Carotenoids are essential for nodule development in soybean.
Abstract
Legume-Rhizobium spp. symbiosis requires signaling between the symbiotic partners and differential expression of plant genes during nodule development. Previously, we cloned a gene encoding a putative β-carotene hydroxylase (GmBCH1) from soybean (Glycine max) whose expression increased during nodulation with Bradyrhizobium japonicum. In this work, we extended our study to three GmBCHs to examine their possible role(s) in nodule development, as they were additionally identified as nodule specific, along with the completion of the soybean genome. In situ hybridization revealed the expression of three GmBCHs (GmBCH1, GmBCH2, and GmBCH3) in the infected cells of root nodules, and their enzymatic activities were confirmed by functional assays in Escherichia coli. Localization of GmBCHs by transfecting Arabidopsis (Arabidopsis thaliana) protoplasts with green fluorescent protein fusions and by electron microscopic immunogold detection in soybean nodules indicated that GmBCH2 and GmBCH3 were present in plastids, while GmBCH1 appeared to be cytosolic. RNA interference of the GmBCHs severely impaired nitrogen fixation as well as nodule development. Surprisingly, we failed to detect zeaxanthin, a product of GmBCH, or any other carotenoids in nodules. Therefore, we examined the possibility that most of the carotenoids in nodules are converted or cleaved to other compounds. We detected the expression of some carotenoid cleavage dioxygenases (GmCCDs) in wild-type nodules and also a reduced amount of zeaxanthin in GmCCD8-expressing E. coli, suggesting cleavage of the carotenoid. In view of these findings, we propose that carotenoids such as zeaxanthin synthesized in root nodules are cleaved by GmCCDs, and we discuss the possible roles of the carotenoid cleavage products in nodulation.
Legume-Rhizobium spp. symbiosis results in the formation of the root nodule, in which rhizobia fix atmospheric nitrogen. Nodule development requires diverse events, such as Nod factor synthesis in the rhizobia, perception of the Nod factor on plant roots by receptor-like kinases, endocytosis of rhizobia into plant cells, and so on (Stacey et al., 2006; Oldroyd et al., 2011; Singh and Parniske, 2012). Sequential expression of numerous plant genes occurs during nodulation, contributing to different stages including nitrogen fixation. Arbuscular mycorrhizal (AM) symbiosis exhibits many similarities to the nodulation process (Oldroyd et al., 2009). For example, SymRK, the receptor-like kinase gene, is required for both rhizobial and AM symbioses (Stracke et al., 2002). Similarly, the signal transduction pathways following perception are also in part the same, and the genes common to the two pathways have been referred to as the common symbiosis (SYM) genes (Kistner et al., 2005). These similarities may reflect common mechanisms for host plant cells to respond to symbionts, although the commonality is not globally defined yet.
Plant carotenoids are mostly C40 tetraterpenoid pigments with a series of double bonds (DellaPenna and Pogson, 2006; Lu and Li, 2008). They play essential roles in photosynthesis. The phytohormone abscisic acid (ABA) is synthesized from xanthophylls, oxygenated derivatives of carotenoids. The beneficial effects of carotenoids for human disease prevention and health promotion are well established and are based on their antioxidant activities (Kopsell and Kopsell, 2006; Rao and Rao, 2007; von Lintig, 2010). Metabolic engineering approaches have produced crop plants with enhanced carotenoid contents and improved nutritional value (Giuliano et al., 2008). For example, enhancement of β-carotene, provitamin A, by engineering the carotenoid biosynthetic pathway resulted in the development of cv Golden rice (Oryza sativa; Ye et al., 2000; Paine et al., 2005; Ha et al., 2010).
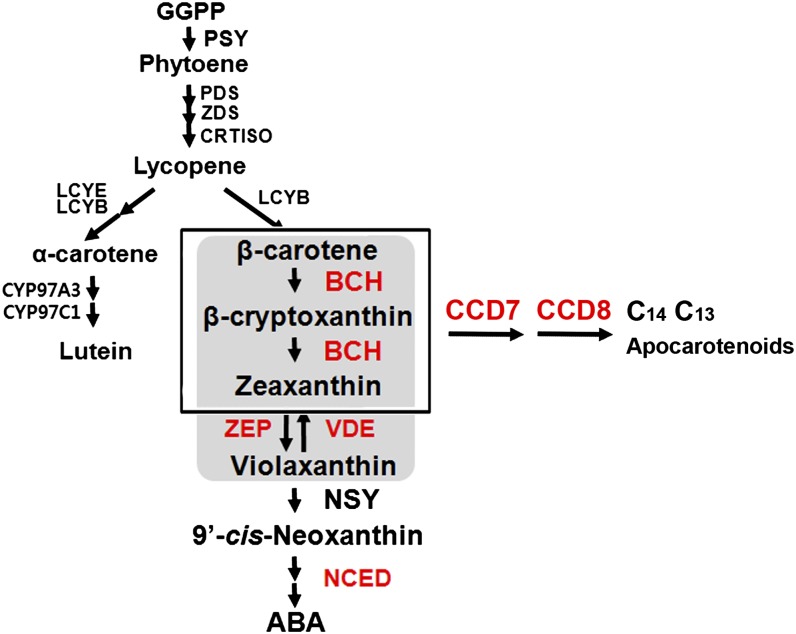
The initial step of carotenoid biosynthesis is the production of phytoene by the enzyme phytoene synthase (Fig. 1; DellaPenna and Pogson, 2006; Cazzonelli and Pogson, 2010). The subsequent activities of desaturases, isomerase, and cyclase convert phytoene into lycopene and further into β-carotene. Xanthophyll synthesis begins with the action of β-carotene hydroxylase (BCH) on β-carotene, producing initially β-cryptoxanthin and thereafter zeaxanthin (Kim et al., 2009). Overexpression of BCH has been found to confer tolerance to light stress (Davison et al., 2002). The subsequent steps catalyzed by zeaxanthin epoxidase (ZEP) and neoxanthin synthase lead to the synthesis of ABA (Takaichi and Mimuro, 1998).
Figure 1.
The biosynthetic pathway of carotenoids in plants. GGPP, Geranylgeranyl diphosphate; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotene isomerase; LCYB, lycopene β-cyclase; CYP97A3 and CYP97C1, cytochrome P450 enzymes; NSY, neoxanthin synthase; LCYE, lycopene ε-cyclase; CRTR-E, ε-carotene hydroxylase. Enzymes in red were examined in this study.
Various carotenoid cleavage dioxygenases (CCDs) catalyze the formation of apocarotenoids with functions as hormones, flavors, and pigments (Auldridge et al., 2006b; Strack and Fester, 2006; Tsuchiya and McCourt, 2009; Walter et al., 2010). Recently, CCD7 and CCD8 were shown to control the synthesis of strigolactones, newly discovered hormones that inhibit shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008; Vogel et al., 2010; Ruyter-Spira et al., 2013). In addition, carotenoid cleavage products have been discovered in plant roots colonized by AM fungi (Strack and Fester, 2006). During AM symbiosis, roots synthesize apocarotenoids at the same time as activating plant genes for carotenoid metabolism. Although RNA interference (RNAi)-mediated inhibition of apocarotenoid synthesis suggests that apocarotenoids are functionally significant (Snowden et al., 2005; Floss et al., 2008), their role in AM symbiosis is unknown.
In a search for genes differentially induced during soybean (Glycine max)-Rhizobium spp. symbiosis, several antioxidant genes, including a gene encoding a putative BCH, were identified. In this report, we describe genes (GmBCHs) encoding a putative BCH whose expression increased in soybean root nodules. Therefore, the biochemical activities of BCHs were investigated. RNAi inhibition of GmBCH expression interfered with nitrogen fixation as well as nodule development. Subsequent analysis of the expression and biochemical activities of GmCCDs in root nodules led us to hypothesize that GmCCD8 could be involved in the synthesis of apocarotenoids from zeaxanthin in these nodules.
RESULTS
Expression of Genes Encoding Putative BCHs in Soybean
We isolated from root nodules of soybean a complementary DNA (cDNA) with strong homology to BCH (GmBCH1), whose expression was higher in nodules than in roots (Fig. 2A; Lee et al., 2005). In a BLAST search with the GmBCH1 sequence against the soybean genome (http://www.phytozome.net/soybean.php; Schmutz et al., 2010), we found several open reading frames encoding products with amino acid sequences highly homologous to that of GmBCH1; they were designated GmBCH2 to GmBCH5 (GenBank accession numbers are as follows: GmBCH1 [AY575953], GmBCH2 [BT093388], GmBCH3 [BT098487], GmBCH4 [JF970190], GmBCH5 [JF970191]). The open reading frames of GmBCH1 and GmBCH2 are very similar to functionally confirmed BCHs (e.g. they have 74% and 75% sequence identity, respectively, to the BCH from coffee [Coffea arabica], CaCRTR-B; Simkin et al., 2008; Supplemental Fig. S1A). They have divergent N-terminal regions, like most previously reported BCHs, but carry four His-containing motifs, in which the spacing of the His residues was conserved (e.g. HXXXXH and HXXHH). The presence of the His residues in these motifs has been confirmed to be essential for the enzymatic activity, as mutagenesis abolishing these His residues resulted in no enzymatic conversion of β-carotene into zeaxanthin (Supplemental Fig. S1A; Bouvier et al., 1998). GmBCH2 appears to possess a plastid transit sequence (see below; Yu et al., 2007), and GmBCH3 has a sequence almost identical to GmBCH1 except for its N-terminal 33 amino acids, so it also contains the above-mentioned motifs common to BCHs. The 5′ untranslated regions of GmBCH1 and GmBCH3 differ. Whereas the GmBCH3 locus could be identified in the present version of the soybean genome database, the unique 5′ region of the GmBCH1 sequence was not detected in the database. Therefore, we attempted to establish the presence of GmBCH1 in the soybean genome by cloning its specific 5′ DNA region (see “Materials and Methods”). A 432-bp DNA fragment, which included the upstream promoter region of GmBCH1, was cloned (Supplemental Fig. S2A), and this 5′ region as well as the coding region of GmBCH1 were also detected by genomic PCR (Supplemental Fig. S2B, lanes 3 and 6). GmBCH4 and GmBCH5 also contain the motifs mentioned above and thus are regarded as additional BCH paralogs (Supplemental Fig. S1A).
