A novel set of HR-related genes and secondary metabolites depends on WRKY transcription factors in tomato.
Abstract
The hypersensitive response (HR) is considered to be the hallmark of the resistance response of plants to pathogens. To study HR-associated transcriptome and metabolome reprogramming in tomato (Solanum lycopersicum), we used plants that express both a resistance gene to Cladosporium fulvum and the matching avirulence gene of this pathogen. In these plants, massive reprogramming occurred, and we found that the HR and associated processes are highly energy demanding. Ubiquitin-dependent protein degradation, hydrolysis of sugars, and lipid catabolism are used as alternative sources of amino acids, energy, and carbon skeletons, respectively. We observed strong accumulation of secondary metabolites, such as hydroxycinnamic acid amides. Coregulated expression of WRKY transcription factors and genes known to be involved in the HR, in addition to a strong enrichment of the W-box WRKY-binding motif in the promoter sequences of the coregulated genes, point to WRKYs as the most prominent orchestrators of the HR. Our study has revealed several novel HR-related genes, and reverse genetics tools will allow us to understand the role of each individual component in the HR.
Unlike animals, plants do not have a specialized adaptive immune system that recognizes and directly attacks and destroys infectious agents. Instead, plants are endowed with a broad range of sophisticated and efficient innate immunity mechanisms that enable them to recognize and restrain a plethora of pathogenic microbes in their natural habitat (Jones and Dangl, 2006). Plants carry so-called pattern recognition receptors that recognize conserved structural molecules of pathogens, referred to as pathogen-associated molecular patterns, leading to pathogen-associated molecular pattern-triggered immunity (PTI). Invading pathogens, in their turn, secrete virulence proteins (so-called effectors) to suppress PTI, leading to effector-triggered susceptibility. As a response to this, plants have evolved resistance proteins that recognize these effectors, rendering them avirulence factors (Avrs), and subsequently mediate effector-triggered immunity. In fact, PTI and effector-triggered immunity function in a similar way, and both types of immunity are based on the activation of immune receptors that trigger a level of defense that is sufficient to stop colonization by the pathogen (DeYoung and Innes, 2006; Jones and Dangl, 2006; Postel and Kemmerling, 2009; Dodds and Rathjen, 2010; Thomma et al., 2011). The innate immune response of plants often culminates in the hypersensitive response (HR). This particular response, which is considered to be one of the hallmarks of the immunity of plants to pathogens, involves programmed cell death (PCD) and occurs at the site of pathogen entry, resulting in efficient containment of the pathogen (Lam et al., 2001).
The interaction between tomato (Solanum lycopersicum) and the biotrophic extracellular fungus Cladosporium fulvum is a model pathosystem that is characterized as a typical gene-for-gene interaction, in which C. fulvum resistance proteins (Cf) of tomato mediate the specific recognition of Avrs of the fungus (de Wit and Joosten, 1999; Rivas and Thomas, 2005; Thomma et al., 2005). In this interaction, the HR is one of the distinctive responses of resistant tomato plants to avirulent strains of the fungus. In most studied pathosystems, pathogen infection is nonsynchronous, rendering the chronological ordering of defense-related postinfection events in resistant plants at the molecular, biochemical, metabolic, and cytological levels difficult (Morel and Dangl, 1997). To obtain insight into the global transcriptome and metabolome reprogramming that occurs in tomato when mounting the HR, we studied tomato plants that express both a Cf resistance gene and the matching Avr gene from the fungus, obtained by crossing Cf-expressing tomato with an Avr-expressing tomato line. In such Cf/Avr “dying seedlings” (DS), the induction of the HR can be suppressed at an elevated temperature and high relative humidity (RH; 33°C and 100% RH). A subsequent shift to normal growth conditions (20°C and 70% RH) initiates a synchronized and systemic induction of the HR in the seedlings (de Jong et al., 2002; Gabriëls et al., 2006; Stulemeijer et al., 2007). In contrast, inoculation of Cf-expressing tomato plants with C. fulvum carrying the matching Avr gene results in an HR that is restricted to only a limited number of cells and that is not synchronized by far, which hampers research toward direct and small local effects. Hence, the DS enable us to synchronize and amplify the responses associated with the HR.
Here, we performed transcriptome and metabolome profiling of DS expressing both Cf-4 and Avr4, as compared with the parental lines expressing either Cf-4 or Avr4, at various time points after the shift to the permissive condition of 20°C and 70% RH. Unbiased transcriptome- and metabolome-wide approaches allow investigation of the behavior and relationships between different elements of a particular biological system (Schneider and Collmer, 2010). With the right tools, these approaches allow visualization of the underlying biological processes, leading to a better insight into the biology of the host cells while the plant is mounting a defense to the invading pathogen (Bolton et al., 2008a; van Baarlen et al., 2008; van Esse et al., 2008, 2009; Hanssen et al., 2011). We used the Affymetrix Tomato Genome Array to study changes in the global gene expression profiles in the DS as compared with their parental lines. In addition, we employed untargeted metabolomics, using gas chromatography (GC)-time-of-flight (TOF) and Liquid Chromatography (LC)-photo diode array (PDA)-quadrupole time-of-flight (QTOF) mass spectrometry (MS), for profiling polar primary metabolites (PPM) and semipolar secondary metabolites (SPSM) of the plant material, respectively. We analyzed and explored the data sets with various bioinformatics and statistics tools to identify major, system-wide transcriptome and metabolome reprogramming events associated with mounting of the HR. We found that the DS undergo a vast and coordinated reprogramming of their transcriptome within several hours after the shift to permissive conditions, on the one hand to induce the HR and on the other hand to control it. We revealed specific interrelationships of various metabolites associated with the HR and a critical role of energy homeostasis during mounting of the HR. Furthermore, we uncover major regulators and regulatory mechanisms of the HR in these tomato plants mounting a synchronous and systemic immune response.
RESULTS
HR-Associated Global Changes in Gene Expression in Tomato Seedlings
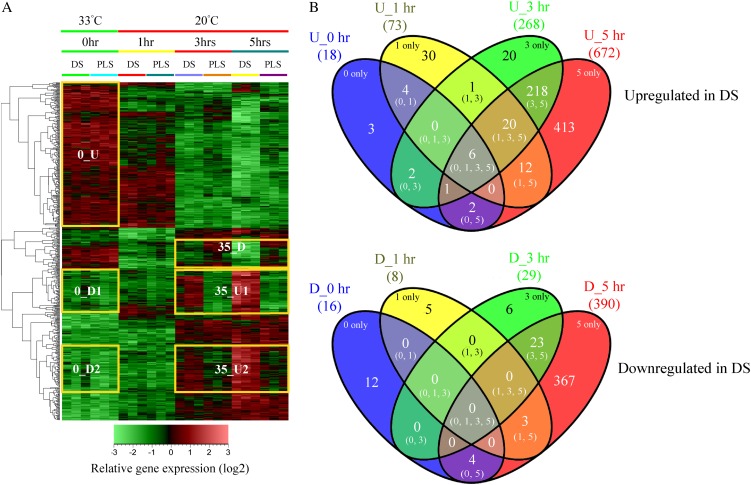
The hierarchical cluster analysis (HCA) presented in Figure 1A provides an overview of the relative expression profiles of the genes that were interrogated by the Affymetrix tomato microarrays in response to transferring the seedlings to environmental conditions permissive for HR. For each time point, the profiles of three independent biological replicates are shown. Before HR induction (0 h), the overall gene expression profile of the DS compared with a 1:1 mixture of their parental lines (PLS; see “Materials and Methods”) is very similar. At this stage, in both the DS and PLS, the majority of the genes with relatively high expression appear to be associated with the initially relatively high temperature of 33°C, as their expression decreases within 1 h after the shift to 20°C (Fig. 1A, yellow box 0_U). This cluster represents a large number of genes, including some of the major marker genes for heat stress, such as genes encoding heat shock proteins, heat stress-associated proteins, heat shock protein-binding proteins, DNAJ heat shock proteins, and universal stress proteins. Unlike the greater part of the genes in this cluster, which are further down-regulated at 3 and 5 h, most of the heat shock proteins show up-regulation at 5 h after the temperature shift in the DS when compared with the PLS (Supplemental Table S1).
Figure 1.
Transcriptome profiles of the Cf-4/Avr4 DS and their PLS before (0 h) and 1, 3, and 5 h after the temperature shift that initiates the HR in the DS. A, HCA of the transcriptome profile. For each time point, the columns labeled “DS” or “PLS” represent gene expression data obtained in three independent replicate experiments. Marked changes in the gene expression profiles of the DS and the PLS at 0 h and at 3 and 5 h after the temperature shift are indicated by the yellow-boxed gene sets. At 0 h, an additional gene set selection was performed to group genes based on their expression profiles. 0_U, Up-regulated at 0 h; 0_D, down-regulated at 0 h; 35_D, down-regulated at 3 and 5 h; 35_U, up-regulated at 3 and 5 h. The bar at bottom shows the color code for the relative change in gene expression of log2-transformed and autoscaled data. B, Venn diagram showing the temporal distribution of unique and overlapping genes differentially regulated upon comparison of the averages between the three biological replicates of the DS and those of the PLS for the different time points. The top panel shows genes up-regulated (U) in the DS compared with the PLS, and the bottom panel shows genes down-regulated (D) in the DS compared with the PLS. Numbers in parentheses represent the number of differentially regulated genes that are unique or overlapping at time points 0, 1, 3, and/or 5 h after the temperature shift.
Upon shifting the plants from 33°C to 20°C, within 1 h, moderate changes in gene expression were observed, which for the greater part involve slight up-regulation of sets of genes in the DS compared with the PLS. This up-regulation becomes much more pronounced at 3 and 5 h after the temperature shift (Fig. 1A, yellow boxes 35_U1 and 35_U2). In contrast, gene set 35_D is down-regulated in the DS compared with the PLS. Figure 1B provides an overview of the numbers of genes that are either significantly up- or down-regulated in the DS compared with the PLS at 0 h (33°C/100% RH) and the different time points after the shift to 20°C/70% RH, based on the averages between the three biological replicates for the individual time points. From the 10,219 genes represented on the tomato genome array, a total of 1,152 genes (11.3%) show differential regulation (fold change [FC] > 2 and P < 0.05), at least under one of the conditions analyzed. Of these genes, 732 are up-regulated and 420 are down-regulated in the DS when compared with the PLS, representing about 7.2% and 4.1% of the total number of genes represented on the array, respectively (Fig. 1B).
The rescue temperature of 33°C, in combination with 100% RH, efficiently suppresses the expression of the majority of the defense-related genes (Fig. 1A, yellow boxes 0_D1 and 0_D2; Gabriëls et al., 2006; Stulemeijer et al., 2007). However, in the DS, there are six genes that consistently show a higher expression compared with the PLS at all time points (Fig. 1B, top panel [0, 1, 3, 5]). These six genes encode a basic endochitinase, an acidic 26-kD endochitinase, a Gly-rich protein, a photoassimilate-responsive protein, a disease resistance-responsive family protein, and aquaporin NIP1.1, which have all been reported to be involved in the response of plants to biotic stress (Margolles-Clark et al., 1996; Bienert et al., 2007; Hong et al., 2007; Gomes et al., 2009; Mangeon et al., 2010). This indicates that in the DS, some level of defense is already induced before the temperature shift without the execution of the HR. One hour after the shift to 20°C, 81 genes show differential regulation in the DS compared with the PLS, of which the majority (90%) are up-regulated (Fig. 1B, top panel, U_1 h [73 genes], and bottom panel, D_1 h [eight genes]). About 41% (30 genes) of these up-regulated genes are unique for this time point (Fig. 1B, top panel, 1 only), whereas a subset of 20 genes that show early up-regulation continue to exhibit higher expression levels at later time points (Fig. 1B, top panel [1, 3, 5]). In the DS, the overlapping and unique sets of genes that are differentially expressed at 3 and 5 h (Fig. 1A, boxes 35_D, 35_U1, and 35_U2) account for about 91% of the total number of differentially regulated genes (Fig. 1B, 3 only, 5 only, and [3, 5]), indicating that defense-related transcriptional reprogramming is significantly intensified at time points later than 1 h after the shift to permissive conditions. Close to 62% of these genes are up-regulated in the DS compared with the PLS, with the majority (413 genes; 63%) being unique for 5 h (Fig. 1B, top panel, 5 only). In addition to a large set of up-regulated genes at 5 h (U_5 h; 672 genes), there are also a large number of down-regulated genes at this time point (397, from which 367 are even unique for 5 h; Fig. 1B, bottom panel, D_5 h and 5 only, respectively).
