binding Activity of TCP15
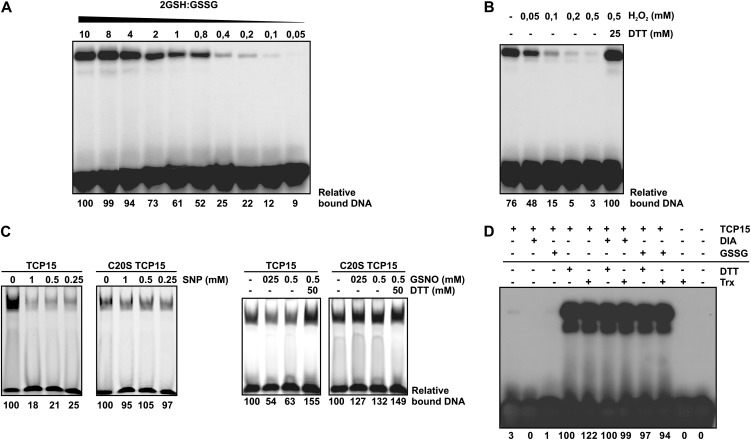
The effect of redox conditions on TCP15 activity was further analyzed by incubating the protein with different proportions of GSH and GSSG. Figure 6A shows that the DNA-binding activity of TCP15 adjusts to the GSH/GSSG ratio of the medium, suggesting that active (reduced) and inactive (oxidized) molecules coexist and interconvert according to redox conditions. This experiment also allowed the calculation of the redox potential for the interconversion, which most likely reflects the potential of the redox-active Cys-20. Fitting the observed curve of DNA-binding activity versus buffer redox potential to the Nernst equation for a two-electron reaction, a midpoint potential of −238.4 ± 1.4 mV at pH 7.5 was obtained. This potential is close to that of the GSH/GSSG pair under these conditions (−269.6 mV; Veine et al., 1998; Bick et al., 2001) and fit in the range of −170 to −330 mV reported for several redox-regulated plant proteins such Rap2.4a, AtbZip16, ABI2, TRXs, and GRXs (Meinhard et al., 2002; Rouhier et al., 2008; Shaikhali et al., 2008, 2012). It can be suggested that redox interconversions may operate under in vivo conditions to modulate the DNA-binding activity of TCP15.
Figure 6.
Cellular redox agents modulate the DNA-binding activity of TCP15. A, TCP15 purified without 2-ME was incubated in buffer at pH 7.5 with different proportions of GSH and GSSG, as indicated. The total concentration of glutathione was kept constant at 10 mm. After incubation, the DNA-binding activity of TCP15 was analyzed by EMSA. B, TCP15 purified without 2-ME was incubated in the presence of different concentrations of H2O2, as indicated, during 1 h at 25°C. After incubation, the DNA-binding activity of TCP15 was analyzed by EMSA. Treatment with DTT, where indicated, was performed during 1 h after the treatment with H2O2. C, TCP15 and its mutant at position 20 of the TCP domain (purified without 2-ME) were incubated in the presence of the NO-producing agents SNP and GSNO at the indicated concentrations during 1 h as specified in “Materials and Methods.” After incubation, the DNA-binding activity of the proteins was analyzed by EMSA. Reversion of the effect of GSNO was assayed by incubating samples with DTT during an additional 20 min. D, TCP15 dialyzed in the absence of reducing agents as described in “Materials and Methods” was incubated in the presence of diamide (DIA), GSSG, or DTT. Alternatively, the protein was incubated with 13 ng µL−1 purified thioredoxin and thioredoxin reductase in the presence of 0.55 mm NADPH (Trx). Where indicated, the treatments with DTT or the thioredoxin system were applied to protein previously oxidized with diamide or GSSG. After the respective treatments, the DNA-binding activity of TCP15 was analyzed by EMSA. Experiments were repeated at least three times with similar results.
To evaluate if, apart from the GSH/GSSG pair, other redox agents that are normally present or produced within cells influence the DNA-binding activity of TCP15, we assayed the effect of hydrogen peroxide (H2O2), thioredoxin, and nitric oxide (NO). Incubation with H2O2 produced a concentration-dependent decrease in the DNA-binding activity of TCP15 (Fig. 6B). Similar results were observed after treatment with the NO-producing agents sodium nitroprusside (SNP) and S-nitrosoglutathione (GSNO; Fig. 6C). C20S-TCP15 was considerably less sensitive to these treatments, indicating that nitrosylation of Cys-20 is mainly responsible for the inactivation of TCP15. The modifications provoked by H2O2 and NO do not permanently inactivate the protein, since the effects of these compounds were reversed after treatment with DTT. Activation of the oxidized form of TCP15 was also achieved after treatment with thioredoxin and thioredoxin reductase in the presence of NADPH (Fig. 6D). The thioredoxin system was effective with TCP15 oxidized with either diamide or GSSG. It can be suggested that this system may also operate in vivo to modulate the activity of class I TCP proteins.