Figure 2.
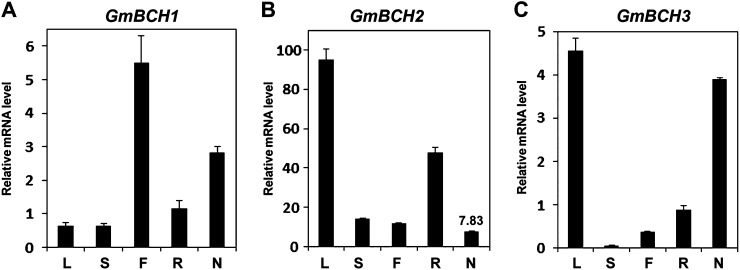
Expression of the soybean BCHs GmBCH1, GmBCH2, and GmBCH3 in soybean tissues, including 27-d-old nodules. Transcript levels were determined by real-time RT-PCR and normalized with the geometric mean of three reference genes (GmELF1b, GmActin2/GmActin7, and Ubiquitin; Vandesompele et al., 2002). Data are representative of three independent experiments. Error bars represent sd (n = 3). L, Leaf; S, stem; F, flower; R, root; N, nodule.
We investigated the expression of the GmBCHs in various tissues by real-time reverse transcription (RT)-PCR. To optimize the PCR, we examined critical aspects of the primers (Supplemental Fig. S3). The expression of GmBCH2 was higher than that of GmBCH1 and GmBCH3 in most tissues, particularly in leaf, and relatively low in nodules, whereas expression of GmBCH1 and GmBCH3 was high in leaf and flower and noticeable in root nodules (Fig. 2). Expression of GmBCH4 and GmBCH5 was high in leaf and quite low in roots and nodules. These results generally match with the RNA-Seq Atlas data (Libault et al., 2010; Severin et al., 2010; Supplemental Table S1). GmBCH3 expression in the transcriptomic data might actually reflect the expression of both GmBCH1 and GmBCH3, because the coding regions of both genes are almost identical and GmBCH1 is not identified in the current version of the soybean genome database (Supplemental Table S1).
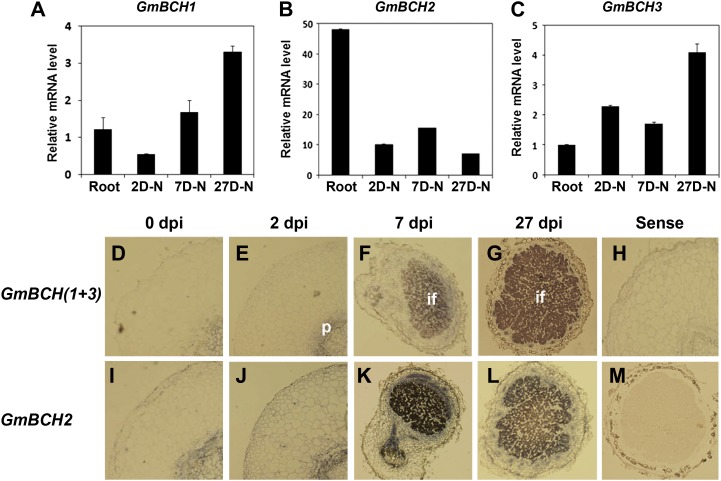
In order to examine GmBCH expression during nodulation, we performed real-time RT-PCR and in situ hybridization. For the in situ hybridization, specific regions of each gene were used to make probes for GmBCH1/GmBCH3 and GmBCH2 (Supplemental Figs. S1 and S4, A and B; see “Materials and Methods”). The probes were checked for hybridization specificity by whole-mount in situ hybridization to young leaves (Supplemental Fig. S4, C and D). The relative expression levels of GmBCH1/GmBCH3 and GmBCH2 obtained from whole-mount in situ hybridization in young leaves were found to be comparable to those from the real-time RT-PCR in Figure 2, implying that the GmBCH probes are not likely to cross hybridize. Sections from nodules at different stages of development were hybridized with a probe for GmBCH1/GmBCH3 together, as they have almost identical DNA sequences. GmBCH1 and GmBCH3 were found to be induced as nodules matured, suggesting that both are involved in the nodulation process (Fig. 3, A, C, and D–G). Their expression was seen in the root pericycle in the early stages of nodulation and became high in the central infected zones in the early and mature nodule stages. GmBCH2 was also expressed during nodulation, especially strongly in 7-d-old nodules (Fig. 3, B and I–L). Its expression was also strong in the root pericycle. The GmBCH expression levels during nodulation were confirmed by the fluorometric measurement of GUS activities using transgenic roots and nodules expressing GmBCH2/GmBCH3 promoter (1.6-kb upstream regions of each)-GUS fusions (Supplemental Fig. S5). These observations could mean that expression of the putative BCH genes, especially GmBCH1, GmBCH2, and GmBCH3, may be involved in nodulation.
Figure 3.
Expression of GmBCHs during nodulation. A to C, Expression of GmBCH1 (A), GmBCH2 (B), and GmBCH3 (C) during nodule development. Transcript levels were determined by real-time RT-PCR and normalized. Expression levels are shown as means and sd of three independent experiments. 2D-N, Two-day-old nodule; 7D-N, 7-d-old nodule; 27D-N, 27-d-old nodule. D to M, In situ hybridization of GmBCHs during nodule development. Sections from 0 (roots), 2, 7, and 27 d post inoculation (dpi) were hybridized with antisense GmBCH(1+3) (D–G) and GmBCH2 (I–L) riboprobes. Sections from 0 (roots) and 27 dpi were hybridized with sense GmBCH(1+3) (H) and GmBCH2 (M) riboprobes as negative controls. if, Infected region; p, Pericycle.
Enzymatic Activities of GmBCHs
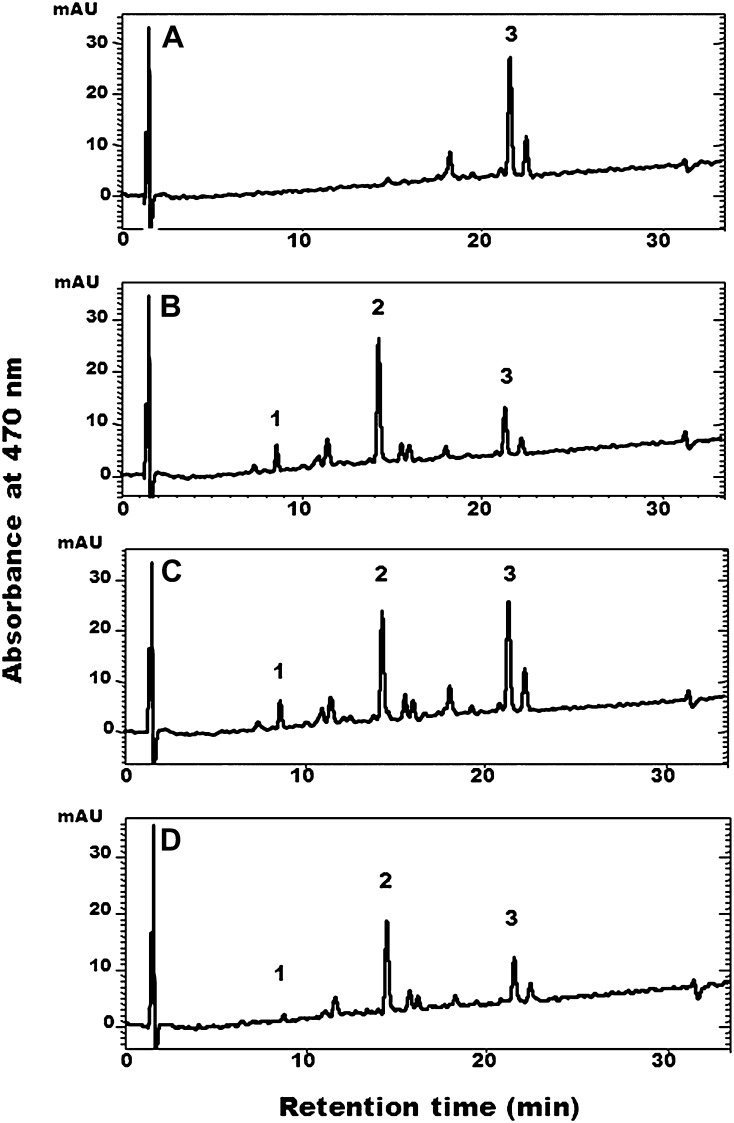
The similarity of GmBCH1, GmBCH2, and GmBCH3 to BCHs of other plants prompted us to test whether they were functionally active in converting β-carotene into zeaxanthin. Escherichia coli carrying pACCAR16ΔcrtX for the production of β-carotene (Misawa et al., 1990) was transformed with GmBCH1 or GmBCH2 cDNA cloned in pUC19 (pGmBCH1 or pGmBCH2) or with pUC19 alone as a negative control. While HPLC analysis of the carotenoids extracted from E. coli transformed with pUC19 alone identified only β-carotene (Fig. 4A), E. coli transformants harboring the GmBCH genes yielded β-cryptoxanthin and zeaxanthin, as defined by their retention times and the absorption spectra of the eluents, as well as their relative molecular masses measured by liquid chromatography/mass spectrometry (MS; Fig. 4, B and C). This indicates that GmBCH1 and GmBCH2 convert β-carotene into β-cryptoxanthin and further into zeaxanthin (see “Discussion”). In addition, enzymatic activity of GmBCH3 was confirmed by expressing it in E. coli (Fig. 4D).