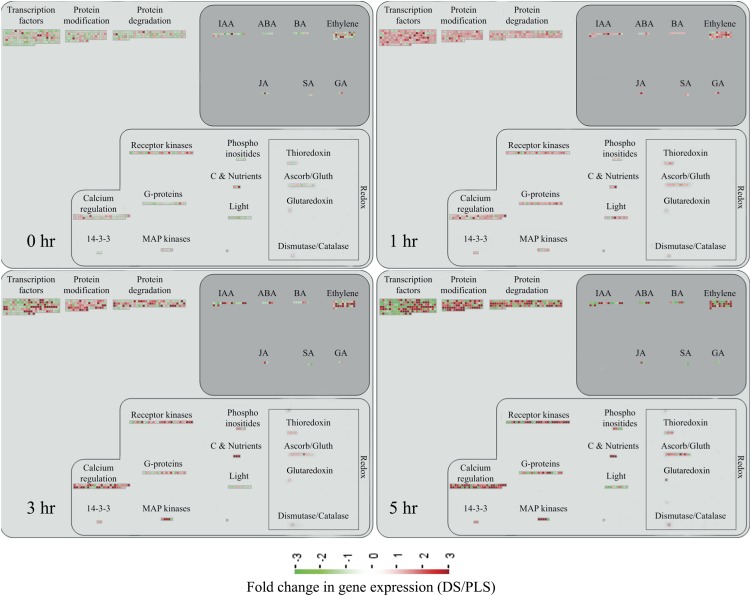
As visualized by MapMan (Thimm et al., 2004), the massive reprogramming in the DS upon initiation of the HR involves the transcriptional activation of genes encoding receptor-like kinases, G-proteins, mitogen-activated protein (MAP) kinases, transcription factors, and genes involved in calcium regulation, redox pathways, hormone biosynthesis, protein modification, and protein degradation (Fig. 2). Cytoplasmic receptor-like kinase (RLK)-encoding genes are the most prominent among the up-regulated RLKs. This group also includes genes encoding Leu-rich repeat-containing RLKs, lysin motif-containing RLKs, Domain of Unknown Function26-containing RLKs, wall-associated kinase, and an RLK10-like protein. Particularly at 3 and 5 h, these genes show significantly higher expression in the DS (Fig. 2, 3 hr and 5 hr).
Figure 2.
Visualization of the transcriptional reprogramming occurring in the DS as compared with their PLS at 0, 1, 3, and 5 h after the temperature shift that induces the HR in the DS. Each panel represents an individual time point and depicts the expression profile of genes (represented by the colored squares) involved in the indicated biological processes. The bar at bottom shows the color codes for the FC in gene expression in the DS compared with the PLS. The map does not represent all differentially regulated genes, as only genes that can be related to specific biological processes are displayed. ABA, Abscisic acid; BA, benzyladenine; IAA, indole-3-acetic acid; JA, jasmonic acid; SA, salicylic acid.
Overrepresentation Analysis of Differentially Regulated Gene Sets
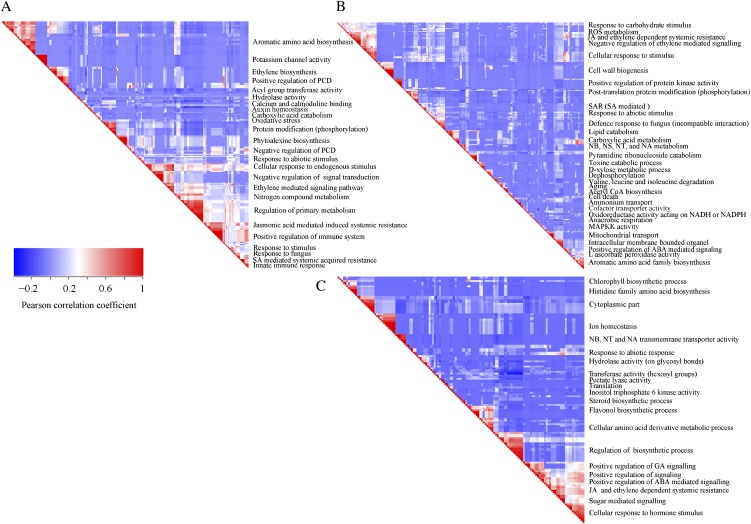
Overrepresentation analysis (ORA) of genes within the up- and down-regulated categories (Fig. 1B) allows making a temporal dissection of the different processes that occur upon mounting of the HR. ORA on gene sets that showed differential regulation at 3 and 5 h after initiating the HR produced a vast list of significantly enriched or underrepresented Gene Ontology (GO) subcategories. To visualize these lists of categories and to reduce the high level of redundancy, we built a similarity matrix using the (gene × GO subcategory) matrix, which is an output of GeneTrail software (Backes et al., 2007). In the (gene × GO subcategory) matrix, a differentially regulated gene gets a score of 1 when it belongs to a particular functional category; if not, its score is 0. Subsequently, the resulting (0, 1) matrix was used to produce a set of manageable “GO subcategory metaclusters” by comparing two GO subcategories for the characteristic presence of a gene. In such an analysis, the high redundancy level of the GO annotation is significantly reduced (for details, see Supplemental Materials and Methods S1). In the panels that are shown, redundant GO subcategories that have high correlation are indicated in red and fall along the diagonal axis of the similarity matrix (Fig. 3).
Figure 3.
ORA of the differentially regulated genes in the DS when compared with the PLS at 3 and 5 h after the temperature shift that induces the HR in the DS. A similarity matrix forms a rectangle that is a mirror image of two identical triangles; therefore, only the right triangle is shown. The red triangles along the long sides of the triangles represent clusters of highly similar GO subcategories, whereas the blue blocks represent clusters of unrelated subcategories. A, Similarity matrices (GO subcategories × GO subcategories) of the overlapping genes differentially expressed at time points 3 and 5 h. B, Up-regulated genes unique for time point 5 h. C, Down-regulated genes unique for time point 5 h. ABA, Abscisic acid; JA, jasmonic acid; NA, nucleic acid; NB, nucleobase; NS, nucleoside; NT, nucleotide; ROS, reactive oxygen species; SA, salicylic acid; SAR, systemic acquired resistance. The strength of the correlation is indicated by the color bar on the left: red, strong correlation; blue, not correlated. For details, see Supplemental Materials and Methods S1.
ORA on the set of genes that show early transcriptional activation upon HR induction (Fig. 1B, top panel, U_1 h; 73 genes), revealed some significantly overrepresented GO subcategories, such as Val, Leu, and Ile degradation, oxidoreductase activity, response to chemical stimulus, secondary metabolism, and response to stimulus (Supplemental Table S2). At time points 3 and 5 h, there is massive transcriptional reprogramming in the DS. ORA on the 218 overlapping up-regulated genes at these time points (Fig. 1B, top panel, [3, 5]) resulted in 226 significantly enriched GO subcategories, and a similarity matrix analysis on these subcategories produced 25 distinct metaclusters (Fig. 3A). Interestingly, functional categories associated with signal perception, signal transduction, and activated defense responses are among these metaclusters. Examples are sets of up-regulated genes associated with aromatic amino acid biosynthesis (Phe and Tyr), ethylene biosynthesis and signaling, calcium/calmodulin-dependent signaling, protein modification (phosphorylation), phytoalexin biosynthesis, and positive as well as negative regulation of PCD (Fig. 3A).
Time point 5 h is characterized by the presence of two large differentially regulated gene sets, one up-regulated in the DS (Fig. 1, A, sets 35_U1 and 35_U2, and B, top panel, 5 only; 413 genes) and one down-regulated in the DS (Fig. 1, A, set 35_D, and B, bottom panel, 5 only; 367 genes). ORA on these two sets of genes revealed a significant enrichment for a number of GO subcategories: 32 and 22 major GO subcategory metaclusters were identified in the up-regulated and down-regulated categories, respectively (Fig. 3, B and C). In the up-regulated gene set, some of the significantly overrepresented GO subcategories are anaerobic respiration, catabolic processes like lipid catabolism and pyrimidine ribonucleoside catabolism, cell wall biogenesis, systemic acquired resistance, mitochondrial transport, positive as well as negative regulation of defense-related processes, aging, and cell death. Typically, these categories are indicative for the activation of processes demanding large amounts of energy and the activation of corresponding cellular processes aimed to meet this energy demand. Among the down-regulated genes, genes involved in chlorophyll and flavonol biosynthesis, ion homeostasis (related to Ca2+ homeostasis), hydrolase activity, response to abiotic stress, and positive regulation of signaling and translation are all significantly overrepresented.
Network Analysis Indicates Strong Association between Catabolic Processes, and Carbon Backbone- and Energy-Demanding Biosynthesis-Related Processes, during Activation of the HR
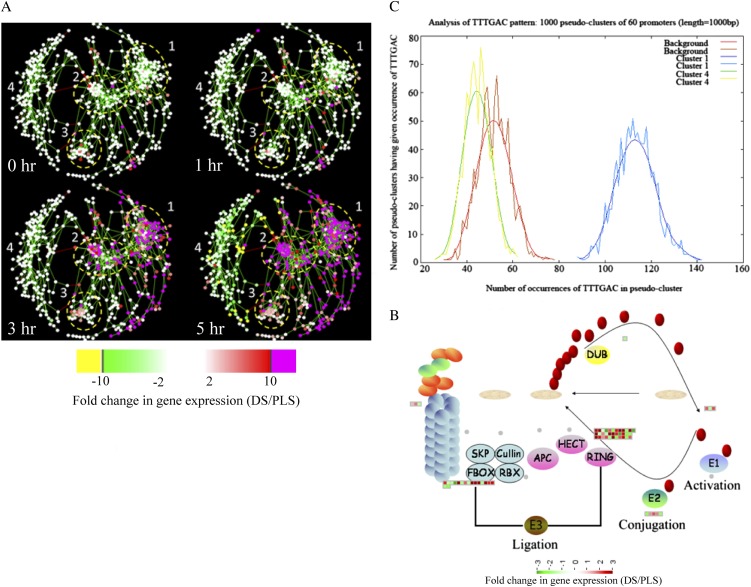
By using Cytoscape software (Shannon et al., 2003) and the plugin Expression Correlation Network (http://www.baderlab.org/Software/ExpressionCorrelation), a coexpression network analysis was performed on all 1,152 genes that are differentially regulated between the DS and the PLS under at least one of the conditions that was analyzed. By using the Cytoscape plugin Module Identification In Networks (MINE; Rhrissorrakrai and Gunsalus, 2011), four clusters of strongly associated genes were identified within the coexpression network (Fig. 4A). Clusters 1, 2, and 3 mostly harbor up-regulated genes, whereas cluster 4 consists of mostly down-regulated genes (Fig. 4A). The expression profile of each gene cluster is shown in Supplemental Figure S1. In some cases, there is substantial variation in the expression levels of the individual genes belonging to one cluster when the individual replicates for the same conditions are compared. The within-sample variation (variation between biological replicates) at early time points is high, resulting in lower numbers of significantly regulated genes (Figs. 1B and 4A; Supplemental Fig. S1). This variation is much less at time points 3 and 5 h, resulting in high numbers of significantly regulated genes.