TCP15-Dependent Transcription Is Sensitive to Oxidative Stress Conditions in Vivo
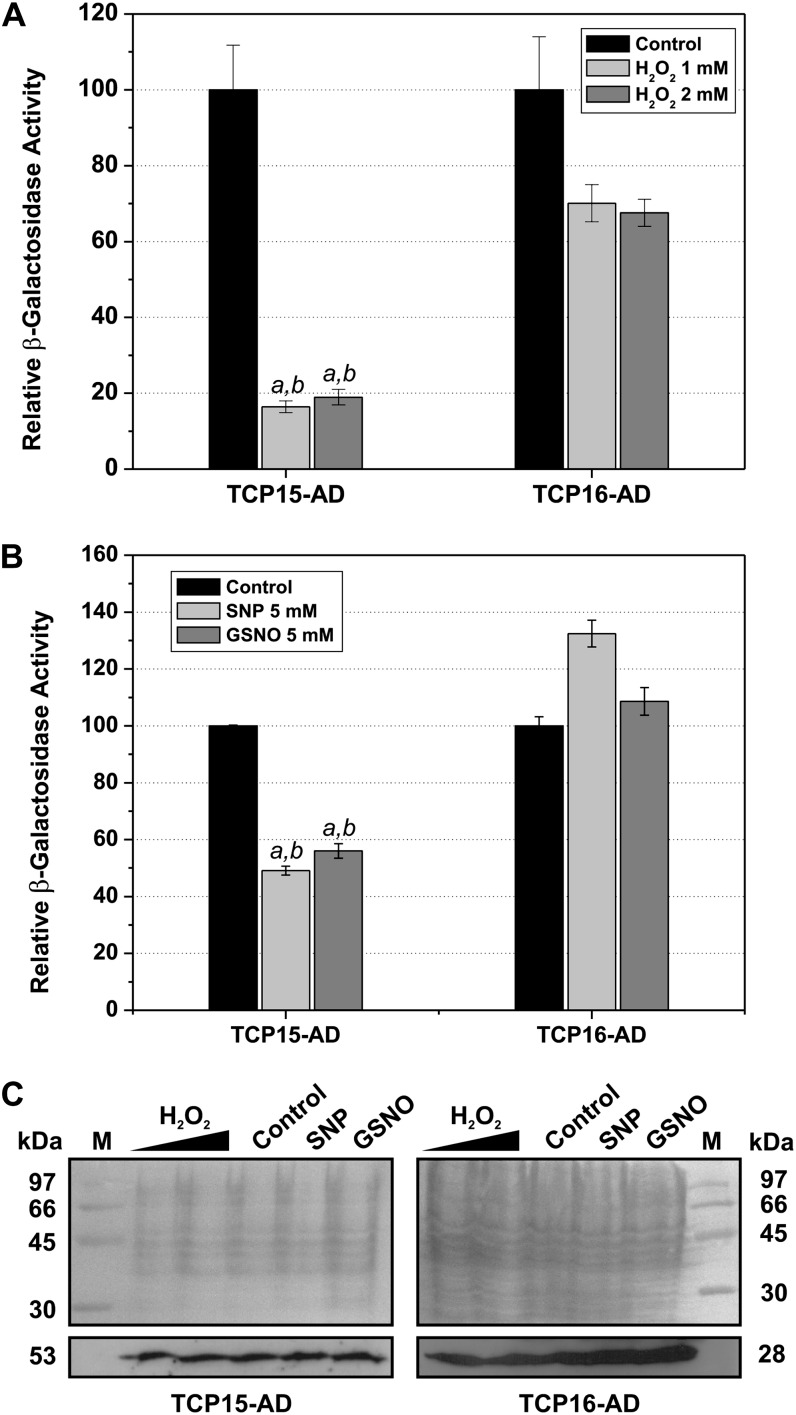
Redox conditions within cells can be modulated by treatment with agents that produce oxidative stress (Delaunay et al., 2000; Jiang et al., 2006). To analyze the effect of cellular oxidative stress on class I TCP protein action, we employed a yeast (Saccharomyces cerevisiae) one-hybrid assay in which the transcription of a lacZ reporter gene was under the control of TCP-binding sites. Expression of TCP15 fused to the GAL4 activation domain (TCP15-AD) in these cells induced the reporter gene, as deduced from β-galactosidase activity measurements (Fig. 7). Treatment of cells with either 1 or 2 mm H2O2 produced an approximately 80% inhibition of reporter gene expression after 4 h of incubation (Fig. 7A). Western-blot analysis of TCP15-AD protein levels indicated that H2O2 treatment barely affected the expression of the fusion protein (Fig. 7C), suggesting that the decrease in reporter gene expression is probably due to inactivation of the protein. In agreement with this, inhibition by the redox agent was considerably lower (about 30%) when a similar experiment was performed with TCP16, which is not sensitive to redox conditions (Fig. 7A). The small inhibition caused by H2O2 treatment on TCP16-dependent expression most likely reflects a redox-sensitive step of gene expression not related to TCP protein action. In fact, this was correlated with a decrease in TCP16-AD protein levels after H2O2 treatment (Fig. 7C). We do not know why this decrease is apparent in cells expressing TCP16-AD and not in cells expressing TCP15-AD. According to western blots and β-galactosidase activity measurements, TCP16-AD seems to be expressed at higher levels than TCP15-AD, and this may reveal a limiting step that is sensitive to oxidative stress. In any case, the significant differences observed between TCP15- and TCP16-dependent reporter gene expression are indicative of a differential sensitivity of TCP15 and TCP16 to changes in redox conditions within cells, most likely due to the presence of Cys-20 in the former.
Figure 7.
Oxidative stress and NO affect TCP15-dependent transcription. β-Galactosidase activity of yeast cells that contain the lacZ reporter gene under the control of the TCP15 or TCP16 target sequence (Viola et al., 2012) and express fusions of the corresponding proteins to the GAL4 activation domain was measured after incubation in the presence or absence of H2O2 or NO-producing agents. A, Increase in β-galactosidase activity after a 4-h incubation in the absence (Control) or presence of H2O2. Values refer to the increase in activity observed in the absence of the redox agent (100%). B, A similar experiment as in A, in which yeast cells were treated with SNP or GSNO during 4 h. Values in A and B represent means ± sd of three independent measurements. Letters indicate significant differences with respect to the control (a) or to the same treatment applied to the TCP16-AD-expressing strain (b; P < 0.01). C, Western-blot analysis of TCP protein levels in control and treated cells. Protein extracts from equivalent amounts of cells as indicated by optical density measurements were subjected to SDS-PAGE, transferred to an appropriate membrane, and either stained with Ponceau S (top panels) or revealed with an antibody against the hemagglutinin tag, present in the fusion proteins (bottom panels). Numbers indicate molecular masses in kD. Experiments were repeated twice with similar results.
In a similar way, treatment of yeast cells with SNP and GSNO produced a decrease in reporter gene expression in the case of cells expressing TCP15 but was ineffective in cells expressing TCP16 (Fig. 7B). Since these treatments did not affect TCP protein levels (Fig. 7C), this result most likely suggests that SNP and GSNO produce the inactivation of TCP15, but not TCP16, in vivo, probably through NO-dependent modification.