Figure 4.
HPLC analysis of carotenoids extracted from β-carotene-producing E. coli transformed with GmBCH1, GmBCH2, or GmBCH3. pACCAR16ΔcrtX-containing E. coli harboring pUC19 alone as a negative control (A) or harboring pGmBCH1 (B), pGmBCH2 (C), or pGmBCH3 (D) was used for carotenoid analysis. Peak 1, Zeaxanthin; peak 2, β-cryptoxanthin; peak 3, β-carotene. mAU, Milliabsorbance units.
Subcellular Localization of GmBCHs
We predicted the subcellular locations of GmBCH1, GmBCH2, and GmBCH3 using ChloroP version 1.1 (http://www.cbs.dtu.dk.services/ChloroP/). GmBCH2 and GmBCH3 were predicted to contain a chloroplast transit peptide at its N terminus, as expected (amino acids 1–47), whereas GmBCH1 was not. To determine the location of the enzymes experimentally, GmBCH1, GmBCH2, and GmBCH3 cDNAs were fused with GFP, a reporter gene, under the control of the cauliflower mosaic virus (CaMV) 35S promoter, and the resulting constructs were introduced into Arabidopsis (Arabidopsis thaliana) protoplasts. As predicted, the GFP signals from the GmBCH2-GFP and GmBCH3-GFP fusions were mainly found in plastids, whereas those of the GmBCH1-GFP fusion appeared to be cytosolic (Fig. 5, A–D). Deletion of the putative cleavage site in the transit peptide in the N-terminal region of GmBCH2 resulted in cytosolic localization of the product (Supplemental Fig. S6A). Moreover, addition of the transit peptide of GmBCH2 to the N terminus of GmBCH1 resulted in localization of most of the GmBCH1 to plastids, suggesting that the cytosolic localization of GmBCH1 is not an artifact of overexpression (Supplemental Fig. S6B).
Figure 5.
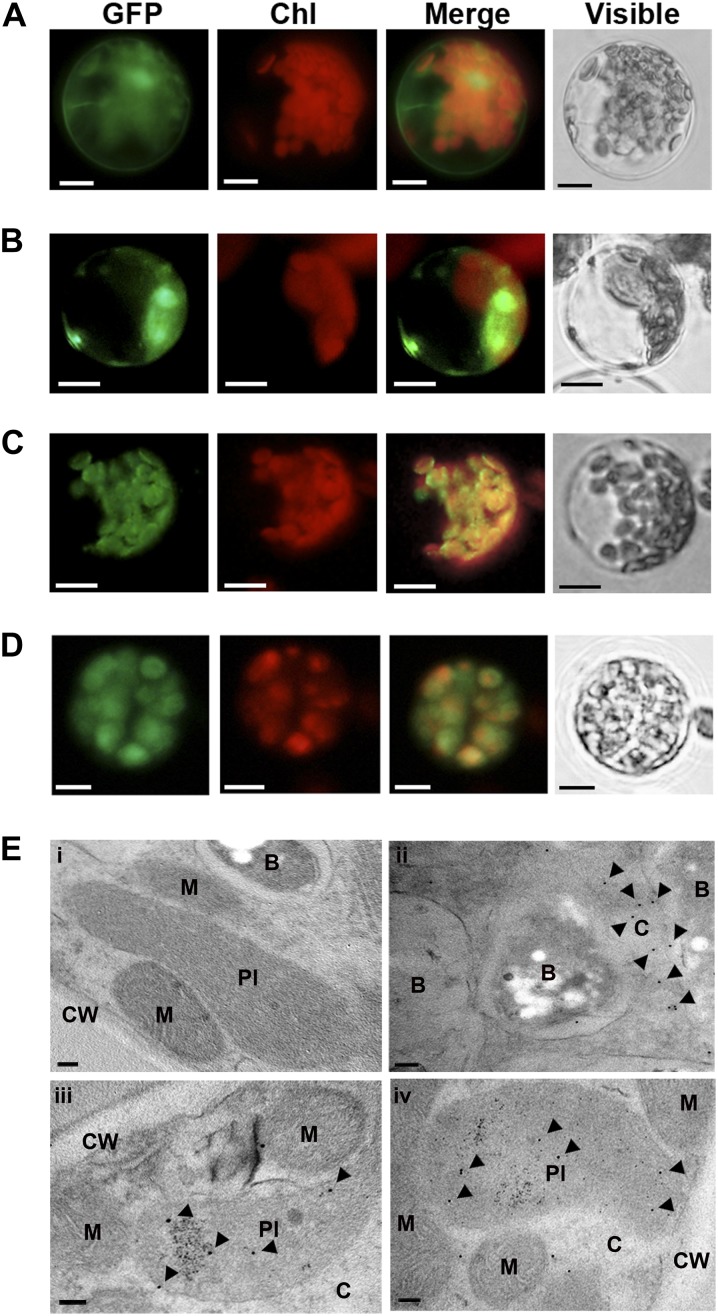
Subcellular localization of GmBCHs in Arabidopsis protoplasts and soybean nodules. A to D, Arabidopsis protoplasts were transfected with control vector (35S-GFP; A), GmBCH1-GFP (B), GmBCH2-GFP (C), or GmBCH3-GFP (D). Bars = 10 μm. E, GmBCHs were localized by EM immunogold labeling with an anti-GFP antiserum in soybean nodules transformed with empty vector (pCAMBIA3301; i), GmBCH1-GFP (ii), GmBCH2-GFP (iii), or GmBCH3-GFP (iv). Gold particles (10 nm) are indicated by arrowheads. B, Bacteroids; C, cytoplasm; CW, cell wall; M, mitochondria; Pl, plastid. Bars = 100 nm.
The subcellular locations of the GmBCHs were further examined by electron microscopic (EM) immunogold labeling with an anti-GFP antiserum and ultrathin sections of soybean nodules expressing GmBCH1-GFP, GmBCH2-GFP, or GmBCH3-GFP. Immunogold particles were detected in the cytosol of nodules expressing GmBCH1-GFP and in the plastids of nodules expressing GmBCH2-GFP and GmBCH3-GFP (Fig. 5E), which is consistent with the localizations reported in Arabidopsis protoplasts as well as the predictions obtained using ChloroP version 1.1. We used the exact binomial test in order to determine whether gold particles were preferentially located in a specific organelle (Conover, 1971). For GmBCH1, more than 78% of the gold particles were found in the cytosol (P = 0.0478). For GmBCH2/GmBCH3, more than 64%/70% of the gold particles were found in the plastid (P = 0.0492/0.0329, respectively). These results suggest that GmBCH2 and GmBCH3 function in the plastids of root nodules, while GmBCH1 resides in the cytosol, unlike other BCHs. It is not clear how a cytosolic BCH can participate in carotenoid metabolism.
Drastically Reduced Nitrogen Fixation in Nodules Expressing RNAi against GmBCHs
To see if GmBCH1/GmBCH3 were essential for nodule growth, we made an RNAi construct against them and subcloned it downstream of the leghemoglobin (Lbc3) promoter. This RNAi construct contained a part of the N-terminal region of GmBCH1 and GmBCH3 that includes the 5′ untranslated region of GmBCH3 (Supplemental Fig. S7). The sequence of the RNAi construct was 99% identical to GmBCH1 and GmBCH3 and had significant identity to GmBCH4 but not to GmBCH2 or GmBCH5. The resulting cassette, GmBCH(1+3)-RNAi, was introduced into pCAMBIA1304, which harbors 35S-GUS (the CaMV 35S promoter fused to GUS) as a reporter. This construct was introduced into Agrobacterium rhizogenes to generate transgenic hairy roots (Lee et al., 2005).
The formation of root nodules was markedly reduced on the GmBCH(1+3)-RNAi hairy roots (Fig. 6A), and similar defective nodulation was observed when a transgenic hairy root formed along with a nontransgenic (GUS-negative) hairy root on the same plant (Fig. 6B). In the meantime, there appeared to be no difference in root growth between the transgenic and nontransgenic hairy roots. The impairment of nodulation on GmBCH(1+3)-RNAi hairy roots resulted in lower nodule weight, as measured in 24 GmBCH(1+3)-RNAi plants and 19 controls (Fig. 6C). When we checked the expression levels of GmBCH1 and GmBCH3 in the transgenic nodules, we found that these varied; hence, as representatives with different nodule weights, we chose RNAi plants 2, 20, and 21 for further expression analysis. RNAi plants 20 and 21, as expected, had significantly reduced expression of GmBCH1 and GmBCH3 together with decreased nodule weights (RNAi type 2), whereas GmBCH1 and GmBCH3 expression was less affected in the RNAi plants, with almost the same nodule weights as the control, such as RNAi plant 2 (RNAi type 1; Fig. 6D). Although the relationship between the expression levels of GmBCH(1+3) and nodule weight is only shown for the three representative transgenic plants (i.e. 2, 20, and 22) in Figure 6D, we actually examined the expression levels and nodule weights in 12 transgenic plants and four controls.
Figure 6.