Figure 4.
Coexpression analysis of genes differentially regulated during the HR and identification of central regulators in the regulatory network. A, Coexpression networks, representing all differentially regulated genes that have a correlation coefficient of 0.92 or greater, at 0, 1, 3, and 5 h after the temperature shift that induces the HR in the DS. Shown is an output of Cytoscape software that was used to build a coexpression network of all 1,152 genes that are significantly differentially regulated between the DS and the PLS under at least one of the conditions analyzed. The dots represent individual genes (the “nodes”), and the length of the connecting lines (the “edges”) between them is a measure of their coexpression. Short edges indicate strong coexpression, whereas long edges indicate weak coexpression. Four clusters of strongly coregulated genes were identified. The bar at bottom shows the color code for the FC in gene expression. B, MapMan representation of genes involved in the proteasome pathway, depicting differentially regulated genes at 5 h after the temperature shift involved in ubiquitin activation (E1), ubiquitin conjugation (E2), and ligation of ubiquitin to proteins that are targeted for degradation (E3). SKP, Cullin, and FBOX together constitute the SCF complex. RBX (for ring box protein), APC (for anaphase-promoting complex), HECT (for homology to E6-AP C terminus), and RING (for really interesting new gene) are different protein complexes in the E3 ubiquitin ligase family. DUB, Deubiquitinating enzyme. Red ovals represent individual ubiquitin proteins. C, POBO output interface indicating a “good” motif with spread sample distribution. In this case, the test set (all cluster 1 genes) and the background (the promoter sequences of all genes of Arabidopsis) as well as the cluster 4 gene set (genes with an opposite expression pattern to the test set) are well separated. This indicates a significant difference in the occurrence of the motif searched for among these clusters (in this case, TTTGAC, the W-box motif). “Bad” motifs are characterized by uniform sample distribution, indicating an absence of variation in the occurrence of the motif (cluster 4 and background).
We used Web-based tools from the Tomato Functional Genomics Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/array/GO_analysis.cgi) and AtCOECiS (http://bioinformatics.psb.ugent.be/ATCOECIS/; Vandepoele et al., 2009; Fei et al., 2011) to compute GO enrichment analysis on each cluster. The coexpression network analysis, in combination with the GO enrichment analyses, revealed a strong association between genes involved in protein degradation (encoding proteins involved in proteasome-related pathways) and cell death with genes involved in the final step of Phe, Tyr, and hydroxycinnamic acid amide (HCAA) biosynthesis. These genes are predominantly present in cluster 1 of Figure 4A, together with many genes implicated in the HR, such as genes encoding amino acid transporters, MAP kinases, a number of WRKY transcription factors, and genes involved in the calcium and calmodulin signaling network. Furthermore, cluster 1 contains genes encoding receptor-like protein kinases, protein kinases and glycosyl hydrolases, genes involved in phospholipid signaling, the jasmonic acid- and ethylene-mediated signaling pathways, oxidative stress, and a number of genes of unknown function (Fig. 4A, cluster 1; Supplemental Table S3, cluster 1 genes and cluster 1 GO). Genes involved in Ca2+-related signaling are also among the core components of cluster 1. This is evident from the presence of a large number of genes involved in calcium sensing and genes encoding other proteins involved in Ca2+-related signaling. These genes include Glutamate decarboxylase2 (GAD2), Diacylglycerol kinase, genes encoding phosphatases, Phospholipase D β1, MAPK3, genes encoding a number of protein kinases, and calmodulin-binding WRKY transcription factors (Pappan et al., 1997; Snedden and Blumwald, 2000; Snedden and Fromm, 2001; Yang and Poovaiah, 2003; Yap et al., 2003; Du et al., 2009; Leivar et al., 2011; Supplemental Table S3, cluster 1 genes). Unlike genes playing a role in the shikimate pathway, which represents the early steps in aromatic amino acid biosynthesis (Maeda and Dudareva, 2012), genes encoding enzymes involved in the final steps of aromatic amino acid biosynthesis, such as Arogenate dehydratase4 (ADT4), ADT6, and Prephenate dehydratase1, show massive up-regulation (more than 35-fold) in expression in the DS as compared with the PLS (Supplemental Table S3, cluster 1 genes, aromatic amino acid biosynthesis). Most of the genes encoding enzymes of the shikimate pathway that are involved in the generation of chorismate for the biosynthesis of aromatic amino acids (Phe, Trp, and Tyr) only show a slight transcriptional up-regulation and predominantly localize in cluster 3 (Fig. 4A). These genes include Chorismate synthase1, Chorismate mutase1 (CM1), CM3, Phospho-2-dehydro-3-deoxyheptonate aldolase2, and 5-Enolpyruvylshikimate-3-phosphate synthase (Supplemental Table S3, cluster 3 genes).
At 5 h after the induction of the HR, MapMan-based mapping of the differentially regulated genes clearly illustrates the induction of genes associated with the ubiquitin-dependent protein degradation pathway (Fig. 4B). In the regulatory network, these genes are mainly localized in cluster 1 (Fig. 4A). Here, one of the main differences between the DS and the PLS lay in the final step of ubiquitination (E3), which involves the ligation of ubiquitin to proteins that are targeted for degradation, with considerably higher related gene expression in the DS (Fig. 4B, RING and FBOX; Supplemental Table S3, cluster 1 genes). There are no clear differences between the DS and PLS concerning the expression of genes involved in processes like ubiquitin activation (E1), whereas at 5 h in the DS, there is a slight, but significant, down-regulation (about 2-fold) of the E2-related bin representing genes involved in ubiquitin conjugation (Fig. 4B).
Network Analysis Indicates WRKY Transcription Factors as Prominent Master Regulators of Transcriptional Reprogramming during the HR
To identify common transcriptional regulators of genes for which the expression profiles are highly similar (Supplemental Fig. S1), we studied the presence of conserved regulatory motifs in the promoter sequences of genes belonging to the same clusters in Figure 4A. One of the significantly overrepresented motifs present in the promoter of genes belonging to cluster 1 (Fig. 4A) is TGACC/T, the so-called W-box, a potential binding site for WRKY transcription factors (Eulgem, 2005). This W-box motif dominates the gene cluster (Supplemental Table S4; Supplemental Fig. S2), as 56 out of the 78 genes present in cluster 1 contain such a TGACC/T motif. However, the Y-box motif (redox-dependent transcription activation) and other motifs of unknown function are also among the significantly overrepresented motifs (Supplemental Table S4A). Validation by the Web-based tool POBO (http://ekhidna.biocenter.helsinki.fi/poxo/pobo/) confirmed the significant enrichment of the W-box motif in the promoters of the coregulated gene cluster of the test set (all cluster 1 genes), while the background (the promoter sequences of all Arabidopsis [Arabidopsis thaliana] genes) and the second input (the promoter sequences of all genes of cluster 4) have a similar, but low, occurrence of the motif (Fig. 4C; Supplemental Table S4A). Interestingly, in cluster 1, there are a number of genes present that are annotated as WRKY transcription factors themselves (Supplemental Table S3, cluster 1 genes, WRKY). This suggests that there is a network of WRKYs that orchestrates the transcriptome reprogramming related to the induction and amplification of processes leading to the HR.
Cluster 2 harbors genes involved in ethylene biosynthesis and ethylene-triggered signaling, transferases involved in secondary metabolism, genes involved in lipid signaling, and others (Supplemental Table S3, cluster 2 genes and cluster 2 GO). For this cluster, POBO validation revealed that binding motifs associated with MYB transcription factors are abundant in the promoter sequences (Supplemental Table S4A). Interestingly, in this cluster, there is a gene present with high sequence identity to Arabidopsis MYB78 (AtMYB78; Supplemental Table S3, cluster 2 genes). Cluster 3 represents genes involved in the early steps of aromatic amino acid biosynthesis, and the promoter sequences of these genes show enrichment of an abscisic acid-responsive element, which is the abscisic acid response element-like binding site motif (Supplemental Table S4A).
Cluster 4 is characterized by the presence of genes that are involved in photosynthesis, the biosynthesis of flavonoids, ion homeostasis, circadian rhythm, and others (Fig. 3C; Supplemental Table S3, cluster 4 genes and cluster 4 GO). One of the fascinating features of this cluster is that there are five genes present that encode unknown proteins and show an inverse expression pattern from the rest of the group. These genes might be involved in the coordinated repression of other genes within this cluster (Supplemental Fig. S1; Supplemental Table S3, cluster 4 genes). Although the promoter sequences of the genes belonging to cluster 4 contain mostly unknown overrepresented motifs, some of the identified motifs that were validated by POBO include AG (MADS) and SORLIP4 (phytochrome A-regulated gene expression; Supplemental Table S4A).
Validation of Gene Expression in C. fulvum-Inoculated Tomato and Enhanced Ethylene Production by the DS
We validated our findings concerning up-regulated genes in the DS model system by checking their actual regulation in the interaction between C. fulvum and tomato by performing quantitative reverse transcription (qRT)-PCR on 10 selected genes from clusters 1 and 2, harboring most of the HR-related genes. The selected genes are involved in ethylene biosynthesis, calmodulin binding, oxylipin biosynthesis, proteolysis, transcription factor activity, aromatic amino acid biosynthesis, secondary metabolite (HCAA) biosynthesis, and signal transduction (kinase activity) and are categorized into different GO subcategories either directly or indirectly linked with plant defense. We inoculated the PLS that were used to generate the DS (MM-Cf0:Cf-4, resistant, and MM-Cf0:Avr4, susceptible) with a strain of C. fulvum secreting Avr4, and gene expression levels, relative to mock-treated plants, were determined at 6 and 10 d after inoculation (Supplemental Fig. S3A). The relative expression levels of the selected genes in the C. fulvum-tomato interaction are consistent with the microarray results (Supplemental Fig. S3B), showing an up-regulation in the resistant plants as compared with the susceptible ones at an early stage of infection (6 d after inoculation).
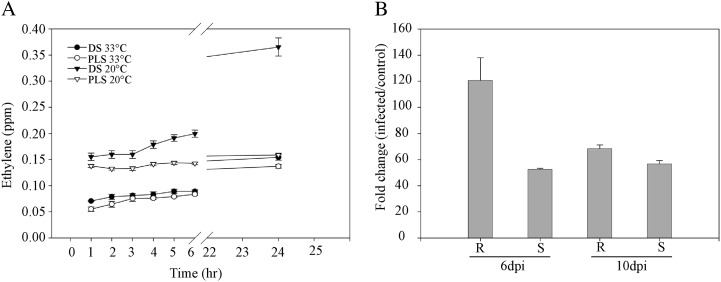
To determine whether the increased expression of genes that encode proteins involved in ethylene production (Figs. 2–4, cluster 2; Table I) also leads to increased ethylene production, we measured the ethylene emission of both the DS and the PLS before and after the temperature shift. Instead of shifting the plants to 20°C and 70% RH, plants were now shifted to 20°C and 100% RH as the plants were kept in closed jars to analyze their head space. Upon rescue at 33°C and 100% RH, there was a slow release of ethylene, both by the DS and PLS (Fig. 5A). Under these particular experimental conditions, lowering the temperature to 20°C did not result in a swift visible HR, indicating that the 100% RH in the jars is sufficient to suppress this response, at least for a period of 24 h. Interestingly, although the development of a visual HR is suppressed, a marked increase in ethylene emission was observed at 4 h after shifting the seedlings to 20°C and 100% RH, as the DS started to emit significantly higher amounts of ethylene than the PLS (Fig. 5A). After 24 h at 20°C and 100% RH, the DS had emitted about three times more ethylene than the PLS.
Table I. Expression profiles of genes involved in ethylene biosynthesis and signaling.