TCP Protein Activity Is Modulated by Treatment of Plants with H2O2
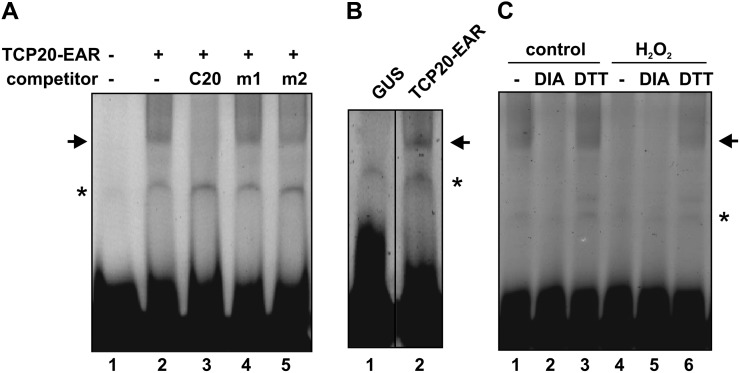
To analyze if redox changes in the external medium act to modulate the activity of TCP transcription factors within the plant, we employed a transient expression approach in which TCP20 fused to the ERF-associated amphiphilic repression (EAR) domain was expressed under the control of the 35S cauliflower mosaic virus (CaMV) promoter (35S::TCP20-EAR). After transformation, TCP20 protein activity was determined by electrophoretic mobility shift assays (EMSAs) using total protein extracts prepared from transformants and an oligonucleotide that contains the binding sequence GTGGGACCGG, preferred by TCP20 according to binding site selection experiments reported earlier (Viola et al., 2011). Figure 8A shows that specific retarded complexes were observed in plants transformed with 35S::TCP20-EAR. Formation of these complexes was abolished by a 100-fold molar excess of the same, unlabeled oligonucleotide but not by similar amounts of oligonucleotides containing changes in the TCP20-binding sequence (Fig. 8A, lanes 3–5). In addition, these complexes were not present when an extract of plants transformed with pBI121 (that expresses GUS from the 35S CaMV promoter) was used (Fig. 8B, lane 1), suggesting that they indeed arise from TCP20-EAR binding to DNA. Complex formation was abolished by previous treatment of the extract with diamide (Fig. 8C, lane 2), showing that the protein expressed in plants is sensitive to oxidation. Treatment of plants with H2O2 during 1 h before protein extraction produced a pronounced decrease in the formation of protein-DNA complexes (Fig. 8C, lane 4). Complex formation was recovered by treatment of the extract with DTT (Fig. 8C, lane 6). In fact, the amount of protein-DNA complex was similar in samples treated with DTT, regardless of the previous treatment with or without H2O2, suggesting that the inclusion of the oxidizing agent affected the activity, rather than the expression, of the protein, most likely through the oxidation of redox-sensitive residues. We conclude that the DNA-binding activity of TCP transcription factors can be modulated within the plant by manipulating the redox conditions of the environment.
Figure 8.
In vivo redox sensitivity of class I TCP transcription factors. Arabidopsis plants were transformed with a construct that expresses TCP20-EAR under the control of the 35S CaMV promoter. Thirty-six hours after transformation, total protein extracts were prepared and their DNA-binding activities were analyzed by EMSA using a labeled oligonucleotide with the TCP20 target sequence GTGGGACCGG (Viola et al., 2011). A, The DNA-binding specificity of proteins present in extracts from plants transformed with 35S::TCP20-EAR was tested by competition with a 100-fold molar excess of unlabeled oligonucleotides containing the TCP20 target site (C20), the same site with mutations at positions 3 and 8 (m1, GTAGGACTGG), or an oligonucleotide with an unrelated target site (m2, CAATAATTGA). B, EMSA using extracts from plants transformed with 35S::TCP20-EAR or with pBI121, which expresses GUS from the 35S CaMV promoter, as a control. C, EMSA using extracts from plants transformed with 35S::TCP20-EAR treated with either water (control) or 10 mm H2O2 during 1 h immediately before extract preparation. Extracts were treated with 10 mm diamide (DIA), 50 mm DTT, or water (–) during 1 h before EMSA analysis. Arrows indicate the positions of a retarded band observed exclusively in extracts from plants transformed with the TCP20-expressing construct, while asterisks indicate an unspecific retarded band. The experiment was repeated twice with similar results.
TCP-Dependent Expression of CUC1 Is Modulated by Redox Treatment
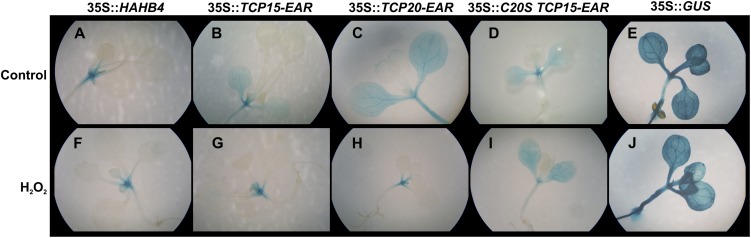
We have previously shown that expression in plants of a repressor form of TCP15 causes the induction of the boundary-specific gene CUC1 in vegetative and floral tissues (Uberti Manassero et al., 2012). Figure 9B and Table I show that transient transformation of a CUC1 reporter line with a construct that expresses TCP15-EAR from the 35S CaMV promoter produces an extension of the CUC1 expression domain, usually restricted to meristematic regions (Takada et al., 2001), to cotyledon lamina. This was also true for constructs that express TCP20-EAR and C20S-TCP15-EAR (Fig. 9, C and D) but not for a construct that expresses the HD-Zip transcription factor Hahb4, used as a control (Fig. 9A; P < 0.0001). The effect of the expression of TCP15 and TCP20 was almost completely abolished by previous treatment of transformed plants with H2O2 (Fig. 9, G and H; Table I; P < 0.0001). H2O2 treatment, however, was ineffective in the case of plants transformed with C20S-TCP15-EAR (Fig. 9I; Table I; P = 1.000), suggesting that H2O2 sensitivity is related to the presence of Cys at position 20 of the TCP domain. The results show that modification of redox conditions affects the function of class I TCP transcription factors that contain Cys-20 within the plant.
Figure 9.