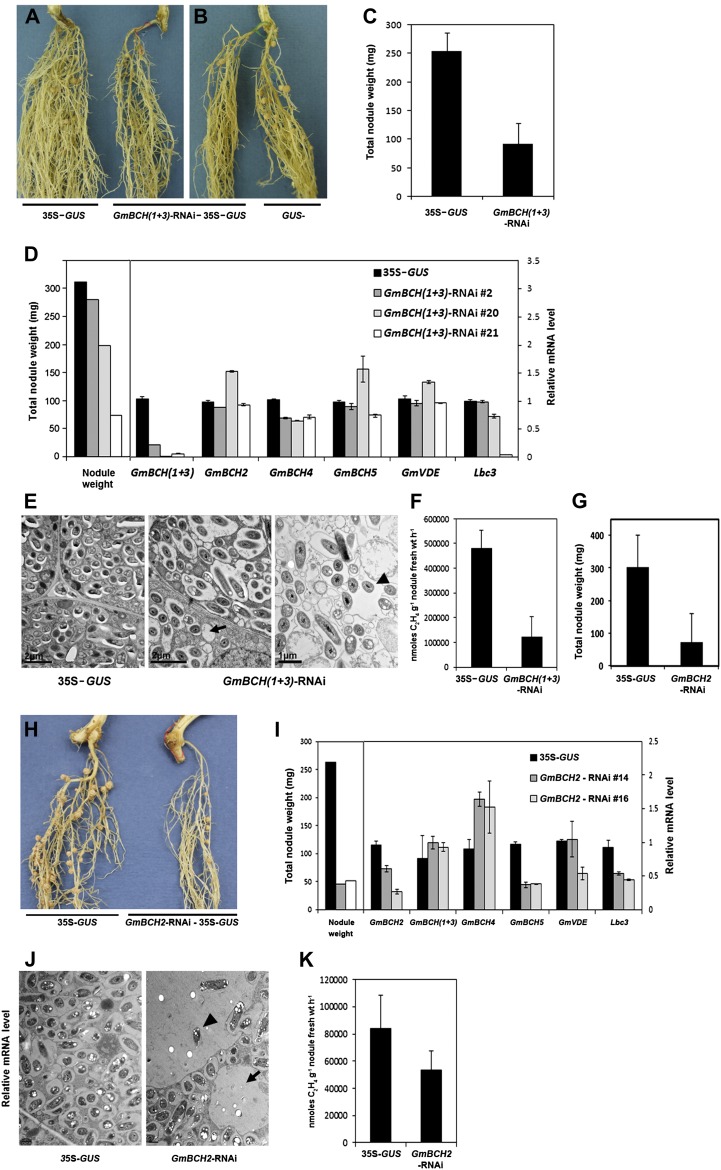
Nodule development on transgenic hairy roots expressing an RNAi construct against both GmBCH1 and GmBCH3. A, Nodules formed on transgenic hairy roots containing pCAMBIA1304 alone (control; 35S-GUS; left plant) or the GmBCH(1+3)-RNAi cloned in pCAMBIA1304 (right plant). After GUS assay, only GUS-positive (transgenic) hairy roots were inoculated with B. japonicum (K599). These experiments were repeated three times, and representative results are shown. In each experiment, five to seven plants were used per construct. B, A GUS-negative (untransformed control) hairy root is shown on the right. C, Total nodule weights (mg) of control (35S-GUS) and GmBCH(1+3)-RNAi plants were measured and are shown as means and sd of three independent experiments. D, Transcript levels are shown in nodules from a transgenic control plant (35S-GUS) and three representative transgenic plants: GmBCH(1+3)-RNAi 2, GmBCH(1+3)-RNAi 20, and GmBCH(1+3)-RNAi 21. RNAi plant 2 had almost the same nodule weight as the control, RNAi plant 20 had a reduced nodule weight, and RNAi plant 21 had a drastically reduced nodule weight. Transcript levels of GmBCH(1+3), GmBCH2, GmBCH4, GmBCH5, GmVDE, and Lbc3 were determined by real-time RT-PCR in controls (35S-GUS) and three GmBCH(1+3)-RNAi nodules and normalized. Expression levels are shown as means and sd of three independent experiments. E, EM images of 27-d-old control (35S-GUS) and GmBCH(1+3)-RNAi plants. GmBCH(1+3)-RNAi nodules often contained empty vesicles (arrow) and bacteroids outside the symbiosomes (arrowhead). F, Nitrogenase activities were measured by the acetylene reduction assay, and data are averaged from three independent experiments. G, Total nodule weights (mg) of control (35S-GUS) and GmBCH2-RNAi plants were measured and are shown as means and sd of three independent experiments. H, Nodules formed on transgenic hairy roots containing pCAMBIA1304 alone (control; 35S-GUS; left plant) or the GmBCH2-RNAi cloned in pCAMBIA1304 (right plant). After GUS assay, nodules were formed as in A. I, Transcript levels in nodules from one transgenic control plant (35S-GUS) and two differentially repressed transgenic plants (GmBCH2-RNAi 14 and GmBCH2-RNAi 16) are shown. Transcript levels of GmBCH2, GmBCH(1+3), GmBCH4, GmBCH5, GmVDE, and Lbc3 were determined by real-time RT-PCR and normalized. Expression levels are shown as means and sd of three independent experiments. J, EM images of 27-d-old control (35S-GUS) and GmBCH2-RNAi nodules. GmBCH2-RNAi nodules often contained empty vesicles (arrow) and bacteroids outside the symbiosomes (arrowhead). K, Nitrogenase activities were measured by the acetylene reduction assay as in F.
To establish the statistical significance of the RNAi plant groups (i.e. control versus RNAi type 1 versus RNAi type 2), we conducted an ANOVA with Tukey’s multiple comparison procedure and found significant differences in nodule weight between control and RNAi type 2 as well as between RNAi type 1 and RNAi type 2 (Supplemental Fig. S8A); there was also a significant difference in the expression of GmBCH(1+3) between these pairs (Supplemental Fig. S8B; Maritz, 1981). In addition, using all 16 plants making up the three groups, we examined the correlation between gene expression level and nodule weights based on the Spearman rank correlation coefficient. This yielded a strong positive correlation of 0.76 (P = 0.001; Supplemental Fig. S8C). Meanwhile, GmBCH4 expression, which was quite low in root nodules, was somewhat affected in most of the RNAi plants. The expression of GmBCH2 as well as GmBCH5 was not altered in any of the plants (Fig. 6D). These observations imply that the effect of the RNAi on GmBCH transcript levels was specific to GmBCH1 and GmBCH3. Expression of GmVDE, a gene contributing positively to the accumulation of zeaxanthin from the opposite direction of GmBCH in the xanthophyll cycle (Fig. 1; Cazzonelli and Pogson, 2010), was also unaffected, indicating that silencing of the GmBCHs in the RNAi nodules was not compensated, at least at the transcription level, by the induction of GmVDE (Fig. 6D). In agreement with the observed retardation of nodule development, on the other hand, expression of Lbc3, which encodes leghemoglobin, an oxygen carrier for symbiosis with rhizobia that is one of the hallmark genes for the development of nitrogen-fixing nodules (Ott et al., 2005), was decreased in RNAi plants 20 and 21 (Fig. 6D). In addition, acetylene reduction assays showed that nitrogen-fixing ability was also lower in the RNAi nodules (Fig. 6F). The RNAi nodules examined by electron microscopy contained empty vesicles rather than symbiosomes with rhizobia in about 60% of the cells examined. In addition, the rhizobia were not even enclosed by symbiosome membranes in about 5% of the cells of the RNAi nodules with strongly repressed GmBCH(1+3) (Fig. 6E; Supplemental Fig. S9). These data indicate that the expression of GmBCH1 and/or GmBCH3 may be essential for nodule development.
Expression of GmBCH2 was prominent during nodulation; hence, we also silenced GmBCH2 in nodules using the leghemoglobin promoter-driven RNAi approach. When we compared nine GmBCH2-RNAi plants with eight control plants, we found that the reduction in GmBCH2 expression resulted in decreased nodule weight and nitrogenase activity (Fig. 6, G–K). GmBCH5 expression was also reduced in the GmBCH2-RNAi nodules, probably due to its strong homology with GmBCH2, while the expression of GmBCH1 and GmBCH3 was not affected and GmBCH4 expression was somewhat increased (Fig. 6I). About 65% of the infected cells examined contained empty vesicles, and most of them exhibited the presence of bacteroids outside symbiosomes (Fig. 6J). It is not clear whether the phenotypic difference observed by electron microscopy of GmBCH(1+3)-RNAi nodules and GmBCH2-RNAi nodules indicates their different roles in nodulation. In addition, the expression of GmBCH4 and GmBCH5 was also observed in nodules, albeit at a low level (Supplemental Fig. S1, B and C). Taken together, these results suggest that the GmBCHs, comprising GmBCH1 to GmBCH5, may be essential for nodule development.
Decreased Expression of the Putative Zeaxanthin Epoxidase Gene during Nodulation
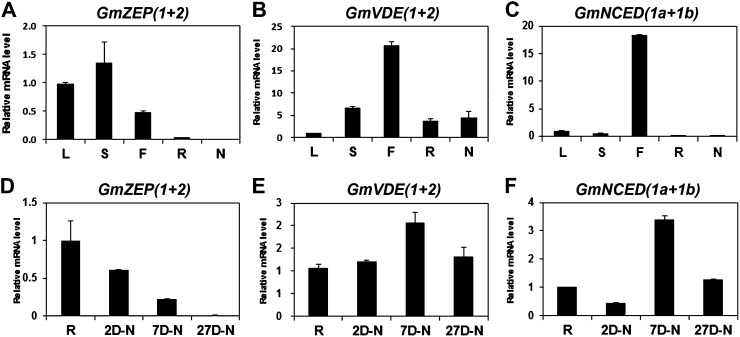
The impairment of nodulation in the RNAi hairy roots suggested to us that a product of BCH action, or some other derivatives of the carotenoid metabolic pathway, plays an important role in nodulation. This led us to examine the expression of genes encoding two enzymes of the xanthophyll cycle: ZEP and violaxanthin deepoxidase (VDE). Since we identified three GmZEPs (Supplemental Fig. S1D) and two GmVDEs (Supplemental Fig. S1E) in the soybean genome sequence, we examined their expression. Actually, we came across a few more DNA sequences homologous to the ZEP and VDE genes in other plants, but they showed only partial identity and were not further studied (Supplemental Fig. S1, D and E). The primers used to examine the expression of GmZEPs and GmVDEs were designed using DNA sequences highly conserved among other plants. The transcript levels of both genes were considerably lower in roots and nodules than in the aerial parts of the plant, such as leaves, stems, and flowers (Fig. 7, A and B; Supplemental Fig. S10). As nodules matured, GmZEP expression decreased and was almost undetectable in mature 27-d-old nodules (Fig. 7D). Expression of the GmVDEs remained relatively constant throughout nodule development (Fig. 7E). In addition, we examined the expression of two soybean 9-cis-epoxycarotenoid dioxygenase1 (NCED1) orthologs (GmNCED1a and GmNCED1b; Supplemental Fig. S1F) and found that their expression was high in flowers but quite low in roots and nodules, although expression in 7-d-old nodules was a little higher than in other stages of nodulation (Fig. 7, C and F). The expression levels of GmZEPs, GmVDEs, and GmNCED1s generally matched with those in the soybean RNA-Seq Atlas, especially expression in the aerial parts of the soybean (Libault et al., 2010; Severin et al., 2010; Supplemental Table S1). These expression data appear to indicate that GmZEPs, GmVDEs, and GmNCED1s do not play major roles, if any, in further carotenoid metabolism after BCH during nodulation, although we cannot rule out the possibility that their protein levels are higher than suggested by their transcript levels.