Expression levels are given as FC in gene expression in the DS as compared with the PLS at 0, 1, 3, and 5 h after the temperature shift that induces the HR in the DS. CTR1, Constitutive triple response1; EIN4, Ethylene insensitive4; ERF, Ethylene response factor; ESE1, Ethylene and salt inducible1; ETR1, Ethylene response1; MBF1C, Multiprotein-binding protein1C.
| Probe Set Identifier | Arabidopsis Identifier | Gene Name |
FC (DS/PLS) |
|||
|---|---|---|---|---|---|---|
| 0 h | 1 h | 3 h | 5 h | |||
| Les.3662.1.S1_at | At1g01480 | ACS2 | 5 | 2 | 26 | 101 |
| LesAffx.3059.1.S1_at | At3g23220 | ESE1 | −1 | 1 | 17 | 56 |
| Les.3769.1.S1_at | At4g11280 | ACS6 | −2 | 2 | 13 | 31 |
| Les.3575.1.S1_at | At3g23240 | ERF1 | 4 | 3 | 13 | 31 |
| Les.36.1.S1_at | At3g04580 | EIN4 | 3 | 2 | 23 | 29 |
| Les.274.1.S1_at | At5g61600 | ERF104 | −1 | 2 | 18 | 22 |
| LesAffx.51274.1.S1_at | At5g47230 | ERF5 | −1 | −1 | 36 | 20 |
| Les.3551.1.S1_at | At3g24500 | MBF1C | −1 | −2 | 2 | 20 |
| Les.3818.1.S1_at | At3g23240 | ERF1 | 1 | 1 | 4 | 20 |
| Les.3573.1.S1_at | At5g47220 | ERF2 | 2 | 2 | 3 | 8 |
| Les.4140.1.S1_at | At4g17500 | ERF1 | −2 | 2 | 4 | 7 |
| Les.3041.1.S1_at | At1g50640 | ERF3 | −1 | 2 | 3 | 3 |
| Les.3679.1.S1_at | At3g23240 | ERF1 | 2 | 3 | 2 | 2 |
| Les.3500.1.S1_at | At1g66340 | ETR1 | −1 | 1 | −1 | −3 |
| Les.31.1.S1_s_at | At5g03730 | CTR1 | 1 | 2 | 1 | 3 |
Figure 5.
Ethylene emission and ACS2 gene expression of the seedlings and C. fulvum-inoculated tomato, respectively. A, Ethylene emission patterns of the DS and their PLS upon rescue at 33°C and 100% RH and after a shift to 20°C and 100% RH. B, Relative expression levels of the ACS2 gene in resistant (R; MM-Cf0:Cf-4) and susceptible (S; MM-Cf0:Avr4) parental lines at 6 and 10 d post inoculation (dpi) with C. fulvum. Bars represent expression of the gene (FC) in inoculated plants relative to the mock-inoculated resistant and susceptible PLS.
To verify whether ethylene-induced signaling processes are also activated when tomato is infected with C. fulvum, we monitored the expression of 1-Aminocyclopropane-1-carboxylate synthase2 (ACS2), which encodes one of the rate-limiting enzymes in ethylene biosynthesis, in the resistant and susceptible parental lines at 6 and 10 d after inoculation with the fungus. In line with our observations in the DS, in resistant tomato, ACS2 expression was strongly up-regulated at 6 d post inoculation when compared with the mock-inoculated plants and was about 2.4 times higher than in the inoculated susceptible plants. ACS2 continued to exhibit a higher expression level up to 10 d post inoculation when compared with the susceptible C. fulvum-inoculated parental line (Fig. 5B).
The Dynamics of the Metabolome of the DS, as Compared with the PLS, Upon Mounting of the HR
To further validate the observed HR-associated transcriptional reprogramming in the DS upon mounting of the HR and to determine its effect on the overall metabolome of these plants, we performed untargeted metabolite profiling of both PPM and SPSM of the DS and the PLS using GC-TOF-MS and LC-PDA-QTOF-MS, respectively. We detected a total of 132 different PPM and 250 different SPSM, of which 42 and 144, respectively, show differential accumulation (FC > 1.5, P < 0.05) in the DS, as compared with the PLS, at least under one of the conditions analyzed. Compared with most of the secondary metabolites, for most of the organic acids and some other primary metabolites, the FC in accumulation is reproducible but relatively low. Hence, the cutoff value was lowered to an FC of 1.5 in order to retain the organic acids in the analysis (Supplemental Table S5).
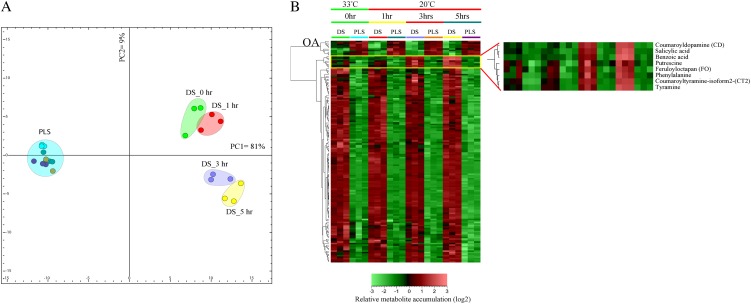
Based on the differentially accumulating metabolites, we computed a discriminant function analysis (DFA) and performed HCA to investigate the temporal accumulation patterns of these metabolites in response to triggering of the HR (Fig. 6A). The first component (PC1) of the DFA already explains more than two-thirds (81% variance) of the variation in the entire data set. PC1 predominantly reflects an inherent genotype-dependent difference in metabolite content (DS versus PLS), even before the temperature shift that induces the HR in the DS (Fig. 6A). The presence of such an inherent difference suggests that, although at 0 h (33°C/100% RH) the differences between the DS and PLS at the transcriptional level are relatively low (Fig. 1), this condition apparently does not fully suppress the Cf-4/Avr4-triggered responses, thereby already causing a substantial difference at the level of the metabolome (see below). Some of the metabolites that contribute to the separation of the PLS and DS along the first principal component (PC1) include amino acids, alkaloids, some HCAAs, benzenoids, flavonoids, and other unidentified metabolites that accumulate to higher amounts in the DS already at 0 h. Furthermore, there is a strong depletion of organic acids, and of some flavonoids and several unidentified metabolites in the DS, at this time point (Supplemental Table S5).
Figure 6.
Metabolome reprogramming in the DS as compared with their PLS. Shown are the metabolome profiles, comprising all differentially regulated PPM and SPSM, of the DS as compared with the PLS at 0, 1, 3, and 5 h after the temperature shift. A, Principal component analysis depicting metabolome reprogramming in the DS as compared with the PLS. PC1 and PC2 represent principal components 1 and 2, respectively. B, HCA of the PPM and SPSM profiles. The bar at bottom shows the color code for the relative change in metabolite accumulation of log2-transformed and autoscaled data. The enlarged part of the heat map at right indicates the aromatic amino acid-derived metabolites that differentially accumulate in the DS at time points 3 and 5 h after the temperature shift. OA, Clade containing most of the organic acids that are differentially down-regulated in the DS.
In line with the overall changes in the transcriptome of the DS, there is further reprogramming of the metabolome during mounting of the HR in these plants. This is mainly reflected by the second principal component (9% variance) that primarily indicates the time-dependent accumulation of HR-associated metabolites that is typical for the DS (Fig. 6A). In the DS, this group of metabolites shows higher accumulation at time points 3 and 5 h after the temperature shift. All of them are aromatic amino acid-derived metabolites, including benzoic acid (14-fold), salicylic acid (26-fold), tyramine (10-fold), dopamine (5-fold), and the HCAAs coumaroyltyramine-isoform 1 (75-fold) and coumaroyldopamine (116-fold; Fig. 6B; Supplemental Table S5), and this massive increase reflects the full activation of HR-related defense responses. There is also a clear positive precursor-final product relationship among these metabolites. Other HCAAs, such as feruloyloctapan, coumaroyltyramine-isoform 1 and -isoform 2, feruloyltyramine-isoform 1 and -isoform 2, and the glycosyl forms of most of these HCAAs, showed higher accumulation both before and after the temperature shift that induces the HR in the DS (Supplemental Table S5A). Moreover, in comparison with other groups of metabolites (amino acids, polyamines, alkaloids, flavonoids, and benzenoids), the HCAAs showed by far the highest accumulation in the DS (Supplemental Table S5). In the DS, out of the top 20 metabolites showing an increase with an FC of over 15, 11 metabolites represent HCAAs and their glycosyl forms (Supplemental Table S5A).
DISCUSSION
In this study, we have used large-scale “omics” technologies to assess the overall temporal transcriptome and metabolome reprogramming associated with mounting of the HR in tomato. For this, we used DS, expressing both the Cf-4 resistance gene and the matching Avr4 effector, as a model to study the resistance response of tomato to C. fulvum. Our analysis revealed that the induction and execution of the HR is the outcome of the synergistic or antagonistic effects of a number of intertwined cellular and biochemical processes.
Major HR-Associated Signal Transduction and Metabolic Pathways
Our metabolome analysis, combined with the ORA on the differentially regulated genes, clearly shows the involvement of both ethylene and salicylic acid in intensifying the defense responses at later time points after initiating the HR by shifting the DS to permissive conditions (Figs. 2, 3, A and B, 5, and 6B; Table I; Supplemental Table S5). The gene S-Adenosyl-l-methionine carboxyl methyltransferase, which is involved in the conversion of salicylic acid to methyl salicylate, was slightly up-regulated at time points 0 and 1 h and showed dramatic reduction in expression (FC = 5) in the DS at later time points (3 and 5 h). This might be a way to maintain high levels of salicylic acid during the HR (Fig. 2, SA). Other genes, such as MAPK4, NPR1, GRX480, and DOX1, which are involved in salicylic acid-mediated signaling, also showed up-regulation. However, these genes could not be mapped in Figure 2. Ethylene plays an important role in the activation of different forms of PCD, including the HR (Lund et al., 1998; Ciardi et al., 2000, 2001; van Loon et al., 2006; Mur et al., 2009). The well-controlled ethylene release by the DS over time only after HR initiation confirms the robustness of the DS system and matches the observed increase in ethylene-related gene expression (Fig. 2). Ethylene has been shown to be required for the induction of N-Hydroxycinnamoyl-coenzyme A:tyramine N-hydroxycinnamoyl transferase (THT) gene expression and the accumulation of HCAAs during the defense of tomato against the bacterial pathogen Pseudomonas syringae (Zacarés et al., 2007; Lopez-Gresa et al., 2011). In the latter studies, exogenous application of ethylene led to an increase in the expression of the THT genes in tomato, whereas application of the ethylene biosynthesis inhibitor AVG reduced the release of ethylene after P. syringae infection and, consequently, suppressed the expression of the THT genes. In potato (Solanum tuberosum), tomato, and tobacco (Nicotiana tabacum), THTs play a pivotal regulatory role in the biosynthesis of HCAAs by catalyzing the transfer of hydroxycinnamic acids from the respective CoA esters to tyramine, dopamine, octamine, and various other amines (Guillet and De Luca, 2005; Kang et al., 2006).