Modulation of CUC1 expression by class I TCP proteins is affected by H2O2 treatment. An Arabidopsis GUS reporter line for CUC1 expression was transformed with constructs that express fusions of class I TCP proteins to the EAR repressor domain under the control of the 35S CaMV promoter or with constructs that express the HD-Zip transcription factor Hahb4 (Dezar et al., 2005) or GUS as a control. Twenty-four hours after transformation, plants were treated with either water (Control) or 4 mm H2O2 during 12 h and then used to analyze GUS expression by histochemistry. About 40 transformed plants were analyzed for each condition. Representative images of the patterns obtained most frequently in each case are shown. A quantitative evaluation of the data are shown in Table I. The experiment was repeated twice with similar results.
Table I. Effect of H2O2 treatment on TCP protein-dependent CUC1 expression.
n = number of plants.
| Constructa | H2O2 Treatment | Total | CUC1 Patternb | Extended Patternc | P (Treated versus Untreated)d |
|---|---|---|---|---|---|
| n | n | n | |||
| HAHB4 | No | 35 | 35 | 0 | 1.000 |
| Yes | 31 | 31 | 0 | ||
| TCP15-EAR | No | 39 | 4 | 35 | <0.0001 |
| Yes | 40 | 29 | 11 | ||
| TCP20-EAR | No | 36 | 5 | 31 | <0.0001 |
| Yes | 39 | 37 | 2 | ||
| C20S-TCP15-EAR | No | 31 | 2 | 29 | 1.000 |
| Yes | 38 | 2 | 36 |
Construct used for transformation. Expression was under the control of the 35S CaMV promoter. bExpression in the meristematic region. No or very low expression was seen in cotyledons. cExpression in cotyledon lamina (more than 50% of surface). dChange in expression pattern after H2O2 treatment. Two-tailed P values were calculated using Fisher’s exact test.
DISCUSSION
Current evidence indicates that TCP transcription factors appeared during the development of Streptophyta and that the family has considerably expanded in angiosperms, which usually contain more than 20 members (Navaud et al., 2007). All species analyzed contain two classes of TCP proteins that differ in the structure of the TCP domain and DNA-binding characteristics (Kosugi and Ohashi, 2002; Viola et al., 2012). Analysis of the consensus amino acid sequences of members of both classes shows the presence of residues that are highly conserved in all TCP proteins and others that are conserved only in one of the classes. Conservation most likely indicates a relevant role of these residues in TCP protein function. A conserved residue is Cys-20, which is present in more than 94% of class I TCP proteins. Our results indicate that Cys-20 is able to undergo redox interconversions or modifications that affect the DNA-binding properties of class I TCP proteins.
Cys-20 Is Probably Located at the Dimer Interface in Class I TCP Proteins
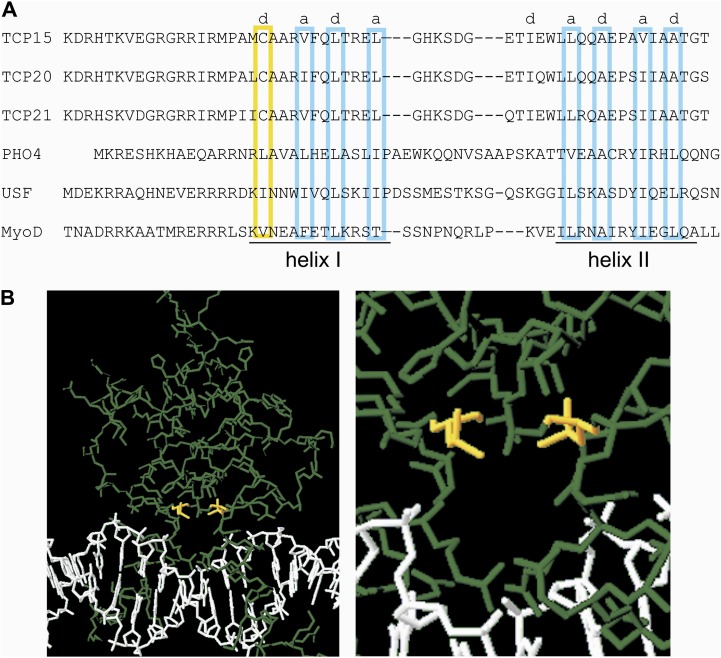
The structure of the TCP domain has been deduced through comparisons with the bHLH domain, present in eukaryotic transcription factors (Aggarwal et al., 2010). Unfortunately, the sequence similarity between the class I TCP and the bHLH domains is too low to obtain a reliable model of the first based on known structures of the second. In bHLH proteins, the dimerization (HLH) domain is composed of two helices separated by a disordered loop, and contacts are established between hydrophobic helical residues present at positions a and d of successive heptads. Assuming that the dimerization domain of TCP proteins has a similar structure, as suggested by Cubas et al. (1999) and Aggarwal et al. (2010), we aligned the dimerization domains of TCP15, TCP20, and TCP21 with those of the bHLH proteins PHO4, USF, and MyoD (Ellenberger et al., 1994; Ferré-D’Amaré et al., 1994; Shimizu et al., 1997) in such a way that hydrophobic residues of helices I and II show a similar arrangement (Fig. 10A). This analysis indicated that Cys-20 of the class I TCP domain, present at the beginning of helix I, is probably located at position d of the first heptad (Fig. 10A). Residue d1 is usually hydrophobic and faces the dimer interface in bHLH proteins (Ellenberger et al., 1994). Accordingly, it can be assumed that Cys-20 also faces the dimer interface in TCP proteins. The crystal structure of the bHLH domain of PHO4 bound to DNA, taken from Shimizu et al. (1997), is shown in Figure 10B. It can be observed that Leu residues at position 16 of the respective monomers, equivalent to Cys-20 of the TCP domain, are located at a close distance (4.8 Å), facing each other. Similar observations can be made in other bHLH-DNA complex structures (Ellenberger et al., 1994; Ferré-D’Amaré et al., 1994). Modeling of a PHO4 protein with Cys instead of Leu-16 yields a distance of 5.9 Å, which is too long for disulfide bond formation (usually around 3.5 Å). However, it can be envisaged that subtle changes in structure may produce a suitable conformation for this reaction to occur. This suitable conformation may form when the protein is not bound to DNA due to increased flexibility. Once the disulfide bond is formed, the protein may be locked in a conformation unsuitable for efficient DNA binding, thus explaining the redox-dependent inactivation observed in this study. In addition, the close proximity of the Cys residues to DNA may explain the fact that their oxidation or modification by redox or alkylating agents affects DNA binding and why the protein is considerably less sensitive to oxidation or modification when bound to DNA. Additionally, the dimer bound to DNA may be locked in a conformation that precludes disulfide bond formation, because the distance between Cys-20 of both monomers may be too large for this reaction to occur.