Figure 7.
Expression of GmZEPs, GmVDEs, and GmNCED1s in soybean. A to C, Expression of GmZEPs, GmVDEs, and GmNCED1s in different tissues. RNA was extracted from different tissues, including 27-d-old nodules of soybean. D to F, Expression of GmZEPs, GmVDEs, and GmNCED1s during nodule development. RNAs were extracted from roots and 2-, 7-, and 27-d-old nodules, and transcript levels were determined by real-time RT-PCR and normalized. Expression of GmZEP1 and GmZEP2 was examined simultaneously using primers for DNA regions of high identity, while the expression of GmZEP3, a gene with low overall homology to other GmZEPs, was measured separately and is shown in Supplemental Figure S10. Expression of GmVDE1 and GmVDE2 was also examined simultaneously using primers for the DNA regions of high identity. Expression of GmNCED1a and GmNCED1b was also examined simultaneously using primers for the DNA regions of high identity. Data are from three independent experiments. L, Leaf; S, stem; F, flower; R, root; N, nodule; 2D-N, 2-d-old nodule; 7D-N, 7-d-old nodule; 27D-N, 27-d-old nodule.
Expression of CCDs in Soybean Root Nodules
We hypothesized that zeaxanthin might be synthesized in root nodules by BCHs and converted to other carotenoids, although not via xanthophyll to ABA, because ZEP expression was very low (Fig. 7D). Therefore, we extracted and quantified carotenoids from root nodules. Contrary to our expectation, carotenoids including zeaxanthin were almost undetectable in root nodules (data not shown). This raised the question of what biochemical reactions occur in root nodules subsequent to zeaxanthin production by the BCHs. Since carotenoid cleavage products have been found in roots infected with mycorrhizal fungi and are regarded as important in AM symbiosis (Strack and Fester, 2006), we reasoned that the carotenoids synthesized in root nodules might be depleted if CCDs were active. The white-colored petals of Chrysanthemum spp. express high levels of CmCCD4a, with the result that no carotenoid can be detected (Ohmiya et al., 2006), and RNAi-mediated suppression of CmCCD4a expression was found to lead to the accumulation of carotenoids and yellow petal color, confirming the relationship between the amount of carotenoid and the expression of CmCCD4a. Thus, we considered the possibility that the conversion of carotenoids into apocarotenoids by CCDs might explain our failure to detect carotenoids in root nodules.
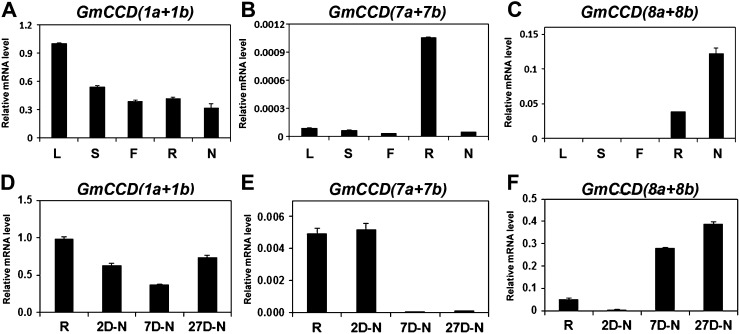
We tested this possibility by examining the expression of CCDs in root nodules of soybean. We chose to examine the expression of CCD1, CCD7, and CCD8 in nodules and excluded CCD4 due to its primary expression in aerial tissues. From the soybean genome sequence (http://www.phytozome.net/soybean.php), we obtained the DNA sequences of putative versions of CCD7 and CCD8, which are reported to cleave carotenoids. The deduced amino acid sequences of CCD7 and CCD8 (designated GmCCD7 and GmCCD8 hereafter) were very similar to previously reported CCD7 and CCD8 sequences and were named GmCCD7a, GmCCD7b, GmCCD8a, and GmCCD8b (Supplemental Fig. S1, G and H). In addition, we also encountered putative GmCCD1a and GmCCD1b sequences and examined their expression (Supplemental Fig. S1I). We found a few more DNA sequences, but their deduced amino acid sequences were only partially identical to previously reported CCDs and not studied further (Supplemental Fig. S1, G and H). Using appropriate primers, the expression of GmCCD7s and GmCCD8s proved to be relatively high in roots and nodules, whereas GmCCD1 expression was seen in all tissues, in agreement with previous data (Libault et al., 2010; Severin et al., 2010; Fig. 8, A–C; Supplemental Table S1). Although the expression of GmCCD7s was rather low, it, as well as that of GmCCD8s, appeared to be induced upon rhizobial infection (Fig. 8, E and F).
Figure 8.
Expression of GmCCDs in soybean. A to C, Expression of GmCCD1s, GmCCD7s, and GmCCD8s in different tissues. RNA was extracted from various tissues, including 27-d-old nodules of soybean. D to F, Expression of GmCCD1s, GmCCD7s, and GmCCD8s during nodulation. RNAs were extracted from roots and 2-, 7-, and 27-d-old nodules, and transcript levels were determined by real-time RT-PCR and normalized. Expression levels are shown as means of three independent experiments. L, Leaf; S, stem; F, flower; R, root; N, nodule; 2D-N, 2-d-old nodule; 7D-N, 7-d-old nodule; 27D-N, 27-d-old nodule.
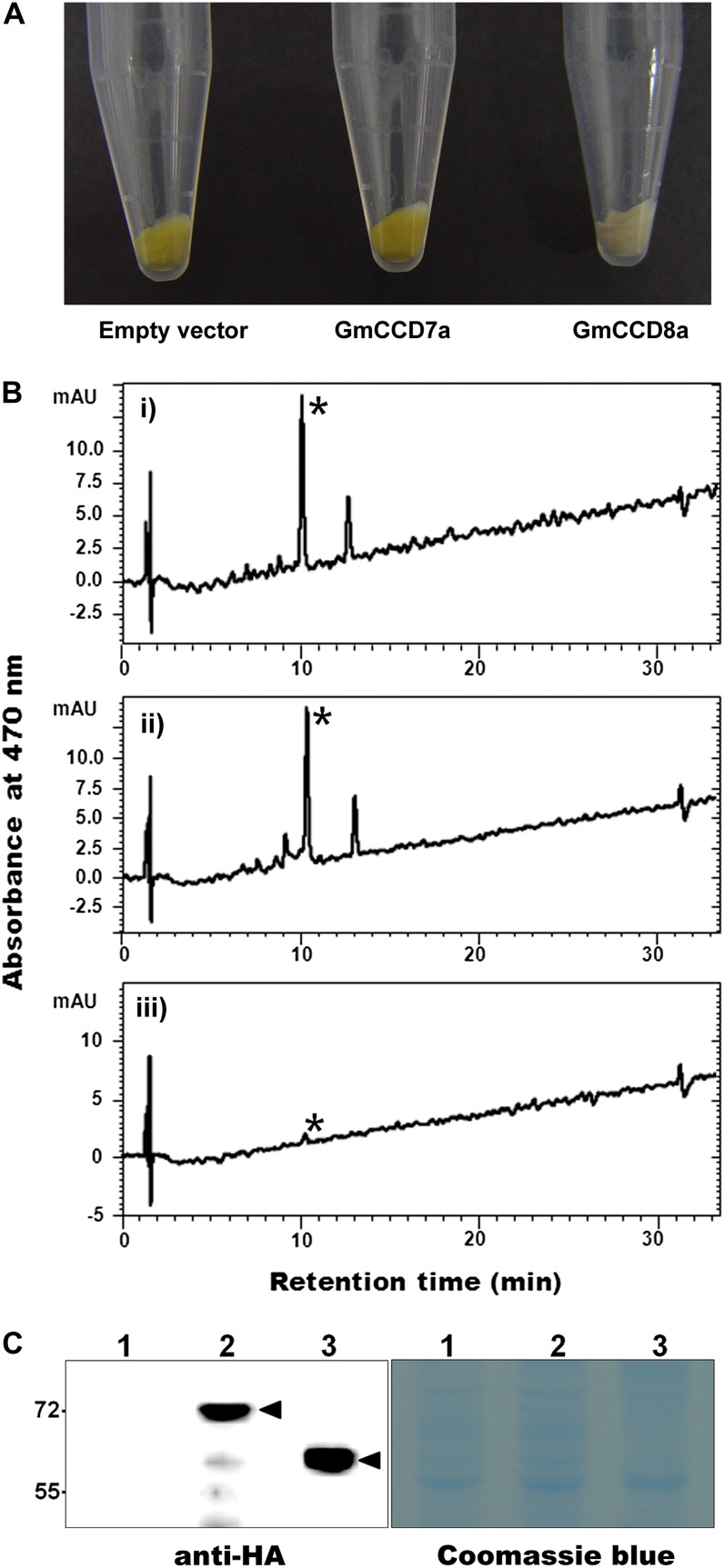
Therefore, we tested whether GmCCD7/GmCCD8 could actually cleave zeaxanthin. E. coli transformed with zeaxanthin biosynthetic genes cloned in the vector pACYC184 (pAC-zeaxanthin) produced zeaxanthin, as shown by HPLC analysis as well as by the yellow color of colonies (Fig. 9, A and B). GmCCD7a-expressing E. coli was also yellow, and a protein of the expected size corresponding to GmCCD7 (70 kD) was detected by SDS-PAGE (Fig. 9C), but lysates gave a strong peak of zeaxanthin, implying that the zeaxanthin produced was not cleaved. In contrast, E. coli transformed with the GmCCD8a-expressing plasmid was no longer yellow while producing a protein of the expected mass (60 kD). This recombinant E. coli strain gave only a very small zeaxanthin peak (Fig. 9, A and B, asterisks). The integrity of GmCCD7a was also tested by another E. coli functional assay, in which the β-carotene-cleaving activity of GmCCD7 was examined and compared with other CCDs in E. coli carrying the plasmid pACCAR16ΔcrtX that enables the accumulation of β-carotene (Supplemental Fig. S11). β-Carotene-cleaving activity was consistently detected in GmCCD7a-expressing E. coli, although it was about 3-fold lower than that of an AtCCD7-expressing E. coli (Supplemental Fig. S11, C and D). Moreover, the apparent gap in the activity between GmCCD7a and the other two CCDs in this assay (Supplemental Fig. S11, C–E) could be further reduced if the relative protein expression level of these proteins was taken into account, as measured by western blots of these cell extracts (Supplemental Fig. S11A). Therefore, it seems likely that both GmCCD7a and GmCCD8a are functional enzymes involved in β-carotene metabolism in soybean nodules. These data also imply that zeaxanthin is cleaved by GmCCD8a but not by GmCCD7a, although we cannot exclude the possibility that GmCCD7a synthesized in root nodules may have cleavage activity on zeaxanthin. Taken together, our observations suggest that carotenoids such as zeaxanthin are synthesized but are cleaved by GmCCD8 during soybean nodule development.