The changes in the transcriptome and metabolome profiles of the DS revealed the activation of both primary and secondary metabolism during mounting of the HR (Figs. 3 and 6). Aromatic amino acids (Phe and Tyr) and various aromatic secondary metabolites derived from these amino acids represent a large portion of the HR-associated metabolites (Fig. 3, A and B; Supplemental Table S5). Phe and Tyr, in combination with various amines, are the primary precursors for the induced HCAA biosynthesis during the defense of plants against pathogens and wounding (Facchini et al., 2002; von Roepenack-Lahaye et al., 2003; Walters, 2003; López-Gresa et al., 2011). At 3 and 5 h, hydroxycinnamic acid-derived compounds, such as benzoic acid (14-fold increase), salicylic acid (26-fold increase), and many HCAAs (20- to 116-fold increase), showed massive accumulation in the DS (Fig. 6B; Supplemental Table S5). The increase in HCAA levels correlates with an up-regulation of the THT genes, as evidenced from the transcriptome of both the DS model and the C. fulvum-infected resistant tomato lines (Supplemental Fig. S3, THT; Supplemental Table S7). In the DS, most of the induced HCAAs have tyramine and dopamine as a backbone. In line with this, tyramine (10-fold increase), dopamine (6-fold increase), and their precursor Tyr (5-fold increase) were present at higher levels in the DS than in the PLS (Supplemental Table S5B). Extraction of HCAAs that are present in the form of higher complexes in the plant cell walls is less likely with the extraction technique that we used (see “Materials and Methods”). However, we detected various HCAAs in the leaf extracts of the DS. This might indicate either that there is a massive production of HCAAs, exceeding their rate of incorporation into the cell wall, or some of the HCAAs in their free form have additional functions, besides cell wall fortification. The increase in the amount of HCAAs is considered to be an indicator of the host plant mounting a barrier against the invading pathogen. Accumulation of HCAAs in the cell wall, for example, ensures a durable barrier against pathogens by reducing the cell wall digestibility by hydrolytic enzymes of the pathogen and/or by directly inhibiting further proliferation of the pathogen (Grandmaison et al., 1993; von Röpenack et al., 1998; Facchini et al., 1999). Moreover, the resulting fortified, and less permeable, cell wall ensures exclusion of the pathogen from accessing plant nutrients and water. In support of an additional function, HCAAs like coumaroyldopamine (116-fold) and coumaroyltyramine (CT; 75-fold) have been reported as novel induced HCAAs upon colonization of resistant tomato plants by P. syringae and were shown to have both antioxidant and antimicrobial activities (Zacarés et al., 2007). In conclusion, both our transcriptional network and metabolome analysis indicate that the HCAAs constitute a key class of metabolites associated with mounting of the HR in tomato.
Energy Homeostasis and Its Major Regulatory Mechanisms during the HR
During rescue of the DS at 33°C and 100% RH, only a small number of genes (34) are differentially regulated in these plants when compared with the PLS (Fig. 1, A and B, 0 h). Still, there already is a marked difference in their metabolome when compared with the PLS, including metabolites associated with the HR (Fig. 6; Supplemental Table S5). This is most likely explained by the activation of various defense-associated responses already at 0 h (which is after 2 weeks of rescue), albeit to a low level and without the execution of the HR and without an increased production of ethylene (Fig. 5A). This suggests that the activation of Cf-4-mediated responses is not completely blocked at elevated temperature and humidity, resulting in a gradual change in the overall metabolome of the DS during the 2 weeks of rescue before initiating the HR by the temperature and humidity shift. It should be noted that in an earlier experiment, using highly sensitive complementary DNA (cDNA)-amplified fragment length polymorphism analysis, it was found that at 0 h, already 70% of all 442 Avr4-responsive tomato genes that were selected were differentially expressed when compared with the PLS at that time point (Gabriëls et al., 2006). This observation supports our findings concerning the “leakiness” of the DS model. Indeed, in addition to the massively altered metabolome at 0 h, the DS are smaller in size than the PLS (de Jong et al., 2002), which is in line with the theory that claims reduction in growth to be a consequence of carbon limitation caused by the increased biosynthesis of defense-related metabolites. This results in a downward adjustment of growth to a level that can be supported by the new level of carbon availability (Smith and Stitt, 2007).
In an incompatible C. fulvum-tomato interaction, once fungal ingress has been resisted and defense responses have again been down-regulated, the plant restores its normal metabolism in order to bring the energy expenditure back to the original situation (Heil and Baldwin, 2002; Bolton, 2009). However, in the DS, the introgression of the fungal Avr4 gene, in combination with the Cf-4 resistance gene, leads to a continuous systemic triggering of the defense machinery, resulting in an amplification of the responses that occur only locally in Cf-4 tomato challenged with C. fulvum expressing Avr4. The associated massive transcriptome and metabolome reprogramming that we observed in the DS costs energy and requires reducing power and carbon skeletons. Disruption of the ion homeostasis (Fig. 3C, mainly at 5 h) reflects the low energy status of the DS, as ATP is normally used to generate the proton-motive force to regulate the transport of ions, such as Ca2+ across the plasma membrane, and ATP depletion is one of the main causes for the inability of a system to maintain its ion homeostasis (Chanson, 1993; Knickerbocker and Lutz, 2001; Arpagaus et al., 2002). Another marker reflecting ATP imbalance in the system is the up-regulation of Adenylate Kinase1 (ADK1) in the DS (3-fold increase as compared with the PLS) at 5 h. ADK1 plays a crucial role in maintaining the right equilibrium between ADP and ATP levels (Hardie et al., 2012).
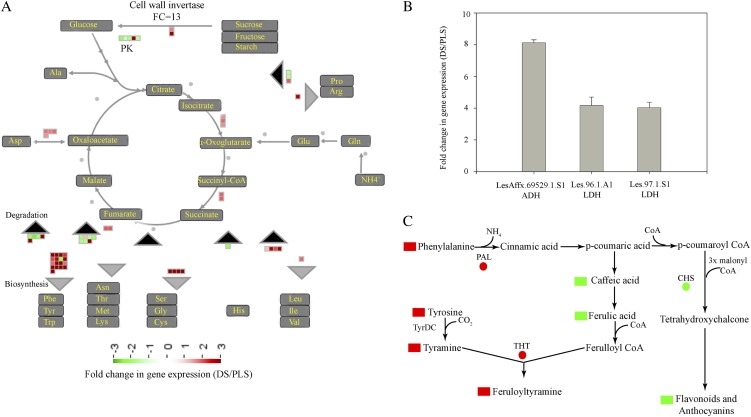
To accommodate the massive metabolome reprogramming, the DS activate various alternative processes generating energy and carbon skeletons. These processes can be categorized as follows: (1) a switch to catabolism; (2) preferential diversion of precursors toward specific biochemical pathways; and (3) increased anaerobic respiration. The first process involves an increased hydrolysis or oxidation of carbohydrates, proteins, nucleic acids, and lipids. Increased hydrolysis of carbohydrates (specifically Suc) into Glc, for example, is caused by transcriptional activation of the gene encoding cell wall invertase (CWI), which is more than 13-fold up-regulated at 5 h in the DS (Fig. 7A). This ensures a continuous supply of Glc for the energy cycle as well as for generating carbon skeletons required for the synthesis of primary metabolites, such as aromatic amino acids (Fig. 7A). The importance of CWI in generating carbohydrates that are catabolized upon the activation of defense responses is shown by RNA interference-mediated knockdown of CWI in tobacco leaves. Lowered CWI expression resulted in compromised defense responses and a delay in the actual hypersensitive cell death (Essmann et al., 2008). Another form of alternative energy generation includes lipid- and ribonucleoside-related catabolic processes (Fig. 3B, lipid and pyrimidine ribonucleoside catabolism). Upon ribonucleoside catabolism, the pentose moiety of the nucleosides serves as a carbon and energy source (Tozzi et al., 2006). Activation of lipid-related catabolic processes was also prominent during mounting of the HR (Fig. 3B). Under carbohydrate starvation conditions, the respiratory demand for carbon skeletons derived from fatty acid β-oxidation increases, thereby triggering induction of the fatty acid breakdown pathway (Graham and Eastmond, 2002).
Figure 7.
Alternative energy and carbon skeleton generation strategies of tomato DS undergoing massive transcriptome and metabolome reprogramming. A, Cell wall invertase-mediated generation of Glc from monosaccharides, disaccharides, and polysaccharides to maintain a continuous supply for the tricarboxylic acid cycle and thereby for amino acid biosynthesis. The gray and black rectangles represent biosynthesis and degradation processes, respectively. B, Expression levels of alcohol dehydrogenase (ADH) and LDH genes in the DS as compared with their PLS at 5 h. C, Redirection of common precursors, such as Phe and Tyr, toward targeted metabolite pathways (red and green shapes represent up-regulation and down-regulation of a gene [circles] or a metabolite [rectangles], respectively). CHS, Chalcone synthase; PAL, Phe ammonia lyase; PK, pyruvate kinase; TyrDC, Tyr decarboxylase.
It is well established that PCD is an active process that aborts cells, tissues, or organs during stress and/or developmental processes, accompanied by reallocation of the recycled nutrients to new sinks (Jones, 2001; Ameisen, 2002; Kuriyama and Fukuda, 2002). Amino acids derived from proteolysis, especially during acute nutrient restriction, are an important source of carbon skeletons (Vierstra, 1993; Vabulas and Hartl, 2005; Vabulas, 2007). The increased accumulation of aromatic amino acids (such as Phe and Tyr) in the DS is partly explained by the moderate activation of genes encoding enzymes involved in the shikimate pathway, which is an early step in aromatic amino acid biosynthesis (Fig. 4A, cluster 3; Supplemental Table S3, cluster 3 genes). In addition to this, a strong transcriptional activation of genes encoding enzymes involved in the final steps of the biosynthesis of these aromatic amino acids also occurs (Fig. 4A, cluster 1; Supplemental Table S3, cluster 1 genes). Remarkably, the latter genes show a high level of coregulation with genes involved in the protein degradation pathway (Fig. 4A, cluster 1; Supplemental Table S3, cluster 1 genes; Supplemental Fig. S3). Such a strong association suggests that protein degradation might function as an alternative pathway for the generation of aromatic amino acids required for the biosynthesis of secondary metabolites, such as benzoic acid, salicylic acid, alkaloids, and the HCAAs. Another example of a switch to catabolism for the generation of energy and carbon skeletons is the activation of genes involved in the degradation of branched amino acids (Leu, Ile, and Val; Fig. 7A; Supplemental Table S2). The catabolism of Leu, Ile, and Val generates acetyl-CoA, which in the tricarboxylic acid cycle generates energy, a process widely observed during starvation and stress (Aubert et al., 1996; Fujiki et al., 2001; Taylor et al., 2004; Kochevenko et al., 2012). Our data suggest that the highly induced hydroxycinnamic acid-derived secondary metabolite biosynthesis pathways are for the greater part responsible for the alterations in aromatic amino acids and carbon skeleton fluxes. Moreover, the depletion of organic acids (Fig. 6B) in the DS suggests that these compounds are precursors for many of the primary (aromatic amino acids) and secondary metabolites. Hence, their depletion can be explained by the massive accumulation of the respective primary and secondary metabolites in the DS.
The second way by which the plant deals with the low-carbon skeleton and energy supplies is the preferential diversion of common precursors toward specific metabolic pathways. This is exemplified by suppression of the flavonoid biosynthesis pathway through the down-regulation of Chalcone synthase gene expression, thereby increasing the availability of precursors like Phe and Tyr for the HCAA biosynthetic pathway (Figs. 3C, flavonol biosynthesis, and 7C), accompanied by up-regulation of the THT genes (Supplemental Table S7). Similarly, preferential redirection of common flavonoid precursors toward the biosynthesis of HCCAs in onion (Allium cepa) epidermis upon infection by Botrytis cinerea was described (McLusky et al., 1999).
As a third way to obtain energy, the DS activate anaerobic energy production (fermentation) pathways, which can be traced by transcriptional activation of l-Lactate dehydrogenase (LDH) and Alcohol dehydrogenase genes (Stulemeijer et al., 2009; Fig. 7C) and the γ-aminobutyrate (GABA) shunt, evident from the high expression of GAD2 and the accumulation of GABA (Supplemental Table S3, cluster 1 genes; Supplemental Table S5B). In hypoxic conditions, pyruvate is broken down by LDH to form lactate and NAD+, giving a boost in NAD+ for glycolytic anaerobic energy generation (Bolton, 2009). Under particular energy-demanding conditions, like the systemic activation of defense responses, pyruvate is produced in high amounts and pyruvate dehydrogenase activity becomes a limiting step in converting pyruvate into acetyl-CoA. Hence, the GABA shunt provides a means to utilize the excess of pyruvate for energy production (Bolton et al., 2008b).