Figure 10.
Cys-20 is probably located at the dimer interface and near DNA in the class I TCP-DNA complex. A, Alignment of the HLH regions of class I TCP proteins and the bHLH proteins PHO4, USF, and MyoD. Positions a and d (boxed) are occupied by hydrophobic residues and are located at the dimer interface. Cys-20 is located at position d1 (boxed in yellow) in class I TCP proteins. B, The structure of the PHO4 bHLH domain bound to DNA (Shimizu et al., 1997). Leu-16, located at position d1 of helix I in both monomers, is shown in yellow. The right panel shows a detail of the region around Leu-16. Structures were visualized using Swiss-PdbViewer (http://www.expasy.org/spdbv/).
Redox Agents Modulate TCP Protein Function in Vivo
The fact that several redox agents that are present within cells influence the DNA-binding properties of class I TCP proteins suggests that redox interconversions may influence TCP protein function in vivo. Dramatic changes in cellular redox potential have been observed under oxidative stress caused by incubation of tissues or cells with H2O2 (Jiang et al., 2006; Meyer et al., 2007). Accordingly, treatment of yeast cells with H2O2 significantly affected the ability of TCP15 to induce the expression of a reporter gene located under the control of the TCP15 target site. This strongly suggests that TCP15 function is affected by oxidative stress, most likely through modification of its redox-active Cys, since the action of TCP16, which contains Val-20, was barely affected under similar conditions. H2O2 triggers an oxidative stress response in yeast that involves the redox-dependent activation of the Yap1 transcription factor (Delaunay et al., 2000). Yap1 activates the expression of antioxidant enzymes that help to reestablish normal redox conditions within cells (Lee et al., 1999). Our results suggest that TCP15 may be inactivated by oxidation under the same conditions that promote the activation of Yap1.
We also showed that the DNA-binding activity of TCP transcription factors can be modulated by direct treatment of plants with H2O2. It has been shown previously that H2O2 treatment produces changes in redox potential within plant cells (Jiang et al., 2006; Meyer et al., 2007). Our results suggest that these redox changes influence the activity of class I TCP transcription factors within the plant. In agreement with this, we have observed that the induction of the boundary-specific gene CUC1 by repressor forms of class I TCP proteins is affected by treatment with a redox agent and that this is dependent on the presence of Cys at position 20 of the TCP domain. This indicates that redox-dependent modifications of TCP protein activity have an impact on downstream events regulated by these transcription factors.
TCP Proteins May Participate in Redox-Dependent Developmental Processes
The exact role of many class I TCP proteins is not currently known, but their functions have been associated with the regulation of development, probably through changes in cell proliferation or growth (Martín-Trillo and Cubas, 2010; Uberti Manassero et al., 2013). Since there is abundant evidence that redox agents modulate several aspects of plant development, from root meristem maintenance to flower and embryo development (May et al., 1998; Vernoux et al., 2000; Foreman et al., 2003; Xing et al., 2005; Cairns et al., 2006; Carol and Dolan, 2006; Pasternak et al., 2008), it is possible that some of these effects may be exerted through the modulation of TCP protein activity. AtTCP20 and other class I proteins interact with the promoters of the mitotic cyclin CYCB1;1, ribosomal protein genes, and genes involved in mitochondrial biogenesis, whose expression is linked to cell proliferation (Trémousaygue et al., 2003; Li et al., 2005; Welchen and Gonzalez, 2005; Giraud et al., 2010). AtTCP14 and AtTCP15 influence cell proliferation, endoreduplication, and the expression of cell cycle-regulated genes (Tatematsu et al., 2008; Kieffer et al., 2011; Li et al., 2012). It has been shown that GSH is required for cell cycle progression and that it is localized to the nucleus at early stages of cell proliferation in plants (Pellny et al., 2009; Diaz-Vivancos et al., 2010). An increase in GSH in the nucleus may serve, among other functions, to modulate the activity of transcription factors involved in cell cycle progression. Class I TCP proteins may be among the proteins that respond to these changes in redox conditions and serve as intermediates in the redox regulation of cell proliferation. An additional role of the modification of class I TCP proteins by redox agents may be the protection of these proteins from permanent inactivation under oxidative stress conditions, considering that disulfide bond formation and modification by GSH, H2O2, or NO are reversible processes.
In addition, it is becoming increasingly evident that plants respond to chronic stress conditions through the modification of developmental programs, a phenomenon known as stress-induced morphogenic response (Potters et al., 2007, 2009). Reactive oxygen species and auxin are thought to be major players in this response. Since some class I TCP proteins have been shown to influence auxin homeostasis (Uberti Manassero et al., 2012), redox modification of the activity of these proteins may provide a link between redox conditions, auxin metabolism, and developmental responses to stress. The identification of targets of TCP proteins that are related to redox metabolism or whose expression is altered by redox conditions will probably lead to the establishment of the molecular mechanisms involved in this link.
CONCLUSION
In conclusion, we have determined that class I TCP proteins that contain Cys at position 20 of the TCP domain are sensitive to redox conditions of the environment. Since a majority of class I proteins contain Cys-20, our results can be extended to most members of this class. Inactivation of TCP proteins by oxidation may result from the formation of disulfide bonds or the modification of Cys-20, located at the dimer interface near the DNA-binding surface of the protein, making these proteins attractive candidates as mediators of the effects of redox conditions on several aspects of plant development.