Figure 9.
Functional assays of GmCCD7 and GmCCD8 in E. coli. A, Expression of GmCCD7a (middle) or GmCCD8a (right) in E. coli strains that carry pAC-zeaxanthin and accumulate zeaxanthin. A zeaxanthin-accumulating E. coli strain with empty vector alone (left) served as a negative control. B, HPLC analysis of carotenoids extracted from zeaxanthin-accumulating E. coli cells expressing GmCCD7a (ii) or GmCCD8a (iii) or with empty vector (i). The zeaxanthin peak is indicated by the asterisks. mAU, Milliabsorbance units. C, Proteins from zeaxanthin-accumulating E. coli cells expressing HA-GmCCD7 (lane 2) and HA-GmCCD8 (lane 3) or with empty vector (lane 1) were isolated (right panel) and immunoblotted with hemagglutinin (HA) antibody (left panel). Proteins of the expected sizes corresponding to GmCCD7 (70 kD) and GmCCD8 (60 kD) were detected. Immunodetected bands are indicated by arrowheads. [See online article for color version of this figure.]
DISCUSSION
We have investigated the significance of carotenoid metabolism in nodule development. This work was prompted by the isolation of a gene encoding BCH as a gene differentially expressed in root nodules (Lee et al., 2005). GmBCH1, GmBCH2, and GmBCH3 expression was found to increase during nodulation and was especially localized to the infected region of nodules (Figs. 2 and 3). In addition, RNAi-mediated repression of these genes resulted in the retardation of nodule development, including impairment in symbiosome formation (Fig. 6; Supplemental Fig. S9). In the RNAi nodules, nitrogen fixation appeared to be damaged, judging from nitrogenase assays as well as the expression of lbc3, an essential gene for nitrogen fixation. We have thus presented evidence, to our knowledge for the first time, that carotenoid metabolism by GmBCHs is required for proper nodule development.
We confirmed that the GmBCHs we isolated from root nodules encoded active enzymes. cDNAs were expressed in E. coli harboring a vector for producing β-carotene (Misawa et al., 1990), and HPLC analyses of E. coli expressing the GmBCHs revealed the accumulation of zeaxanthin and also of large amounts of β-cryptoxanthin (Fig. 4). Similar results have been reported from functional assays in E. coli (Yu et al., 2007), while the expression of other BCH orthologs resulted in the synthesis of more zeaxanthin than cryptoxanthin (Sun et al., 1996; Galpaz et al., 2006). The synthesis of β-cryptoxanthin, that is, the asymmetric addition of hydroxyl groups to the β-end group of β-carotene, could be due to a slightly different conformation of the BCHs in E. coli or the failure of the BCHs to form a stable dimer (Sun et al., 1996; Yu et al., 2007). In any case, the results confirm that the putative GmBCHs are, indeed, functional BCHs.
To our surprise, we failed to detect any carotenoids in root nodules. To account for this, we tested the possibility that all the carotenoids were cleaved to synthesize ABA, since ABA is able to coordinate some aspects of nodulation (Ding et al., 2008). However, the expression of putative GmZEPs and putative GmNCED1s was found to be quite low in nodules (Fig. 7, A and C; Supplemental Fig. S10A), and furthermore, GmZEP expression became almost undetectable in mature nodules (Fig. 7D; Supplemental Fig. S10B), while the expression of putative GmVDEs in root nodules was similar to the level in roots (Fig. 7E). In fact, nodule number is decreased by ABA treatment (Suzuki et al., 2004), and a Lotus japonicus mutant with reduced endogenous ABA exhibited enhanced nodulation and nitrogen fixation (Tominaga et al., 2009). Since ABA was suggested to be a negative regulator of nodulation, it is possible that its concentration is not high in effective nodules. However, we cannot exclude the possibility that a certain amount of carotenoid is metabolized to ABA during specific stages of nodulation.
Despite its high sequence homology with other plant CCD8s (Supplemental Fig. S1H), GmCCD8a apparently exhibited activity on both β-carotene and zeaxanthin in our E. coli functional assay, contradicting the current view of substrate specificity and the proposed role for CCD8 in the sequential cleavage reactions of C40 carotenoids (Alder et al., 2008; Walter et al., 2010). On the other hand, this result is consistent with an earlier report that showed direct cleavage activity of AtCCD8 on a few C40 carotenoids (Auldridge et al., 2006a). In addition, it was reported that CCD8 interfered with carotenoid biosynthesis when it was overexpressed in E. coli (Alder et al., 2008). Therefore, our results here, together with the previous data of Auldridge et al. (2006a), appear to suggest that more in-depth studies on the reaction catalyzed by CCD8, including the nature of the substrate and the cleavage product, are needed. Before drawing a conclusion that GmCCD8a is able to cleave zeaxanthin, it will be necessary to identify the cleavage products generated in the assay. The data in Figure 9 also show that GmCCD7a did not alter the HPLC profile of zeaxanthin-producing E. coli. Since recombinant AtCCD7 has broad substrate specificity and cleaves C40 carotenoids, including β-carotene, into C27 and C13 apocarotenoids (Booker et al., 2004; Schwartz et al., 2004) and GmCCD7 cleaved β-carotene, albeit less efficiently than AtCCD7 (Supplemental Fig. S11), we cannot exclude the possibility that the GmCCD7 synthesized in root nodules can cleave diverse C40 carotenoids, including zeaxanthin. Based on these observations, we propose that GmCCD7, and possibly GmCCD8, cleave C40 carotenoids and that the cleavage products are further cleaved by GmCCD8 (Alder et al., 2008) inside nodule tissue.
The carotenoids synthesized inside the chloroplasts of leaves play essential roles in photosynthesis. On the other hand, those present in flowers, fruits, or roots are not needed for photosynthesis and accumulate in special subcellular compartments such as chromoplasts and cytoplasmic lipid vesicles. For example, a β-carotene oxygenase in a unicellular green alga (Haematococcus pluvialis) was localized to the lipid vesicles outside plastids (Grünewald et al., 2001). In our study, both in silico prediction and actual experiment showed that GmBCH2 and GmBCH3 were present in plastids, whereas GmBCH1 was present, unexpectedly, in the cytosol (Fig. 5). Interestingly, the localization of GmBCH1 in the cytosol seems to be closely associated with a short stretch of N-terminal sequences present only in GmBCH1, in addition to the putative transit-peptide sequence that is present in all of the plastidial GmBCH isoform sequences. Perhaps adding this sequence makes the transit peptide nonfunctional. Although no corresponding locus to GmBCH1 was found in the soybean genome database (http://www.phytozome.net), we have been able to clone GmBCH1 repeatedly by RT-PCR and also could isolate its 0.45-kb upstream sequences from cv Williams 82, from which the soybean sequence database was generated, as well as cv Sinpaldal 2, which has been used as the material of this study, by DNA walking (Supplemental Fig. S2). Therefore, it is likely that GmBCH1 may be present in an unsequenced gap of the present soybean genome sequence. The substrate of GmBCH1 in the cytosol of infected cells, and the significance of its cytosolic location for the symbiotic interaction between soybean and Rhizobium spp., need to be studied in the future.