Major Regulators and Regulatory Mechanisms of the HR
Our data depict the HR as an active process, regulated at the transcriptional, posttranscriptional, translational, and posttranslational levels, to maintain the cellular homeostasis of the system (Figs. 2–4 and 7). The robustness of the system is maintained through tightly regulated activation and repression of defense signaling. For example, enhanced activation of 2C-type protein phosphatases that are associated with the regulation of MAPK4 and MAPK6 and potassium channels (Lee et al., 2007; Schweighofer et al., 2007) can be mentioned as one of the major repression mechanisms of defense signaling during mounting of the HR (Fig. 3B, dephosphorylation; Supplemental Table S6). Another example is repression of the ethylene-induced signaling pathway through the transcriptional activation of repressors (RTE1 and CTR1; Huang et al., 2003; Dong et al., 2010) or lowered expression of signal receptors and transducers, such as EIN2 (Kieber et al., 1993; Supplemental Table S6). Furthermore, repression of PCD is enforced through the down-regulation of positive regulators of cell death, such as LSD1-like1 (Epple et al., 2003), and activation of the immune repressor RIN4 (Afzal et al., 2011; Supplemental Table S6). Next to this, genes encoding glutathione S-transferases (class III) that function as protectants against oxidative stress and as potent detoxifiers of toxins are transcriptionally activated (Fig. 3, A and B; Supplemental Table S6; Marrs, 1996; Kilili et al., 2004). In addition to its role in protein turnover, ubiquitin-dependent protein degradation also regulates defense through the targeted destruction of proteins involved in intensifying defense signaling (Santner and Estelle, 2009; Vierstra, 2009).
As in the DS massive and highly coordinated transcriptional changes are observed, it is anticipated that master regulators are involved. Some of the major transcription factors implicated in the regulation of the overall transcriptome and metabolome of the plant include WRKYs, AP2/ERFs, bZIPs, and MYBs (Daniel et al., 1999; Singh et al., 2002; Eulgem and Somssich, 2007; Lippok et al., 2007; Oñate-Sánchez et al., 2007; Naoumkina et al., 2008; Rushton et al., 2010). Despite the presence of all these, and other, transcription factors in the different hubs of the network (Fig. 4, A and B; Supplemental Tables S3 and S4), WRKY transcription factors in particular seem to play a critical role in orchestrating the HR-related transcriptome changes of the DS. This observation is strengthened by the fact that a W-box motif, to which WRKY transcription factors bind, is present in the promoter sequence of 72% of the genes that are implicated to have an essential role in the HR (Supplemental Table S4B) and that fall within the same cluster as these transcription factors themselves (Fig. 4, cluster 1; Supplemental Table S3, cluster 1 genes). The presence of two genes encoding MAP kinases highly homologous to MAPK3 in the same cluster supports the work that showed some WRKYs to be directly phosphorylated by MAP kinases, thereby inducing phytoalexin biosynthesis (Mao et al., 2011). Our findings strengthen the role of WRKYs in plant defense and defense-related secondary metabolite reprogramming. Such a strong association of differentially regulated defense-related genes and the presence of a shared regulatory motif in their promoter region takes the assumption of “guilt by association” one step further. Furthermore, this coexpression analysis should allow us to predict the role of yet uncharacterized genes such as Les.113.1.S1_at, which shows 106-fold up-regulation in the DS and locates to similar positions in the network as some of the well-characterized gene groups (Supplemental Table S3, genes in each cluster listed as “no hit found”). Moreover, most of the genes that we found in our cluster analysis have not been characterized in tomato, although some of them are annotated based on the presence of functional domains and their sequence similarity with other plant genes. Hence, our analysis suggests that these novel genes are also involved in the HR. This approach shows that, based on a robust and synchronized inducible system, it is possible to extract valuable information from the complex biological events that take place during the response of resistant plants to pathogens.
Currently, we are performing functional analysis of a number of genes that we have selected based on our transcriptional network analysis and for which a role in defense is further supported by our metabolome data. Such studies will pave the way to better control and optimize pathways involved in the HR, which will eventually contribute to enhanced resistance of crop plants to pathogens.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tomato (Solanum lycopersicum) DS were generated by crossing transgenic tomato cv Moneymaker (MM)-Cf0 expressing Avr4 (Cf0:Avr4) to MM-Cf0 tomato transgenic for the Hcr9-4D (Cf-4) resistance gene (Cf0:Cf-4; de Jong et al., 2002; Gabriëls et al., 2006; Stulemeijer et al., 2007). Pretreatment of the seeds before sowing was carried out as described (Stulemeijer et al., 2007). Ten seeds were sown per pot of 7 × 7 × 8 cm, and these were kept in a transparent closed box for 7 d (until the cotyledons had emerged) at 25°C, under a 16-h/8-h light/dark regime, in an incubator (Elbanton). Plants were rescued at 33°C and 100% RH for a period of 2 weeks, after which samples of the Cf-4/Avr4 DS and, as control, a 1:1 mixture of the Cf0:Cf-4 and Cf0:Avr4 PLS that were crossed to generate the Cf-4/Avr4 DS were collected at 0, 1, 3, and 5 h after initiation of the HR by a transfer of the plants to 20°C and 70% RH. Samples consisted of cotyledons and just appearing primary leaves and were immediately frozen, ground to a fine powder in liquid nitrogen, and stored at −80°C until further use. Four pots, each containing 10 seedlings, were considered as an experimental unit, and three independent biological replicates were generated for the DS and PLS at the individual time points.
To validate gene expression profiles observed in the DS and PLS, the Cf0:Avr4 (susceptible) and Cf0:Cf-4 (resistant) parental tomato lines were also inoculated with race 5 of C. fulvum secreting Avr4 (De Wit, 1977). Four biological replicates were performed, also including mock-treated susceptible and resistant plants, and leaflets were harvested at 6 and 10 d after inoculation with the fungus.
Microarray Experiments
Total RNA was extracted and purified from the collected tomato seedling material using the NucleoSpin RNA/protein kit (Macherey-Nagel). RNA labeling, hybridization of the microarrays (Affymetrix Tomato Genome Arrays; GeneChip Tomato Genome; Affymetrix), and data extraction were performed by Service XS according to standard protocols provided by the manufacturer. For this, the RNA concentration was determined with the Nanodrop (type ND-1000), and RNA quality was assessed with the RNA 6000 Nano Labchip kit (Agilent Technologies). Biotin-labeled complementary RNA was synthesized from 2 µg of total RNA using Affymetrix one-cycle target labeling and control agents (900493; Affymetrix). GeneChips were hybridized with 20 µg of fragmented biotin-labeled copy RNA, followed by automated washing and staining, and they were then scanned with an Affymetrix scanner type G7. Affymetrix GCOS software was used to convert the raw data to CEL files, and data analysis was performed by packages from the Bioconductor project (Gentleman et al., 2004) implemented in Genemath XT version 2.12 (Applied Maths). The raw array data (CEL files) were normalized using the GeneChip Robust Multiarray Averaging probe summarization algorithm, applying the empirical Bayes approach (Wu et al., 2004). For each time point, three independent biological replicates were performed.
RNA Isolation and qRT-PCR Gene Expression Analysis
RNA was isolated from 100 mg of tomato leaf tissue using 1 mL of Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The total RNA was treated with DNAse-I Amplification Grade (Invitrogen) and purified with an RNeasy Mini Kit (Qiagen). RNA concentration and integrity were examined, and 1 µg of RNA was used for the reverse transcription reaction, using the iScript Select cDNA Synthesis Kit (Bio-Rad Laboratories), in a final volume of 20 µL. qRT-PCR was performed in triplicate reactions, with the same cDNA pool, in the presence of fluorescent dye (iQ SYBR GREEN Super Mix 2*) using an iCycler iQ instrument (Bio-Rad Laboratories) with specific primer pairs (Supplemental Table S8). The constitutively expressed Actin gene from tomato (Solyc10g080500.1.1) was used as a reference, and the expression levels of other genes were determined relative to this reference.
Extraction and GC-TOF-MS Analysis of PPM
PPM were extracted following the procedure described by Lisec et al. (2006) with minor modifications. Briefly, 50 mg (fresh weight) of frozen leaf tissue was homogenized in an Eppendorf tube and extracted with 700 µL of methanol (100%), precooled at −20°C, and spiked with ribitol (0.2 mg mL−1) as an internal standard. The sample was then incubated for 10 min at 70°C and centrifuged at 21,000g for 10 min. The supernatant was transferred to a new Eppendorf tube, and 375 µL of chloroform (precooled at −20°C) and 750 µL of water (precooled at 4°C) were added. The mixture was centrifuged at 21,000g for 10 min, and 200 µL of the supernatant was dried in a vacuum concentrator without heating. Online derivatization was performed with methoxyamination reagent (methoxyamine hydrochloride [20 mg mL−1 pyridine]) and N-methyl-N-trimethylsilyltrifluoroacetamide using a CTC CombiPal injection and pipetting robot.
A mixture (5 µL) containing a series of n-alkanes (C10–C34) was injected into each sample for the calculation of the retention index. A 2-µL aliquot of each derivatized sample was injected with a gradient of 6°C s−1 from 70°C to 240°C, using an Optic3 injector (ATAS) in split ratio of 1:20, into an Agilent 6890N gas chromatograph equipped with a 40-m × 0.25-µm (i.d.) fused-silica capillary column with a chemically bonded 0.25-µm DB 5-MS stationary phase (J&W Scientific). The injector temperature was kept at 270°C, the septum purge flow rate was 20 mL min−1, and the purge was turned on after 60 s. The gas flow rate through the column was 1 mL min−1, and the column temperature was held at 70°C for 2 min, then increased by 10°C min−1 to 310°C, and held there for 5 min. The column effluent was introduced into the ion source of a Pegasus III TOF mass spectrometer (Leco). The transfer line and the ion source temperatures were 270°C and 200°C, respectively. Ions were generated by a 70-eV electron beam at an ionization current of 2.0 mA, and compounds were detected at a scanning range of 20 spectra s−1 and recorded in the mass range 50 to 600 mass-to-charge ratio. The acceleration voltage was turned on after a solvent delay of 295 s. The detector voltage was 1,400 V.
The resulting chromatograms were processed in an untargeted manner using the MetAlign software (Lommen, 2009). Extraction and reconstitution of compound mass spectra were performed according to the method described before (Tikunov et al., 2012). The internal standard (ribitol) was used to normalize the reconstituted metabolites over the samples. Identification of metabolites was performed using NIST-MS Search and retention indices using the Golm Metabolite database as a reference library (http://gmd.mpimp-golm.mpg.de/download/), and further confirmation was obtained through the injection of authentic standards.
Extraction and LC-QTOF-MS Analysis of SPSM
Extraction and LC-QTOF-MS analysis of SPSM were performed according to the protocols described by De Vos et al. (2007) with slight modifications. Briefly, 50 mg of ground frozen leaf tissue was extracted with 150 µL of a methanol (99.875%)-formic acid (0.125%) mixture (v/v) and sonicated for 15 min. The mixture was centrifuged at 20,000g for 10 min, and the supernatant was filtered using a 0.45-µm inorganic membrane filter (Anotop 10; Whatman), fitted onto a disposable syringe, and transferred into a 150-µL insert in a 1.8-mL glass vial.