MATERIALS AND METHODS
Cloning, Expression, and Purification of Recombinant Proteins
The complete coding regions of Arabidopsis (Arabidopsis thaliana) TCP15 and TCP21 as well as shorter forms of TCP20 (amino acids 1–157) and TCP16 (amino acids 1–80) were cloned in frame with MBP in expression vector pMAL-c2 (New England Biolabs) as described (Viola et al., 2011, 2012). A TCP15 mutant containing Ser at position 20 of the TCP domain (C20S-TCP15) was constructed by overlap extension mutagenesis (Silver et al., 1995) using complementary oligonucleotides with the desired mutations (5′-CCTGCCATGTCTGCTGCACGTG-3′ and 5′-CACGTGCAGCAGACATGGCAGG-3′). All constructs were checked by DNA sequencing. For expression, Escherichia coli cells bearing the corresponding plasmids were grown and induced as described previously (Viola et al., 2011). Purification of recombinant proteins was performed as indicated by the manufacturers of the pMAL-c2 system, except that 2-mercaptoethanol (2-ME) was omitted from the purification buffer. Purified proteins (greater than 95% as judged by Coomassie Brilliant Blue staining of denaturing polyacrylamide gels) were used for the assays. Protein amounts were measured as described by Sedmak and Grossberg (1977). DNA-binding assays were performed with the proteins fused to MBP. Controls made with proteins obtained after cleavage with factor Xa indicated that the MBP moiety does not affect the behavior of the recombinant proteins. In the case of recombinant TCP15 and its mutant, several products of smaller size than the full-length proteins were observed by SDS-PAGE. According to western-blot analysis using antibodies against MBP (Supplemental Fig. S1), these products are truncated forms of the proteins containing MBP and portions of TCP15. Some of these products may be active toward DNA binding, thus explaining the presence of more than one DNA-protein complex in EMSAs, especially when exposed during prolonged periods of time (Figs. 1C, 2A, and 6D).
E. coli thioredoxin1 (encoded by the trxA gene) was expressed from plasmid pET-32a(+) (Novagen) and purified by nickel affinity chromatography. E. coli thioredoxin reductase (encoded by the trxB gene) was expressed from plasmid pTrR301 and purified as described by Mulrooney (1997).
Treatment of Proteins with Redox Agents and NO Donors
When indicated, purified proteins were dialyzed overnight at 4°C in 20 mm Tris-HCl (pH 7.4), 200 mm NaCl, and 1 mm EDTA. Treatments with redox agents were performed in the same buffer during 1 h at 25°C. Reagents were dissolved in the same buffer. Incubations with the NO donor SNP were performed for 1 h under white light to produce the photolysis of SNP. In the case of GSNO, incubations were performed on ice and in the dark. Incubation with NEM was also performed in darkness. Diamide, DTT, GSH, GSSG, NEM, and SNP were purchased from Sigma. GSNO was prepared by nitrosation under acid conditions as described previously (Gordge et al., 1998). Briefly, equal volumes of GSH (200 mm in 50 mm HCl) and sodium nitrite (200 mm) were incubated on ice for 30 min. GSNO was stabilized by the addition of 1 mm EDTA, and its concentration was estimated by A334 using a molar absorption coefficient of 0.96 mm−1 cm−1. The reagent was prepared fresh each day and kept on ice in the dark until use.
Electrophoresis of Proteins
Nonreducing SDS-PAGE was performed essentially as described by Laemmli (1970), except that 2-ME was omitted from the loading buffer. Samples (1 µg of MBP-TCP fusion protein) were preincubated at room temperature in 20 mm Tris-HCl (pH 7.4), 200 mm NaCl, and 1 mm EDTA plus the indicated additions, mixed with loading buffer, boiled during 5 min, and loaded onto a 12% (w/v) polyacrylamide gel. After electrophoresis, gels were stained with Coomassie Brilliant Blue.
Western-Blot Analysis
For western-blot analysis, proteins were separated through SDS-PAGE and then transferred to Hybond-ECL (GE Healthcare Life Sciences) membranes. Membranes were stained with Ponceau S and subsequently probed with antibodies against MBP (New England Biolabs), in the case of purified recombinant proteins, or the hemagglutinin tag (Invitrogen), in the case of extracts from yeast (Saccharomyces cerevisiae) cells, at a dilution of 1:1,000, and developed with anti-mouse or anti-rabbit immunoglobin conjugated with horseradish peroxidase using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Gene Cloning and Plant Transformation
TCP20 was expressed in plants fused to the EAR repressor domain. For this purpose, TCP20 coding sequences spanning nucleotides 7 to 456 were amplified with specific primers, fused to a double-stranded oligonucleotide with EAR domain coding sequences, and cloned in the binary vector pBI121 under the control of the 35S CaMV promoter, as described previously for TCP15 (Uberti Manassero et al., 2012). The construct to express full-length C20S-TCP15-EAR was obtained in a similar way. Constructs were introduced into either wild-type Arabidopsis plants or a stably transformed line that expresses the uidA (GUS) reporter gene under the control of the relevant CUC1 regulatory regions (Spinelli et al., 2011) by transient transformation, essentially as described by Li et al. (2009) with modifications described by Uberti Manassero et al. (2012). Histochemical GUS assays were performed 36 h after transformation as described by Hull and Devic (1995). Total protein extracts were prepared from plants 36 h after transformation. For this purpose, frozen tissue (0.2 g) was ground to powder under liquid N2 and homogenized for 1 min in 0.5 mL of lysis buffer containing 50 mm HEPES-KOH, pH 7.5, 100 mm KCl, 5 mm EDTA, 10% glycerol, 0.1% Triton X-100, 5% polyvinylpyrrolidone, and 1 mm phenylmethylsulfonyl fluoride. Extracts were centrifuged at 14,000g and 4°C during 20 min. Protein in the supernatant was quantified as described by Sedmak and Grossberg (1977).