Carotenoid cleavage products such as β-ionone or dihydroactinidiolide are synthesized in conditions of stress, being involved in plant protection mechanisms (Bouvier et al., 2005). It is thought that the synthesis of carotenoids and their cleavage products promotes symbiosis between plants and the arbuscular mycorrhiza (Strack and Fester, 2006; Walter et al., 2010). However, since little attention has been paid to the presence of (apo)carotenoids in root nodules, it remains unclear what role the former plays in root nodule symbiosis, given that our work points to the presence of apocarotenoids as well as carotenoids in the nodules. Since nodulation and nitrogen fixation were severely inhibited in the GmBCH-RNAi root nodules (Fig. 6) and the expression of GmBCHs and GmCCDs was induced during nodulation, the biosynthesis of carotenoids and presumably apocarotenoids appears to play a significant role in nodulation. A possible role of carotenoid cleavage products is to protect the infected cells from oxidative stress. The symbiosomes enclosing rhizobia must produce tremendous amounts of reactive oxygen species, since a nitrogen-fixing infected cell contains about 20,000 rhizobia. Alternatively, apocarotenoids may act as signaling molecules during the maturation of nodules. A further possibility is that C13 and C14 apocarotenoids are essential for rhizobial symbiosis, as proposed for AM symbiosis (Walter et al., 2010). While there seems to be no clear indication which apocarotenoid(s) is effective in AM symbiosis, identification of the apocarotenoids present in root nodules may offer a key to understanding the establishment and functioning of legume-Rhizobium spp. symbiosis.
MATERIALS AND METHODS
Plants, Rhizobia, and Growth Conditions
Soybean (Glycine max ‘Sinpaldal 2’) seeds were sterilized and grown in darkness on moist, absorbent paper at 28°C for 3 d. Three-day-old seedlings were inoculated with rhizobia (Bradyrhizobium japonicum ‘USDA110’), transferred to sterilized vermiculites, and grown at 28°C for 1 month. Tissues harvested from soybean were frozen immediately in liquid nitrogen and stored at −70°C until used for RNA extraction. For real-time PCR, mature leaves (fully expanded), stems, flowers (including flower buds and mature flowers), roots, and the mature nodule (27 d old) were collected separately.
Gene Isolation and Vector Construction
A partial cDNA clone of GmBCH2 (BE607999) was identified by a BLAST search at the National Center for Biotechnology Information EST database. Because this clone did not contain the 5′ end of the open reading frame, RACE PCR was performed to recover the missing 5′ DNA sequence, and the coding sequence was extended using the CapFishing kit (Seegene). Full-length first-strand cDNA synthesized with oligo(dT)-ACP was amplified using the primers listed in Supplemental Table S2. The degenerate primers for GmZEP and the primers for the full-length cDNA are shown in Supplemental Table S2 as well. To generate the constructs for bacterial expression, the coding regions of the GmBCHs were amplified with Pfu DNA polymerase (Corebio; Supplemental Table S2). Full-length cDNAs encoding the putative CCD7 and CCD8 in soybean were obtained from the soybean genome sequence (http://www.phytozome.net/soybean.php), and full-length GmCCD7 and GmCCD8 cDNAs were amplified from nodule RNA by RT-PCR (Supplemental Table S2). The resulting amplified products were cloned into pUC19 to make pGmBCH1, pGmBCH2, pGmBCH3, p3HA-GmCCD7, and p3HA-GmCCD8, and their sequences were confirmed by DNA sequencing.
A DNA fragment including the 5′ region as well as the coding region of GmBCH1 was isolated from soybean genomic DNA using the DNA Walking SpeedUP Premix Kit (Seegene). PCR was performed with an adaptor provided in the kit and the following gene-specific primers: GmBCH1 primer, 5′-GAGAGTGTTTGTGTTCGCCTGCG-3′; second nested GmBCH1 primer, 5′-AGTAAGGAATGTGATGATCCC-3′; third nested GmBCH1 primer, 5′-CTATCCCCCATGAAGCGAATGCC-3′. The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced.
To make GmBCH promoter-GUS fusions, the 5′ upstream sequences of GmBCH2 and GmBCH3 were identified in the soybean genome (http://www.phytozome.net/soybean.php) as shown in Supplemental Figure S5. The 1.5-kb 5′ upstream regions of GmBCH3 and GmBCH2 were amplified by PCR with Pfu DNA polymerase (Corebio) using the primers shown in Supplemental Table S2 and cloned upstream of GUS in pCAMBIA3301. Fluorometric assays of GUS activity were performed as described by Jefferson et al. (1987) with modifications.
Real-Time RT-PCR
Total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen), and cDNAs were synthesized with Moloney murine leukemia virus reverse transcriptase (Promega) after treatment with DNase I to remove contaminating genomic DNA. One microliter of first-strand cDNA was used as a template, and the primers used are listed in Supplemental Table S2. Real-time RT-PCR was performed with SYBR Green PCR Master Mix (Takara) using a Rotor-Gene 3000 (Corbett Research) and the ABI Prism 5700 sequence detection system. All RT-PCR transcript levels were normalized with the geometric mean of three reference genes (GmELF1b, GmActin2/GmActin7, and Ubiquitin; Vandesompele et al., 2002).
In Situ RNA Hybridization
In situ RNA hybridization was performed according to Oh et al. (2001). Nodules were harvested 2, 7, and 27 d after rhizobial inoculation. Each nodule was processed by microtechniques, hybridized with digoxigenin-labeled antisense and sense RNA probes under standard conditions, and washed with low-stringency and high-stringency buffers for longer than under standard conditions. Hybridization stringency was established by the washing steps in order to avoid cross hybridization. The probes for GmBCH(1+3) and GmBCH2 were made using the N-terminal regions of the GmBCHs, which have no significant sequence similarity to each other (Supplemental Fig. S4); we used a 400-bp region for the GmBCH(1+3) probe and a 300-bp region for the GmBCH2 probe, as indicated in Supplemental Figure S1A. Whole-mount in situ hybridization of soybean leaves was performed with the GmBCH(1+3) and GmBCH2 probes according to Weigel and Glazebrook (2002), and images from whole-mount in situ hybridization were quantified with ImageJ (National Institutes of Health) as described by Ubuka and Bentley (2009).
HPLC Analysis of Carotenoids
To measure their activities, pGmBCH1, pGmBCH2, pGmBCH3, and pUC19 as a negative control were introduced into Escherichia coli JM109 carrying pACCAR16ΔcrtX, which expresses genes for the production of β-carotene (Misawa et al., 1990). E. coli transformants were grown overnight at 28°C in 2 mL of Luria-Bertani liquid medium containing 50 μg mL−1 ampicillin and 35 μg mL−1 chloramphenicol. The overnight cultures were used to inoculate 50 mL of Luria-Bertani medium with the same antibiotics. After 3 h, 0.1 mm isopropylthio-β-galactoside was added, and the E. coli was further incubated in darkness at 28°C for 72 h. For the expression of GmCCD7 or GmCCD8, p3HA-GmCCD7 or p3HA-GmCCD8 was introduced into E. coli (BL21) containing a carotenoid-producing construct (pAC-zeaxanthin or pACCAR16ΔcrtX). The transformants were grown as described above and incubated for 24 h after adding isopropylthio-β-galactoside. Carotenoid cleavage activity was inferred from the absence of accumulating carotenoids (i.e. the absence of yellow color).
Cell pellets of E. coli cultures were resuspended in 80% acetone and concentrated. After redissolving in methanol, 10-μL samples were used for HPLC. Assays were performed in ambient conditions using a Prostar 230 ternary gradient pump, a Prostar 430 autosampler, and a Prostar 335 photodiode array detector (Varian). Separation was carried out on a 4.6- × 150-mm carotenoid column (YMC Co.) with a particle size of 3 mm. The mobile phase consisted of solvent A (methanol:tert-butyl methyl ether, 10:90, v/v) and solvent B (water:methanol, 5:95, v/v). A linear gradient was used (10% solvent A at 0 min, 65% solvent A at 40 to 45 min, 95% solvent A at 45 to 50 min). The flow rate was maintained at 1 mL min−1, and the chromatographic profile was recorded at 470 nm. MS data for the carotenoids were obtained using the 1200L liquid chromatography/MS apparatus (Varian). MS conditions were as follows: Atmospheric Pressure Chemical Ionization positive ion mode; mass range, mass-to-charge ratio of 200 to 800; corona current, 2.0 μA; nebulizing gas pressure (nitrogen), 60 p.s.i.; drying gas (nitrogen) flow rate, 4 L min−1; drying gas temperature, 300°C.
Subcellular Localization of GmBCHs Using Arabidopsis Protoplasts and EM Immunogold Labeling
To make GFP fusion constructs, the full-length cDNAs of the GmBCHs were amplified by PCR with Pfu DNA polymerase (Corebio) using primers (Supplemental Table S2) containing XbaI and BamHI sites and fused in frame to GFP. To make a truncated GmBCH2-GFP, a part of GmBCH2 (corresponding to amino acids 48–314) was used. To make a fusion of GmBCH1-GFP with the transit peptide of GmBCH2, we used a region of GmBCH2 corresponding to amino acids 1 to 47. Protoplasts isolated from Arabidopsis (Arabidopsis thaliana) were transfected by the polyethylene glycol method as described by Yoo et al. (2007). After 16 h of incubation, fluorescence was examined with a fluorescence microscope.
To make the transgenic plants expressing GmBCH1-GFP, GmBCH2-GFP, or GmBCH3-GFP for EM immunogold labeling, fusions of GmBCHs-GFP under the control of the CaMV 35S promoter were inserted between the HindIII/EcoRI sites of pCAMBIA3301. Transgenic nodules were produced according to Lee et al. (2005). Immunoelectron microscopic studies were performed according to Lin et al. (2011). Sections of 27-d-old nodule on copper grids were labeled with anti-GFP rabbit antibody (Abcam) and then with 10-nm gold-conjugated goat anti-rabbit antibody (Abcam). The sections were viewed in a JSM-1200EX II transmission electron microscope (JEOL).