The LC-PDA-QTOF-MS platform consisted of a Waters Alliance 2795 HT HPLC system equipped with a Luna C18(2) precolumn (2.0 × 4 mm) and an analytical column (2.0 × 150 mm, pore size of 100 Å, particle size of 3 µm; Phenomenex) connected to an Ultima V4.00.00 QTOF mass spectrometer (Waters, MS Technologies). Degassed solutions of formic acid in ultrapure water (1:1,000, v/v; eluent A) and formic acid in acetonitrile (1:1,000, v/v; eluent B) were pumped into the HPLC system at 190 µL min−1, and the gradient was linearly increased from 5% to 35% eluent B over a 45-min period, followed by 15 min of washing and equilibration of the column. The column, sample, and room temperatures were kept at 40°C, 20°C, and 20°C, respectively. Ionization was performed using an electrospray ionization source, and masses were detected in negative mode. A collision energy of 10 eV was used for full-scan LC-MS in the range of mass-to-charge ratio 100 to 1,500. For LC-MS/MS, a collision energy of 35 eV was applied. Leu enkephaline, (M – H)− = 554.2620, was used for online mass calibration (lock mass) using a separate spray inlet. The chromatograms were aligned using the MetAlign (accurate mass option) software package (Lommen, 2009). Extraction and reconstitution of compound mass spectra were performed according to the method described by Tikunov et al. (2012).
The identification of the secondary metabolites was based on their detected accurate masses and their corresponding retention times recorded for tomato fruit, as presented in the metabolome tomato database (Iijima et al., 2008). We used different public databases (Dictionary of Natural Products [http://dnp.chemnetbase.com] and KNApSAcK [http://kanaya.aist-nara.ac.jp/KNApSAcK/]) to putatively identify some of the metabolites that are not present in the metabolome tomato database (Moco et al., 2006). For confirmation of the identity of these metabolites, we performed three-stage fragmentation of the parent ion using a Thermo LC-LTQ-Obitrap Fourier transform mass spectrometry system in high-mass resolution mode (van der Hooft et al., 2011). The identification levels were given according to the proposed minimum reporting standards for chemical analysis, the Metabolomics Standards Initiative (Sumner et al., 2007). The identities of the HCAAs trans-feruloyltyramine (FT) and trans-CT present in total extracts of the DS were confirmed by using synthetic trans-FT and trans-CT as standards. For this, trans-FT and trans-CT were synthesized according to López-Gresa et al. (2011), and chromatograms, UV absorption spectra, and MS/MS spectra of the FT and CT standards and an extract of the DS obtained at 5 h after the temperature shift were compared (Supplemental Fig. S4).
Ethylene Measurements by GC Coupled to Flame Ionization Detection
Ethylene was measured using GC coupled to flame ionization detection on a Focus gas chromatograph (Thermo Scientific), equipped with a flame ionization detector and an RT QPLOT (15 m × 0.53 mm [i.d.]) column (Restek), using helium as the carrier gas. First, the seedlings were kept for 24 h in 1,000-mL air-tight glass jars at 33°C and 100% RH, and 2-mL samples were taken regularly from the head space for analyses. Then, the plants were transferred to 20°C and 100% RH, and again, 2-mL air samples were collected over a period of 24 h. Samples were stored in air-tight syringes and were injected in the gas chromatograph using a 500-µL injection loop.
Statistical Analysis and Bioinformatics
For both transcriptome and metabolome studies, three independent biological replicates were performed and analyzed. Differentially expressed genes were filtered based on the following criteria: expression FC > 2 and P < 0.05, based on two-tailed Student’s t tests on the expression values of the DS and PLS at individual time points. The FC in gene expression was calculated by subtracting the expression value (log2) of the PLS (control) from the DS (treatment). The same procedure was employed to filter both PPM and SPSM. Here, we reduced the FC cutoff value to 1.5, in order to include most of the tightly regulated primary metabolites in the analysis, and we corrected for the false discovery rate.
About 80% to 90% of the genes on the tomato Affymetrix genome array are of unknown function. Therefore, to annotate them, the BLAST output of these genes from the Tomato Functional Genomics Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/array/download_annotation.cgi) was used. In this output list, the potential Arabidopsis (Arabidopsis thaliana) orthologs and their respective protein reference sequence identifiers were included. Subsequently, this combined information was used to perform ORA, aimed at identifying significantly enriched functional GO subcategories of genes that are differentially regulated during the HR. For this analysis, GeneTrail, a Web-based tool that compares the frequency of differentially regulated genes belonging to a certain functional category in the test set with that of a reference set (in this case, all genes of Arabidopsis), was used. Separate computation of the ORA was done for the up- and down-regulated gene categories using the sets of genes that have unique or overlapping expression patterns between the different time points. To reduce the number of redundant functional categories describing the same biological process, a similarity matrix was built based on the output of GeneTrail using R software (Supplemental Materials and Methods S1). In addition to GeneTrail, we used other Web-based tools like Tomato Functional Genomics Database GO enrichment analysis (http://ted.bti.cornell.edu/cgi-bin/TFGD/array/GO_analysis.cgi) and AtCOECiS (http://bioinformatics.psb.ugent.be/ATCOECIS/; Vandepoele et al., 2009; Fei et al., 2011) to compute GO enrichment analysis of gene clusters that are selected from our coexpression network analysis.
Genemath XT software (Applied Maths) was used for DFA and HCA. For HCA, Pearson’s correlation coefficients were used to calculate the distance or similarity between two entries, and the resulting clusters were summarized using a complete linkage algorithm. To compare the expression values, both for the transcriptome and the metabolome profiles, the raw values of each sample were log transformed and autoscaled by the use of the average as an offset and the sd as scale (raw value average [offset]/sd [scale]).
To gain more insight into the interaction of differentially regulated genes at the systems level, a regulatory network was built using Cytoscape (version 2.6; Shannon et al., 2003) and the Expression Correlation Network plugin (http://www.baderlab.org/Software/ExpressionCorrelation). Network construction was based on a weighed gene-gene network, in which an edge weight for two genes is given based on the similarity of their expression profiles. A similarity matrix was computed using Pearson’s correlation coefficients and was then converted into a sparse network by connecting each gene to its nearest neighbors with a strict similarity cutoff score of 0.92. This type of computation is based on selecting a gene and determining its nearest neighbors in the expression space within the defined distance cutoff value. The Cytoscape plugin MINE was used to identify the major hubs where pathways converge. MINE performs a type of clustering that follows a guilt-by-association approach (Rhrissorrakrai and Gunsalus, 2011).
We used Tair Motif Finder (http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp) to identify potential nucleotide patterns (hexamers) in the promoter regions (a 1-kb stretch upstream of the open reading frame) of the coregulated genes. The Web-based tool POBO (Kankainen and Holm, 2004) was used to validate the result from the Tair Motif Finder. MapMan (Thimm et al., 2004) was used to visualize the expression pattern of genes that fall into different functional groups.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression profiles of the different gene clusters identified in the coexpression network analysis of differentially regulated genes during mounting of the HR in the DS, as shown in Figure 4A.
Supplemental Figure S2. Venn diagram showing the number and distribution of genes containing the sequence TTTGAC, interrogated for the presence of either a C or a T, following the core sequence TGAC.
Supplemental Figure S3. qRT-PCR analysis of the expression of genes, selected from the coexpression network analysis of the DS, in leaves of C. fulvum-inoculated tomato.
Supplemental Figure S4. Confirmation of the identities of trans-FT and trans-CT present in total extracts of dying seedlings obtained at 5 h after the temperature shift using synthesized trans-FT and trans-CT as standards.
Supplemental Table S1. Expression profiles of genes encoding heat shock proteins in the DS compared with the PLS.
Supplemental Table S2. ORA of significantly up-regulated genes in the DS 1 h after the temperature shift.
Supplemental Table S3. Description of the genes that cluster in groups of strongly associated genes that were identified by coexpression network analysis using Cytoscape, and output of the computation of the GO enrichment analysis on each cluster.
Supplemental Table S4. The Arabidopsis Information Resource statistical regulatory element motif analysis of genes present in clusters 1, 2, 3, and 4 of the coexpression network presented in Figure 4A.
Supplemental Table S5. Metabolites showing differential accumulation in the DS as compared with the PLS.
Supplemental Table S6. Expression profile of genes involved in the repression of defense signal transduction and protection against oxidative damage during mounting of the HR.
Supplemental Table S7. Expression profile of genes involved in the biosynthesis of HCAAs during mounting of the HR.
Supplemental Table S8. Nucleotide sequences of primers used for qRT-PCR gene expression analysis.
Supplemental Materials and Methods S1. Reduction of the redundancy of GO subcategories obtained from GeneTrail.
Supplementary Material
Acknowledgments
We are indebted to José María Bellés (Instituto de Biología Molecular y Celular de Plantas) for providing hydroxycinnamic acid amide standards. We thank Anna Undas and Francel Verstappen (Laboratory of Plant Physiology, Wageningen University), Bert Schipper (Plant Research International), and Arjen Peppel and Uulke van Meeteren (Horticultural Supply Chains Group, Wageningen University) for the technical help we received during primary and secondary metabolite analysis and with ethylene measurements. We are grateful for the technical advice we obtained from Chris Maliepaard (Plant Breeding, Wageningen University) and Bert Essenstam (Unifarm, Wageningen University and Research Center) on multivariate analysis of both the metabolome and transcriptome data. The excellent plant care by Bert Essenstam is highly appreciated. We are grateful for all technical and intellectual support we received from the metabolomics groups of the Laboratory of Plant Physiology at Wageningen University and of Plant Research International.