DNA-Binding Assays
For EMSAs, purified recombinant proteins (50–500 ng, depending on the amount of DNA used) were incubated with double-stranded DNA (0.3–0.6 ng, 30,000 cpm) generated by hybridization of the complementary oligonucleotides 5′-AATTCAGATCTGTGGGCCCACGAG-3′ and 5′-GATCCTCGTGGGCCCACAGATCTG-3′ (binding sites underlined) and labeled with [α-32P]dATP by filling in the 3′ ends using the Klenow fragment of E. coli DNA polymerase I. Alternatively, one of the oligonucleotides was 5′ end labeled with 6-FAM. Binding reactions (20 µL) containing, in addition to protein and labeled DNA, 20 mm HEPES (pH 7.5), 50 mm KCl, 2 mm MgCl2, 0.5 mm EDTA, 0.5% Triton X-100, 1 µg of poly(dI-dC), and 10% glycerol were incubated on ice for 20 min, supplemented with 2.5% Ficoll, and immediately loaded onto a running gel (5% acrylamide, 0.08% bis-acrylamide in 0.5× TBE plus 2.5% glycerol; 1× TBE is 90 mm Tris-borate, pH 8.3, 2 mm EDTA). The gel was run in 0.5× TBE at 20 mA for 2 h and dried prior to autoradiography or directly scanned for fluorescence. Quantitative analysis was performed with a Typhoon (GE Healthcare) scanner. EMSAs using protein extracts from transformed plants were performed in a similar way, except that 10 µg of protein extract and complementary oligonucleotides containing the preferred binding sequences of TCP20 (5′-AATTCAGATCTGTGGGACCGGGAG-3′ and 5′-GATCCTCCCGGTCCCACAGATCTG-3′; Viola et al., 2011) were used. For competition, complementary oligonucleotides with changes in the TCP20 binding site (m1, 5′-AATTCAGATCTGTAGGACTGGGAG-3′ and 5′-GATCCTCCCAGTCCTACAGATCTG-3′; m2, 5′-AATTCAGATCTCAATAATTGAGAG-3′ and 5′-GATCCTCTCAATTATTGAGATCTG-3′) were used.
One-Hybrid Analysis in Yeast
Yeast strains carrying the TCP15 or TCP16 binding sequence inserted into the genome in front of the lacZ reporter gene (Viola et al., 2012) were transformed with constructs that express fusions of the TCP proteins to the GAL4 activation domain in plasmid pGADT7 (Clontech). Transformed cells at an optical density at 600 nm of 0.3 were incubated either in the absence or presence of H2O2, SNP, or GSNO. After incubation, specific β-galactosidase activity was assayed as described by Reynolds et al. (1997) using o-nitrophenylgalactoside as substrate. In addition, an amount of cells corresponding to an optical density at 600 nm of 7.5 was disrupted using the urea/SDS protein extraction method (Printen and Sprague, 1994). Extracts from these cells were analyzed through SDS-PAGE followed by western blotting to detect the expression of the fusion proteins as described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Western-blot analysis of recombinant MBP-TCP15 and its mutant using anti-MBP antibodies.
Supplemental Figure S2. Gel-filtration analysis of recombinant MBP-TCP20.
Supplementary Material
Acknowledgments
We thank Javier Palatnik (Instituto de Biología Celular y Molecular de Rosario) for sending us the CUC1 reporter line and Vanesa Piattoni (Instituto de Agrobiotecnología del Litoral) for help with gel-filtration studies.
Glossary
- HLH
helix-loop-helix
- bHLH
basic helix-loop-helix
- MBP
maltose-binding protein
- GSSG
oxidized glutathione
- DTT
dithiothreitol
- GSH
reduced glutathione
- NEM
N-ethylmaleimide
- H2O2
hydrogen peroxide
- NO
nitric oxide
- SNP
sodium nitroprusside
- GSNO
S-nitrosoglutathione
- CaMV
cauliflower mosaic virus
- EMSA
electrophoretic mobility shift assay
- 2-ME
2-mercaptoethanol
References
- Aggarwal P, Das Gupta M, Joseph AP, Chatterjee N, Srinivasan N, Nath U. (2010) Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick JA, Setterdahl AT, Knaff DB, Chen Y, Pitcher LH, Zilinskas BA, Leustek T. (2001) Regulation of the plant 5'-adenylyl sulfate reductase by oxidative stress. Biochemistry 40: 9040–9048 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. (2006) Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Dolan L. (2006) The role of reactive oxygen species in cell growth: lessons from root hairs. J Exp Bot 57: 1829–1834 [DOI] [PubMed] [Google Scholar]
- Comelli RN, Gonzalez DH. (2007) Conserved homeodomain cysteines confer redox sensitivity and influence the DNA binding properties of plant class III HD-Zip proteins. Arch Biochem Biophys 467: 41–47 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Delaunay A, Isnard AD, Toledano MB. (2000) H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19: 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezar CA, Gago GM, Gonzalez DH, Chan RL. (2005) Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res 14: 429–440 [DOI] [PubMed] [Google Scholar]
- Diaz Vivancos P, Wolff T, Markovic J, Pallardó FV, Foyer CH. (2010) A nuclear glutathione cycle within the cell cycle. Biochem J 431: 169–178 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Fass D, Arnaud M, Harrison SC. (1994) Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev 8: 970–980 [DOI] [PubMed] [Google Scholar]
- Fairman R, Beran-Steed RK, Anthony-Cahill SJ, Lear JD, Stafford WF, III, DeGrado WF, Benfield PA, Brenner SL. (1993) Multiple oligomeric states regulate the DNA binding of helix-loop-helix peptides. Proc Natl Acad Sci USA 90: 10429–10433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR, Pognonec P, Roeder RG, Burley SK. (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J 13: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V, Wagner R, Braütigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T. (2005) Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem 280: 5318–5328 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J. (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DH, Welchen E, Attallah CV, Comelli RN, Mufarrege EF. (2007) Transcriptional coordination of the biogenesis of the oxidative phosphorylation machinery in plants. Plant J 51: 105–116 [DOI] [PubMed] [Google Scholar]
- Gordge MP, Hothersall JS, Noronha-Dutra AA. (1998) Evidence for a cyclic GMP-independent mechanism in the anti-platelet action of S-nitrosoglutathione. Br J Pharmacol 124: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Luan S. (2003) Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol 132: 1149–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine GF, Hernandez JM, Grotewold E. (2004) Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J Biol Chem 279: 37878–37885 [DOI] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. (2009) In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks LM, Cahoon RE, Bonner ER, Rivard RS, Sheffield J, Jez JM. (2007) Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell 19: 2653–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull GA, Devic M (1995) The beta-glucuronidase (gus) reporter gene system. Gene fusions: spectrophotometric, fluorometric, and histochemical detection. In H Jones, ed, Methods in Plant Molecular Biology: Plant Gene Transfer and Expression Protocols. Humana Press, Totowa, NJ, pp 125–141 [DOI] [PubMed] [Google Scholar]