Generation of Transgenic Root Nodules
To make GmBCH(1+3)-RNAi and GmBCH2-RNAi constructs, a 230 bp-fragment targeting both GmBCH1 and GmBCH3 and a 180 bp-fragment of GmBCH2 were amplified by PCR with Pfu DNA polymerase (Corebio) using the primers shown in Supplemental Table S2. The amplified fragments were inserted into the HindIII/XbaI and XhoI/KpnI sites of pKANNIBAL (Wesley et al., 2001). The GmBCH(1+3)-RNAi construct was transferred into the binary plasmid pCAMBIA1304, and the resulting plasmid was introduced into Agrobacterium rhizogenes (K599) by the freeze-thaw method. Hairy roots emerging after infection with the agrobacteria were examined for GUS expression in order to identify the transgenic hairy roots, and only one transgenic hairy root in each plant was spared to be used for nodulation, removing all the others (Lee et al., 2005).
Acetylene Reduction Assay
Ethylenes produced per g (fresh weight) of nodules were determined as described previously (Oh et al., 2001).
Transmission Electron Microscopy
Nodule specimens (approximately 1 × 3 mm2 with 1-mm-thick underlying tissues) from transgenic roots containing the GmBCH(1+3)-RNAi construct were excised with a razor blade and processed as reported previously (Kim, 2008). After metal staining, the sections were examined with a transmission electron microscope (JEM-1010; JEOL) operated at an accelerating voltage of 80 kV.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of the amino acid sequences of BCHs, ZEPs, VDEs, NCED1s, CCD7s, CCD8s, and CCD1s.
Supplemental Figure S2. The upstream region of GmBCH1 isolated by DNA walking.
Supplemental Figure S3. Assessment of the PCR efficiency of primer sets for GmBCHs.
Supplemental Figure S4. Assessment of probes for in situ hybridization.
Supplemental Figure S5. Expression of GmBCH2-GUS and GmBCH3-GUS in transgenic soybean nodules measured by fluorometric assay of GUS.
Supplemental Figure S6. Subcellular localization of N-terminally deleted GmBCH2 and a fusion of GmBCH1 with the transit peptide of GmBCH2.
Supplemental Figure S7. DNA sequences used in preparing for GmBCH(1+3)-RNAi and GmBCH2-RNAi constructs.
Supplemental Figure S8. Statistical analyses of nodule weight and gene expression in GmBCH(1+3)-RNAi nodules.
Supplemental Figure S9. EM analysis of GmBCH(1+3)-RNAi nodules.
Supplemental Figure S10. Expression of GmZEP3 in soybean.
Supplemental Figure S11. Functional assay of GmCCD7 and GmCCD8 in β-carotene-accumulating E. coli.
Supplemental Table S1. RNA-Seq expression data for soybean genes in various tissues.
Supplemental Table S2. Primers used for gene cloning and gene expression by real-time RT-PCR.
Supplementary Material
Acknowledgments
We thank Drs. Norihiko Misawa and Francis X. Cunningham, Jr., for generously providing pACCAR16ΔcrtX and pAC-zeaxanthin, respectively.
Glossary
- AM
arbuscular mycorrhizal
- ABA
abscisic acid
- BCH
β-carotene hydroxylase
- ZEP
zeaxanthin epoxidase
- CCDs
carotenoid cleavage dioxygenases
- RNAi
RNA interference
- cDNA
complementary DNA
- RT
reverse transcription
- MS
mass spectrometry
- CaMV
cauliflower mosaic virus
- EM
electron microscopic
- VDE
violaxanthin deepoxidase
References
- Alder A, Holdermann I, Beyer P, Al-Babili S. (2008) Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochem J 416: 289–296 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ. (2006a) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, McCarty DR, Klee HJ. (2006b) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Isner JC, Dogbo O, Camara B. (2005) Oxidative tailoring of carotenoids: a prospect towards novel functions in plants. Trends Plant Sci 10: 187–194 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Keller Y, d’Harlingue A, Camara B. (1998) Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim Biophys Acta 1391: 320–328 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Conover WJ (1971) Practical Nonparametric Statistics. John Wiley & Sons, New York, pp 97–104 [Google Scholar]
- Davison PA, Hunter CN, Horton P. (2002) Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418: 203–206 [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GE. (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20: 2681–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DS, Schliemann W, Schmidt J, Strack D, Walter MH. (2008) RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol 148: 1267–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J. (2006) A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA. (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26: 139–145 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Grünewald K, Hirschberg J, Hagen C. (2001) Ketocarotenoid biosynthesis outside of plastids in the unicellular green alga Haematococcus pluvialis. J Biol Chem 276: 6023–6029 [DOI] [PubMed] [Google Scholar]
- Ha SH, Liang YS, Jung H, Ahn MJ, Suh SC, Kweon SJ, Kim DH, Kim YM, Kim JK. (2010) Application of two bicistronic systems involving 2A and IRES sequences to the biosynthesis of carotenoids in rice endosperm. Plant Biotechnol J 8: 928–938 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Smith JJ, Tian L, Dellapenna D. (2009) The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol 50: 463–479 [DOI] [PubMed] [Google Scholar]
- Kim KW. (2008) Visualization of micromorphology of leaf epicuticular waxes of the rubber tree Ficus elastica by electron microscopy. Micron 39: 976–984 [DOI] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopsell DA, Kopsell DE. (2006) Accumulation and bioavailability of dietary carotenoids in vegetable crops. Trends Plant Sci 11: 499–507 [DOI] [PubMed] [Google Scholar]
- Lee MY, Shin KH, Kim YK, Suh JY, Gu YY, Kim MR, Hur YS, Son O, Kim JS, Song E, et al. (2005) Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol 139: 1881–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Lin WL, Dickson DW, Sahara N. (2011) Immunoelectron microscopic and biochemical studies of caspase-cleaved tau in a mouse model of tauopathy. J Neuropathol Exp Neurol 70: 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Li L. (2008) Carotenoid metabolism: biosynthesis, regulation, and beyond. J Integr Plant Biol 50: 778–785 [DOI] [PubMed] [Google Scholar]
- Maritz JS (1981) Distribution-Free Statistical Methods. Chapman & Hall, New York, p 217 [Google Scholar]
- Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K. (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172: 6704–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HS, Son O, Chun JY, Stacey G, Lee MS, Min KH, Song ES, Cheon CI. (2001) The Bradyrhizobium japonicum hsfA gene exhibits a unique developmental expression pattern in cowpea nodules. Mol Plant Microbe Interact 14: 1286–1292 [DOI] [PubMed] [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in Chrysanthemum petals. Plant Physiol 142: 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Harrison MJ, Paszkowski U. (2009) Reprogramming plant cells for endosymbiosis. Science 324: 753–754 [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, et al (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23: 482–487 [DOI] [PubMed] [Google Scholar]
- Rao AV, Rao LG. (2007) Carotenoids and human health. Pharmacol Res 55: 207–216 [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H (2013) The biology of strigolactones. Trends Plant Sci 18: 72–83 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon S, Schlueter J, Ma J, Mitros T, Nelson W, Hyten D, Song Q, Thelen J, Cheng J, et al. (2010) Genome sequence of the paleopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Loewen MC. (2004) The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J Biol Chem 279: 46940–46945 [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Moreau H, Kuntz M, Pagny G, Lin C, Tanksley S, McCarthy J. (2008) An investigation of carotenoid biosynthesis in Coffea canephora and Coffea arabica. J Plant Physiol 165: 1087–1106 [DOI] [PubMed] [Google Scholar]
- Singh S, Parniske M. (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15: 444–453 [DOI] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G, Libault M, Brechenmacher L, Wan J, May GD. (2006) Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol 9: 110–121 [DOI] [PubMed] [Google Scholar]
- Strack D, Fester T. (2006) Isoprenoid metabolism and plastid reorganization in arbuscular mycorrhizal roots. New Phytol 172: 22–34 [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Sun Z, Gantt E, Cunningham FX., Jr (1996) Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271: 24349–24352 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Akune M, Kogiso M, Imagama Y, Osuki K, Uchiumi T, Higashi S, Han SY, Yoshida S, Asami T, et al (2004) Control of nodule number by the phytohormone abscisic acid in the roots of two leguminous species. Plant Cell Physiol 45: 914–922 [DOI] [PubMed] [Google Scholar]
- Takaichi S, Mimuro M. (1998) Distribution and geometric isomerism of neoxanthin in oxygenic phototrophs: 9′-cis, a sole molecular form. Plant Cell Physiol 39: 968–977 [Google Scholar]
- Tominaga A, Nagata M, Futsuki K, Abe H, Uchiumi T, Abe M, Kucho K, Hashiguchi M, Akashi R, Hirsch AM, et al (2009) Enhanced nodulation and nitrogen fixation in the abscisic acid low-sensitive mutant enhanced nitrogen fixation1 of Lotus japonicus. Plant Physiol 151: 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, McCourt P. (2009) Strigolactones: a new hormone with a past. Curr Opin Plant Biol 12: 556–561 [DOI] [PubMed] [Google Scholar]
- Ubuka T, Bentley GE. (2009) Identification, localization, and regulation of passerine GnRH-I messenger RNA. J Endocrinol 201: 81–87 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T, Simkin AJ, Goulet C, Strack D, Bouwmeester HJ, et al (2010) SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J 61: 300–311 [DOI] [PubMed] [Google Scholar]
- von Lintig J. (2010) Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu Rev Nutr 30: 35–56 [DOI] [PubMed] [Google Scholar]
- Walter MH, Floss DS, Strack D. (2010) Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232: 1–17 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 212–214 [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I. (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu B, Lydiate DJ, Schäfer UA, Hannoufa A. (2007) Characterization of a beta-carotene hydroxylase of Adonis aestivalis and its expression in Arabidopsis thaliana. Planta 226: 181–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.