Glossary
- PTI
pathogen-associated molecular pattern-triggered immunity
- HR
hypersensitive response
- PCD
programmed cell death
- DS
dying seedlings
- RH
relative humidity
- PPM
polar primary metabolites
- SPSM
semipolar secondary metabolites
- GC
gas chromatography
- TOF
time-of-flight
- QTOF
quadrupole time-of-flight
- LC
liquid chromatography
- HCA
hierarchical cluster analysis
- PLS
parental lines
- FC
fold change
- MAP
mitogen-activated protein
- ORA
overrepresentation analysis
- GO
Gene Ontology
- HCAA
hydroxycinnamic acid amide
- qRT
quantitative reverse transcription
- DFA
discriminant function analysis
- cDNA
complementary DNA
- GABA
γ-aminobutyrate
- MS
mass spectrometry
- FT
feruloyltyramine
- MINE
Module Identification In Networks
References
- Afzal AJ, da Cunha L, Mackey D. (2011) Separable fragments and membrane tethering of Arabidopsis RIN4 regulate its suppression of PAMP-triggered immunity. Plant Cell 23: 3798–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameisen JC. (2002) On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ 9: 367–393 [DOI] [PubMed] [Google Scholar]
- Arpagaus S, Rawyler A, Braendle R. (2002) Occurrence and characteristics of the mitochondrial permeability transition in plants. J Biol Chem 277: 1780–1787 [DOI] [PubMed] [Google Scholar]
- Aubert S, Alban C, Bligny R, Douce R. (1996) Induction of β-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett 383: 175–180 [DOI] [PubMed] [Google Scholar]
- Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. (2007) GeneTrail: advanced gene set enrichment analysis. Nucleic Acids Res 35: W186–W192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert GP, Møller ALB, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK, Jahn TP. (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Bolton MD. (2009) Primary metabolism and plant defense: fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Bolton MD, Kolmer JA, Xu WW, Garvin DF. (2008a) Comparative transcript profiling of Lr1- and Lr34-mediated leaf rust resistance in wheat. Phytopathology 98: S24. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Kolmer JA, Xu WW, Garvin DF. (2008b) Lr34-mediated leaf rust resistance in wheat: transcript profiling reveals a high energetic demand supported by transient recruitment of multiple metabolic pathways. Mol Plant Microbe Interact 21: 1515–1527 [DOI] [PubMed] [Google Scholar]
- Chanson A. (1993) Active-transport of proton and calcium in higher-plant cells. Plant Physiol Biochem 31: 943–955 [Google Scholar]
- Ciardi JA, Tieman DM, Jones JB, Klee HJ. (2001) Reduced expression of the tomato ethylene receptor gene LeETR4 enhances the hypersensitive response to Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 14: 487–495 [DOI] [PubMed] [Google Scholar]
- Ciardi JA, Tieman DM, Lund ST, Jones JB, Stall RE, Klee HJ. (2000) Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol 123: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Lacomme C, Morel J-B, Roby D. (1999) A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J 20: 57–66 [DOI] [PubMed] [Google Scholar]
- de Jong CF, Takken FLW, Cai X, de Wit PJGM, Joosten MHAJ. (2002) Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol Plant Microbe Interact 15: 1040–1049 [DOI] [PubMed] [Google Scholar]
- De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD. (2007) Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc 2: 778–791 [DOI] [PubMed] [Google Scholar]
- De Wit PJGM. (1977) A light and scanning-electron microscopic study of infection of tomato plants by virulent and avirulent races of Cladosporium fulvum. Eur J Plant Pathol 83: 109–122 [Google Scholar]
- de Wit PJGM, Joosten MHAJ. (1999) Avirulence and resistance genes in the Cladosporium fulvum-tomato interaction. Curr Opin Microbiol 2: 368–373 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Innes RW. (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dong C-H, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C. (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem 285: 40706–40713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy ASN, Poovaiah BW. (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Epple P, Mack AA, Morris VR, Dangl JL. (2003) Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci USA 100: 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann J, Bones P, Weis E, Scharte J. (2008) Leaf carbohydrate metabolism during defense: intracellular sucrose-cleaving enzymes do not compensate repression of cell wall invertase. Plant Signal Behav 3: 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG. (2002) Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot 80: 577–589 [Google Scholar]
- Facchini PJ, Yu M, Penzes-Yost C. (1999) Decreased cell wall digestibility in canola transformed with chimeric tyrosine decarboxylase genes from opium poppy. Plant Physiol 120: 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z, Joung J-G, Tang X, Zheng Y, Huang M, Lee JM, McQuinn R, Tieman DM, Alba R, Klee HJ, et al. (2011) Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res 39: D1156–D1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanabe A. (2001) Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett 499: 161–165 [DOI] [PubMed] [Google Scholar]
- Gabriëls SH, Takken FL, Vossen JH, de Jong CF, Liu Q, Turk SC, Wachowski LK, Peters J, Witsenboer HM, de Wit PJGM, et al. (2006) cDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol Plant Microbe Interact 19: 567–576 [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80–R80.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes D, Agasse A, Thiébaud P, Delrot S, Gerós H, Chaumont F. (2009) Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim Biophys Acta 1788: 1213–1228 [DOI] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ. (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41: 156–181 [DOI] [PubMed] [Google Scholar]
- Grandmaison J, Olah GM, Van Calsteren MR, Furlan V. (1993) Characterization and localization of plant phenolics likely involved in the pathogen resistance expressed by endomycorrhizal roots. Mycorrhiza 3: 155–164 [Google Scholar]
- Guillet G, De Luca V. (2005) Wound-inducible biosynthesis of phytoalexin hydroxycinnamic acid amides of tyramine in tryptophan and tyrosine decarboxylase transgenic tobacco lines. Plant Physiol 137: 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen IM, van Esse HP, Ballester AR, Hogewoning SW, Parra NO, Paeleman A, Lievens B, Bovy AG, Thomma BPHJ. (2011) Differential tomato transcriptomic responses induced by Pepino mosaic virus isolates with differential aggressiveness. Plant Physiol 156: 301–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Baldwin IT. (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Hong W, Xu YP, Zheng Z, Cao JS, Cai XZ. (2007) Comparative transcript profiling by cDNA-AFLP reveals similar patterns of Avr4/Cf-4- and Avr9/Cf-9-dependent defence gene expression. Mol Plant Pathol 8: 515–527 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, Suzuki T, Suzuki H, Okazaki K, Kitayama M, et al. (2008) Metabolite annotations based on the integration of mass spectral information. Plant J 54: 949–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM. (2001) Programmed cell death in development and defense. Plant Physiol 125: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kang S, Kang K, Chung GC, Choi D, Ishihara A, Lee D-S, Back K. (2006) Functional analysis of the amine substrate specificity domain of pepper tyramine and serotonin N-hydroxycinnamoyltransferases. Plant Physiol 140: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Holm L. (2004) POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res 32: W222–W229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kilili KG, Atanassova N, Vardanyan A, Clatot N, Al-Sabarna K, Kanellopoulos PN, Makris AM, Kampranis SC. (2004) Differential roles of tau class glutathione S-transferases in oxidative stress. J Biol Chem 279: 24540–24551 [DOI] [PubMed] [Google Scholar]
- Knickerbocker DL, Lutz PL. (2001) Slow ATP loss and the defense of ion homeostasis in the anoxic frog brain. J Exp Biol 204: 3547–3551 [DOI] [PubMed] [Google Scholar]
- Kochevenko A, Araújo WL, Maloney GS, Tieman DM, Do PT, Taylor MG, Klee HJ, Fernie AR. (2012) Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits. Mol Plant 5: 366–375 [DOI] [PubMed] [Google Scholar]
- Kuriyama H, Fukuda H. (2002) Developmental programmed cell death in plants. Curr Opin Plant Biol 5: 568–573 [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411: 848–853 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li LG, Cheong YH, Pandey GK, Lu GH, Buchanan BB, Luan S. (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Antolín-Llovera M, Ferrero S, Closa M, Arró M, Ferrer A, Boronat A, Campos N. (2011) Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 23: 1494–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippok B, Birkenbihl RP, Rivory G, Brümmer J, Schmelzer E, Logemann E, Somssich IE. (2007) Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol Plant Microbe Interact 20: 420–429 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lommen A. (2009) MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem 81: 3079–3086 [DOI] [PubMed] [Google Scholar]
- Lopez-Gresa MP, Torres C, Campos L, Lison P, Rodrigo I, Belles JM, Conejero V. (2011) Identification of defence metabolites in tomato plants infected by the bacterial pathogen Pseudomonas syringae. Environ Exp Bot 74: 216–228 [Google Scholar]
- Lund ST, Stall RE, Klee HJ. (1998) Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Mangeon A, Junqueira RM, Sachetto-Martins G. (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolles-Clark E, Harman GE, Penttila M. (1996) Enhanced expression of endochitinase in Trichoderma harzianum with the cbh1 promoter of Trichoderma reesei. Appl Environ Microbiol 62: 2152–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA. (1996) The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 127–158 [DOI] [PubMed] [Google Scholar]
- McLusky SR, Bennett MH, Beale MH, Lewis MJ, Gaskin P, Mansfield JW. (1999) Cell wall alterations and localized accumulation of feruloyl-3′-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant J 17: 523–534 [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CHR. (2006) A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol 141: 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL. (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ 4: 671–683 [DOI] [PubMed] [Google Scholar]
- Mur LA, Lloyd AJ, Cristescu SM, Harren FJ, Hall MA, Smith AR. (2009) Biphasic ethylene production during the hypersensitive response in Arabidopsis: a window into defense priming mechanisms? Plant Signal Behav 4: 610–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina MA, He X, Dixon RA. (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Anderson JP, Young J, Singh KB. (2007) AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol 143: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappan K, Qin W, Dyer JH, Zheng L, Wang X. (1997) Molecular cloning and functional analysis of polyphosphoinositide-dependent phospholipase D, PLDbeta, from Arabidopsis. J Biol Chem 272: 7055–7061 [DOI] [PubMed] [Google Scholar]
- Postel S, Kemmerling B. (2009) Plant systems for recognition of pathogen-associated molecular patterns. Semin Cell Dev Biol 20: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Rhrissorrakrai K, Gunsalus KC. (2011) MINE: module identification in networks. BMC Bioinformatics 12: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Thomas CM. (2005) Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu Rev Phytopathol 43: 395–436 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Schneider DJ, Collmer A. (2010) Studying plant-pathogen interactions in the genomics era: beyond molecular Koch’s postulates to systems biology. Annu Rev Phytopathol 48: 457–479 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Blumwald E. (2000) Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J 24: 317–326 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Fromm H. (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151: 35–66 [DOI] [PubMed] [Google Scholar]
- Stulemeijer IJS, Joosten MHAJ, Jensen ON. (2009) Quantitative phosphoproteomics of tomato mounting a hypersensitive response reveals a swift suppression of photosynthetic activity and a differential role for hsp90 isoforms. J Proteome Res 8: 1168–1182 [DOI] [PubMed] [Google Scholar]
- Stulemeijer IJS, Stratmann JW, Joosten MHAJ. (2007) Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol 144: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L, Amberg A, Barrett D, Beale M, Beger R, Daykin C, Fan TM, Fiehn O, Goodacre R, Griffin J, et al. (2007) Proposed minimum reporting standards for chemical analysis. Metabolomics 3: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH. (2004) Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Nürnberger T, Joosten MHAJ. (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, van Esse HP, Crous PW, de Wit PJGM. (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol Plant Pathol 6: 379–393 [DOI] [PubMed] [Google Scholar]
- Tikunov YM, Laptenok S, Hall RD, Bovy A, de Vos RC. (2012) MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL. (2006) Pentose phosphates in nucleoside interconversion and catabolism. FEBS J 273: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Vabulas RM. (2007) Proteasome function and protein biosynthesis. Curr Opin Clin Nutr Metab Care 10: 24–31 [DOI] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU. (2005) Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310: 1960–1963 [DOI] [PubMed] [Google Scholar]
- van Baarlen P, van Esse HP, Siezen RJ, Thomma BPHJ. (2008) Challenges in plant cellular pathway reconstruction based on gene expression profiling. Trends Plant Sci 13: 44–50 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hooft JJJ, Vervoort J, Bino RJ, Beekwilder J, de Vos RCH. (2011) Polyphenol identification based on systematic and robust high-resolution accurate mass spectrometry fragmentation. Anal Chem 83: 409–416 [DOI] [PubMed] [Google Scholar]
- van Esse HP, Fradin EF, de Groot PJ, de Wit PJGM, Thomma BPHJ. (2009) Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol Plant Microbe Interact 22: 245–258 [DOI] [PubMed] [Google Scholar]
- van Esse HP, Van’t Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, Vervoort J, de Wit PJGM, Thomma BPHJ. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Geraats BPJ, Linthorst HJM. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (1993) Protein-degradation in plants. Annu Rev Plant Physiol Plant Mol Biol 44: 385–410 [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- von Roepenack-Lahaye E, Newman MA, Schornack S, Hammond-Kosack KE, Lahaye T, Jones JDG, Daniels MJ, Dow JM. (2003) p-Coumaroylnoradrenaline, a novel plant metabolite implicated in tomato defense against pathogens. J Biol Chem 278: 43373–43383 [DOI] [PubMed] [Google Scholar]
- von Röpenack E, Parr A, Schulze-Lefert P. (1998) Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J Biol Chem 273: 9013–9022 [DOI] [PubMed] [Google Scholar]
- Walters DR. (2003) Polyamines and plant disease. Phytochemistry 64: 97–107 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Yang T, Poovaiah BW. (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8: 505–512 [DOI] [PubMed] [Google Scholar]
- Yap KL, Yuan T, Mal TK, Vogel HJ, Ikura M. (2003) Structural basis for simultaneous binding of two carboxy-terminal peptides of plant glutamate decarboxylase to calmodulin. J Mol Biol 328: 193–204 [DOI] [PubMed] [Google Scholar]
- Zacarés L, López-Gresa MP, Fayos J, Primo J, Bellés JM, Conejero V. (2007) Induction of p-coumaroyldopamine and feruloyldopamine, two novel metabolites, in tomato by the bacterial pathogen Pseudomonas syringae. Mol Plant Microbe Interact 20: 1439–1448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.