- Jiang K, Schwarzer C, Lally E, Zhang S, Ruzin S, Machen T, Remington SJ, Feldman L. (2006) Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol 141: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee J, Godon C, Lagniel G, Spector D, Garín J, Labarre J, Toledano MB. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem 274: 16040–16046 [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Park E, von Arnim AG, Nebenführ A. (2009) The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li B, Dong AW. (2012) The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant 5: 270–280 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J. (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- May M, Vernoux T, Leaver C, Van Montagu M, Inze D. (1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J Exp Bot 49: 649–667 [Google Scholar]
- Meinhard M, Rodriguez PL, Grill E. (2002) The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta 214: 775–782 [DOI] [PubMed] [Google Scholar]
- Meyer AJ. (2008) The integration of glutathione homeostasis and redox signaling. J Plant Physiol 165: 1390–1403 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Mulrooney SB. (1997) Application of a single-plasmid vector for mutagenesis and high-level expression of thioredoxin reductase and its use to examine flavin cofactor incorporation. Protein Expr Purif 9: 372–378 [DOI] [PubMed] [Google Scholar]
- Nag A, King S, Jack T. (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E. (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O, Dabos P, Carnus E, Tremousaygue D, Hervé C. (2007) TCP transcription factors predate the emergence of land plants. J Mol Evol 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Noctor G, Gomez L, Vanacker H, Foyer CH. (2002) Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J Exp Bot 53: 1283–1304 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pasternak M, Lim B, Wirtz M, Hell R, Cobbett CS, Meyer AJ. (2008) Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J 53: 999–1012 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Pellny TK, Locato V, Vivancos PD, Markovic J, De Gara L, Pallardó FV, Foyer CH. (2009) Pyridine nucleotide cycling and control of intracellular redox state in relation to poly (ADP-ribose) polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol Plant 2: 442–456 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Jansen MAK. (2009) Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ 32: 158–169 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Palme KJ, Jansen MAK. (2007) Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci 12: 98–105 [DOI] [PubMed] [Google Scholar]
- Printen JA, Sprague GF., Jr (1994) Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138: 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Lundblad V, Dorris D, Keavency M (1997) Yeast vectors and assays for expression of cloned genes. In F Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology. Greene Publishing and Wiley-Interscience, New York, pp 13.6.1–13.6.6 [Google Scholar]
- Rouhier N, Lemaire SD, Jacquot JP. (2008) The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu Rev Plant Biol 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Sedmak JJ, Grossberg SE. (1977) A rapid, sensitive, and versatile assay for protein using Coomassie Brilliant Blue G250. Anal Biochem 79: 544–552 [DOI] [PubMed] [Google Scholar]
- Serpa V, Vernal J, Lamattina L, Grotewold E, Cassia R, Terenzi H. (2007) Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem Biophys Res Commun 361: 1048–1053 [DOI] [PubMed] [Google Scholar]
- Shaikhali J, Heiber I, Seidel T, Ströher E, Hiltscher H, Birkmann S, Dietz KJ, Baier M. (2008) The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol 8: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikhali J, Norén L, de Dios Barajas-López J, Srivastava V, König J, Sauer UH, Wingsle G, Dietz KJ, Strand Å. (2012) Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. J Biol Chem 287: 27510–27525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. (1997) Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J 16: 4689–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J, Limjoco T, Feinstone S (1995) Site-specific mutagenesis using the polymerase chain reaction. In MA Innis, DH Gelfand, JJ Sninsky, eds, PCR Strategies. Academic Press, San Diego, pp 179–188 [Google Scholar]
- Spinelli SV, Martin AP, Viola IL, Gonzalez DH, Palatnik JF. (2011) A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol 156: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starovasnik MA, Blackwell TK, Laue TM, Weintraub H, Klevit RE. (1992) Folding topology of the disulfide-bonded dimeric DNA-binding domain of the myogenic determination factor MyoD. Biochemistry 31: 9891–9903 [DOI] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. (2012) The ArabidopsisO-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Takeda T, Amano K, Ohto MA, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C. (2006) RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol Biol 61: 165–177 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E. (2008) Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J 53: 42–52 [DOI] [PubMed] [Google Scholar]
- Tavakoli N, Kluge C, Golldack D, Mimura T, Dietz KJ. (2001) Reversible redox control of plant vacuolar H+-ATPase activity is related to disulfide bridge formation in subunit E as well as subunit A. Plant J 28: 51–59 [DOI] [PubMed] [Google Scholar]
- Terrile MC, París R, Calderón-Villalobos LI, Iglesias MJ, Lamattina L, Estelle M, Casalongué CA. (2012) Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J 70: 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trémousaygue D, Garnier L, Bardet C, Dabos P, Hervé C, Lescure B. (2003) Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33: 957–966 [DOI] [PubMed] [Google Scholar]
- Tron AE, Bertoncini CW, Chan RL, Gonzalez DH. (2002) Redox regulation of plant homeodomain transcription factors. J Biol Chem 277: 34800–34807 [DOI] [PubMed] [Google Scholar]
- Uberti Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH. (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Uberti Manassero NG, Viola IL, Welchen E, Gonzalez DH. (2013) TCP transcription factors: architectures of plant form. Biomol Concepts 4: 111–127 [DOI] [PubMed] [Google Scholar]
- Veine DM, Arscott LD, Williams CH Jr. (1998) Redox potentials for yeast, Escherichia coli and human glutathione reductase relative to the NAD+/NADH redox couple: Enzyme forms active in catalysis. Biochemistry 37: 15575–15582 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Reinheimer R, Ripoll R, Manassero NG, Gonzalez DH. (2012) Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J Biol Chem 287: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH. (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435: 143–155 [DOI] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. (2005) Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2: evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol 139: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. (2006) Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiol 141: 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt H, Thomas RM. (1997) The self-association of basic helix-loop-helix peptides. Prog Colloid Polym Sci 107: 115–121 [Google Scholar]
- Xing S, Rosso MG, Zachgo S. (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.